Abstract
Background
The androgen regulated probasin (PB) promoter has been used extensively to target transgenes to the prostate in transgenic mice; however, limited data exists on the mechanism that dictates prostate-specific gene expression. Tissue-specific gene expression involves synergistic effects among transcription factors associated in a complex bound to cis-acting DNA elements.
Methods
Using comprehensive linker scan mutagenesis, enzyme mobility shift and supershift assays, chromatin immunoprecipitation and transgenic animal studies, we have extensively characterized the prostate specific PB promoter.
Results
We identified a series of non-receptor transcription factors that are bound to the prostate-specific rat PB promoter. These factors include several ubiquitously distributed proteins known to participate in steroid receptor-mediated transcription. In addition, we identified two tissue-specific DNA elements that are crucial in directing prostate-specific PB expression, and confirmed the functional importance of both elements in transgenic animal studies. These two elements are functionally interchangeable and can be bound by multiple protein complexes, including the forkhead transcription factor FoxA1, a “pioneer factor” that has a restricted distribution to some cells type that are ectoderm and endoderm in origin. Using transgenic mice we further demonstrate that the minimal PB promoter region (−244/−96 bp) that encompasses these tissue-specific elements results in prostate-specific gene expression in transgenic mice, contains androgen receptor and FoxA1 binding sites as well as ubiquitous transcription factor binding sites.
Conclusion
We propose that these sequence-specific DNA-binding proteins, including tissue-restricted and ubiquitous factors, create the first level of transcriptional control, which responds to intracellular pathways that directs prostate-specific gene expression.
Keywords: prostate-specific, androgen receptor, FoxA1, c-jun, NFI, Oct-1
INTRODUCTION
Transcriptional regulation of gene expression involves a synergistic effect among the basal transcriptional machinery, proximal sequence-specific activators, cofactors, long-range enhancer-promoter interactions, chromatin remodeling complexes, as well as the signal transduction cascades (1). In addition to alternative splicing and DNA rearrangement, transcriptional synergy has long been proposed as an important mechanism to achieve organism diversity. Comparative genome analysis implies that a progressively more elaborate regulation of gene expression rather than the creation of new genes is responsible for the complexity of an organism, and that a constrained organization of metazoan enhancers is essential for the precise patterns of gene expression during development (2). Thus, a greater elaboration of cis-regulatory DNA sequences, and an increased complexity in the multi-protein transcription complexes, could be two pervasive mechanisms that lead to increased physiological and behavioral complexity during evolution (1).
A complex eukaryote differentially transcribes approximately 20-25,000 genes in precise spatial and temporal patterns to maintain cell specificity and homeostasis. It is widely accepted that the cell accomplishes this by employing combinatorial subsets of basal transcription factors, ubiquitous DNA binding factors or cofactors, signal-responsive or tissue-specific activators and/or coactivators. A small grouping of these factors could lead to an exponentially larger number of regulatory decisions (1,3,4). The basis for such transcriptional synergy stems from the highly sophisticated cis-regulatory DNA sequences seen in higher and complex eukaryotes. However, the rapidly expanding cofactor and chromatin remodeling complexes might be critical for integrating the complex cis-regulatory information.
Androgen receptor signaling is a specialized signaling pathway that plays pivotal roles in prostatic differentiation and function. In addition, transcriptional synergy applies to nuclear receptors, and the functional flexibility seen in members of this family is largely due to the principle of cooperativity. For instance, combinatorial gene regulation by AR has been proposed to involve differential recruitment of diverse co-regulatory complexes as well as the differential interaction with various intracellular pathways (5).
The rat probasin (PB) gene is an androgen regulated, prostate epithelial cell specific gene whose expression is associated with differentiation. The PB promoter has been used in numerous gene targeting studies (6-13) to drive prostate-specific gene expression and PB directed Cre is used to conditionally inactivate genes (14,15). In the PB proximal promoter, two strong androgen receptor binding sites (ARBSs) (−236/−223 and −140/−117 bp) and two weak ARBSs (−209/−196 and −107/−93 bp) were identified (21;22), and these ARBSs function together with the surrounding sequences to control androgen-dependent PB activation. Thus, a DNA fragment (−244 to −96 bp) has been termed the Androgen Responsive Region (ARR) (23;24). More recently, an artificial PB promoter ARR2PB, which contains an extra copy of ARR (−244/−96 bp) fused in front of the −286/+28 bp proximal promoter, targeted high levels of reporter genes into both prostate cell lines and transgenic mouse prostate (11). While these data indicate that critical prostate-specific information is contained within a 314 bp DNA fragment, reduction of the DNA fragment to a minimal prostate-specific sequence that defines key transcription factors involved has not yet been reported.
We have recently reported that two forkhead binding sites are present in the PSA promoter, and both can be bound by Forkhead Box A1 (FoxA1; formerly termed hepatocyte nuclear factor-3α) (16). Furthermore, we have shown that the expression of FoxA1 is essential for normal murine prostate development and differentiation (17). Functional FoxA1 binding sites were further identified in other prostatic enhancers across species, and some of these sites were positioned immediately adjacent to AREs, suggesting a well-conserved organization of cis-acting elements (16,18,19). In this report, we further characterized the PB −286/+28 promoter in both cell culture and transgenic mice in order to identify additional transcription factors and investigate the mechanisms of the androgen-regulated prostate-specific gene expression.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Thirty-one substitutive linker mutations of the −286PB promoter, named from m1 to m31, were generated by sequential replacement of the 10 bp sequence (5′-CTCGAGTACT-3′), using PCR based site-directed mutagenesis as described by Chen et al., (20). All final PCR products were subcloned into pGEM-Teasy vector (Promega Corp., Madison, WI), and verified with sequencing analysis. DNA sequencing was performed (by DNA Sequencing Core Laboratory, Vanderbilt-Ingram Cancer Center, Vanderbilt University Medical Center) as described in the Thermo sequence fluorescent-labeled primer cycle sequencing kit (Perkin Elmer Inc., Boston, MA). All mutant and wild type −286PB promoters were inserted upstream of the Luc reporter gene in pGL3-basic vector (Promega Corp., Madison, WI). Same approaches were used to construct the mtOct-1, mtC-jun, mtNFI, TS1/TS1, TS2/TS2, TS2/TS1, and four TS2 mutants (mt1, mt2, mt3, and mt4) expression vectors.
The −286PBCAT, ARR2PBCAT, and delARR2PBCAT reporter plasmids were described before (11). Two mutants of −286PBCAT, mtTS1/−286PBCAT and mtTS2/−286PBCAT, were generated by replacing wt −286PB promoter with the m5 and m18 substitutive linker mutations of −286PB promoter, respectively. The ARR2TKCAT was generated by replacing the endogenous −96 to −28 bp PB promoter with thymidine kinase (TK) promoter in ARR2PBCAT.
Cell Culture and Transient Transfection Assays
The human prostate carcinoma cell lines PC-3, LNCaP, the monkey kidney cell line COS-1, the human breast carcinoma cell line MCF-7, and the human pancreas carcinoma cell line PANC-1 were obtained from American Type Culture Collection (Manassas, VA). PC-3 cells were maintained in RPMI medium, 10% FBS and 250nM DEX and the other cell lines were cultured as recommended by ATCC.
The additional over-expression of rat AR in LNCaP was used to increase the overall detectable response of an AR driven promoter. The endogenous mutant AR expressed by LNCaP cells (T868A) binds to the androgen response elements in the same manner as the wild type AR except that the point mutation in the ligand binding domain allows for a extended ligand specificity now including the anti-androgen, flutamide but not by bicalutamide. Activation of the wild type AR and AR(T877A) results in similar mobility, nuclear distribution, and transcriptional activity of both ARs (21). Therefore, the mutant human AR of the LNCaP cells should not interfere with the rat AR that was also expressed in these cells. Transient transfection assays were performed using lipofectin reagent according to the manufacturer procedure (Life Technologies Inc., Gaithersburg, MD). Briefly, cells were plated at an initial density of 2~3 × 104/well in 24-well tissue culture plates and grown to roughly 70% confluence (~36 hour incubation). Then, 0.25 μg testing construct, 0.25 μg rat AR expression vector (rAR) or rat GR expression vector (rGR) (21), 0.0125 μg pRL-CMV (Promega Corp., Madison, WI) were mixed with serum-free MEM medium in tube A (25 μl/well). 4μl lipofectin were mixed with 21 μl serum-free MEM medium in tube B (25μl/well), and allow to standing at room temperature for 40 minutes. Two solutions in tube A and B were combined, mixed gently, and incubated at room temperature for 12-15 minutes, then 0.2 ml/well MEM medium with 10% FBS (stripped serum) were added to the DNA/lipofectin mixture. Aspirate medium from the wells and add 0.25ml of above solution to each well. The cells were incubated with the DNA/lipofectin mixture for 6 hours, followed by removal of the medium and addition of 0.5 ml/well medium with or without steroids for 24 hours. The cells were harvested by removing the medium, washing the cells once with phosphate-buffered saline, and incubating with 100 μl passive lysis buffer (Promega Corp., Madison, WI) for 1 hour at room temperature. Both firefly and Renilla Luciferase activity were determined in a lumicounter (LUM/star, BMG LabTechnologies, Inc., Durham, NC) by using the dual-Luciferase reporter assay system (Promega Corp., Madison, WI) to control for transfection efficiency. The cells transfected with CAT reporter constructs were harvested by removing the medium, washing the cells once with phosphate-buffered saline, and adding 100 μl lysis buffer (0.1 M Tris, pH 7.8, 0.1% triton X-100). The CAT activity was determined by the two-phase flour diffusion assay using [3H]acetylcoenzyme (5.9 Ci/mmol, Amersham Pharmacia Biotech) as the substrate (22).
Establishment of PBCAT Transgenic Mouse Lines
All animal studies were conducted in accordance with the principles and procedures outlined by Vanderbilt University Council on Animal Care. All the PBCAT transgenic mice were generated by microinjection of the DNA into the male pronucleus of a fertilized oocyte by the Transgenic Core/ES Cell Shared Resource of the Vanderbilt-Ingram Cancer Center. Animals were housed in standard lighting (12 h of light, 12 h of darkness) and allowed food and water ad libitum. Transgenic animals were identified by polymerase chain reaction (PCR)-based screening using isolated mouse tail DNA as described previously (15). The primer pairs used in PCR screening were the PB forward primer 5′-TAGCATCTTGTTCTTAGTCTT-3′ and the CAT reverse primer 5′-CAACGGTGGTATATCCAGTG-3′. All transgenic mouse lines were maintained as the B6D2F1 mouse strain.
CAT Assays on PBCAT Transgenic mice
To measure the levels of CAT reporter gene expression in transgenic mice, tissue samples including anterior (AP), dorsal (DP), lateral (LP) and ventral prostate (VP), brain, spleen, thymus, heart, lung, liver, kidney, testes, muscle, adrenals, epididymis, seminal vesicle, uterus, ovary, and oviduct were microdissected; Proteins were extracted by homogenization in lysis buffer (0.1 M Tris, pH 7.8, 0.1% triton X-100); And protein concentrations were determined by protein assay (Bio-Rad Laboratories, Hercules, CA). The CAT assays were performed as described before (22).
