SUMMARY
Draft genome sequences for Schistosoma japonicum and S. mansoni are now available. The schistosome genome encodes ~13000 protein-encoding genes for which the functions of few are well understood. Nonetheless, the new genes represent potential intervention targets, and molecular tools are being developed to determine their importance. Over the past 15 years, noteworthy progress has been achieved towards development of tools for gene manipulation and transgenesis of schistosomes. A brief history of genetic manipulation is presented, along with a review of the field with emphasis on reports of integration of transgenes into schistosome chromosomes.
Keywords: Schistosomes, genetic manipulation, transgenesis, chromosome integration, germ line, retrovirus, murine leukaemia virus, pseudotyped gammaretrovirus, transposon, piggyBac
INTRODUCTION
Schistosomes are considered the most important of the human helminth infections in terms of morbidity and mortality. More than 200 million people are infected with schistosomes and a further 800 million are at risk of schistosomiasis in >75 countries in tropical and sub-tropical latitudes. Treatment and control of schistosomiasis rely on the anthelmintic drug praziquantel, but there is concern that drug resistance will appear. New interventions, including vaccines, drugs and diagnostics, are needed for this neglected tropical disease (Hotez et al. 2008; Brindley et al. 2009, and references therein).
Draft genome sequences for Schistosoma japonicum and S. mansoni were reported recently, a landmark event that ushered in the post-genomic era for schistosomiasis (Schistosoma japonicum Genome & Functional Analysis Consortium, 2009; Berriman et al. 2009; Han et al. 2009). Despite the abundant new datasets, functional analysis of target genes to underpin new interventions for schistosomiasis requires routine approaches for both reverse and forward genetics. To date, functional genomics approaches beyond conventional RNA interference have not been available for schistosomes although reporter plasmids and RNAs have been introduced to several developmental stages (e.g. Davis et al. 1999; Wippersteg et al. 2002b, 2005; Mourão et al. 2009; Krautz-Peterson et al. 2010; Stefanic et al. 2010). Functional genomics including somatic and germline transgenesis are desirable because these techniques facilitate validation of essential genes/gene products to be targeted with drugs or vaccines (e.g. van Ooij et al. 2008; Homann et al. 2009; Langridge et al. 2009; Buguliskis et al. 2010). This review addresses genetic manipulation of schistosomes. More specifically, this review focuses on reports of genetic manipulation of schistosomes dealing with approaches targeting integration of transgenes into schistosome chromosomes.
BRIEF HISTORY OF GENETIC MANIPULATION IN SCHISTOSOMES
Advances with genetic manipulation of parasitic helminths including schistosomes have been reviewed (Grevelding, 2006; Brindley and Pearce, 2007; Kalinna and Brindley, 2007; Alrefaei et al. 2011; Mann et al. 2011). In brief, transgenesis of schistosomes was pioneered by Davis and co-workers who bombarded adult stages of S. mansoni with mRNA-encoding firefly luciferase and a luciferase-encoding plasmid (Davis et al. 1999). Subsequently, Grevelding and colleagues undertook a series of studies employing particle bombardment of S. mansoni stages using plasmids co-precipitated on gold beads. The plasmids encoded fluorescent reporter proteins and were driven by promiscuous (e.g. HSP70) or tissue-specific gene promoters (e.g. cathepsin F) from schistosomes (Wippersteg et al. 2002a, b, 2003, 2005; Rossi et al. 2003; Dvorak et al. 2010).
Heyers and colleagues introduced plasmid DNA (coated on gold beads) into miracidia, sporocysts and adults of S. mansoni by particle bombardment (Heyers et al. 2003). The plasmid construct encoded green fluorescent protein (GFP) under control of the S. mansoni HSP70 (heat shock protein 70 kDa) promoter and termination elements (Wippersteg et al. 2002b). The bombarded larvae and adults expressed GFP, and the transformed miracidia penetrated and established in the intermediate snail host Biomphalaria glabrata. Gold particles were detected in the germ balls of parasites in snail tissue, indicating feasibility of returning transformed parasites to the developmental cycle, a step expected to be useful for establishing lines of transgenic schistosomes.
