Abstract
A major challenge in allogeneic bone marrow (BM) transplantation is overcoming engraftment resistance to avoid the clinical problem of graft rejection. Identifying gene pathways that regulate BM engraftment may reveal molecular targets for overcoming engraftment barriers. Previously, we developed a mouse model of BM transplantation that utilizes recipient conditioning with non-myeloablative total body irradiation (TBI). We defined TBI doses that lead to graft rejection, that conversely are permissive for engraftment, and mouse strain variation with regards to the permissive TBI dose. We now report gene expression analysis, using Agilent Mouse 8x60K microarrays, in spleens of mice conditioned with varied TBI doses for correlation to the expected engraftment phenotype. The spleens of mice given engrafting doses of TBI, compared with non-engrafting TBI doses, demonstrated substantially broader gene expression changes, significant at the multiple testing-corrected P < .05 level and with fold change ≥ 2. Functional analysis revealed significant enrichment for a down-regulated canonical pathway involving B-cell development. Genes enriched in this pathway suggest that suppressing donor antigen processing and presentation may be pivotal effects conferred by TBI to enable engraftment. Regardless of TBI dose and recipient mouse strain, pervasive genomic changes related to inflammation was observed and reflected by significant enrichment for canonical pathways and association with upstream regulators. These gene expression changes suggest that macrophage and complement pathways may be targeted to overcome engraftment barriers. These exploratory results highlight gene pathways that may be important in mediating BM engraftment resistance.
Keywords: Bone marrow, graft rejection, gene expression
INTRODUCTION
Transplantation of allogeneic bone marrow (BM) and peripheral blood stem cells is curative for many hematologic malignancies and non-malignant disorders of hematopoiesis (Gluckman 2009; Thomas and Blume 1999). Pre-transplant recipient conditioning with chemotherapy or irradiation is required, however, to avoid the problem of graft rejection (Barrett 2000). Many preparative regimens in clinical use are effective for achieving donor engraftment, but have numerous toxicities and contribute substantially to post-transplant morbidity and mortality (Bacher et al. 2009). Thus, understanding barriers that resist BM engraftment is central to successful allogeneic hematopoietic cell transplantation.
These barriers are difficult to study because multiple cell types, molecular pathways, and resistance mechanisms act in concert to reject donor BM cells. Both host natural killer cells and T-cell lymphocytes can mediate immune resistance to engraftment (Shizuru et al. 2010). Elimination of donor BM by these immune cells rely on cell killing mechanisms that can involve granzyme, perforin, or Fas ligand-mediated death pathways (Zimmerman et al. 2005a). Key cytolytic pathways have not been identified, however, because using targeted knockout rodent models to neutralize each one at a time, or even several simultaneously, fail to eliminate engraftment barriers (Baker et al. 1995; Komatsu et al. 2003; Scheffold et al. 2005; Zimmerman et al. 2005b). In addition, non-immune mechanisms related to the requirement for creating BM “space” in recipients, reflecting effects on hematopoietic stem cell niche interactions, contribute to engraftment resistance (Schofield 1978). These redundancies in mechanisms causing BM rejection have confounded efforts to study engraftment barriers using traditional cellular and immunologic techniques.
Total body irradiation (TBI) has long been used in preparative regimens for allogeneic BM transplantation in both the clinical setting as well as in experimental rodent models (Barrett 1982; Reddy et al. 2008). Previously, we developed an MHC-matched mouse model of BM allograft rejection where the minimum TBI dose permissive for engraftment varied between two mouse strains (Cao et al. 2009). We had used this TBI dose as trait variance for a quantitative genetic study and identified, by linkage analysis, a trait locus on mouse chromosome 16 termed Bmgr5. This trait locus regulated BM engraftment by conferring dominant susceptibility alleles with large effects in regards to the engraftment phenotype.
Our non-myeloablative BM transplant model is also highly suitable for forward genetic studies based on global transcriptome profiling. It defined a range of TBI doses that accurately and reliably correlate with BM engraftment and mouse strain variation with regards to the permissive TBI dose. Thus, systematic experimental grouping according to the TBI dose, and the recipient mouse strain, could be used to assign gene expression profiles that are derived from mice expected to engraft as opposed to those expected to reject. The aim of the present study was to use Agilent 8x60K microarrays to investigate differential gene expression induced by TBI in spleens of these recipient mice. Such an evaluation may provide insight into key gene pathways that mediate the permissiveness for engraftment enabled by recipient conditioning with TBI.
MATERIALS AND METHODS
Animal facilities and equipment
All work with mice was conducted in accordance with procedures outlined in the NIH Guide to Care and Use of Experimental Animals. Experimental protocols were approved by the University of Utah Institutional Animal Care and Use Committee. All mice were bred and maintained at the Animal Resource Center at the University of Utah, Salt Lake City, Utah. Total body irradiation (TBI) was delivered using an X-RAD 320 Biological Irradiator (Precision X-Ray, North Branford, CT).
Allogeneic BM transplantation
BM was harvested from AKR/J (H2k, Thy1.1) donor mice following CO2 asphyxiation and transplanted at a dose of 1 × 107 per recipient mice via retro-orbital injection. BALB.K (H2k, Thy1.2) and B10.BR (H2k, Thy1.2) recipient mice were conditioned with non-myeloablative TBI delivered in a single fraction on day 0. The TBI dose was 300, 400, or 500 cGy, depending on the recipient mouse strain. Engraftment in recipient mice was evaluated at 6 and 8 weeks post-transplantation. T cell chimerism was assessed by flow cytometry analysis using monoclonal antibodies against Thy1.1 (Ox-7) expressed by donor mice and Thy1.2 (53-2.1) expressed by recipient mice as previously described (Cao et al. 2009).
Gene expression microarray
Genome wide gene expression profiling was performed using total RNA isolated from spleens of BALB.K and B10.BR mice at rest and 24 hours after TBI (300 and 400 cGy for BALB.K, or 300 and 500 cGy for B10.BR). Whole spleens were collected in the Trizol Reagent (Life Technologies, Grand Island, NY) and homogenized with a rotor-stator device for RNA extraction using the RNeasy Mini Kit (Qiagen, Valencia, CA) with on-column DNase treatment. One color Cy3 RNA labeling and array hybridization to Agilent SurePrint G3 8x60K Mouse Gene Expression Arrays (Agilent Technologies, Santa Clara, CA) were performed by the Huntsman Cancer Institute Microarray Core Facility (University of Utah, Salt Lake City, UT). All microarray data have been deposited in the NCBI Gene Expression Omnibus database (Accession Number GSE43310).