Electrophoretic Mobility Shift Assays
Nuclear extract for PC-3 cells was prepared as described by Dignam et al (23). Nuclear extracts for LNCaP and MCF-7 cells were purchased from Geneka Biotechnology, Inc. (Montreal, Quebec, Canada) and the Hela nuclear extract was purchased from Promega Corp. (Madison, WI). All oligonucleotides for EMSAs were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). The probes were radiolabeled with γ-[32p] dATP using the T4 polynucleotide kinase according to the manufacture’s instructions (New England Biolabs, Beverly, MA) and purified by 15% PAGE. One nanogram [20,000 disintegration per minute (dpm)] of the probe was incubated on ice for 10 min with different amount of nuclear extracts, 0.5 μg poly (dI-dC), and buffer D [10 mM HEPES pH 7.9, 7.5 mM MgCl2, 50 mM KCl, 0.1 mM EDTA, 10% (v/v) glycerol, 0.5 mM phenylmethysulfonide fluoride (PMSF) and 0.5 mM dithiothreitol (DTT)] in a total volume of 20 μl. In oligoncleotide competitions, 50 to 200-fold molar excess of cold, double-stranded oligonucleotide were added to the incubation mixtures. In supershift analyses, antibodies were added after the binding reaction and incubated for an additional 20 min on ice. All supershift antibodies to C-fos, C-jun, NFI, and Oct-1 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The concentration of antibody in each EMSA reaction was 0.2 μg/μl. The protein-DNA complexes were separated on a 0.75-mm nondenaturing PAGE containing acrylamide/bisacrylamide (20/1) 5% (v/v), 0.5 X TBE. Gels were run at room temperature in 1 X TBE and dried before being autoradiographed.
ChIP
ChIP assays were performed as described previously (16). Neo-Tag cells were either treated with 10−8 M DHT or continued to grow in the androgen-depleted medium. After 24 h of treatment, cells were washed with PBS and cross-linked with 1% formaldehyde at 37 C for 10 min. Cells were scraped into conical tube, pelleted for 4 min at 2000 rpm at 4 C, resuspended in SDS lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8.1), 1x proteinase inhibitor cocktail] for 200 μl per 106 cells, and sonicated with four to five sets of 10-sec pulses at an 80% maximum power (Fisher Sonic Dismembrator, model 50; Fisher Scientific, Pittsburgh, PA). After centrifugation for 10 min, supernatants were collected and diluted 1:10 in ChIP dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), 167 mM NaCl], followed by preclearing for 30 min with 3 μg of sonicated salmon sperm DNA with protein A agarose (80 μl of 50% slurry in 10 mM Tris-HCl, 1 mM EDTA). IP was performed overnight at 4 C with specific antibodies. Protein A agarose (60 μl) with salmon sperm DNA was added for 1 h with rotation to collect the complex. Beads were sequentially washed for 5 min each with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 150 mM NaCl), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 500 mM NaCl), LiCl wash buffer (0.25 M LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl) and twice with TE buffer (10 mM Tris-HCl, 1 mM EDTA). The complex was eluted twice with 250 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and eluates were pooled. The formaldehyde cross-linking was reversed by adding 20 μl 5 M NaCl and incubating for 6–8 h at 65 C. Eluates were incubated for an additional 1 h at 45 C with 2 μl of 10 mg/ml Proteinase K. DNA was extracted using QIAquick Spin Column (Qiagen) and 3–5 μl of extracted DNA was used in each PCR amplification. Primers (TCCCAGTTGGCAGTTGTACACAGA and CACCAACATCTATCTGATTGGAGGA) used encompassed transcription factor binding sites on the large rat PB promoter. ChIP experiments were repeated three times.
RESULTS
Seven Cis-acting DNA Elements within the −286 PB are Critical for PB Gene Expression
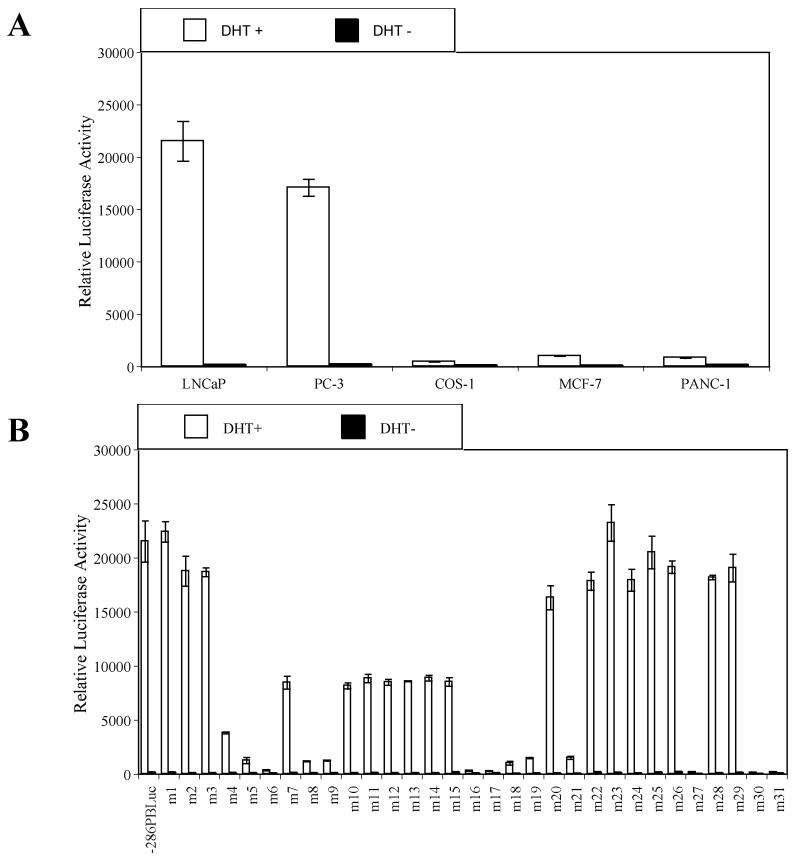
In order to identify cis-acting DNA elements within the proximal PB promoter required for androgen dependent prostate-specific gene expression, we performed a comprehensive linker mutation analysis with the sequential introduction of 10 bp (5′-CTCGAGTACT-3′) replacement mutations covering the entire −286/+28 PB sequences. Thirty-one PB mutants (m1-m31) were generated. Individual PB mutants and a wild type (wt) −286/+28 PB control were subcloned into the pGL3-basic vector. The resulting reporter constructs were co-transfected with a rat AR expression vector (rAR) into prostatic (LNCaP and PC-3), and non-prostatic (COS-1, MCF-7, and PANC-1) cell lines, and luciferase activity was measured in the presence and absence of dihydrotestosterone (DHT). DHT induction of the wt − 286/+28 PB promoter activities was approximately 135-fold and 83-fold in prostatic LNCaP and PC-3 cells, respectively, whereas DHT induced the same wt PB promoter in non-prostatic COS-1, MCF-7 and PANC-1 was only 3.7-fold, 10.2-fold, and 5.5-fold, respectively (Fig. 1 A).
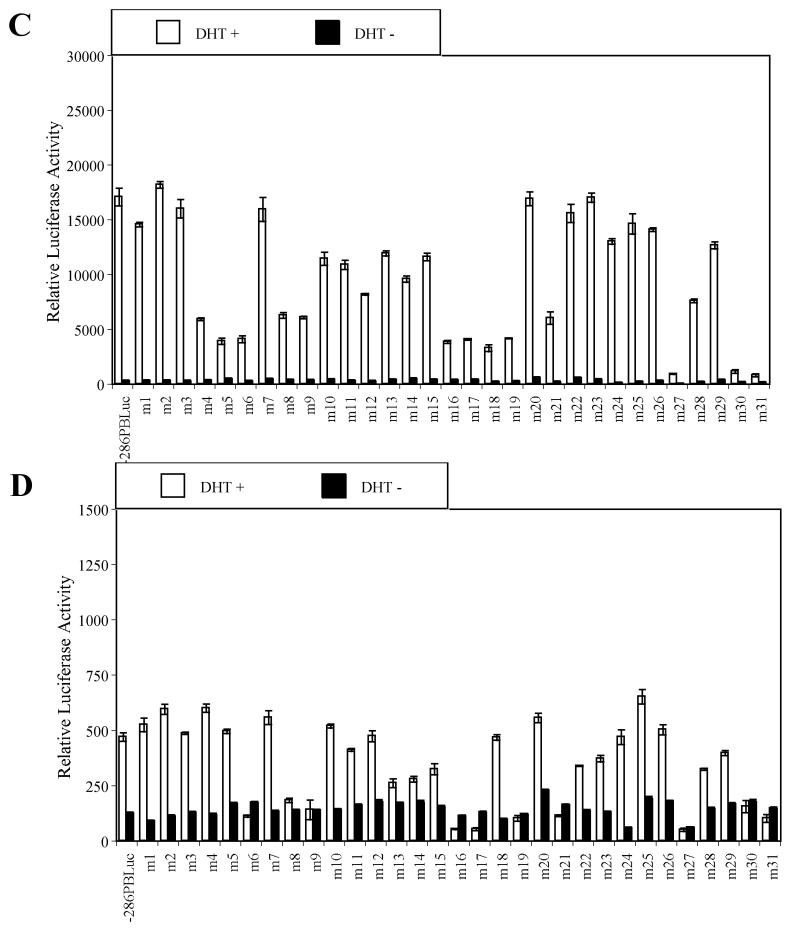
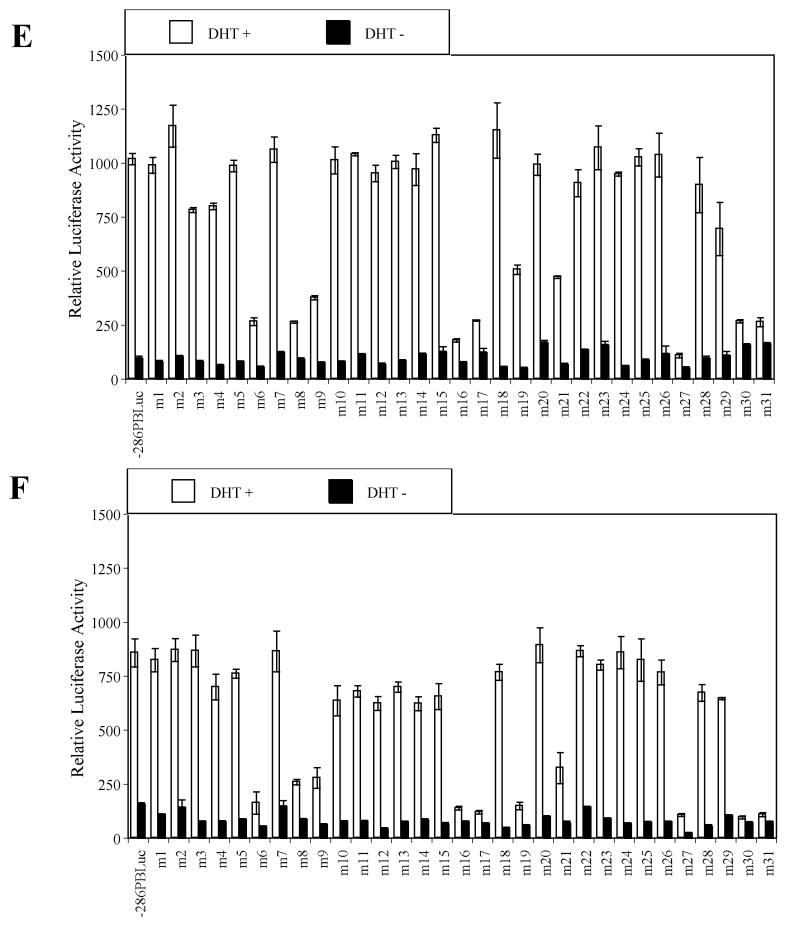
Fig. 1.