Correnti and Pearce (2004) demonstrated that square wave electroporation could introduce reporter genes into schistosomes. Subsequently, the technique has found wide acceptance for introduction of plasmids, long dsRNA, siRNA, virions and other reporters into all three major species of human schistosomes, S. mansoni (e.g. Morales et al. 2008; Dvorak et al. 2010), S. japonicum (Zhao et al. 2008) and S. haematobium (Rinaldi, unpublished). Square wave electroporation has been successfully employed to introduce nucleic acids into eggs, miracidia, sporocysts, schistosomules and adult developmental stages of schistosomes, frequently using a single pulse of 125 volts for 20 milliseconds, in 4 mm gap pathway cuvettes (e.g. Correnti et al. 2005; Faghiri and Skelly, 2009; Kines et al. 2010). Moreover, electroporation has also been employed to develop a method to assess for the presence of an active RNAi pathway by silencing the exogenous reporter gene, firefly luciferase. This straightforward approach offers investigators a means to study the presence of a functional RNAi pathway in less well known parasites and/or to detect the activity of non-conventional interfering molecules such as short hairpin RNAs in schistosomes (Rinaldi et al. 2008; Ayuk et al. 2011).
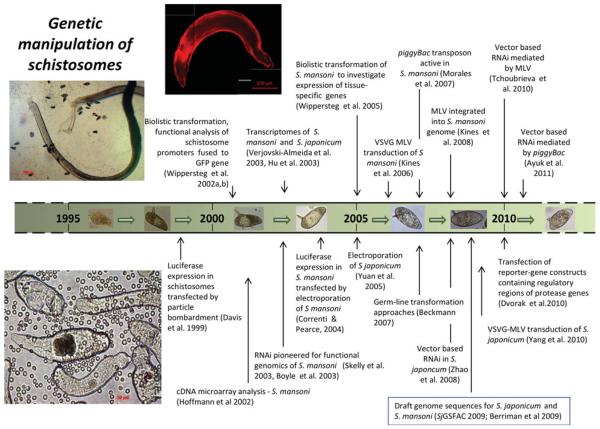
Findings reported by Grevelding and colleagues are notable in relation to the introduction of transgenes into the germline of schistosomes (Grevelding, 2006; Beckmann et al. 2007). Plasmids (both super-coiled and linear) encoding GFP were introduced into miracidia by particle bombardment, after which the transformed miracidia infected Biomphalaria glabrata snails by the natural route. Investigation of the cercariae (F0 generation) shed from the snails and adult worms from hamsters infected with the cercariae and of eggs (F1) from the rodents revealed the presence of the GFP transgene. Miracidia harvested from eggs obtained from the livers of the hamsters were used for snail infections and the resulting cercariae were employed to infect hamsters to derive subsequent schistosome generations, F2 and F3. Molecular analyses of F2 and F3 cercariae or adults failed to detect transgenes. Nonetheless, the findings demonstrated that the transgenes were passed from one developmental stage to the next within one generation and, furthermore, from one generation to the next. Since the germ cells are considered to be the only invariable cell type in the developmental cycle of the schistosome, the investigators concluded that the transgenes were present in the germline and their germline-transformation approach had succeeded. Loss or instability of the transgene before (non-Mendelian) inheritance by the F2 progeny likely occurred because transgenes had not integrated into the schistosome genome, a phenomenon well known with extrachromosomal arrays of transgenes in Caenorhabditis elegans (see Semple et al. 2010). Fig. 1 provides a time line - over the past 15 years - of the pioneering and key advances in the genetic manipulation of schistosomes.
Fig. 1.
Schematic time line of advances on genetic manipulation of schistosomes. Key events are noted, with pioneering or key reports cited.
SCHISTOSOME TRANSGENESIS WITH INTEGRATION COMPETENT VECTORS
Although approaches to genetic manipulation of schistosomes with non-integration competent vectors have been informative, there are major advantages to genomic integration of transgenes including Mendelian inheritance, sustained transgene activity and transgene-vectored RNA interference (see Giordano-Santini and Dupuy, 2011). Several classes of integration competent vectors enjoy utility in functional genomics and gene therapy for a spectrum of eukaryotes. These include transposons, gammaretroviruses, lentiviruses and recombinase systems (e.g. see Mates et al. 2007; Damasceno et al. 2010; Turan et al. 2011). Several of these are now being actively investigated for utility in integrating transgenes into the schistosome genome (Alrefaei et al. 2011).
