Abstract
Objectives
MicroRNA (miRNA)-related single-nucleotide polymorphisms (SNPs) in miRNA processing machinery genes can affect cancer risk, treatment efficacy, and patient prognosis. A miR-SNP of rs11077 located in the 3′ untranslated region (3′ UTR) of the miRNA processing machinery gene XPO5 was examined in 112 advanced non-small-cell lung cancer (NSCLC) patients to evaluate its association with cancer outcome.
Materials and methods
The miR-SNP was genotyped with ligase detection reaction method. Survival curves were calculated using the Kaplan-Meier method, and multivariate survival analysis was performed using a Cox proportional hazards model.
Results
The AC genotype of rs11077, which carries C or A allele, was significantly associated with a better chemotherapy response (P = 0.001). In addition, rs11077 was independently associated with overall survival in advanced NSCLC patients through multivariate analysis (relative risk 0.457; 95% confidence interval: 0.251–0.831; P = 0.010).
Conclusion
rs11077 was associated with chemotherapy response and survival of advanced NSCLC patients. The analysis of miR-SNPs in miRNA processing machinery genes can help identify patient subgroups that are at high risk for poor disease outcomes.
Keywords: NSCLC, miR-SNP, survival, XPO5, rs11077, chemotherapy response
Introduction
Lung cancer is the most frequent cancer and one of the leading causes of cancer-related deaths worldwide, accounting for 30% of all cancer-related deaths.1,2 Non-small-cell lung cancer (NSCLC) accounts for 89% of all lung cancers, and about one-third of NSCLC patients are diagnosed with locally advanced stage disease.3,4 Despite aggressive treatment for these patients, the prognosis of this cancer is still poor, with a 5-year survival rate of approximately 10% and a median survival time of 16–18 months.5,6 Chemotherapy with platinum-based combinations including NP (vinorelbine plus cisplatin), TP (paclitaxel plus cisplatin), and GP (gemcitabine plus cisplatin) regimens, for no more than six-treatment course, is the standard first-line therapy for advanced-stage NSCLC. Many clinical factors, such as tumor stage, metastasis, sex, and weight loss, are predictors of prognosis for NSCLC patients.7 Pharmacogenomic studies focusing on the relationship between chemotherapy sensitivity and molecular markers have linked genomics information to therapy evaluation, and a recent study has also indicated that gene modulation can influence drug response.8
MicroRNAs (miRNAs) are RNA molecules measuring ~22 nucleotides in length that act as post-transcriptional regulators of mRNA expression.9,10 More than 700 miRNAs have been identified in humans, and these miRNAs are responsible for regulating the expression of at least 30% of protein-coding genes.11 A growing body of evidence suggests that miRNAs play important roles in a broad range of biological processes, such as embryonic development; cellular differentiation, proliferation, and apoptosis; cancer development; and insulin secretion.9,10 During miRNA processing, long primary transcripts of miRNAs (pri-miRNAs) are processed in the nucleus by the RNase III Drosha and are transported to the cytoplasm by the nuclear transport factor exportin-5 (XPO5) and RAN. In the cytoplasm, RNase III Dicer and transactivation-responsive RNA-binding protein (TRBP) mediate pre-miRNA processing to release a 21-bp dsRNA. The RNA-induced silencing complex (RISC), including GEMIN3 and GEMIN4, will select one strand as the mature miRNA and guide mature miRNAs to their target mRNA sites.9,12–16 miRNA-related single-nucleotide polymorphisms (miR-SNPs), defined as single-nucleotide polymorphisms (SNPs) in the miRNA genes, miRNA binding site, and miRNA processing machinery, can modulate miRNA and target gene expression. As a result, miR-SNPs may affect cancer risk, treatment efficacy, and patient prognosis.8,17–19
The pharmacogenomic analysis of miRNA and treatment efficacy represents a novel research field in which to evaluate and improve drug efficacy in cancer therapy. A miR-SNP of rs11077 located in the 3′ untranslated region (3′ UTR) of the miRNA processing gene XPO5 is associated with chemotherapy response in metastatic colon cancer, as well as recurrence in resected NSCLC.8,18 XPO5 is found in the nuclear membrane, mediates the transport of pre-miRNA, and adjusts miRNA expression. Knockdown XPO5 expression leads to reduced miRNA levels.20 In the present study, we evaluated the predictive power of this SNP regarding chemotherapy efficacy and overall survival of advanced NSCLC patients.