Microarray data analysis
RNA samples for each individual microarray was pooled from 3 male mice. For each experimental group, microarrays were then performed in triplicate for statistical analysis. Raw microarray signal data was extracted using Agilent Feature Extraction software and loaded for processing in Agilent GeneSpring GX 11 software. Log-transformed signal intensity values were globally normalized using the percentile shift algorithm, shifting to the 75th percentile of each sample, for per chip normalization. This was followed by baseline shifting to the median of all samples for per gene normalization. Statistical significance in gene expression was determined using P-values calculated from the unpaired Student’s T-test. All microarray-related P-values reported in this study are corrected for multiple testing using the Benjamini-Hochberg False Discovery Rate method (B-H P-values). Of significantly differentially expressed RNA (B-H P-value < .05), only those with greater than or equal to a 2-fold increase or decrease in expression compared to controls were used for functional analyses.
Functional analysis of gene expression data
Pathway analysis of gene expression changes was performed using Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, Redwood City, CA). Significant enrichment for genesets in IPA-curated canonical pathways was determined using Fisher’s exact test P-values with multiple testing adjustments according to the Benjamini-Hochberg False Discovery Rate method. Additionally, a ratio is calculated from the number of molecules, from our gene expression data, that overlap with the pathway divided by the total number of molecules that map to the canonical pathway.
IPA Upstream Regulator Analysis was used to predict which upstream regulators are activated or inhibited to explain the pattern of significant up/down-regulation of gene expression. A Fisher’s exact test P-value was used to assess for significant overlap between known targets of each upstream regulator, manually curated in the IPA database, and observed gene expression changes in our dataset. A Z-score algorithm was used to evaluate the known targets of upstream regulators in addition to the expected direction of change in gene expression. Statistical significance was conferred at the Z-score ≥ 2.0 level.
Flow cytometry
Single-cell suspensions from spleens were blocked with normal mouse serum and stained with anti-B220 (RA3-6B2) PerCP and andi-CD11c (HL3) APC as described (Gu et al. 2013). Flow cytometry data were collected on a FACSCalibur instrument (BD Biosciences, San Jose, CA).
Real-time PCR
Total RNA from spleen was reverse transcribed with the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA.) for real-time PCR using the LightCycler 480 SYBR Green I Master reagent on a LightCycler 480 instrument (Roche Diagnostics, Indianapolis, IN). PCR cycling parameters were as follows: initial incubation at 95°C for 10 min, 45 cycles of 95°C for 10 s, 65°C for 20 s, and 72°C for 30 s. Primer sequences are provided in Supplemental Table 1. Relative quantitation was performed by the comparative cycle threshold method using β-Actin as internal controls (Schmittgen and Livak 2008).
Statistical analysis
Statistical analysis was performed using SPSS 13.0 software. Differences in the proportions of engrafting mice and the means of real-time PCR relative expression values was determined, respectively, using the Fisher’s exact test and Student’s T-test. Statistical significance was conferred at the P < .05 level using two-tailed P-values. Venn-diagrams were constructed using BioVenn (Hulsen et al. 2008).
RESULTS
Allogeneic BM engraftment phenotype
Prior to collecting samples for microarray analysis, the engraftment phenotype in BALB.K and B10.BR recipients was reassessed. Recipient mice were conditioned with a non-myeloablative dose of TBI and transplanted with allogeneic BM from AKR/J donors. Donor engraftment was defined as the detection of donor T-cells at both 6 and 8 weeks post-transplant by flow cytometry of peripheral blood. As shown in Table 1, and in congruence with prior observations, all BALB.K mice engrafted with AKR/J BM when conditioned with TBI 400 cGy but not with TBI 300 cGy (Cao et al. 2009). Thus, for this strain 400 cGy is the engrafting TBI (E-TBI) dose while 300 cGy is the non-engrafting TBI (NE-TBI) dose. TBI 400 cGy was not permissive for BM engraftment in B10.BR mice (0/7, 0%), however, which required TBI 500 cGy for reliable engraftment. Engrafted BALB.K and B10.BR mice demonstrate comparable levels of donor T-cell chimerism and representative engrafting mice were confirmed to achieve multi-lineage donor engraftment in B-cell and myeloid cells by RT-PCR (data not shown). These results confirm a stable allogeneic BM engraftment phenotype suitable for global gene expression analysis.
Gene expression in mice conditioned with TBI
Gene expression profiling was performed using total RNA from spleens of BALB.K and B10.BR mice 24 hours after TBI without donor BM injection. Host immune cells in secondary lymphoid spaces are thought to represent the largest barrier to allogeneic BM engraftment (Shizuru et al. 2010). The cell types involved are diverse and include natural killer cells, T-cells, their corresponding antigen presenting cells, and other effector cells such as macrophages. Thus, we selected bulk splenocyte populations as the source of RNA for global gene expression analysis. Determining the optimal time point after TBI to harvest spleens for RNA extraction was more difficult. Assays based on spleen cell proliferation or progenitor cell assays, both of which measure early rejection, suggest that BM allograft rejection can occur during the first 5 days after transplantation (Baker et al. 1995; Davenport et al. 1995). Key transcriptional events in recipients induced by TBI permitting engraftment is likely to occur during this time period. Meanwhile, the early biochemical events that begin with TBI-induced DNA double-strand breaks and that then progress to recruitment of DNA damage sensors, chromatin remodeling, and ultimately to downstream transcription repression and induction occurs during the first few hours (Friedl et al. 2012; Xu and Price 2011). Our own prior studies in spleens of mice treated with TBI showed that apoptotic splenocytes, detected by propidium iodide staining, arise within 24 hours after irradiation and near total viable cell depletion is achieved within 72 hours (Cao et al. 2009). As a final consideration, the trafficking of transplanted donor BM stem cells likely involve early trafficking to spleen, as suggested by studies based on in vivo bioluminescence imaging studies (Cao et al. 2004). Thus, we chose to evaluate transcripts isolated from spleens of recipient mice 24 hours after TBI, without prior BM injections to avoid potentially confounding donor RNA.
The gene expression analysis was performed using Agilent G3 8x60K microarrays and the results are summarized in Table 2. We performed single color Cy3 labeling and used BALB.K and B10.BR mice not given TBI as control samples for data analysis. Nearly 6000 probe features are differentially expressed between un-irradiated BALB.K compared with B10.BR mice, at a significance level of B-H P-value < .05 and with fold change ≥ 2.0. This widespread degree of differential gene expression is consistent with published observations regarding mouse strain-dependent transcriptional regulation (Su et al. 2008). A similar extent of global expression changes was observed after treatment with E-TBI doses, which were 400 cGy for BALB.K and 500 cGy for B10.BR, when compared with un-irradiated same-strain controls. Substantially less extensive gene expression changes were observed with mice irradiated with NE-TBI doses, which was TBI 300 cGy for both BALB.K and B10.BR mice.