Retroviruses
Both simple and complex retroviruses (family Retroviridae) are widely employed in functional genomics and gene therapy biotechnologies (e.g. see Hannon and Rossi, 2004; Petrus et al. 2010; Sliva and Schnierle, 2010). The simplex retroviruses include the genus Gammaretrovirus which includes the murine leukaemia virus (MLV). Complex retroviruses include the genus Lentivirus which includes the primate pathogens, HIV and SIV. Attributes of retroviruses that advance their appeal as gene transfer vectors include self-reliant infectious nature, ability to integrate into the chromosomes of the infected cell, potential to be modified to increase host cell range and availability of numerous constructs from commercial sources and academic colleagues.
For safety, retroviral vector systems are usually employed in two components – the retroviral vector, which does not encode viral proteins and the retrovirus packaging cell line, which provides the viral proteins necessary for vector transfer. Infectious, but replication incompetent virions are released from transfected packaging cells. The virus can infect target cells but cannot produce new virions after integration into host chromosome because the integrated provirus does not encode viral proteins (Miller, 1992). The restricted host-cell range of retroviral vectors limits their use for stable gene transfer in eukaryotic cells. To overcome this latter limitation, Burns and colleagues pioneered to use of vesicular stomatitis virus glycoprotein (VSVG) pseudotyped murine leukaemia virus (MLV)-derived vectors in which the retroviral envelope glycoprotein is replaced by the glycoprotein (G) of the rhabdovirus, vesicular stomatitis virus (VSV) (Burns et al. 1993). VSVG is able to bind to phospholipids on membranes of eukaryotic cells at large, endowing the VSVG pseudotyped virion with a potentially very broad range of target tissues and cells (Mastromarino et al. 1987; Emi et al. 1991). VSVG pseudotyped virions can infect non-mammalian cells including fish cell lines that are ordinarily refractory to infection because they do not express a cognate receptor for the envelope (surface) protein ligands of the wild type virions.
In our laboratory it has been well established that the infectious replication incompetent MLV retrovirus pseudotyped with VSVG can transduce S. mansoni leading to integration of retroviral transgenes into schistosome chromosomes (Kines et al. 2006, 2008, 2010; Rinaldi et al. 2011). This was notable given it was thought that evolutionary blocks would constrain the utility of MLV in non-mammalian taxa (Dirks and Miller, 2001). The MLV-derived vector pLNHX was modified to include reporter genes (firefly luciferase or green fluorescent protein) under the control of an endogenous schistosome gene promoter – the RNA polymerase II (Pol II) schistosome actin gene promoter or the RNA polymerase III (Pol III) Spliced Leader (SL) RNA gene promoter. Constructs and the plasmid encoding VSVG were employed to transfect GP2-293 packaging cells modified to express the MLV gag and pol genes (Mann et al. 2008). Eggs, primary sporocysts, schistosomules and adult stages of S. mansoni have been successfully transduced with the VSVG pseudotyped MLV virions. Two-colour immunofluorescence, Southern hybridization and RT-PCR confirmed successful transduction of the schistosomes by this gammaretrovirus. Furthermore, an anchored PCR (retrotransposon-anchored PCR, RAP) approach that employs primers specific for multi-copy endogenous mobile genetic elements interspersed in the schistosome genome was successfully deployed to locate integration junctions of transgenes in the genome of S. mansoni, definitively establishing the presence of proviral MLV transgenes integrated into schistosome chromosomes (Kines et al. 2008). In terms of promoters, schistosome actin, HSP70 and spliced leader (SL) gene promoters, as well as the 5′-LTR of MLV, all were found to drive transgene expression in viriontransduced schistosomes (Kines et al. 2006, 2008).