Materials and methods
Tissue specimens and DNA extraction
Blood samples were collected at the Fourth Hospital of Hebei University, Shijiazhuang, People’s Republic of China from NSCLC patients who received chemotherapy treatment in the Department of Respiratory Medicine between 2001 and 2010. The genomic DNA was immediately extracted using the Wizard Genomic DNA extraction kit (Promega Corporation, Fitchburg, WI, USA) and stored at −20°C. All procedures were supervised and approved by the Human Tissue Research Committee at the hospital, and informed consent was obtained from all participants.
Genotyping of miR-SNPs
The miR-SNP rs11077 of the miRNA processing gene XPO5 was genotyped using the ligase detection reaction method with the forward primer 5′-GAATCTGGTCACCTGATGGGA-3′ and reverse primer 5′-GTGCCTGAGTGGACCTTGAG-3′ to amplify the DNA fragments flanking miR-SNPs based on the NCBI SNP database (http://www.ncbi.nlm.nih.gov/snp/). Polymerase chain reaction was performed using a PCR Master Mix Kit according to the manufacturer’s instructions (Promega). The ligation was performed using the different probes S1 5′- GTACCTCCAAGGACCAGGGCTGGGA-3′ or S2 5′-TTTGTACCTCCAAGGACCAGGGCTGGGC-3′ matched to the alleles of miR-SNPs. The S1 or S2 probe was ligated with S3 5′-AGTCTTTAGTGCTAACATCCCCTTT-3′ downstream of the SNP site, and the ligated products were separated using the ABI PRISM Genetic Analyzer 3730XL (Applied Biosystems; Life Technologies, Carlsbad, CA, USA). Polymorphisms were confirmed based on a 3-bp length difference in the ligated products.
Statistical analysis
The χ2 test was used to analyze dichotomous values of clinical characteristic frequencies or SNP frequencies among the different overall response rate (ORR) groups. Survival curves were calculated using the Kaplan–Meier method, and comparisons between the curves were made using the log-rank test. Multivariate survival analysis was performed using a Cox proportional hazards model. All of the statistical analyses were performed using the SPSS software package (v 18.0; IBM Corporation, Armonk, NY, USA). A P-value of <0.05 was considered to be statistically significant.
Results
A total of 116 patients enrolled in this study were reviewed every 3 months for 2 years by letter or telephone interview. One patient was lost in the first year and two in the second year. The remaining 112 patients, including 27 stage III and 85 stage IV patients, were analyzed using Kaplan–Meier analysis. The relationships between the 2-year survival data and clinical characteristics were compared using the log-rank test in advanced NSCLC patients. Sex, age, TNM classification, smoking status, and histology were not statistically associated with overall survival through univariate analysis (Table 1).
Table 1.
Analysis of clinical characteristics associated with overall survival and chemotherapy response in advanced NSCLC patients
| Characteristics | No of cases | 2-year survival rate (%) | ORR P-value | OS P-value |
|---|---|---|---|---|
| Sex | 0.527 | 0.438 | ||
| Male | 74 | 17.6 | ||
| Female | 38 | 26.3 | ||
| Age (years) | 0.396 | 0.798 | ||
| ≤60 | 52 | 19.2 | ||
| >60 | 60 | 21.7 | ||
| TNM classification | 0.067 | 0.788 | ||
| III | 27 | 11.1 | ||
| IV | 85 | 23.5 | ||
| Smoking | 0.658 | 0.313 | ||
| Yes | 51 | 23.5 | ||
| No | 61 | 18.0 | ||
| Histology | 0.301 | 0.906 | ||
| SQ | 34 | 17.6 | ||
| AC | 78 | 21.8 | ||
| Chemotherapy regimen | 0.195 | 0.504 | ||
| GP | 53 | 15.1 | ||
| TP | 28 | 25.0 | ||
| NP | 31 | 25.8 |
Abbreviations: AC, adenocarcinoma; GP, gemcitabine plus cisplatin; NP, vinorelbine plus cisplatin; NSCLC, non-small-cell lung cancer; ORR, overall response; OS, overall survival; SQ, squamous cell carcinoma; TNM, tumor node metastasis; TP, paclitaxel plus cisplatin.