Identification of key cis-acting DNA elements within the –286PB promoter. Prostatic cell lines and non-prostatic cell lines were tranfected with the –286PB Luc reporter constructs together with the AR and Renilla luciferase expression vectors and relative Luc activities were determined in the absence or presence of 10−8 M DHT as described in Materials and Methods. The reporter gene activities were measured in at least three determinations ± SD, and all experiments were adjusted for transfection efficiency. The luciferase activities are expressed as relative luciferase light units/min/μg protein. (A), the wild type −286PB promoter activities on different cells are shown. The reporter gene activities of the wild type and mutant – 286PB promoters are shown in (B), LNCaP cells; (C), PC-3 cells; (D), COS-1 cells; (E), MCF-7 cells; and (F), PANC-1 cells.
As ligand-bound AR binding to ARBS-1 and ARBS-2 is essential for −286PB Luc promoter activity following androgen treatment, reporter activity of m6 (−236/−227 bp), m16 (−136/−127 bp), m17 (−127/−117 bp) mutant constructs was almost completely abolished in all tested cell lines (Fig. 1B-F), and the same was true for the m27 (−26/−17 bp) mutant construct. Very low promoter activity was detected in both prostatic and non-prostatic cell lines transfected with m30 (+5/+14 bp) and m31 (+15/+24 bp) mutant constructs, likely due to the disruption of the transcription initiation site (Fig. 1B-F).
Importantly, the linker mutation analysis revealed that the promoter activities of m8 (−216/−207 bp), m9 (−206/−197 bp), m19 (−106/−97 bp), and m21 (−86/−77 bp) mutant constructs was dramatically reduced in all cell lines tested (Fig. 1B-F). In contrast to the mutations that affected PB promoter activity in all tested cell lines, mutations in Androgen Receptor Binding Site-1 (ARBS-1) or ARBS-2 suppressed promoter activity to around 40% of the wt control in LNCaP cells, but had less effect in other cells (Fig. 1B-F). Most intriguingly, the promoter activities of m4 (−256/−247 bp), m5 (−246/−237 bp), and m18 (−116/−107 bp) were decreased to 19.6%, 6.8%, and 4.7% in LNCaP cells and 40.6%, 28.7%, and 22.2 % in PC-3 cells, however these mutations had almost no effect in non-prostatic cells (Fig. 1B-F). In summary, these findings indicate that regions within the PB promoter altered in m8 (−216/−207 bp), m9 (−206/−197 bp), m19 (−106/−97 bp), and m21 (−86/−77 bp) constructs contain non cell specific, essential cis-regulatory sequences, while regions altered in the m4 (−256/−247 bp), m5 (−246/−237 bp), and m18 (−116/−107 bp), as well as the previously identified AR binding sites ARBS-1 (−236/−225) or ARBS-2 (−135/−121) contain prostate-specific cis-regulatory elements.
Oct-1, C-jun, and NFI Bind to the −286 PB Promoter
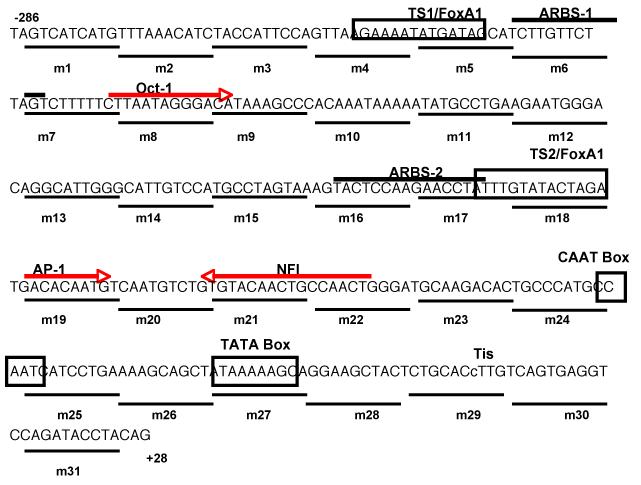
In order to identify potential trans-acting proteins that bind to PB regulatory sequence, transcription factor database searches were performed using web-based search engines TESS (www.cbil.upenn.edu/tess) and TRANSFAC (www.cbrc.jp/research/db/TFSEARCH.html). Putative binding sites for Oct-1 (−218 to − 206 bp), AP-1 (−108 to −96 bp) and NFI (−88 to −72 bp) were predicted to be contained in these key cis-acting elements (Fig. 2). To determine if transcription factor binding site sequences for Oct-1, c-jun (AP-1), and NFI are functional, site-directed mutagenesis of the wt −286 PB Luc reporter construct was used to disable the Oct-1, c-jun, and NFI binding sites by generating mt-Oct-1, mt-c-jun, and mt-NFI reporter plasmids (Fig. 3A). These mutants can no longer bind Oct-1, c-jun, and NFI as confirmed in EMSA assays (see Fig. 4). Co-transfection of the individual mutant with a rAR expression vector into LNCaP cells followed by DHT treatment revealed reporter activity of the mutant-Oct-1, c-jun, and NFI reporter plasmids was 11.8%, 15.9%, and 13.8% compared to the wild-type −286 PB Luc control (Fig. 3B). These observations demonstrate that specific Oct-1, c-jun, and NFI binding sites are important for full −286 PB promoter activity.
Fig. 2.
The locations of the key cis-acting DNA elements within –286PB are shown. TS1, TS2, TATA box, and a putative CATT box are boxed. The potential Oct-1, C-jun, and NF-1 binding sites in –286PB promoter are marked with big arrows. The directions of the arrows indicate the sense or the antisense strand that matched the consensus sequence. ARBS-1 and ARBS-2 are up-lined. The lowercase c (small arrow) is the transcription initiation site (Tis) of the PB gene.
Fig. 3.
Mutational analysis of Oct-1, C-jun, and NFI binding sites within the –286PB promoter. (A), Wild type PB promoter sequences containing underlined putative Oct-1, C-jun, and NFI motifs were mutated (uppercase characters) as indicated in mtOct-1, mtC-jun, and mtNFI. The consensus Oct-1, C-jun, and NFI sequences are shown as cOct-1, cC-jun, and cNFI. (B), LNCaP cells were transiently transfected with wt –286PBLuc, mtOct-1, mtC-jun, or mtNFI reporter constructs together with the AR and Renilla luciferase expression vectors and relative Luc activities were determined in the absence or presence of 10−8 M DHT as described in Materials and Methods. The reporter gene activities were measured in at least three determinations ± SD, and all experiments were adjusted for transfection efficiency. The luciferase activities are expressed as relative luciferase light units/min/μg protein.
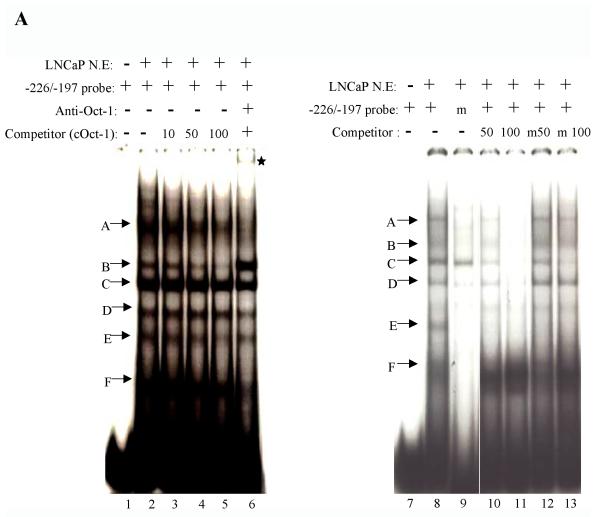
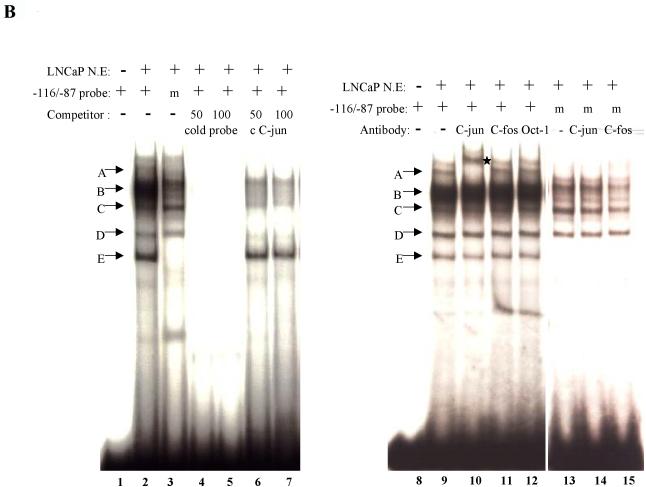
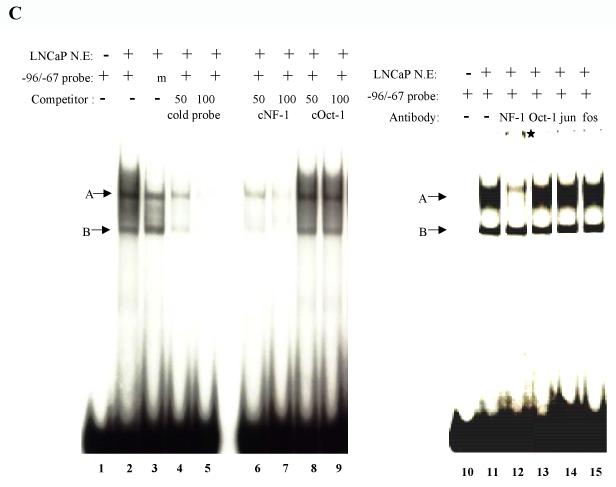
Fig. 4.
Characterization of the putative Oct-1, C-jun, and NFI binding sites within the –286PB promoter. (A), LNCaP nuclear extracts (10 μg) were incubated with radiolabeled probes corresponding to the wild type or mutated Oct-1 binding sites (−226/−197 bp) in the absence or presence of unlabeled competitor oligonucleotides. The competitors are consensus Oct-1 fragment, wild type Oct-1 binding site, and mutant version of the Oct-1 binding site. Arrows indicate the specific protein-DNA complexes. Complexes A, D, E and F were specifically eliminated or reduced; however, the complex B was enhanced when anti-Oct-1 antibody was added into reaction. Asterisk indicates a supershifted band with anti-Oct-1 antibody. (B), LNCaP nuclear extracts (10 μg) were incubated with radiolabeled probes corresponding to the wild type or mutated C-jun binding sites (−116/−87 bp) in the absence or presence of unlabeled competitor oligonucleotides. The competitors are consensus C-jun fragment, wild type C-jun binding site. Arrows indicate the specific protein-DNA complexes. Complexes A, B, C and D are specifically eliminated or reduced by a consensus C-jun competitor. The protein-DNA complex A was supershifted upon addition of anti-C-jun antibody. Asterisk indicates the supershifted band with anti-C-jun antibody. (C), LNCaP nuclear extracts (10 μg) were incubated with radiolabeled probes corresponding to the wild type or mutated NFI binding sites (−96/−67 bp) in the absence or presence of unlabeled competitor oligonucleotides. The competitors are unlabeled consensus NFI, consensus Oct-1, wt NFI binding site. Two specific protein-DNA complexes are marked as A and B. Both complexes A and B are inhibited upon addition of unlabeled consensus NFI probe. The complex A was supershifted when anti-NFI antibody was included in the reaction. Asterisk indicates a supershifted band with anti-NFI antibody.