It is likely that the schistosome cells transduced by the virions were frequently tegumental and/or intestinal cells (Mann et al. 2008). However, we have also fragmented adult worms into several pieces before exposing the (still visibly motile) fragments to virions, which resulted in increased density of transgenes integrated into the schistosome chromosomes (Rinaldi et al. 2011). Nonetheless, in order for heritable transmission to occur, germline transduction would have to have taken place. The schistosome egg represents an advantageous developmental stage at which to direct transgenes (Kines et al. 2010; Mann et al. 2011). Accordingly, we proceeded to transduce schistosome eggs with VSVG-pseudotyped MLV facilitated by electroporation. Square wave electroporation was more effective in delivering VSVG-pseudotyped MLV into schistosome eggs than passive soaking. Quantitative PCR (qPCR) analysis revealed that schistosome eggs electroporated with virions had several fold more copies of provirus than eggs simply soaked in virions (Kines et al. 2010). These findings highlight the potential of the schistosome egg as a target into which to deliver chromosomal integration competent transgenes, aiming to establish germline transgenesis in schistosomes.
VSVG-pseudotyped MLV has been employed to transfer transgenes into S. japonicum; Yang et al. (2010) transduced schistosomules (perfused from rabbits) with retroviral transgene encoding human telomerase reverse transcriptase (hTERT). RT-PCR, in situ hybridization immunohistochemistry and immunoblot analysis determined that S. japonicum can be successfully transduced with VSVG-pseudotyped MLV and that the MLV vector can transport sizeable genes as cargo – the hTERT gene was ~3.5 kb in length (Yang et al. 2010). These findings also suggested the tantalizing possibility of using the hTERT transgene to immortalize cells from schistosome tissues, utilizing the oncogenic potential of hTERT to establish schistosome cell lines.
Transposons
Transposons are naturally occurring mobile genetic elements that move by a cut-and-paste mechanism; they are flanked by inverted terminal repeat (ITR) sequences and mobilized by a transposase encoded by their single open reading frame. There are ~20 superfamilies of these Class II mobile genetic elements, with member species widespread throughout eukaryote phyla (Feschotte and Pritham, 2007; Yuan and Wessler, 2011). Several are known from schistosomes including examples of the Merlin and CACTA groups (Berriman et al. 2009). Transposons can frequently mobilize in species phylogenetically distant from where they were first isolated, a facility which has been harnessed in functional genomics and experimental gene therapy (Plasterk et al. 1999; Ivics et al. 2009). Accordingly, it is feasible that exogenous transposons might also mobilize in schistosomes and thereby supply integration competent vectors for functional genomics of schistosomes. Several well-studied transposons including piggyBac, Hermes and mariner, are transpositionally active in planarians (Gonzalez-Estevez et al. 2003). In evolutionary terms, it is notable that host-parasite interactions play a key role in the horizontal transfer of transposons across phyla (Gilbert et al. 2010).
The piggyBac transposon is used widely in functional genomics and experimental gene therapy (Gonzalez-Estevez et al. 2003; Balu et al. 2005; Wilson et al. 2007). This transposon was isolated from the genome of a moth. It is a short inverted terminal repeat element of 2.5 kb in length with ITRs of 13 bp in length and a single open reading frame encodings the transposase. piggyBac exhibits precise excision upon transposition and affinity for TTAA target sites (Fraser et al. 1985, 1996; Cary et al. 1989; Elick et al. 1996). Recently, it has been determined that piggyBac is also active in schistosomes. Morales and colleagues examined whether the piggyBac transposon could deliver reporter transgenes into the genomes of S. mansoni (Morales et al. 2007). Linearized piggyBac donor plasmid carrying the firefly luciferase gene as reporter cargo under the control of schistosome gene promoters – actin (pXL-BacII-SmAct-Luc) or HSP70 (pXL-BacII-SmHSP70-Luc) – was introduced together with mRNA encoding the piggyBac transposase into cultured schistosomules by square wave electroporation. Activity of the helper transposase mRNA was confirmed by hybridization of genomic DNA from the transformed schistosomes to a luciferase gene probe. The hybridization signals indicated that the piggyBac transposon had integrated into numerous sites within schistosome chromosomes. Integration events were recovered using an anchored PCR approach employing several endogenous mobile genetic elements from the schistosome genome as anchors, which revealed characteristic piggyBac TTAA footprints in the vicinity of several protein encoding genes, annotated as adenylosuccinate lyase, glutathione peroxidase 1 and glutathione S transferase, as well as loci near endogenous mobile genetic elements including Boudicca and SR2. These findings provided the first direct evidence of somatic transgenesis of schistosomes, or indeed of any parasitic helminth. They demonstrated the transpositional activity of piggyBac in schistosomal tissues, expanding the host range of piggyBac to the digenetic trematodes. Very recently, we reported that vector-based RNAi activity driven by transgene cargo carried by the piggyBac vector pXL-BacII (Ayuk et al. 2011) – see below.