Association between clinical characteristics and rs11077 with chemotherapy response
Chemotherapy response was defined according to the Response Evaluation Criteria in Solid Tumors.21 The ORR was determined by comparing patients with a complete response (CR) or partial response (PR) to patients with stable disease (SD) or progressive disease (PD). Using an intent-to-treat analysis, the ORR was 18%, with a 0.9% CR and 17.1% PR. In addition, 65% of NSCLC patients achieved SD and 17% of patients developed PD (Table 2). The relationships between ORR and clinical characteristics were analyzed with the χ2 test. The clinical characteristics included sex, age, TNM classification, smoking status, and histology. Only TNM classification was associated with treatment response at borderline difference levels (P = 0.067).
Table 2.
Multivariate analysis of prognostic factors associated with overall survival in NSCLC patients with Cox proportional hazards model
| Factors | Relative risk | 95% CI | P-value |
|---|---|---|---|
| Sex | 0.606 | 0.359–1.023 | 0.061 |
| Age | 0.979 | 0.660–1.452 | 0.915 |
| TNM classification | 0.809 | 0.498–1.314 | 0.391 |
| Smoking | 1.735 | 1.041–2.894 | 0.035 |
| Chemotherapy regimen | 0.862 | 0.683–1.089 | 0.213 |
| Histology | 0.962 | 0.615–1.504 | 0.864 |
| rs11077 genotype | 0.457 | 0.251–0.831 | 0.010 |
Abbreviations: CI, confidence interval; NSCLC, non-small-cell lung cancer; TNM, tumor node metastasis.
One hundred twelve NSCLC patients and 80 healthy controls were genotyped for rs11077 polymorphisms with ligase detection reaction method. The rs11077 CC (carrying two homozygous C alleles), AC (carrying heterozygous A and C alleles), and AA (carrying two homozygous A alleles) genotype frequencies in the control samples were one, 14, and 65, respectively, which was similar to the genotype frequencies in NSCLC patients (0, 18, and 94 for CC, AC, and AA, respectively). The C allele was the minor frequency allele. No statistically significant association was detected between cancer risk and the distribution of the rs11077 polymorphism in the 112 NSCLC patients and 80 healthy controls. We subsequently assessed the relationship between rs11077 and the overall survival of these NSCLC patients. The NSCLC patients were divided into two groups on the basis of their rs11077 genotype, and their overall survival curves were plotted using the Kaplan–Meier method. The 2-year survival rates of AC and AA patients were 33.3% and 18.1%, respectively. A significant difference in overall survival was found between the two groups (Figure 1). The patients with the AC allele exhibited a significantly longer survival time compared to AA patients (P = 0.007).
Figure 1.
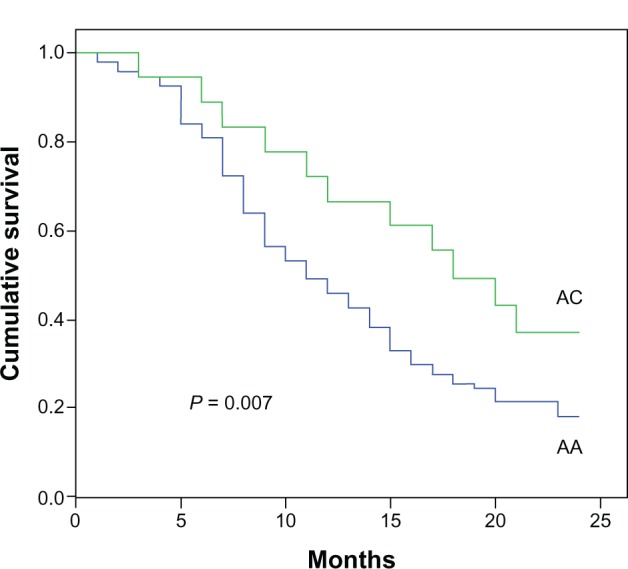
Genotype of rs11077 and their association with NSCLC survival.