Table 2.
Differentially expressed genes (B-H P-value < .05) in spleen 24 hours after TBI.
| Fold change ≥ 2.0, No. | Permissive for allogeneic BM engraftment | |||
|---|---|---|---|---|
|
| ||||
| Total Probes a | Probe feature
|
|||
| Genes | LincRNA | |||
|
|
|
|||
| BALB.K vs. B10.BR | 5,918 | 2,837 | 1,299 | NA |
| BALB.K | ||||
| TBI 300 cGy vs. 0 cGy | 3,485 | 2,268 | 371 | No |
| TBI 400 cGy vs. 0 cGy | 6,170 | 3,715 | 759 | Yes |
| B10.BR | ||||
| TBI 300 cGy vs. 0 cGy | 3,064 | 1,907 | 281 | No |
| TBI 500 cGy vs. 0 cGy | 5,991 | 3,763 | 535 | Yes |
Abbreviations: B-H P-value, Benjamini-Hochberg False Discovery Rate multiple testing corrected P-value; BM, bone marrow; LincRNA, long intergenic noncoding RNA; NA, not applicable; TBI, total body irradiation.
Analysis was performed using Agilent SurePrint G3 Mouse GE 8x60K microarrays, each printed with 55,682 probes corresponding to 39,430 Entrez gene RNAs and 4,622 lincRNAs.
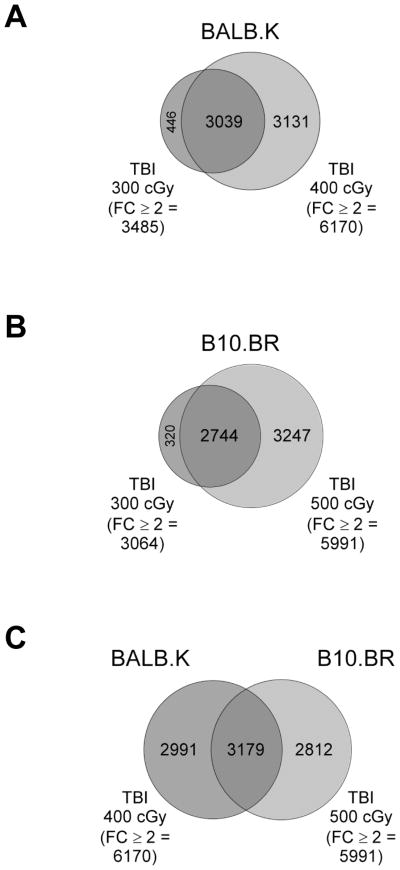
Venn-diagrams of significant microarray results are shown in Fig. 1. For both BALB.K and B10.BR mice, nearly all probe features significantly regulated in mice given NE-TBI doses overlapped with mice given E-TBI doses (Fig. 1A–B). Thus, each mouse strain acquired approximately 3000 additional significant probe features as a result of increasing the irradiation dose from NE-TBI to E-TBI levels. A comparison of genomic changes in BALB.K and B10.BR mice given E-TBI doses, as shown in Fig. 1C, revealed that approximately half of all significant probe features for either strain intersected and are shared between the two mice while the remainders are unique to each without overlap. These results show experimental groupings that will be used to generate genelists from the microarray data for functional analyses.
Fig. 1. Venn-diagrams of microarray results according to mouse strain and TBI dose.
Agilent Mouse 8x60K microarrays were used to analyze gene expression in spleens of control mice not given TBI or 24 hours after TBI at indicated doses. Shown are proportional Venn diagrams displaying overlap of probe features significantly differentially expressed at B-H P-value < .05 and with fold change ≥ 2 in (A) BALB.K mice given TBI 400 cGy (the E-TBI dose) versus TBI 300 cGy (the NE-TBI dose), (B) B10.BR mice given TBI 500 cGy (the E-TBI dose) versus TBI 300 cGy (the NE-TBI dose), and (C) BALB.K mice given TBI 400 cGy versus B10.BR given TBI 500 cGy.
Gene pathway analysis in mice conditioned with engrafting TBI doses
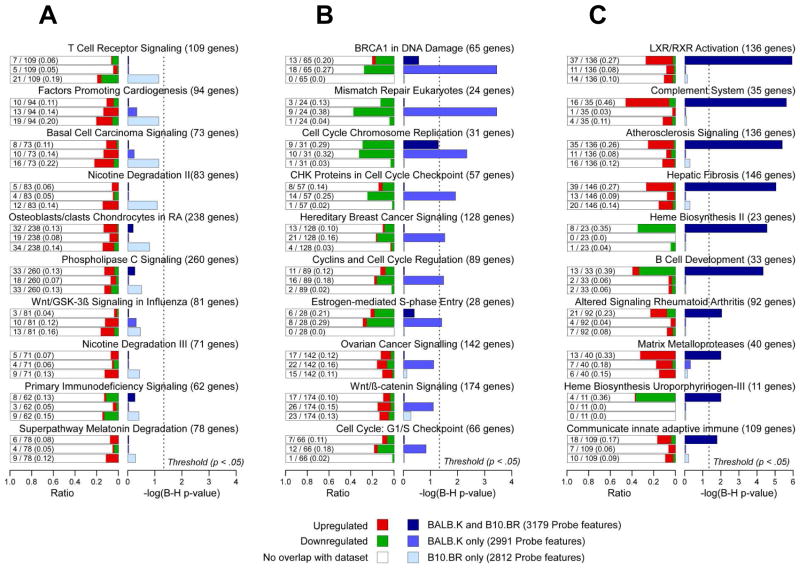
We first performed pathways analysis of gene expression profiles corresponding to BALB.K and B10.BR mice conditioned with E-TBI doses. A genelist of differentially expressed probe features shared by BALB.K and B10.BR mice is provided as Supplemental File 1; for probe features unique to BALB.K mice as Supplemental File 2; and for probe features unique to B10.BR mice as Supplemental File 3. These genelists were used to identify significant enrichment for canonical pathways in the IPA database. As shown in Fig. 2A, the genelist of probe features unique to B10.BR mice had no gene pathways that were significantly enriched after correction for multiple testing. Fig. 2B shows that there were seven canonical pathways significantly enriched in the genelist unique to BALB.K mice. These pathways are all down-regulated and over-represented for genes involved in control of DNA damage repair, cell cycle progression, and apoptosis. None of these pathways were significantly enriched in the genelist of probe features shared by both B10.BR and BALB.K mice or in the one unique to the B10.BR strain alone.
Fig. 2. Canonical pathways enriched in BALB.K and B10.BR mice conditioned with engrafting TBI doses.