Other integration competent vectors
We are unaware of reports of other integrated transgenes in schistosomes beyond the findings with MLV and piggyBac (above). However, it is not unlikely that other retrovirus and transposons, including endogenous schistosome mobile genetic elements, could find utility in genome studies of schistosomes. A potential advantage of endogenous mobile genetic elements is that they may not suffer infection blocks from host restriction/innate immunity factors (e.g. Takeuchi and Matano, 2008; Strebel et al. 2009). DNA site-specific recombinases (SSRs), such as Cre, FLPe and φC31, from bacteriophages of fungi, and other provenances are influential tools for analyzing gene function in vertebrates (e.g. see Bischof and Basler, 2008) and might also be active in schistosomes. Several are now in service for site-specific gene manipulation of Plasmodium falciparum (Adjalley et al. 2010; O'Neill et al. 2011).
In overview, transgenesis mediated by integration competent vectors such as MLV and piggyBac can provide a routine functional genomics platform for forward and reverse genetics of schistosomes. Forward genetics, where specimens displaying a mutant phenotype after insertional (or chemical, etc.) mutagenesis are selected – the `from phenotype to genotype' approach – can now be attempted using transduction of schistosomes (e.g. with MLV or piggyBac) followed by high throughput sequence analysis of the schistosome genome in similar fashion to other pathogens (e.g. Langridge et al. 2009). Further, given that draft schistosome genomes are now available, MLV or piggyBac can be used for reverse genetics. With reverse genetics, functional analysis involves targeting a known gene sequence for inactivation where the function of the gene of interest is then inferred from the resulting phenotype (`from genotype to phenotype' approach) (Boutros and Ahringer, 2008). Conventional RNAi and more recently vector-based RNAi in schistosomes are the reverse genetic tools of choice for reverse genetics, approaches widely used for discovery of targets for experimental drug and/or vaccine development (e.g. Sayed et al. 2006; Mourão et al. 2009; Stefanic et al. 2010).
VECTOR-BASEDRNAi
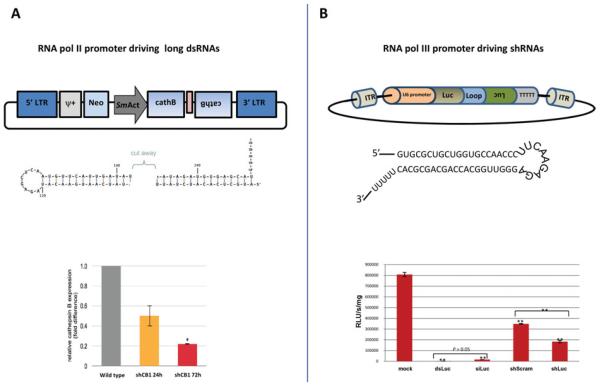
Experimental RNAi works well in schistosomes, in general (see Krautz-Peterson et al. 2010). Skelly and co-workers and Boyle and colleagues first described successful knockdown in S. mansoni (Boyle et al. 2003; Skelly et al. 2003), and since then numerous reports describing endogenous and reporter gene knockdown in S. mansoni and S. japonicum have appeared (e.g. Kumagai et al. 2009; Rinaldi et al. 2009). RNAi is active in S. haematobium (Rinaldi and co-workers, unpublished). However, conventional RNAi by double stranded RNA frequently leads to transient gene silencing and, in addition, may be inaccessible to some developmental stages and/or tissues of schistosomes. In vivo, e.g. vector-based RNAi approaches that lead to integration of transgenes encoding cassettes that express short interfering RNAs can circumvent deficiencies with exogenous RNAi approaches by providing continuous and/or conditional gene silencing (see Sliva and Schnierle, 2010). In brief, these experimental systems frequently employ a gene construct encoding an oligonucleotide of the target siRNA, a short loop domain (~9 residues), followed by the reverse complement of the siRNA, and driven by a Pol III (or Pol II) promoter. The construct can then be introduced into target cells for endogenous expression of