Note: The patients with the AC allele exhibited a significantly longer survival time than AA patients.
Abbreviations: NSCLC, non-small-cell lung cancer; AA, carrying two homozygous A alleles; AC, carrying heterozygous A and C alleles.
We performed a multivariate analysis with the Cox proportional hazards model for clinical characteristics. As shown in Table 2, the rs11077 SNP was identified as an independent predictor of overall survival for advanced NSCLC patients (relative risk 0.457; 95% confidence interval: 0.251–0.831; P = 0.010). Smoking status was also identified as an independent predictor of survival.
The relationship between chemotherapy response and rs11077 genotype was assessed using the χ2 test. As shown in Table 3, the ORR was determined by comparing patients with a CR plus PR to patients with SD plus PD. AA type patients comprised 82 PD or SD patients and 12 CR or PR patients, whereas AC type patients comprised ten PD or SD patients and eight CR or PR patients. AC type NSCLC patients showed better treatment response compared to the AA type patients (P = 0.001).
Table 3.
Relevant association of rs11077 genotype with chemotherapy response
| Genotype | SD/PD (n) | CR/PR (n) | P-value |
|---|---|---|---|
| AA | 64/18 | 0/12 | 0.001 |
| AC | 9/1 | 1/7 |
Abbreviations: CR, complete response; n, number of patients; PD, progressive disease; PR, partial response; SD, stable disease; AA, carrying two homozygous A alleles; AC, carrying heterozygous A and C alleles.
Discussion
The identification of predictive markers for chemotherapy response is a novel field in pharmacogenomics and cancer research. Researching the association of miRNA expression with tumors and chemosensitivity has attracted great interest in cancer research. The altered expression of a number of miRNAs has been to found to predict anticancer treatments.22 In a previous report, rs11077 has been associated with recurrence in postsurgical NSCLC patients.18 In the present study, we report that miR-SNP rs11077 has a predictive value on chemotherapy response and can modulate the lifespan of advanced NSCLC patients. The miR-SNP in the miRNA processing machinery genes of XPO5 are involved in the prognosis of NSCLC.
A mutated and inactive XPO5 resulted in reduced miRNA processing and decreased miRNA target inhibition; the restored XPO5 seemed to act as a tumor suppressor and reversed the impaired export of pre-miRNA.23 The miR-SNP of rs11077 of XPO5 has been associated with the risk of esophageal cancer, as well as the survival in myeloma.19,24 The mechanism for this SNP modifying NSCLC survival remains unclear. This SNP is located in the 3′ UTR of XPO5, and the SNP might affect mRNA stability and alter the expression of XPO5. The fact that the rs11077 CC genotype was associated with reduced Renilla expression in a Renilla luciferase system implies that this SNP could modify XPO5 expression and result in overall expression of miRNA.19 The genes or miRNA binding at this SNP site to mediate XPO5 expression need to be further investigated.
We report for the first time that AC genotype of XPO5 exhibit a better chemotherapy response than that of AA type. The altered XPO5 expression may affect the expression of miRNAs specific for drug metabolism, leading to an altered chemotherapy response and advanced NSCLC survival. miRNA expressional profiles upon altered XPO5 expression need to be performed to identify the candidate miRNAs responsible for chemotherapy response.