Shown are canonical pathways most significantly enriched in genelists of probe features (B-H P-value < .05, and fold change ≥ 2) that are (A) unique to B10.BR, (B) unique to BALB.K, or (C) shared by both BALB.K and B10.BR mice conditioned with TBI doses adequate for BM engraftment. For each canonical pathway, shown are bar plots of the −log10 of Benjamini-Hochberg corrected P-values for each pathway according to genelists with significant probe features unique to B10.R (light blue), unique to BALB.K (medium blue), or shared by both BALB.K and B10.BR (dark blue). Also shown, for each canonical pathway, are bar plots for the ratio of genes present in the indicated genelist divided by the total number of genes in the pathway, with red indicating up-regulated and green indicating down-regulated genes.
The genelist with significant probe features shared by both BALB.K and B10.BR mice, in which both mice were conditioned with E-TBI doses, may be most insightful for identifying key pathways important in mediating permissiveness for BM engraftment. As shown in Fig. 2C, this genelist is significantly overrepresented in seven up-regulated and three down-regulated canonical pathways. A complete list of genes in our genelist that overlap with components for each IPA canonical pathway is provided in Supplemental Table 2. All up-regulated pathways participate in inflammation, and this was reflected by partial overlap between pathway components comprised of a number of pro-inflammatory molecules. For example, as shown in Supplemental Fig. 1, the retinoid and liver X receptor, atherosclerosis signaling, and hepatic fibrosis pathways intersect at group of genes that include Mcsf, the macrophage colony stimulating factor, individual apolipoproteins implicated in macrophage inflammatory responses, and pro-inflammatory cytokines such as interleukin 1 (IL-1), the IL-1 family members IL-18 and IL-33, and their corresponding receptors (Mak et al. 2002; Smith 2011). Also up-regulated are canonical pathways involving alternative pathway complement factors and matrix metalloproteases, both of which have well recognized roles in innate immunity and inflammation (Mastellos et al. 2005; Parks et al. 2004). The complement pathway (−log10 B-H P-value = 5.63, ratio = 0.46), along with the down-regulated B-cell development pathway (−log10 B-H P-value = 4.34, ratio = 0.39), bear special mention in that both the P-value of significant enrichment and the ratio of overlapping pathway molecules are high. Intriguingly, down-regulated genes enriched in the B-cell pathway are known to participate in indirect presentation of alloantigens and T-cell activation (Redfield et al. 2011). These results highlight down-regulated B-cell lymphocyte function and up-regulation of several inflammatory mechanisms as major genomic changes induced by conditioning with E-TBI doses.
Gene expression in BALB.K mice conditioned with engrafting versus non-engrafting TBI doses
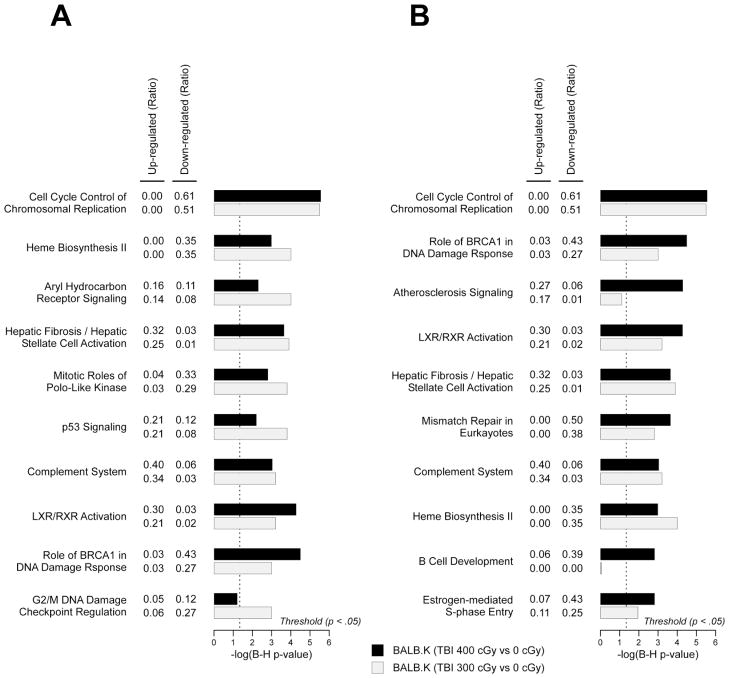
We next analyzed for canonical pathways enriched in BALB.K mice conditioned with E-TBI as compared with NE-TBI doses. A genelist of significantly differentially expressed probe features in BALB.K mice given 300 cGy, the NE-TBI dose, is supplied as Supplemental File 4; and for probe features in BALB.K given 400 cGy, the E-TBI dose, as Supplemental File 5. Among the ten gene pathways most significantly enriched for mice conditioned with NE-TBI doses, shown in Fig. 3A, essentially all are also significantly enriched in mice given E-TBI doses. Among canonical pathways most enriched in mice given E-TBI doses, however, we observed that genes in the B-cell development pathway were not at all significantly induced or repressed if conditioning was given at NE-TBI levels instead (Fig. 3B). These results suggest that down-regulation of B-cell signaling may be a key gene expression change acquired with increasing conditioning intensity from NE-TBI to E-TBI doses.
Fig. 3. Canonical pathways enriched in BALB.K conditioned with engrafting versus non-engrafting TBI doses.
Shown are canonical pathways most significantly enriched in genelists of probe features (B-H P-value < .05, and fold change ≥ 2) from BALB.K mice conditioned with (A) non-engrafting TBI doses, i.e. 300 cGy, or (B) engrafting TBI doses, i.e. 400 cGy. For each canonical pathway, shown are bar plots of the −log10 of Benjamini-Hochberg corrected P-values for each pathway according to genelists with significant probe features unique to BALB.K mice conditioned with 300 cGy (light grey) or 400 cGy (black). Each canonical pathway is also annotated with ratio of up-regulated and down-regulated molecules overlapping with the pathway.