shRNA targeting the gene of interest. The shRNA is processed in the cytoplasm to siRNA (Manjunath and Dykxhoorn, 2010). Both plasmid-based and retroviral (integrating) vectors are widely used for vector-based RNAi procedures, the latter offering long term gene silencing of expression (Couto and High, 2010; Sliva and Schnierle, 2010). Zhao and co-workers pioneered the approach in schistosomes, demonstrating silencing of the expression of the Mago nashi gene in S. japonicum by siRNAs derived from shRNA expressed by mammalian Pol III promoter H1 (Zhao et al. 2008). We recently demonstrated that MLV encoding long hairpin RNAs, ~120 bp long (hpRNA), driven by a RNA polymerase II promoter (S. mansoni actin) targeting S. mansoni cathepsin B in the adult stage of S. mansoni delivered silencing of the protease (Fig. 2, left panel) (Tchoubrieva et al. 2010). On the other hand, in many species including insects, mammals, birds and pathogenic protozoa, Pol III promoter-based DNA vectors have been employed to express small interfering RNA (siRNA) or short hairpin RNA (~21 bp long) (shRNA) (Lambeth et al. 2005; Wakiyama et al. 2005; Wise et al. 2007; Linford et al. 2009). Aiming to establish vector-based RNAi driven by a Pol III promoter, we cloned S. mansoni and human U6 gene promoters (~270 bp) into pLNHX driving shRNA targeting firefly luciferase. We targeted luciferase because the effect of RNAi against luciferase can be readily discerned (in contrast to many endogenous genes) (Rinaldi et al. 2008). Luciferase activity was significantly reduced in worms transduced with piggyBac encoding shLuc (Fig. 2, right) (Ayuk et al. 2011).
Fig. 2.
Vector-based RNA interference (RNAi) in Schistosoma mansoni. Panel A: Vector-based RNAi mediated by long hairpin RNAs (hpRNA) driven by a RNA polymerase II promoter (from the S. mansoni actin gene) carried by a retroviral vector to knock down of the S. mansoni cathepsin B1. Top: schematic representation of retroviral vector construct and dsRNA hairpin. Bottom: knock down of S. mansoni cathepsinB1. Panel B: Vector-based RNAi mediated by short hairpin RNAs (shRNA) driven by a RNA polymerase III promoter (from the S. mansoni U6 gene) carried by the piggyBac donor plasmid to knock down of the exogenous reporter gene firefly luciferase. Top: schematic representation of construct encoding the shRNA. Bottom: knock down of reporter firefly luciferase activity. Adapted from Tchoubrieva et al. (2010) and Ayuk et al. (2011) with permission.
DEVELOPMENTAL STAGES OF SCHISTOSOMES FOR TRANSGENESIS
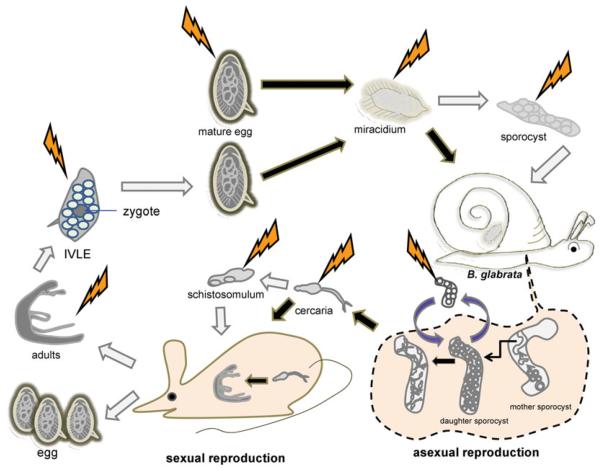
Since developmental cycles of the three major schistosomes of humans can be accomplished using laboratory rodents as the mammalian hosts and laboratory-reared snails as the intermediate hosts (Lewis, 1998), most developmental stages of these schistosomes are theoretically accessible to genetic manipulation (Fig. 3). Moreover, some stages can be cultured ex vivo or in vitro and returned to the snails or mice to continue development (see Mann et al. 2010). On the other hand, discrete stages are differentially accessible to delivery of transgenes using approaches including particle bombardment, square wave electroporation, cationic polymer-based gene delivery, and/or transduction by virions or other infectious agents (Heyers et al. 2003; Beckmann et al. 2007; Mann et al. 2008). Other approaches, such as microinjection, should be of value, as indicated by progress with introduction of transgenes in tape-worms and parasitic nematodes (Spiliotis et al. 2010).