Since the PB reporter is highly expressed in transient transfection assays in LNCaP cells, and regions identified to potentially bind Oct-1, c-jun, and NFI are important for robust PB reporter activity, the putative Oct-1, c-jun, and NFI binding sites within the −286/+28 PB were examined using nuclear extracts on EMSA. As shown in Fig. 4A, the labeled −226 to −197 bp probe produced several protein-DNA complexes (lanes 2 and 8), while a mutant −226 to −197 bp probe (the putative Oct-1 site that was specifically mutated) only formed one protein-DNA complex (lane 9). Protein-DNA complexes termed A, B, and E is weaker or is abolished when an unlabeled consensus Oct-1 oligonucleotide was used as a competitor against the wt probe (Fig. 4A lanes 3-5). All protein-DNA complexes were competed by a 100-fold molar excess of the unlabeled −226 to −197 bp oligonucleotides whereas only complex C and E were competed by 100-fold molar excess of the unlabeled mutated −226 to −197 bp oligonucleotides as a competitor (Fig. 4A lanes 11 and 13). When an anti-Oct-1 antibody was included in the binding reaction, a super shift band was observed with a disruption in complex A. Complex D, E, and F became weaker in intensity, while complex B was enhanced (Fig. 4A, lane 6). Reversely, the −226 to −197 bp cold oligonucleotide effectively competed with the binding of radiolabeled Oct-1 consensus sites in EMSA studies (data not shown). Taken together, these results demonstrated that PB promoter contains a cis-regulatory element for Oct-1 at −226 to −197 bp region.
When a radiolabeled −116 to −87 bp probe, containing the putative c-jun binding site, was incubated with the LNCaP nuclear extracts, several protein-DNA complexes with different mobility were detected (Fig. 4B, lane 2). The binding was reduced or abolished when a radiolabeled mutated −116 to −87 bp probe is incubated with the LNCaP nuclear extracts (Fig. 4B, lane 3). The specificity of protein-DNA interaction was ascertained using competition assay. The shifted bands were abrogated when 50 to 100-fold molar excess unlabeled −116 to −87 bp probe was added to the reaction as a competitor (Fig. 4B, lanes 4 and 5). When cold consensus c-jun oligonucleotides were used as competitors, three complexes were abolished (Fig. 4B, lane 6 and 7). In super shift assay, the anti-c-jun antibody resulted in super-shifted band on the gel (Fig. 4B, lane 10) while no effect was detected when anti-c-fos or anti-Oct-1 antibodies was used (lanes 11 and 12). Furthermore, no super shift was observed when the radiolabeled mutated −116 to −87 bp probe was incubated with LNCaP nuclear extracts (Fig. 4B, lanes 14 and 15). These results demonstrate that −116 to − 87 bp contained an AP-1 binding site.
Incubation of a radiolabeled probe comprising the sequence from −96 to −67 bp with LNCaP nuclear extracts resulted in the appearance of two specific labeled bands A and B (Fig. 4C, lane 2), while when a mutated −96 to −67 bp as a probe, the complex A was reduced while complex B was maintained (Fig. 4C, lanes 2 and 3). Excess of cold −96 to −67 bp sequences competed off the binding of the complexes (Fig. 4C, lanes 4 and 5). The binding of nuclear proteins to the −96 to −67 bp sequence was challenged with excess cold consensus NFI and consensus Oct-1 oligonucleotides. Competition was observed only for the cold NFI oligonucleotides (Fig. 4C, lanes 6 and 7), but not for the Oct-1 oligonucleotides (lanes 8 and 9). To further define the nature of the protein within these protein-DNA complexes, we performed super-shift EMSA. The NFI antibody shifted the complex A to higher region in the gel with a drastic reduction in the original band (Fig. 4C, lane 12); while no effect was observed for anti-Oct-1, anti-C-jun, and anti-C-fos antibodies. (Fig. 4C, lanes 13-16).
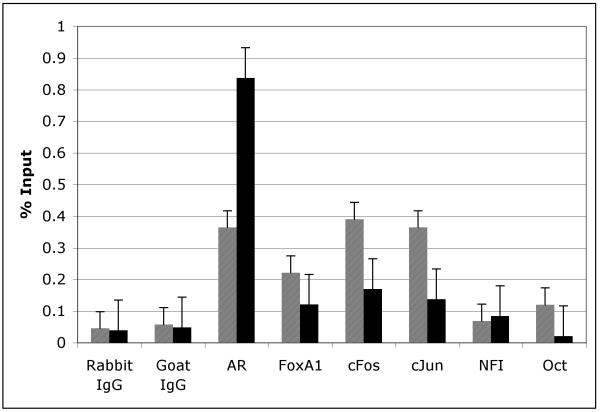
A new mouse prostate cell line, NeoTag, expresses the SV40 Large T antigen driven by a rat PB promoter. This cell line allowed us to perform ChIP assays to investigate the in vivo association of FoxA1, c-Jun, c-Fos, Oct-1, and NFI with the −286 PB promoter in both the presence and absence of DHT (Fig. 5). By ChIP analysis, FoxA1 is constitutively bound and c-Jun, c-Fos, NFI and Oct-1 are bound to the −286 PB promoter both in the presence and absence of DHT (Fig. 5). Taken together, these studies indicate that the AR, FoxA1 c-Jun, c-Fos, Oct-1 and NFI interact with defined cis-regulatory sequence within the PB promoter in vivo.
Fig. 5.
Cis-binding factors interact with the large probasin promoter in vivo. Neo-Tag cells which contain the endogenous large probasin promoter were treated in the presence or absence of DHT for 24 hours, after which ChIP assays were performed as indicated in materials and methods.
TS1 and TS2 share similar biological function on −286 PB promoter
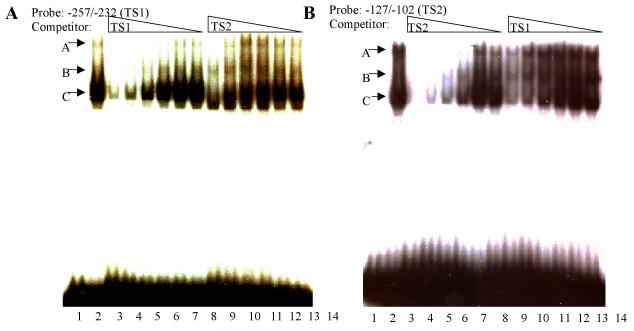
By mutation analysis and testing in cell cultures, m4-m5 and m17-m18 were found to be critical for expression in prostate cells but not in non-prostatic cell lines and the sites were name Tissue Specific 1 and 2 (TS1 and TS2), respectively (Fig. 2). To further characterize these sites, EMSA using nuclear extracts derived from LNCaP cells was conducted using radiolabeled fragments that encompassed the two TS sites: TS1 (probe −257 to −232 bp) or TS2 (probe −127 to −102 bp). As shown in Fig. 6A, three complexes were formed with radiolabeled TS1 in the presence of 20 μg LNCaP nuclear extracts (Fig. 6A, lane 2), as compared to the control in the absence of nuclear extracts (lane 1). Similarly, three complexes were observed when TS2 was used as a probe (Fig. 6B, lane 2). However, a comparison of the highest shifted protein-DNA complex of TS2 (complex A, lane 2, Fig. 6B) appeared as a stronger band compared to that of TS1 (complex A, lane 2, Fig. 6A). The interrelationship between TS1 and TS2 was further explored by comparing their relative binding profiles with the LNCaP nuclear extracts. A fixed amount of the radiolabeled TS1 probe was competed with a series of decreasing concentrations of unlabeled TS1 or TS2 probes in EMSA studies. Cold TS1 (Fig. 6A, lanes 3-8) was a slightly stronger competitor than TS2 in competing labeled TS1 (Fig. 6A, lanes 9-14). When a fixed amount of a radiolabeled TS2 probe incubated with LNCaP nuclear extracts, cold TS2 (Fig. 6B, lanes 3-8) was a slightly stronger competitor than TS1 in competing labeled TS2 (Fig. 6B, lanes 9-14). Most interestingly, these two DNA elements compete reciprocally, suggesting that similar, if not identical, transcription factors bind to both of these tissue-specific elements.
Fig. 6.
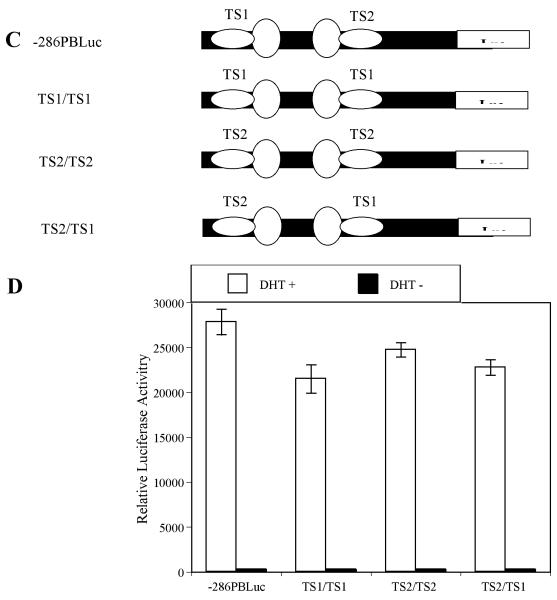
TS1 and TS2 share similar biological function on −286PB promoter. (A) and (B), Similar transcription factors bind to the TS1 and TS2 in LNCaP cells. The radiolabeled PB –257 to –232 bp probe containing the TS1 (A) and PB –127 to –102 bp probe containing the TS2 (B) were incubated with 10 μg of the LNCaP nuclear extracts in the absence or presence of 200×, 100×, 50×, 25×, 12.5× and 6.25× unlabeled TS1 or TS2 competitors. (C), Schematic representation of the wild type – 286PBLuc, TS1/TS1, TS2/TS2, and TS2/TS1 reporter constructs. (D), LNCaP cells were transiently transfected with wild type –286PBLuc, TS1/TS1, TS2/TS2, and TS2/TS1 reporter constructs together with the AR and Renilla luciferase expression vectors and relative Luc activities were determined in the absence or presence of 10−8 M DHT as described in Materials and Methods. The reporter gene activities were measured in at least three determinations ± SD, and all experiments were adjusted for transfection efficiency. The luciferase activities are expressed as relative luciferase light units/min/μg protein.