Real-time PCR
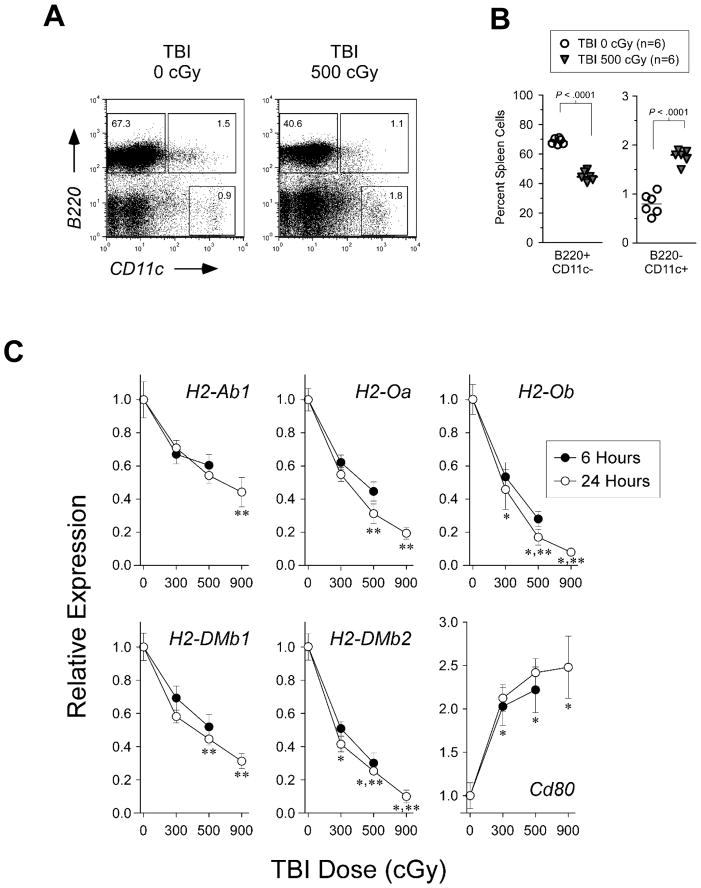
Decreased transcript abundance of genes enriched in the B-cell development pathway may reflect relative TBI-mediated depletion of specific spleen lineage cells rather than down-regulated transcription. Indeed, as shown in Fig. 4A–B, we observed in the spleen of our mice 24 hours after TBI a slight expansion of radioresistant B220-CD11c+ dendritic cells but also a more than one-third reduction in the proportion of B220+CD11c- B-cells and B220+CD11c+ plasmacytoid dendritic cells. Down-regulated genes in the B-cell canonical pathway include those that encode H2-O and H2-M and that we have previously shown to be exclusively expressed in these cell populations (Chen and Jensen 2004; Chen et al. 2006). Real-time PCR demonstrates, however, that the relative expression for two of these genes, H2-Ob and H2-DMb2, was significantly reduced after TBI compared with H2-Ab1 (Fig. 4C). The H2-Ab1 gene encodes the beta chain of the MHC class II molecule and its constitutive expression is also highly restricted to these antigen presenting cells (Glimcher and Kara 1992). Further, for all H2-O and H2-M genes there was a dose-response relationship between transcript repression and increasing TBI dose, such that increasing from NE-TBI to E-TBI levels significantly reduced gene expression in all instances. Relative expression by real-time PCR was increased for CD80, a gene up-regulated in the B-cell pathway, thereby validating the microarray findings. These results suggest that increasing the dose from NE-TBI to E-TBI levels significantly repress gene expression in the B-cell development pathway.
Fig. 4. Flow cytometry and real-time PCR analyses.
(A) Representative FACS plot and (B) dot plot of percent spleen cells gated in B220+CD11c- B-cell and B220-CD11c+ dendritic cell populations from B10.BR mice 24 hours after TBI. (C) Real-time PCR of RNA isolated from spleens of B10.BR mice 6 and 24 hours after TBI for the indicated genes. Shown are the relative expression compared with un-irradiated control. For relative expression 24 hours after TBI, indicated are significant difference compared with H2-Ab1 at each TBI dose level (*, P-value < .05), and significant difference compared with the TBI 300 cGy dose level for each gene (**, P-value < .05).
Upstream regulator of gene expression in mice conditioned with engrafting doses of TBI
IPA upstream regulator analysis examines the known targets of an upstream regulator, which can be a transcription factor, cytokine, chemical, drug, or other molecule. It then compares the targets’ actual direction of change, in a dataset such as our genelists, to expectations derived from the literature and issues a prediction for each upstream regulator based on a z-score calculation. Results of an analysis for upstream regulators of gene expression changes induced by conditioning with E-TBI doses for BALB.K mice is supplied as Supplemental File 6; for B10.BR mice as Supplemental File 7; and for both mice as a summary of the most significant upstream regulators in Table 3. Overall there was remarkable similarity in predicted upstream regulators for both BALB.K and B10.BR mice. In both strains the most significant upstream regulators included Tp53, a transcription factor known to be activated by ionizing radiation, and the IKK kinase complex, a signalsome that activates NF-kappaB in response to both inflammatory cytokines and DNA damage (Mercurio et al. 1997; Rashi-Elkeles et al. 2011). In turn, the predicted upstream inflammatory stimuli in our dataset include innate sensoring of danger signals, such as lipopolysaccharide, and by toll-like receptor 3, a Tp53-inducible sensor of double-stranded DNA. These stimuli are predicted to be ultimately delivered by upstream regulatory cytokine groups that include interferons, tumor necrosis factor, and transforming growth factor-beta (Menendez et al. 2011; Shan et al. 2007). These results show that the gene expression profile in our mice are broadly inflammatory in nature and consistent with expected physiologic responses to ionizing radiation such as conditioning with TBI.
Table 3.
Upstream regulators of gene expression in spleens of mice conditioned with engrafting TBI doses. a
| BALB.K (TBI 400 cGy vs TBI 0 cGy)
|
B10.BR (TBI 500 cGy vs TBI 0 cGy)
|
||||
|---|---|---|---|---|---|
| Regulator | Molecule type | Z-score | Regulator | Molecule type | Z-score |
|
|
|
||||
| ATRA | Chemical | 9.078 | Tp53 | Transcription factor | 7.602 |
| Tp53 | Transcription factor | 8.205 | TGFB1 | Growth factor | 7.576 |
| LPS | Chemical | 8.203 | ATRA | Chemical | 7.116 |
| IKKB | Kinase | 6.845 | IKKB | Kinase | 7.030 |
| TNF | Cytokine | 6.824 | LPS | Chemical | 6.950 |
| Calcitriol | Chemical | 6.536 | Smarcb1 | Transcription factor | 6.632 |
| TLR3 | Membrane receptor | 6.417 | IKKA | Kinase | 6.587 |
| TGFB1 | Growth factor | 6.230 | IFNG | Cytokine | 6.357 |
| Cdkn2a | Transcription factor | 6.225 | IFNA2 | Cytokine | 6.308 |
| IKKA | Kinase | 6.022 | TNF | Cytokine | 5.872 |
| PMA | Chemical | 5.931 | PMA | Chemical | 5.811 |
| IFNG | Cytokine | 5.920 | TLR3 | Membrane receptor | 5.749 |
| Smarcb1 | Transcription factor | 5.904 | Cebpa | Transcription factor | 5.687 |
| Decitabine | Chemical | 5.871 | IFNA | Cytokine group | 5.634 |
| IFNA | Cytokine group | 5.829 | Sp1 | Transcription factor | 5.615 |
Abbreviations: ATRA, all-trans retinoic acid; IFNA, interferon alpha; IFNA2, interferon alpha 2; IFNG, interferon gamma; IKKA, IkappaB kinase alpha; IKKB, IkappaB kinase beta; LPS, lipopolysaccharide; PMA, phorbol myristate acetate; TBI, total body irradiation; TGFB1, transforming growth factor 1; TLR3, toll-like receptor 3; TNF, tumor necrosis factor.