Only one healthy control carried the homozygous CC allele of this miR-SNP. The frequency of the CC allele is dramatically lower in our patients from the Hebei area in China than is reported in the European population (less than 1% versus 21.7%). Several hundred non-Hodgkin lymphoma and esophageal cancer patients were examined for the rs11077 SNP distribution, and the homozygous CC allele had a low frequency of less than 1% (Guo, unpublished data, 2013).
Although miR-SNP studies for miRNA processing machinery genes are at an early stage, our results are encouraging because they indicate that miR-SNPs may have an effect on chemotherapy sensitivity and cancer survival. However, the results from this study require validation in other populations and in laboratory-based functional studies. MicroRNAs have been emphasized as a key factor in patients’ susceptibility to therapeutic response in many complex diseases, including cancer.25
Conclusion
A miR-SNP in the 3′ UTR region of XPO5 was found to be an independent prognostic marker for chemosensitivity and survival in advanced NSCLC patients.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(Suppl 8):S4–S66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 2.Cao C, Zhang YM, Wang R, et al. Excision repair cross complementation group 1 polymorphisms and lung cancer risk: a meta-analysis. Chin Med J (Engl) 2011;124:2203–2208. [PubMed] [Google Scholar]
- 3.Gandara D, Narayan S, Lara PN, Jr, et al. Integration of novel therapeutics into combined modality therapy of locally advanced non-small cell lung cancer. Clin Cancer Res. 2005;11:5057s–5062s. doi: 10.1158/1078-0432.CCR-05-9012. [DOI] [PubMed] [Google Scholar]
- 4.Yin M, Liao Z, Huang YJ, et al. Polymorphisms of homologous recombination genes and clinical outcomes of non-small cell lung cancer patients treated with definitive radiotherapy. PloS One. 2011;6:e20055. doi: 10.1371/journal.pone.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang P, Allen MS, Aubry MC, et al. Clinical features of 5,628 primary lung cancer patients: experience at Mayo Clinic from 1997 to 2003. Chest. 2005;128:452–462. doi: 10.1378/chest.128.1.452. [DOI] [PubMed] [Google Scholar]
- 6.Cullen MH, Billingham LJ, Woodroffe CM, et al. Mitomycin, ifosfamide, and cisplatin in unresectable non-small-cell lung cancer: effects on survival and quality of life. J Clin Oncol. 1999;17:3188–3194. doi: 10.1200/JCO.1999.17.10.3188. [DOI] [PubMed] [Google Scholar]
- 7.Bi N, Yang M, Zhang L, et al. Cyclooxygenase-2 genetic variants are associated with survival in unresectable locally advanced non-small cell lung cancer. Clin Cancer Res. 2010;16:2383–2390. doi: 10.1158/1078-0432.CCR-09-2793. [DOI] [PubMed] [Google Scholar]
- 8.Boni V, Zarate R, Villa JC, et al. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fuorouracil and irinotecan. Pharmacogenomics J. 2011;11:429–436. doi: 10.1038/tpj.2010.58. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA target. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 14.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Z, Wu C, Wang X, Wang C, Zhang R, Shan B. A polymorphism at the miR-502 binding site in the 3′-untranslated region of the histone methyltransferase SET8 is associated with hepatocellular carcinoma outcome. Int J Cancer. 2012;131:1318–1322. doi: 10.1002/ijc.27352. [DOI] [PubMed] [Google Scholar]
- 18.Campayo M, Navarro A, Viñolas N, et al. A dual role for KRT81: a miR-SNP associated with recurrence in non-small-cell lung cancer and a novel marker of squamous cell lung carcinoma. PLoS One. 2011;6:e22509. doi: 10.1371/journal.pone.0022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Larrea CF, Navarro A, Tejero R, et al. Impact of MiRSNPs on survival and progression in patients with multiple myeloma undergoing autologous stem cell transplantation. Clin Cancer Res. 2012;18:3697–3704. doi: 10.1158/1078-0432.CCR-12-0191. [DOI] [PubMed] [Google Scholar]
- 20.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 21.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 22.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 23.Melo SA, Moutinho C, Ropero S, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]