Since similar protein/DNA binding patterns were observed for TS1 and TS2, we next examined whether these two sites are functionally interchangeable. Thus, using site-directed mutagenesis, we generated mutant constructs where TS2 was replaced with TS1 (Fig. 6C, TS1/TS1), or TS1 was replaced with TS2 (TS2/TS2), or the positions of TS1 and TS2 were switched (TS2/TS1). The resulting mutants and wt control were transiently transfected into LNCaP cells, and luciferase activity was compared. Although overall reporter gene activities of the TS1/TS1, TS2/TS2, and TS2/TS1 are 77.2%, 89.5%, and 81.8% of the wt −286 PB Luc control (TS1/TS2), respectively, switching the TS sites did not significantly decrease the androgen-inductivity of PB promoter (Fig. 6D). These data strongly suggested that TS1 and TS2 are functionally interchangeable and bind similar transcriptions factors.
Prostatic cell-specific transcriptional factor(s) binds to the TS1 and TS2
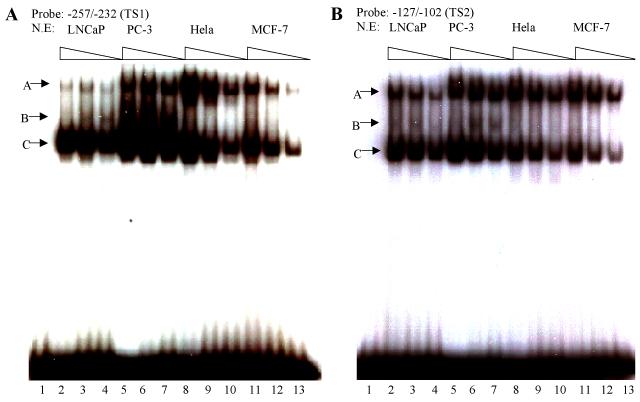
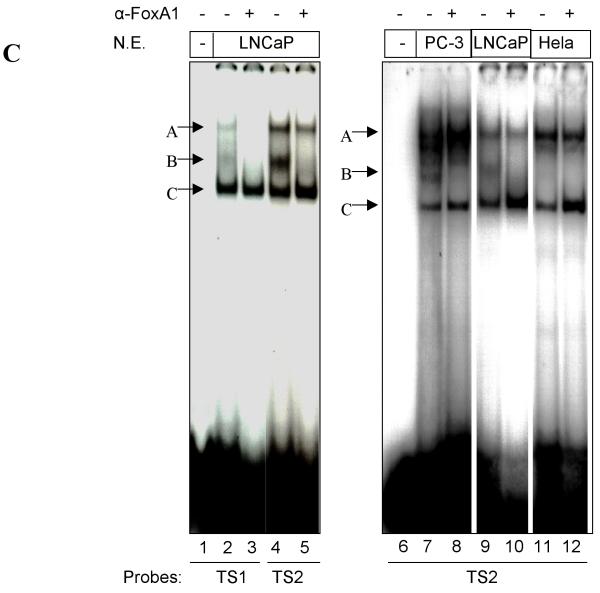
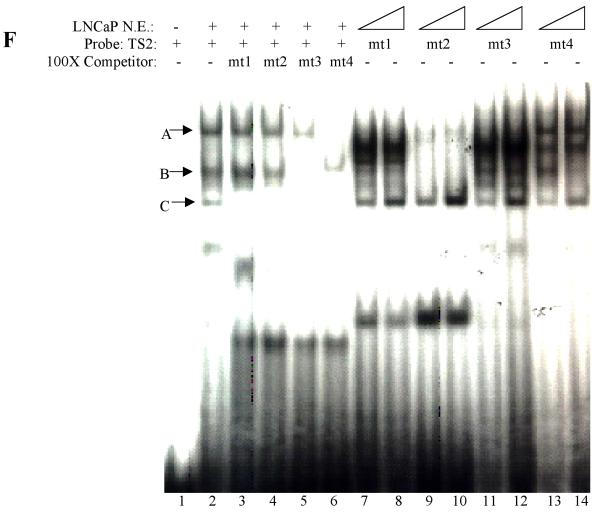
Although prostate-specific transcription factors have not yet been identified, several putative prostate-specific factor DNA binding sites had been described in the Prostatic Binding Protein C3 (PBP C3) gene androgen responsive unit, the PSA enhancer region, the PAP promoter, and in the proximal PB promoter (24-27). To determine the cell-specificity of the proteins that interact with TS1 and TS2, EMSA on radiolabeled TS1 and TS2 was performed by using nuclear proteins extracted from prostatic cell lines (LNCaP and PC-3), and non-prostatic cell lines (Hela, MCF-7). Two protein-DNA complexes (designated as A and C) were detected as binding to TS1 and TS2 in all nuclear extracts tested (Fig. 7A and B) indicating these complexes are not prostate-specific. In contrast, a weak complex (designated as B) was only detected in prostatic LNCaP and PC-3 nuclear extracts but not with non-prostatic Hela and MCF-7 nuclear extracts (Fig. 7A and B), indicating these complexes consist of proteins important for prostate specific TS1 and TS2 activity. EMSA and super-shift assay demonstrated that this transcription factor is FoxA1, as complex B was super-shifted upon the addition of FoxA1 antibody (Fig. 7C).
Fig. 7.
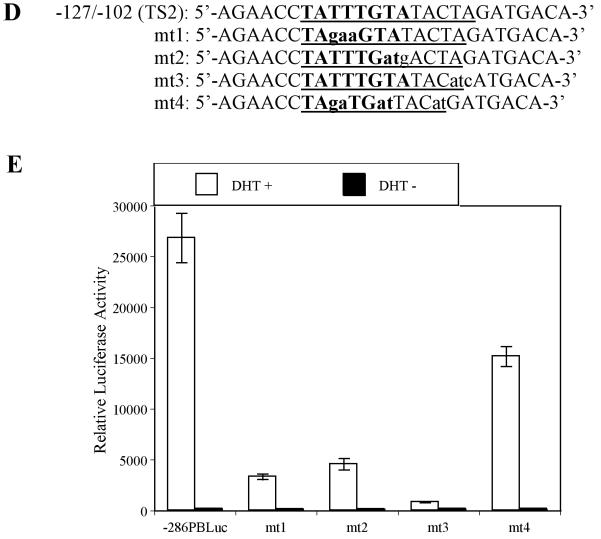
Interaction between cell specific transcription factor(s) and TS1 and TS2 are required for –286PB activation. (A) and (B), Electromobility shift assays of TS1 and TS2 reveal cell specific protein-DNA complexes. The radiolabeled PB –257 to – 232 bp probe containing the TS1 (A) and PB –127 to –102 bp probe containing the TS2 (B) were incubated with 20 μg, 10 μg, and 5 μg of the LNCaP, PC-3, Hela, and MCF-7 nuclear extracts; complexes were resolved on 5% polyacrylimide gels. (C), Radiolabeled TS1 (lanes 2-3) and TS2 probes (lanes 4-5) were used in EMSA with LNCaP nuclear extracts, in the presence (lanes 3 and 5) or absence (lanes 2 and 4) of FoxA1 antibody. Same super shift experiments were performed for PC-3 (lanes 7-8) and Hela cells (lanes 11-12). (D), Wild type PB promoter sequences –127 to –102 bp, containing a FoxA1 motif (bolded) within the TS2 (underlined), was mutated (lowercase characters) as indicated in mt1-mt4. (E), LNCaP cells were transiently transfected with wild type –286PBLuc, and mt1-mt4 reporter constructs together with the AR and Renilla luciferase expression vectors and relative Luc activities were determined in the absence or presence of 10−8 M DHT as described in Materials and Methods. The reporter gene activities were measured in at least three determinations ± SD, and all experiments were adjusted for transfection efficiency. The luciferase activities are expressed as relative luciferase light units/min/μg protein. (F), LNCaP nuclear extracts (10 μg or 20 μg) were incubated with radiolabeled probes corresponding to the wt or mutated TS2 (−127 to −102 bp) in the absence or presence of 100X unlabeled mutated TS2 competitor oligonucleotides; complexes were resolved on 5% polyacrylimide gels.
TS1 and TS2 are required for −286 PB promoter activity in prostatic cells
It had been reported that the promoter activity was significantly reduced in prostatic cells when the TS1 sequences were mutated (16,26,28). To functionally characterize the TS2 element, four TS2 mutant constructs were generated using PCR based site-directed mutagenesis (Fig. 7D). The resulting constructs and wt control were transiently transfected into LNCaP and MCF-7 cells. Luciferase assay revealed that the androgen-induced promoter activities of TS2 mutants were 12.1%, 17.0%, 3.1%, and 52.8% of that measured in wt −286 PB Luc control. This reduction was only observed in LNCaP cells (Fig. 7E) but not in MCF-7 cells (data not shown).
To test whether these TS2 mutations alter the TS2-binding proteins, mutant and wt TS2 were used as probes and were tested in EMSA studies (Fig. 7F). Different binding patterns were observed in these mutant TS2 probes (lanes 7-14) as compared to wt TS2 (lane 2): i) the binding of FoxA1 was abolished for mt1 and mt2 (lanes 7-10); ii) mt3 completely abolished complex A, but not FoxA1 and complex C (lanes 11 and 12); iii) mt4 slightly affected complexes A, B, and C (lanes 13 and 14). Competition assay demonstrated a result consistent with these in vitro binding assays (Fig. 7F, lanes 3-6). Importantly, these results suggested that in addition to FoxA1, unknown transcription factor(s) binds TS2 element.
TS1 and TS2 are Critical in Directing Androgen Dependent Prostate-Specific Gene Expression in Transgenic Mice
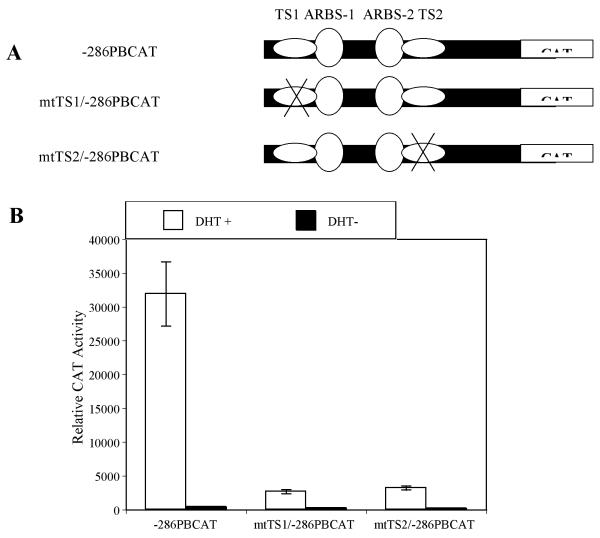
In order to examine the in vivo functional significance of TS1 and TS2, we compared the activities of mutated TS1 and TS2 −286 PB promoter with wt −286 PB promoter using transgenic mice. Previously to test the PB promoter in transgenic mice, the CAT reporter was used (9-11). For that reason the CAT reported was selected for these in vivo studies. PCR based site-directed mutagenesis of the wt − 286 PBCAT was used to generate mtTS1/−286 PBCAT and mtTS2/−286 PBCAT (Fig.8A). The mtTS1 sequence is same as the mt5 used in linker mutation study, while the mtTS2 is same as the mt18 sequence. These constructs were first tested in transient transfection assay before creating transgenic lines. The CAT activities of the mtTS1/−286 PBCAT and mtTS2/−286 PBCAT were only 8.4% and 10.1% of CAT activity of the wt −286 PBCAT in LNCaP cells (Fig. 8B). Three wt −286 PBCAT transgenic founders (2 males and 1 female), 4 mtTS1/−286 PBCAT transgenic founder (2 males and 2 females), and 3 mtTS2/−286 PBCAT transgenic founders (2 males and 1 female) were generated and confirmed later with PCR. As shown in Table 1, CAT activities were detected in the prostate lobes but not in any other tissues in two male wt −286 PBCAT founders. This result strongly demonstrated that −286 PB promoter is sufficient to confer prostatic selection to PB expression. However, low CAT activities were detected in the prostate lobes of one male founder and male offspring of one female founder of the mtTS1/−286 PBCAT, indicting that TS1 site is important, but may not be absolutely required for the prostate-specificity of PB promoter. Importantly, no CAT activity was detected in any prostate lobes from the mtTS2/−286 PBCAT transgenic mice established from three separate founders, suggesting that TS2 site is critical for the prostate-specific activation of the PB promoter. As expected, no CAT activity was found in any other tissues from the mtTS1/−286 PBCAT or mtTS2/−286 PBCAT transgenic mice.