Statistical significance was conferred at the Z-score ≥ 2.0 level
DISCUSSION
Inflammation, macrophage activation, and up-regulation of the complement pathway can be viewed as unintended consequences of TBI conditioning in that they either augment the barrier to engraftment or, worse, contribute to toxicity of the preparative regimen. By contrast, our analysis identified a down-regulated canonical pathway related to B-cell development that instead may be directly beneficial for promoting BM engraftment. Considerable evidence from solid organ allograft models suggests that presentation of donor antigens by host antigen presenting cells, including B-cell lymphocytes, via the indirect pathway of allorecognition can activate alloreactive CD4 T-cells to mediate graft rejection (Redfield et al. 2011). Repression of transcripts encoding H2-M and H2-O in the B-cell development gene pathway suggests one way through which recipient conditioning at E-TBI doses may mitigate this putative BM engraftment resistance mechanism. As we and others have shown, H2-M and H2-O are MHC class II-like molecules co-expressed in the endosomes of mouse B-cells and dendritic cells that, along with their human counterparts HLA-DM and HLA-DO, participate in loading of peptide antigens into the peptide binding groove of MHC class II molecules (Fallas et al. 2007; Gu et al. 2013; Jensen 2007). This forms the MHC-peptide antigen complex that interacts with cognate T-cell receptors to activate responding CD4 T-cells. Reduced expression of H2-M and H2-O may compromise this indirect antigen processing pathway. Specifically, we have demonstrated that H2-O knockout mouse are impaired in presentation of exogenous antigens (Gu et al. 2013) and, further, it has been shown that mice deficient in H2-M have prolonged allograft survival compared with wildtype mice (Felix et al. 2000). Whether these mechanisms contribute to resistance of BM engraftment has not been validated by in vivo engraftment assays. If confirmed, elucidation of how genes in this canonical pathway participate in BM rejection may identify key molecules or cell types that may be targeted for pre-transplant conditioning.
An unresolved question still surrounding BM engraftment barriers is to what extent the initial presentation and recognition of donor antigens trigger downstream aggressive inflammatory and cytotoxic events that eventually result in elimination of donor BM cells (Hayry et al. 1984). Our results suggest that, in the complete absence of donor antigens, gene expression changes induced by even a non-myeloablative preparative regimen are sufficient to generate a robust inflammatory milieu primed to reject donor cells. In both human clinical samples and in rodent models, it has been shown that gut tissue injury in recipients occur as the result of toxicities from the conditioning regimen (Hill et al. 1997; van der Velden et al. 2010). Further, gut injury leads to translocation of lipopolysaccharide across the damaged mucosa into the systemic circulation, thereby initiating release of inflammatory cytokines such as tumor necrosis factor and IL-1. The broad inflammatory signature of the genomic profile observed in our mice conditioned with TBI, with respect to both the associated canonical pathways and predicted upstream regulators, underscores the widespread nature of this inflammatory response.
From our functional analysis of gene expression patterns in mice conditioned with TBI, the up-regulated complement canonical gene pathway stands out as being significantly enriched with regards to both the P-value for significance and ratio of gene overlap. The role of complement factors in BM engraftment resistance is largely unexplored, but has been shown to contribute strongly to rejection of solid organ allografts in mice (Kwan et al. 2012b). Both host T-cells and antigen presenting cells produce complement proteins including C3a and C5a, as well as their respective G-protein-coupled receptors, C3aR and C5aR. In rodent models of organ allografting, complement blockade attenuates T cell activation and delays rejection while increasing complement activation, in recipients deficient for the complement regulatory protein decay-accelerating factor, accelerates allograft rejection (Vieyra et al. 2011). Further, complement activation has been observed after marrow-ablative TBI doses in models of BM transplant associated graft-versus-host disease and to contribute to its pathogenesis (Kwan et al. 2012a). Results from our genomic profiling show that complement factors are also activated in bulk splenocytes after non-myeloablative TBI conditioning. Thus, complement inhibitors may be an effective strategy for overcoming allogeneic BM engraftment barriers.
Another component of innate immunity that may be therapeutically viable relate to recent studies of BM engraftment barriers that implicate a previously unappreciated role for the reticulo-endothelial system, in particular those of macrophages. Host macrophages can clear donor BM hematopoietic stem cells, both autologous and allogeneic, via a mechanism dependent on interactions between SIRPA and CD47 (Jaiswal et al. 2009). The pro-inflammatory genomic profile induced by TBI in our mice includes many gene expression changes that cause macrophage differentiation and activation (Verschoor et al. 2012). Upon inflammation, circulating monocytes extravasate through endotheliuem into tissue towards chemotactic danger signals. Once there they differentiate into macrophages under the influence of macrophage colony stimulating factor, which was up-regulated after TBI in our mice (Delneste et al. 2003). Lipopolysaccharide, an upstream regulator identified in the analysis, is a known activator of tissue-resident macrophages. This process can be further stimulated by apolipoprotein subsets, which we observed to be up-regulated in several inflammatory canonical pathways. Also up-regulated were macrophage derived pro-inflammatory cytokines including tumor necrosis factor, IL-1, IL-18, and IL-33 (Berbee et al. 2010). These findings support therapeutic approaches targeting CD47-mediated phagocytosis of donor BM stem cells to overcome engraftment resistance (Logan et al. 2012).
In summary, our results here provide novel insight into the gene expression profile induced in mice conditioned with TBI as recipients for a BM transplant. We assayed for genomic changes in a tissue representing a secondary lymphoid cell compartment, the spleen, and at a time point, 24 hours after TBI, which is physiologically relevant for studying the BM rejection phenotype. From our functional analysis of the gene expression pattern, we highlighted canonical pathways involved with complement, macrophage, and B-cell function as areas that may be attractive for further interrogation. In our BM transplant model and in the common clinical practice where transplants are performed between HLA-compatible individuals, the preparative regimen is the primary determinant of engraftment. Further studies of findings presented here may therefore prove particularly insightful for better understanding the basis for BM engraftment resistance. For example, characterizing gene expression changes in other tissue sites, such as peripheral blood and BM, or in specific cell types that may participate in cellular rejection may be informative. Potential findings will need to considered in the context of whether deleterious effects for promoting graft-versus-host disease is also conferred (Rowe et al. 2006). Ultimately, this may lead to the design of novel preparative regimens for overcoming engraftment barriers and improved outcome after allogeneic hematopoietic cell transplantation in patients.