Fig. 3.
Cartoon representation of points of the developmental cycle of Schistosoma mansoni amenable to genetic manipulation. Both the mammalian stages involved in sexual reproduction and the snail stages with asexual reproduction are presented. Thunder bolts suggest accessible points of introduction of transgenes into the schistosome e.g. transgene delivery by electroporation, microinjection, etc. Black arrows indicate processes that occur naturally whereas white arrows represent processes that can be manipulated. Dashed line indicates events inside the snail. IVLE, in vitro laid eggs.
Adult worms can be obtained from infected rodents and can be maintaining in culture. `Viable' fragments of worms – obtained by dicing adult schistosomes into several pieces – can be used as a study model as well (Rinaldi et al. 2011); whereas these fragments are not as tractable as primary cell cultures of Echinococcos multilocularis (Spiliotis et al. 2010) or fragments of planarians (Shibata et al. 2010), they do allow access to internal organs and cells of the schistosome. The schistosome egg and the miracidium have desirable attributes for consideration in relation to genetic manipulation, these include the presence of a single celled zygote within the egg-shell upon its release from the blood fluke (Jurberg et al. 2009), favourably high ratio of germ to somatic cells even as it develops and ease of maintenance in vitro. Primary sporocysts transformed from miracidia in vitro are worthy targets for genomic manipulation because this developmental stage can be transplanted into Biomphalaria glabrata snails to establish lines of S. mansoni (Kapp et al. 2003). Schistosomules obtained by mechanical transformation of cercariae shed from snails have been used to investigate the activity of transgenes and/or schistosome gene promoters driving transgenes (Correnti et al. 2005, 2007; Morales et al. 2007).
The `in vitro laid egg' (IVLE) deserves special mention. Pearce and colleagues demonstrated that eggs develop after release from adult schistosomes in vitro (Freitas et al. 2007). Eggs released from the fertilized adult female schistosome can develop in vitro and eventually release viable miracidia, provided that the eggs have been laid soon after the adult worms have been perfused from experimentally infected rodents. (From about 48 hours after perfusion from mice, eggs shed from worms exhibit reduced viability.) Notably, when released from the female worm, the schistosome egg includes a single cell zygote, in which cleavage has yet to take place (Jurberg et al. 2009). Accordingly, introduction of transgenes into this young egg may be able to accomplish germ (and somatic) transgenesis in a developmental stage that seems to be reasonably accessible in the laboratory (Mann et al. 2011).
Finally, it is obvious that the availability of immortalized cell lines would enhance functional genomics investigations (Brindley and Pearce, 2007). Unfortunately, none are yet available. Progress with primary cell cultures in related flatworms (Spiliotis et al. 2010) indicates that cell lines can be established and perhaps progress with transgenesis of schistosomes with oncogenes such as hTERT (Yang et al. 2010) will provide a route forward in this area.