Fig. 8.
Evaluation of androgen-induced activities of the wild type –286PBCAT, mtTS1/−286PBCAT and mtTS2/−286PBCAT constructs in LNCaP cells. (A) Schematic representation of the wild type –286PBCAT, mtTS/1-286PBCAT and mtTS2/−286PBCAT constructs. (B) These constructs were cotransfected with AR and Renilla luciferase expression vectors into LNCaP cells and treated with or without 10−8 M DHT for 24 hours before harvest. Background activities of cell lysates with no DNA transfection were subtracted from the data contained in the experimental group, before the normalization with Renilla luciferase activities. The activity induction in response to hormone treatment was determined by comparing receptor gene activity induced by DHT to the corresponding baseline values in the absence of hormone. Data shown here are a representative from at least three independent experiments in triplicate. Error bars indicate SD values. Results are presented as relative CAT activities dpm/min/mg protein.
Table I.
The CAT activities of the wild type −286PBCAT, mtTS1/−286PBCAT, and mtTS2/−286PBCAT in transgenic mice
| DNA Construct | Founder | Sex | AP | DP | LP | VP |
|---|---|---|---|---|---|---|
| −286PBCAT | A0240 | M | 225 | 1078 | 2863 | 3962 |
| A0451 | F | 0 | 0 | 0 | 0 | |
| A0459 | M | 285 | 837 | 2168 | 4295 | |
|
| ||||||
| mtTS1/−286PBCAT | A0265 | F | 0 | 380 | 846 | 625 |
| A0267 | F | 0 | 0 | 0 | 0 | |
| A0269 | M | 0 | 0 | 0 | 0 | |
| A0279 | M | 154 | 480 | 636 | 1350 | |
|
| ||||||
| mtTS2/−286PBCAT | A0307 | F | 0 | 0 | 0 | 0 |
| A0374 | M | 0 | 0 | 0 | 0 | |
| A0692 | M | 0 | 0 | 0 | 0 | |
The data are represented as CAT activity (dpm/min/mg protein), which was determined in individual male founders. In the female founders, CAT activity was determined in transgenic F1 male offspring. No CAT activity was detected in any other tissue tested. AP=anterior prostate, DP=dorsal Prostate, LP=lateral prostate, VP=ventral prostate.
ARR Sequence (−244 to −96 bp) Contains Sufficient Information for Prostate-Specific Expression of the PB Gene
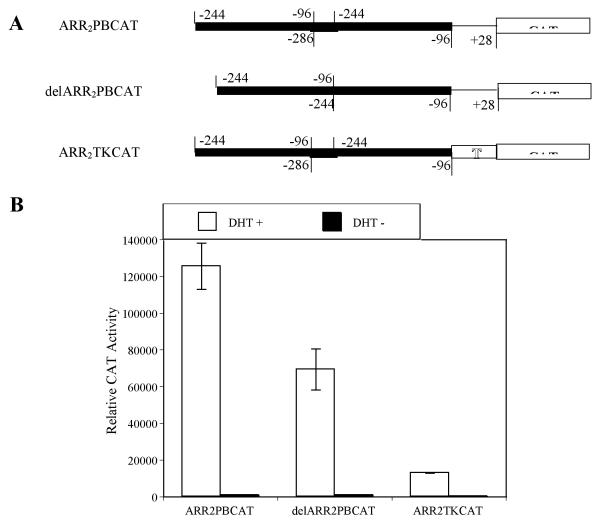
The cis-acting elements involved in androgen dependent prostate-specific PB gene expression were contained within −286 PB promoter as demonstrated in the wt −286 PBCAT transgenic mice studies (Table 1). Deletion analysis has shown that the minimal PB promoter (−53 to +28 bp) does not direct prostatic cell specific expression of a reporter gene in transient transfection assays (unpublished observation). In transgenic mouse studies, the ARR2PB, containing two ARR, was found to be much stronger in prostate-targeting as compared to the proximal PB promoter (11), suggesting that the additional copy of ARR increased the transgene activity in transgenic mice. In order to further narrow down the PB sequence required for prostate-specific expression, two CAT reporter constructs, delARR2PBCAT and ARR2TKCAT (see the material and methods), were used to generate transgenic mice. In the transient transfection assays, the CAT activities of the delARR2PBCAT and ARR2TKCAT were 55.2% and 13.4% of wt ARR2PBCAT in LNCaP cells (Fig. 9). Two ARR2TKCAT transgenic founders (1 males and 1 female) and 7 delARR2PBCAT transgenic founders (6 males and 1 female) were generated and confirmed by PCR. In male offspring, CAT activities were detected in prostate lobes, but not in any other tissues, of the ARR2TKCAT founder and 2 delARR2PBCAT male founders (Table. 2). These data demonstrated that −244 to −96 bp PB promoter is sufficient for the prostate-specific expression of PB gene in transgenic mice.
Fig. 9.
Evaluation of androgen-induced activities of the ARR2PBCAT delARR2PBCAT, and ARR2TKCAT constructs in LNCaP cells. (A) Schematic representation of the ARR2PBCAT delARR2PBCAT, and ARR2TKCAT constructs. (B) These constructs were cotransfected with AR and Renilla luciferase expression vectors into LNCaP cells and treated with or without 10−8 M DHT for 24 hours before harvest. Background activities of cell lysates with no DNA transfection were subtracted from the data contained in the experimental group, before the normalization with Renilla luciferase activities. The activity induction in response to hormone treatment was determined by comparing receptor gene activity induced by DHT to the corresponding baseline values in the absence of hormone. Data shown here is representative for at least three independent experiments in triplicate. Error bars indicate SD values. Results are presented as relative CAT activities dpm/min/mg protein.
Table II.
The CAT activities of the delARR2PBCAT and ARR2TKCAT in transgenic mice
| DNA Construct | Founder | Sex | AP | DP | LP | VP |
|---|---|---|---|---|---|---|
| delARR2PBCAT | A0337 | M | 0 | 0 | 0 | 0 |
| A0338 | M | 0 | 0 | 0 | 0 | |
| A0339 | M | 0 | 0 | 0 | 0 | |
| A0340 | M | 530 | 1824 | 3249 | 2835 | |
| A0344 | M | 842 | 2214 | 22440 | 28062 | |
| A0349 | M | 0 | 0 | 0 | 0 | |
| A0368 | F | 0 | 0 | 0 | 0 | |
|
| ||||||
| ARR2TKCAT | A0421 | M | 493 | 1317 | 8307 | 4925 |
| A0770 | F | 0 | 0 | 0 | 0 | |
The data are represented as CAT activity (dpm/min/mg protein), which was determined in individual male founders. In the female founders, CAT activity was determined in transgenic F1 male offspring. No CAT activity was detected in any other tissue tested. AP=anterior prostate, DP=dorsal Prostate, LP=lateral prostate, VP=ventral prostate.
DISCUSSION
Tissue-specific gene expression is governed by the spatial expression of trans-acting regulatory factors, and the combinatorial action of multiple transcriptional activators and repressors interacting with distinct promoter and enhancer elements (1,3,4,29,30). Transgenic animal studies have determined that the key cis-regulatory elements controlling androgen regulated prostate-specific PB expression were contained within the proximal promoter (11). Using promoter mutagenesis analyses, EMSA assays and transgenic mouse studies, we have performed the most extensive characterization of any regulatory sequence required for tissue specific expression in the murine prostate. Our studies have identified key cis-regulatory sites, including two tissue-specific elements (TS1 and TS2), which are critical for PB tissue-specific activation. Our data demonstrated that the androgen regulated prostate-specific PB expression involves several transcriptional factors (Oct-1, AP-1 and NFI), cell type-limited regulatory proteins (AR and FoxA1) that act in combination, and likely yet to be identified factors.
Functional interactions between Oct-1 and steroid hormone receptors have been well-documented to be crucial for the transcriptional control of steroid hormone-responsive genes (31-35). For instance, Oct-1 is essential for androgen regulated SLP gene expression in vivo (34,35), and is required for mouse mammary tumor virus (MMTV) transactivation (36). Mutagenesis studies demonstrated that the −218/−206 bp DNA fragment in the PB promoter is important for promoter activation in both prostatic and non-prostatic cells. Using EMSA and super shift assays with nuclear extracts from prostate epithelial cell lines, we identified that this element is bound by Oct-1. This result was confirmed by ChIP analysis. The maximal androgen induction of PB promoter in LNCaP cells was greatly reduced when the promoter binding by Oct-1 is disrupted through site-directed DNA mutation. Cross talk between Oct-1 and androgen signaling pathway may preferentially occur through interactions between Oct-1 and AR in a DNA-dependent manner, which leads to the recruitment of SRC-1 (37). It has been documented that Oct-1 can directly interact with steroid receptors through its conserved POU domain (37,38), suggesting a conserved functional mechanism. On the PB promoter, the Oct-1 binding element is located immediately downstream to ARBS-1, suggesting a similarly important functional interaction between Oct-1 and AR holds true for PB gene activation.
The proto-oncoprotein c-jun heterodimerizes with c-fos to form the transcription factor AP-1, which regulates transcription through binding to consensus AP-1 cis-acting elements. Functional cross talk between c-jun and AR has been extensively studied. In a search for interacting partners, AR was found to inhibit the protein-DNA interaction between c-jun and the AP-1 binding element (39). It has been reported that c-jun stimulates transcription via AR through an indirect mechanism, in which c-jun promotes AR-mediated transactivation in the absence of its interaction with c-fos or with the AP-1 DNA site. This positive regulatory effect of c-jun on AR transactivation is dose-dependent and is through a direct interaction with the AR DNA binding domain/hinge region (40). Studies also suggested that c-jun supports AR activity by enhancing AR DNA binding and facilitating the ligand-dependent interaction between AR amino- and carboxyl-terminals (40-42). Thus, c-jun was proposed as a mediator for AR-induced transactivation. However, androgen-induced PSA gene expression was inhibited by 12-O-tetradecanoylphorbol 13-acetate, which elevates AP-1 activity in LNCaP cells. Transient over-expression of c-jun and c-fos also inhibited androgen-induced PSA promoter activity. Reversely, purified AR recombinant proteins also inhibit the formation of c-jun/AP-1 complexes (43). Co-immunoprecipitation experiments suggested such mutual repression of DNA binding is due to a direct interaction between AR and c-jun (43). On the PB promoter, linker mutation, DNA sequence analysis, and ChIP revealed a functional c-jun binding site (−108/−94 bp). This binding site is downstream of ARBS2 and is critical for PB activation, since disruption of c-jun DNA binding resulted in an 84% reduction of total PB androgen-induced activity. Our finding supported the importance of signaling cross talk between androgen and AP-1 pathways in directing PB gene expression, and also suggested that, in the context of PB promoter, the transcriptional activity is modulated by cooperation between AR and c-jun.