Supplementary Material
Table 1.
Allogeneic BM engraftment in BALB. K and B10.BR mice
| Engrafted, No. (%) | % Donor T- cells | P-value a | |
|---|---|---|---|
|
|
|
||
| AKR/J → BALB.K | |||
| TBI 300 cGy | 0 / 5 | 0 ± 0 | .002 |
| TBI 400 cGy | 6 / 6 | 69.3 ± 8.9 | - |
| AKR/J → B10.BR | |||
| TBI 300 cGy | 0 / 5 | 0 ± 0 | .002 |
| TBI 400 cGy | 0 / 7 | 0 ± 0 | .001 |
| TBI 500 cGy | 5 / 5 | 74.4 ± 10.4 | ns |
Abbreviations: BM, bone marrow; ns, not significant; TBI, total body irradiation.
P-value for number of mice engrafted versus BALB.K recipients given TBI 400 cGy.
Acknowledgments
This work was supported in part by grants R01AI076652 (T.M.C.) from the National Institute of Allergy and Infectious Diseases.
Footnotes
ASSESSION NUMBERS: Agilent 8x60K microarrays: GSE43310
AUTHORSHIP CONTRIBUTIONS
Conceived and designed the experiments: TMC, JAS, AT, and XC. Performed the experiments: YW, ST, Q.L, and TMC. Analyzed the data: XC, YW, ST and TMC. Wrote the paper: XC and TMC.
CONFLICTS OF INTEREST DISCLOSURES
The authors declare no competing financial interests.
References
- Bacher U, Klyuchnikov E, Wiedemann B, Kroeger N, Zander AR. Safety of conditioning agents for allogeneic haematopoietic transplantation. Expert Opin Drug Saf. 2009;8:305–15. doi: 10.1517/14740330902918273. [DOI] [PubMed] [Google Scholar]
- Baker MB, Podack ER, Levy RB. Perforin- and Fas-mediated cytotoxic pathways are not required for allogeneic resistance to bone marrow grafts in mice. Biol Blood Marrow Transplant. 1995;1:69–73. [PubMed] [Google Scholar]
- Barrett A. Total body irradiation before bone marrow transplantation: a review. Clin Radiol. 1982;33:131–5. doi: 10.1016/s0009-9260(82)80035-4. [DOI] [PubMed] [Google Scholar]
- Barrett AJ. Conditioning regimens for allogeneic stem cell transplants. Curr Opin Hematol. 2000;7:339–42. doi: 10.1097/00062752-200011000-00003. [DOI] [PubMed] [Google Scholar]
- Berbee JF, Coomans CP, Westerterp M, Romijn JA, Havekes LM, Rensen PC. Apolipoprotein CI enhances the biological response to LPS via the CD14/TLR4 pathway by LPS-binding elements in both its N- and C-terminal helix. J Lipid Res. 2010;51:1943–52. doi: 10.1194/jlr.M006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao TM, Thomas A, Wang Y, Tsai S, Logronio K, Shizuru JA. A chromosome 16 quantitative trait locus regulates allogeneic bone marrow engraftment in nonmyeloablated mice. Blood. 2009;114:202–10. doi: 10.1182/blood-2009-03-208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YA, Wagers AJ, Beilhack A, Dusich J, Bachmann MH, Negrin RS, Weissman IL, Contag CH. Shifting foci of hematopoiesis during reconstitution from single stem cells. Proc Natl Acad Sci U S A. 2004;101:221–6. doi: 10.1073/pnas.2637010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Jensen PE. The expression of HLA-DO (H2-O) in B lymphocytes. Immunol Res. 2004;29:19–28. doi: 10.1385/IR:29:1-3:019. [DOI] [PubMed] [Google Scholar]
- Chen X, Reed-Loisel LM, Karlsson L, Jensen PE. H2-O expression in primary dendritic cells. J Immunol. 2006;176:3548–56. doi: 10.4049/jimmunol.176.6.3548. [DOI] [PubMed] [Google Scholar]
- Davenport C, Kumar V, Bennett M. Rapid rejection of H2k and H2k/b bone marrow cell grafts by CD8+ T cells and NK cells in irradiated mice. J Immunol. 1995;155:3742–9. [PubMed] [Google Scholar]
- Delneste Y, Charbonnier P, Herbault N, Magistrelli G, Caron G, Bonnefoy JY, Jeannin P. Interferon-gamma switches monocyte differentiation from dendritic cells to macrophages. Blood. 2003;101:143–50. doi: 10.1182/blood-2002-04-1164. [DOI] [PubMed] [Google Scholar]
- Fallas JL, Yi W, Draghi NA, O’Rourke HM, Denzin LK. Expression patterns of H2-O in mouse B cells and dendritic cells correlate with cell function. J Immunol. 2007;178:1488–97. doi: 10.4049/jimmunol.178.3.1488. [DOI] [PubMed] [Google Scholar]
- Felix NJ, Brickey WJ, Griffiths R, Zhang J, Van Kaer L, Coffman T, Ting JP. H2-DMalpha(-/-) mice show the importance of major histocompatibility complex-bound peptide in cardiac allograft rejection. J Exp Med. 2000;192:31–40. doi: 10.1084/jem.192.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl AA, Mazurek B, Seiler DM. Radiation-induced alterations in histone modification patterns and their potential impact on short-term radiation effects. Front Oncol. 2012;2:117. doi: 10.3389/fonc.2012.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimcher LH, Kara CJ. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- Gluckman E. Ten years of cord blood transplantation: from bench to bedside. Br J Haematol. 2009;147:192–9. doi: 10.1111/j.1365-2141.2009.07780.x. [DOI] [PubMed] [Google Scholar]
- Gu Y, Jensen PE, Chen X. Immunodeficiency and autoimmunity in h2-o-deficient mice. J Immunol. 2013;190:126–37. doi: 10.4049/jimmunol.1200993. [DOI] [PubMed] [Google Scholar]
- Hayry P, von Willebrand E, Parthenais E, Nemlander A, Soots A, Lautenschlager I, Alfoldy P, Renkonen R. The inflammatory mechanisms of allograft rejection. Immunol Rev. 1984;77:85–142. doi: 10.1111/j.1600-065x.1984.tb00719.x. [DOI] [PubMed] [Google Scholar]
- Hill GR, Crawford JM, Cooke KR, Brinson YS, Pan L, Ferrara JL. Total body irradiation and acute graft-versus-host disease: the role of gastrointestinal damage and inflammatory cytokines. Blood. 1997;90:3204–13. [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, Weissman IL. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–85. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–8. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Mammolenti M, Jones M, Jurecic R, Sayers TJ, Levy RB. Antigen-primed CD8+ T cells can mediate resistance, preventing allogeneic marrow engraftment in the simultaneous absence of perforin-, CD95L-, TNFR1-, and TRAIL-dependent killing. Blood. 2003;101:3991–9. doi: 10.1182/blood-2002-09-2859. [DOI] [PubMed] [Google Scholar]