The NFI family of proteins (44) are known to be important in regulation of several androgen dependent genes (45,46). NFI footprints were identified in the upstream enhancer of the prostate secretory component gene, and in an androgen response unit located in the first intron of the PBP C1 gene (24,46). The in vivo glucocorticoid receptor (GR) induction of MMTV promoter involves a functional synergism between GR and NFI (47). More recently, an unidentified NFI family member(s) was shown to be essential for both GR induced MMTV expression on naked DNA, and the recruitment of the chromatin remodeling enzyme BRG-1 (48) to regulatory sequence. A recent report (18) indicates that NFI binding sites are significantly associated with FoxA1 binding sites in regions of acetylated DNA, and the fact that knockdown of NFI altered endogenous AR target gene expression suggests an important role for NFI family members in endocrine regulated gene expression and chromatin structure. Linker mutagenesis, ChIP, and DNA sequence analysis revealed a critical NFI binding site (−88/−70 bp) in PB promoter. Functional importance of NFI binding was demonstrated by a mutation in NFI site, which resulted in a significant loss of PB promoter activity. This finding is consistent with a previous report showing that the binding of NFI proteins to PB promoter is androgen independent but prostate cell specific (49). However, our transgenic studies demonstrated that −244 to −96 bp of PB sequence is sufficient for the PB gene prostate-specific expression, suggesting that the promoter binding by NFI proteins is required for optimal PB gene activation in prostate cell or tissues, but not absolutely required for the prostatic specific expression of the PB gene.
Importantly, linker mutation and transgenic studies revealed two tissue-specific DNA elements that are fundamentally important for androgen-regulated prostate-specific PB promoter activation. The transgenic mice experiments demonstrated that mutation of TS1 dramatically decreased the transgene expression in prostate lobes while mutation of TS2 almost completely abolished the transgene activities in the transgenic mouse prostate. Even though the activity of the promoter was reduced, prostate-specific activity was maintained. Although it is possible that the site of transgene integration rather then the TS2 mutation prevented transgene expression, it seems unlikely that transgene integration was responsible for the loss of activity since three separate founder lines were established. No transgene activity was detected in any other tissues either in TS1 and TS2 mutant or the wild type −286PBCAT transgenic mice, suggesting that the PB promoter is tightly controlled by prostate-specific transcriptional regulatory complexes present in the prostatic epithelial cells. These data suggests that TS1 and TS2 work cooperatively to control PB gene expression in the prostate, similar to the cooperatively seen between ARBS-1 and ARBS-2 (50).
Both TS1 and TS2 are adjacently positioned to ARBS-1 and ARBS-2, respectively, suggesting a fixed binding site organization may be conserved due to functional importance. The TS1 and TS2 elements are maintained in the PB promoters including the −10,806/+28 and the −426/+28 promoters that have been successfully utilized to generate transgenic mice (9,10). In addition, the ARR2PB promoter duplicates the ARR region, resulting in four ARBS and three tissue-specific elements (one TS1 plus two TS2) that function cooperatively as a strong prostate-specific promoter in vitro and in vivo (25). Interestingly, TS1 and TS2 are interchangeable in biological function; however, wild type conformation of the TS1 and TS2 is required for the optimal PB activity in prostate cells, suggesting that TS1 and TS2 bind similar but not completely identical transcriptional complexes. We have demonstrated that FoxA1, a forkhead transcription factor, is bound to TS1 and TS2 where it directly interacts with the AR (16,51), reviewed in (51). Using TS1 and TS2 probes and nuclear extracts derived from prostate (LNCaP and PC-3) and non-prostate (MCF-7 and Hela) cell lines, we show by EMSA that differences in electrophoretic mobility are attributable to FoxA1 binding to TS1 and TS2 (Fig. 7C). We have recently shown that FoxA1 is expressed in human and mouse prostate epithelium, and that FoxA1 expression is essential for normal prostate development and differentiation (17,51-53). Specifically, we have previously shown that prostates rescued from FoxA1 KO mice show a severely disrupted ductal pattern resembling primitive epithelial cords, with an expansion of the basal cell population, and a lack of epithelial differentiation and probasin expression, as well as other molecular alterations. Interestingly, a putative prostate-specific binding site, close to a strong ARE, has been reported within the PSA core enhancer, and this putative binding site also can bind FoxA1 (25). Adjacently positioned forkhead and AREs also have been identified in several key regulatory regions of other prostatic genes, suggesting a fundamental role for FoxA1 in prostate-specific gene expression (16). Most intriguingly, a direct interaction between FoxA1 and AR has been observed in these promoters, suggesting that both transcription factors cooperatively participate in the expression of these androgen regulated and prostate-specific genes (17). Further, the DNA binding of FoxA1 has been reported to disrupt nucleosome formation thus relieving chromatin-mediated transcriptional repression (54). However, a TS2 mutant with an intact FoxA1 binding site also showed a significant reduction in PB activity in LNCaP cells, indicating that additional TS2-interacting transcription factors are required for the promoter activation. Treatment with androgen recruits AR and AP1 to the probasin promoter while FoxA1, Oct-1 and NFI are constitutively bound. This suggests that FoxA1, Oct-1, and NFI play an architectural role by modifying local chromatin environment to enhance the binding of ligand bound AR and AP1 resulting in a cooperative stimulation of promoter activity. Thus, we propose that tissue-limited transcription factors such as FoxA1 and AR as well as constitutively expressed factors combine to dynamically regulates prostate-specific gene expression (1).
Through cell culture and transgenic animal studies, we demonstrated that although additional flanking DNA sequences are required for maximal promoter activity, the PB ARR (−244 to −96 bp) is sufficient for prostate-specific gene expression. Thus, the PB ARR defines a realistic target size, within which the organization and interaction of the DNA regulatory elements, along with their corresponding transcription factors, can be used to study the mechanism of androgen-dependent prostate-specific gene expression. The strong activity of the composite promoter ARR2PB demonstrates that multiple copies of the ARR create a powerful androgen regulated and prostate-specific promoter. We demonstrate that the TS1 and TS2 sequences (located within the ARR) are important regulatory regions for prostate specific gene expression. Additional critical transcription factors that bind within the TS sequences still remain to be identified.
Acknowledgments
The authors thank Darlene Hancock for help with manuscript preparation. This work was supported by a grant from NIDDK to RJM (R01-DK55748) and a DOD studentship to QS (W81XWH-07-1-0042), and the Integrated Biological Systems Training in Oncology (1 T32 CA119925 PI Stephen Hann), and the Frances Williams Preston Laboratories of the T.J. Martell Foundation. David DeGraff, Ph.D., was supported by the American Cancer Society Great Lakes Division-Michigan Cancer Research Fund Postdoctoral Fellowship (PF-09-285-01-TBE). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
National Institutes of Health Grant R01-DK055748, American Cancer Society Postdoctoral Fellowship PF-09-285-01-TBE, T32 CA119925, and DOD studentship (W81XWH-07-1-0042)
References
- 1.Matusik RJ, Jin RJ, Sun Q, Wang Y, Yu X, Gupta A, Nandana S, Case TC, Paul M, Mirosevich J, Oottamasathien S, Thomas J. Prostate epithelial cell fate. Differentiation. 2008;76(6):682–698. doi: 10.1111/j.1432-0436.2008.00276.x. [DOI] [PubMed] [Google Scholar]
- 2.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424(6945):147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 3.Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science. 2002;295(5556):813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- 4.Matusik RJ, Jin RJ, Sun Q, Wang YQ, Gupta A, Nandana S, Case TC, Paul M, Mirsevich J, Oottamasathien S, Thomas J. Prostate Epithelial Cell Fate. Differentiation. 2008 doi: 10.1111/j.1432-0436.2008.00276.x. In press. [DOI] [PubMed] [Google Scholar]
- 5.Heinlein CA, Chang C. Androgen receptor (AR) coregulators: an overview. Endocr Rev. 2002;23(2):175–200. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 6.Barrios R, Lebovitz R, A.L. W, D.L. W, Matusik R, DeMayo F, M.W. L. RasT24 driven by a probasin promoter induces prostatic hyperplasia in transgenic mice. Transgenics. 1996;2:23–28. [Google Scholar]
- 7.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, Cunha GR, Donjacour AA, Matusik RJ, Rosen JM. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92(8):3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasper S, Sheppard PC, Yan Y, Pettigrew N, Borowsky AD, Prins GS, Dodd JG, Duckworth ML, Matusik RJ. Development, progression, and androgen-dependence of prostate tumors in probasin-large T antigen transgenic mice: a model for prostate cancer. Lab Invest. 1998;78(3):319–333. [PubMed] [Google Scholar]
- 9.Greenberg NM, DeMayo FJ, Sheppard PC, Barrios R, Lebovitz R, Finegold M, Angelopoulou R, Dodd JG, Duckworth ML, Rosen JM, et al. The rat probasin gene promoter directs hormonally and developmentally regulated expression of a heterologous gene specifically to the prostate in transgenic mice. Mol Endocrinol. 1994;8(2):230–239. doi: 10.1210/mend.8.2.8170479. [DOI] [PubMed] [Google Scholar]
- 10.Yan Y, Sheppard PC, Kasper S, Lin L, Hoare S, Kapoor A, Dodd JG, Duckworth ML, Matusik RJ. Large fragment of the probasin promoter targets high levels of transgene expression to the prostate of transgenic mice. Prostate. 1997;32(2):129–139. doi: 10.1002/(sici)1097-0045(19970701)32:2<129::aid-pros8>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Thomas TZ, Kasper S, Matusik RJ. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology. 2000;141(12):4698–4710. doi: 10.1210/endo.141.12.7837. [DOI] [PubMed] [Google Scholar]
- 12.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4(3):223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 13.Klezovitch O, Chevillet J, Mirosevich J, Roberts RL, Matusik RJ, Vasioukhin V. Hepsin promotes prostate cancer progression and metastasis. Cancer Cell. 2004;6(2):185–195. doi: 10.1016/j.ccr.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, Thomas GV, Li G, Roy-Burman P, Nelson PS, Liu X, Wu H. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4(3):209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 15.Wu CT, Altuwaijri S, Ricke WA, Huang SP, Yeh S, Zhang C, Niu Y, Tsai MY, Chang C. Increased prostate cell proliferation and loss of cell differentiation in mice lacking prostate epithelial androgen receptor. Proc Natl Acad Sci U S A. 2007;104(31):12679–12684. doi: 10.1073/pnas.0704940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao N, Zhang J, Rao MA, Case TC, Mirosevich J, Wang Y, Jin R, Gupta A, Rennie PS, Matusik RJ. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol Endocrinol. 2003;17(8):1484–1507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- 17.Gao N, Ishii K, Mirosevich J, Kuwajima S, Oppenheimer SR, Roberts RL, Jiang M, Yu X, Shappell SB, Caprioli RM, Stoffel M, Hayward SW, Matusik RJ. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132(15):3431–3443. doi: 10.1242/dev.01917. [DOI] [PubMed] [Google Scholar]
- 18.Jia L, Berman BP, Jariwala U, Yan X, Cogan JP, Walters A, Chen T, Buchanan G, Frenkel B, Coetzee GA. Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS ONE. 2008;3(11):e3645. doi: 10.1371/journal.pone.0003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132(6):958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, Przybyla AE. An efficient site-directed mutagenesis method based on PCR. Biotechniques. 1994;17(4):657–659. [PubMed] [Google Scholar]
- 21.Farla P, Hersmus R, Trapman J, Houtsmuller AB. Antiandrogens prevent stable DNA-binding of the androgen receptor. J Cell Sci. 2005;118(Pt 18):4187–4198. doi: 10.1242/jcs.02546. [DOI] [PubMed] [Google Scholar]
- 22.Rennie PS, Bruchovsky N, Leco KJ, Sheppard PC, McQueen SA, Cheng H, Snoek R, Hamel A, Bock ME, MacDonald BS, et al. Characterization of two cis-acting DNA elements involved in the androgen regulation of the probasin gene. Mol Endocrinol. 1993;7(1):23–36. doi: 10.1210/mend.7.1.8446105. [DOI] [PubMed] [Google Scholar]
- 23.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Celis L, Claessens F, Peeters B, Heyns W, Verhoeven G, Rombauts W. Proteins interacting with an androgen-responsive unit in the C3(1) gene intron. Mol Cell Endocrinol. 1993;94(2):165–172. doi: 10.1016/0303-7207(93)90165-g. [DOI] [PubMed] [Google Scholar]
- 25.Farmer G, Connolly ES, Jr., Mocco J, Freedman LP. Molecular analysis of the prostate-specific antigen upstream gene enhancer. Prostate. 2001;46(1):76–85. doi: 10.1002/1097-0045(200101)46:1<76::aid-pros1011>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Patrikainen L, Shan J, Porvari K, Vihko P. Identification of the deoxyribonucleic acid-binding site of a regulatory protein involved in prostate-specific and androgen receptor-dependent gene expression. Endocrinology. 1999;140(5):2063–2070. doi: 10.1210/endo.140.5.6696. [DOI] [PubMed] [Google Scholar]
- 27.Zelivianski S, Igawa T, Lim S, Taylor R, Lin MF. Identification and characterization of regulatory elements of the human prostatic acid phosphatase promoter. Oncogene. 2002;21(23):3696–3705. doi: 10.1038/sj.onc.1205471. [DOI] [PubMed] [Google Scholar]
- 28.Kivinen A, Patrikainen L, Kurkela R, Porvari K, Vihko P. USF2 is connected to GAAAATATGATA element and associates with androgen receptor-dependent transcriptional regulation in prostate. Prostate. 2004;59(2):190–202. doi: 10.1002/pros.20015. [DOI] [PubMed] [Google Scholar]
- 29.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92(1):5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 30.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 31.Chandran UR, Warren BS, Baumann CT, Hager GL, DeFranco DB. The glucocorticoid receptor is tethered to DNA-bound Oct-1 at the mouse gonadotropin-releasing hormone distal negative glucocorticoid response element. J Biol Chem. 1999;274(4):2372–2378. doi: 10.1074/jbc.274.4.2372. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez MI, Tovaglieri A, Robins DM. Androgen receptor interactions with Oct-1 and Brn-1 are physically and functionally distinct. Mol Cell Endocrinol. 2002;190(1-2):39–49. doi: 10.1016/s0303-7207(02)00035-7. [DOI] [PubMed] [Google Scholar]
- 33.Kutoh E, Stromstedt PE, Poellinger L. Functional interference between the ubiquitous and constitutive octamer transcription factor 1 (OTF-1) and the glucocorticoid receptor by direct protein-protein interaction involving the homeo subdomain of OTF-1. Mol Cell Biol. 1992;12(11):4960–4969. doi: 10.1128/mcb.12.11.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scarlett CO, Robins DM. In vivo footprinting of an androgen-dependent enhancer reveals an accessory element integral to hormonal response. Mol Endocrinol. 1995;9(4):413–423. doi: 10.1210/mend.9.4.7659085. [DOI] [PubMed] [Google Scholar]
- 35.Scheller A, Scheinman RI, Thompson E, Scarlett CO, Robins DM. Contextual dependence of steroid receptor function on an androgen-responsive enhancer. Mol Cell Endocrinol. 1996;121(1):75–86. doi: 10.1016/0303-7207(96)03854-3. [DOI] [PubMed] [Google Scholar]
- 36.Prefontaine GG, Lemieux ME, Giffin W, Schild-Poulter C, Pope L, LaCasse E, Walker P, Hache RJ. Recruitment of octamer transcription factors to DNA by glucocorticoid receptor. Mol Cell Biol. 1998;18(6):3416–3430. doi: 10.1128/mcb.18.6.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez MI, Robins DM. Oct-1 preferentially interacts with androgen receptor in a DNA-dependent manner that facilitates recruitment of SRC-1. J Biol Chem. 2001;276(9):6420–6428. doi: 10.1074/jbc.M008689200. [DOI] [PubMed] [Google Scholar]
- 38.Prefontaine GG, Walther R, Giffin W, Lemieux ME, Pope L, Hache RJ. Selective binding of steroid hormone receptors to octamer transcription factors determines transcriptional synergism at the mouse mammary tumor virus promoter. J Biol Chem. 1999;274(38):26713–26719. doi: 10.1074/jbc.274.38.26713. [DOI] [PubMed] [Google Scholar]
- 39.Kallio PJ, Poukka H, Moilanen A, Janne OA, Palvimo JJ. Androgen receptor-mediated transcriptional regulation in the absence of direct interaction with a specific DNA element. Mol Endocrinol. 1995;9(8):1017–1028. doi: 10.1210/mend.9.8.7476976. [DOI] [PubMed] [Google Scholar]
- 40.Bubulya A, Wise SC, Shen XQ, Burmeister LA, Shemshedini L. c-Jun can mediate androgen receptor-induced transactivation. J Biol Chem. 1996;271(40):24583–24589. doi: 10.1074/jbc.271.40.24583. [DOI] [PubMed] [Google Scholar]
- 41.Bubulya A, Chen SY, Fisher CJ, Zheng Z, Shen XQ, Shemshedini L. c-Jun potentiates the functional interaction between the amino and carboxyl termini of the androgen receptor. J Biol Chem. 2001;276(48):44704–44711. doi: 10.1074/jbc.M107346200. [DOI] [PubMed] [Google Scholar]
- 42.Wise SC, Burmeister LA, Zhou XF, Bubulya A, Oberfield JL, Birrer MJ, Shemshedini L. Identification of domains of c-Jun mediating androgen receptor transactivation. Oncogene. 1998;16(15):2001–2009. doi: 10.1038/sj.onc.1201697. [DOI] [PubMed] [Google Scholar]
- 43.Sato N, Sadar MD, Bruchovsky N, Saatcioglu F, Rennie PS, Sato S, Lange PH, Gleave ME. Androgenic induction of prostate-specific antigen gene is repressed by protein-protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J Biol Chem. 1997;272(28):17485–17494. doi: 10.1074/jbc.272.28.17485. [DOI] [PubMed] [Google Scholar]
- 44.Murtagh J, Martin F, Gronostajski RM. The Nuclear Factor I (NFI) gene family in mammary gland development and function. J Mammary Gland Biol Neoplasia. 2003;8(2):241–254. doi: 10.1023/a:1025909109843. [DOI] [PubMed] [Google Scholar]
- 45.Darne CH, Morel L, Claessens F, Manin M, Fabre S, Veyssiere G, Rombauts W, Jean CL. Ubiquitous transcription factors NF1 and Sp1 are involved in the androgen activation of the mouse vas deferens protein promoter. Mol Cell Endocrinol. 1997;132(1-2):13–23. doi: 10.1016/s0303-7207(97)00116-0. [DOI] [PubMed] [Google Scholar]
- 46.Verrijdt G, Schoenmakers E, Alen P, Haelens A, Peeters B, Rombauts W, Claessens F. Androgen specificity of a response unit upstream of the human secretory component gene is mediated by differential receptor binding to an essential androgen response element. Mol Endocrinol. 1999;13(9):1558–1570. doi: 10.1210/mend.13.9.0347. [DOI] [PubMed] [Google Scholar]
- 47.Chavez S, Beato M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci U S A. 1997;94(7):2885–2890. doi: 10.1073/pnas.94.7.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hebbar PB, Archer TK. Nuclear factor 1 is required for both hormone-dependent chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter. Mol Cell Biol. 2003;23(3):887–898. doi: 10.1128/MCB.23.3.887-898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeung LH, Read JT, Sorenson P, Nelson CC, Jia W, Rennie PS. Identification and characterization of a prostate-specific androgen-independent protein-binding site in the probasin promoter. Biochem J. 2003;371(Pt 3):843–855. doi: 10.1042/BJ20021816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasper S, Rennie PS, Bruchovsky N, Sheppard PC, Cheng H, Lin L, Shiu RP, Snoek R, Matusik RJ. Cooperative binding of androgen receptors to two DNA sequences is required for androgen induction of the probasin gene. J Biol Chem. 1994;269(50):31763–31769. [PubMed] [Google Scholar]
- 51.DeGraff DJ, Yu X, Sun Q, Mirosevich J, Jin R, Wang Y, Gupta A, Nandana S, Case T, Paul M, Hong-Ying H, Shapiro E, Logan S, Suzuki K, Orgebin-Crist MC, Matusik R. The role of Foxa proteins in the regulation of androgen receptor activity. In: Tindall DJ, Mohler JL, editors. Androgen Action in Prostate Cancer. 2008. [Google Scholar]
- 52.Mirosevich J, Gao N, Gupta A, Shappell SB, Jove R, Matusik RJ. Expression and role of Foxa proteins in prostate cancer. Prostate. 2006;66(10):1013–1028. doi: 10.1002/pros.20299. [DOI] [PubMed] [Google Scholar]
- 53.Mirosevich J, Gao N, Matusik RJ. Expression of Foxa transcription factors in the developing and adult murine prostate. Prostate. 2005;62(4):339–352. doi: 10.1002/pros.20131. [DOI] [PubMed] [Google Scholar]
- 54.Crowe AJ, Sang L, Li KK, Lee KC, Spear BT, Barton MC. Hepatocyte nuclear factor 3 relieves chromatin-mediated repression of the alpha-fetoprotein gene. J Biol Chem. 1999;274(35):25113–25120. doi: 10.1074/jbc.274.35.25113. [DOI] [PubMed] [Google Scholar]