- Kwan WH, Hashimoto D, Paz-Artal E, Ostrow K, Greter M, Raedler H, Medof ME, Merad M, Heeger PS. Antigen-presenting cell-derived complement modulates graft-versus-host disease. J Clin Invest. 2012a;122:2234–8. doi: 10.1172/JCI61019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan WH, van der Touw W, Heeger PS. Complement regulation of T cell immunity. Immunol Res. 2012b;54:247–53. doi: 10.1007/s12026-012-8327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan AC, Weissman IL, Shizuru JA. The road to purified hematopoietic stem cell transplants is paved with antibodies. Curr Opin Immunol. 2012;24:640–8. doi: 10.1016/j.coi.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak PA, Laffitte BA, Desrumaux C, Joseph SB, Curtiss LK, Mangelsdorf DJ, Tontonoz P, Edwards PA. Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors alpha and beta. J Biol Chem. 2002;277:31900–8. doi: 10.1074/jbc.M202993200. [DOI] [PubMed] [Google Scholar]
- Mastellos D, Germenis AE, Lambris JD. Complement: an inflammatory pathway fulfilling multiple roles at the interface of innate immunity and development. Curr Drug Targets Inflamm Allergy. 2005;4:125–7. doi: 10.2174/1568010053622993. [DOI] [PubMed] [Google Scholar]
- Menendez D, Shatz M, Azzam K, Garantziotis S, Fessler MB, Resnick MA. The Toll-like receptor gene family is integrated into human DNA damage and p53 networks. PLoS Genet. 2011;7:e1001360. doi: 10.1371/journal.pgen.1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–6. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–29. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- Rashi-Elkeles S, Elkon R, Shavit S, Lerenthal Y, Linhart C, Kupershtein A, Amariglio N, Rechavi G, Shamir R, Shiloh Y. Transcriptional modulation induced by ionizing radiation: p53 remains a central player. Mol Oncol. 2011;5:336–48. doi: 10.1016/j.molonc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Negrin R, Hill GR. Mouse models of bone marrow transplantation. Biol Blood Marrow Transplant. 2008;14:129–35. doi: 10.1016/j.bbmt.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RR, 3rd, Rodriguez E, Parsons R, Vivek K, Mustafa MM, Noorchashm H, Naji A. Essential role for B cells in transplantation tolerance. Curr Opin Immunol. 2011;23:685–91. doi: 10.1016/j.coi.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe V, Banovic T, MacDonald KP, Kuns R, Don AL, Morris ES, Burman AC, Bofinger HM, Clouston AD, Hill GR. Host B cells produce IL-10 following TBI and attenuate acute GVHD after allogeneic bone marrow transplantation. Blood. 2006;108:2485–92. doi: 10.1182/blood-2006-04-016063. [DOI] [PubMed] [Google Scholar]
- Scheffold C, Scheffold YC, Cao TM, Gworek J, Shizuru JA. Cytokines and cytotoxic pathways in engraftment resistance to purified allogeneic hematopoietic stem cells. Biol Blood Marrow Transplant. 2005;11:1–12. doi: 10.1016/j.bbmt.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- Shan YX, Jin SZ, Liu XD, Liu Y, Liu SZ. Ionizing radiation stimulates secretion of pro-inflammatory cytokines: dose-response relationship, mechanisms and implications. Radiat Environ Biophys. 2007;46:21–9. doi: 10.1007/s00411-006-0076-x. [DOI] [PubMed] [Google Scholar]
- Shizuru JA, Bhattacharya D, Cavazzana-Calvo M. The biology of allogeneic hematopoietic cell resistance. Biol Blood Marrow Transplant. 2010;16:S2–7. doi: 10.1016/j.bbmt.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE. The biological paths of IL-1 family members IL-18 and IL-33. J Leukoc Biol. 2011;89:383–92. doi: 10.1189/jlb.0810470. [DOI] [PubMed] [Google Scholar]
- Su WL, Modrek B, GuhaThakurta D, Edwards S, Shah JK, Kulkarni AV, Russell A, Schadt EE, Johnson JM, Castle JC. Exon and junction microarrays detect widespread mouse strain- and sex-bias expression differences. BMC Genomics. 2008;9:273. doi: 10.1186/1471-2164-9-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas ED, Blume KG. Historical markers in the development of allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 1999;5:341–6. doi: 10.1016/s1083-8791(99)70010-8. [DOI] [PubMed] [Google Scholar]
- van der Velden WJ, Herbers AH, Feuth T, Schaap NP, Donnelly JP, Blijlevens NM. Intestinal damage determines the inflammatory response and early complications in patients receiving conditioning for a stem cell transplantation. PLoS One. 2010;5:e15156. doi: 10.1371/journal.pone.0015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschoor CP, Puchta A, Bowdish DM. The macrophage. Methods Mol Biol. 2012;844:139–56. doi: 10.1007/978-1-61779-527-5_10. [DOI] [PubMed] [Google Scholar]
- Vieyra M, Leisman S, Raedler H, Kwan WH, Yang M, Strainic MG, Medof ME, Heeger PS. Complement regulates CD4 T-cell help to CD8 T cells required for murine allograft rejection. Am J Pathol. 2011;179:766–74. doi: 10.1016/j.ajpath.2011.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10:261–7. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman Z, Jones M, Shatry A, Komatsu M, Mammolenti M, Levy R. Cytolytic pathways used by effector cells derived from recipient naive and memory T cells and natural killer cells in resistance to allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2005a;11:957–71. doi: 10.1016/j.bbmt.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Zimmerman Z, Shatry A, Deyev V, Podack E, Mammolenti M, Blazar BR, Yagita H, Levy RB. Effector cells derived from host CD8 memory T cells mediate rapid resistance against minor histocompatibility antigen-mismatched allogeneic marrow grafts without participation of perforin, Fas ligand, and the simultaneous inhibition of 3 tumor necrosis factor family effector pathways. Biol Blood Marrow Transplant. 2005b;11:576–86. doi: 10.1016/j.bbmt.2005.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.