PERSPECTIVE
We have reviewed advances in functional genomics and transgenesis of schistosomes, focusing on approaches leading to chromosomal integration of transgenes. The retrovirus MLV and the transposon piggyBac have now both been shown to integrate reliably into the chromosomes of S. mansoni and hence both exhibit great potential as vectors to drive functional genomics for schistosomes. However, improvements are needed to establish transgenic schistosomes and protocols. An impediment has been the difficulty of delivering transgenes to the germline. Targeting integration competent transgenes to IVLE may surmount this roadblock (Mann et al. 2011). Other gateways to the schistosome germline include the daughter sporocysts where the germ cells are comparatively massive (see Coustau and Yoshino, 2000). We envisage that advances in technologies which will drive functional genomics forward quickly, including expansion of in vivo RNAi, high-throughput insertional mutagenesis and, hopefully, gains-of-function approaches involving drug selection of transgenic schistosomes. Advances in S. mansoni can be expected to be adapted to the other schistosomes, to the food-borne flukes such as Opisthorchis viverrini, Clonorchis sinensis and Fasciola hepatica, and other helminth parasites at large.
ACKNOWLEDGEMENTS
Support from NIH-NIAID award R01AI072773 is gratefully acknowledged. We thank Tunika Okatcha and Danielle Skinner for critical review of the manuscript.
REFERENCES
- Adjalley SH, Lee MC, Fidock DA. A method for rapid genetic integration into Plasmodium falciparum utilizing mycobacteriophage Bxb1 integrase. Methods in Molecular Biology. 2010;634:87–100. doi: 10.1007/978-1-60761-652-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alrefaei YN, Okatcha TI, Skinner DE, Brindley PJ. Memórias do Instituto Oswaldo Cruz. 2011. Progress with schistosome transgenesis. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuk MA, Suttiprapa S, Rinaldi G, Mann VH, Lee CM, rindley PJ. Schistosoma mansoni U6 gene promoter-driven short hairpin RNA induces RNA interference in human fibrosarcoma cells and schistosomules. International Journal for Parasitology. 2011;41:783–789. doi: 10.1016/j.ijpara.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu B, Shoue DA, Fraser MJ, Jr., Adams JH. High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proceedings of the National Academy of Sciences, USA. 2005;102:16391–16396. doi: 10.1073/pnas.0504679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann S, Wippersteg V, El-Bahay A, Hirzmann J, Oliveira G, Grevelding CG. Schistosoma mansoni: germline transformation approaches and actin-promoter analysis. Experimental Parasitology. 2007;117:292–303. doi: 10.1016/j.exppara.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, Aslett MA, Bartholomeu DC, Blandin G, Caffrey CR, Coghlan A, Coulson R, Day TA, Delcher A, DeMarco R, Djikeng A, Eyre T, Gamble JA, Ghedin E, Gu Y, Hertz-Fowler C, Hirai H, Hirai Y, Houston R, Ivens A, Johnston DA, Lacerda D, Macedo CD, McVeigh P, Ning Z, Oliveira G, Overington JP, Parkhill J, Pertea M, Pierce RJ, Protasio AV, Quail MA, Rajandream MA, Rogers J, Sajid M, Salzberg SL, Stanke M, Tivey AR, White O, Williams DL, Wortman J, Wu W, Zamanian M, Zerlotini A, Fraser-Liggett CM, Barrell BG, El-Sayed NM. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Basler K. Recombinases and their use in gene activation, gene inactivation, and transgenesis. Methods in Molecular Biology. 2008;420:175–195. doi: 10.1007/978-1-59745-583-1_10. [DOI] [PubMed] [Google Scholar]
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nature Reviews Genetics. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Boyle JP, Wu XJ, Shoemaker CB, Yoshino TP. Using RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts. Molecular and Biochemical Parasitology. 2003;128:205–215. doi: 10.1016/s0166-6851(03)00078-1. [DOI] [PubMed] [Google Scholar]
- Brindley PJ, Mitreva M, Ghedin E, Lustigman S. Helminth genomics: the implications for human health. PLoS Neglected Tropical Diseases. 2009;3:e538. doi: 10.1371/journal.pntd.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindley PJ, Pearce EJ. Genetic manipulation of schistosomes. International Journal for Parasitology. 2007;37:465–473. doi: 10.1016/j.ijpara.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Buguliskis JS, Brossier F, Shuman J, Sibley LD. Rhomboid 4 (ROM4) affects the processing of surface adhesins and facilitates host cell invasion by Toxoplasma gondii. PLoS Pathogens. 2010;6:e1000858. doi: 10.1371/journal.ppat.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proceedings of the National Academy of Sciences, USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E, Fraser MJ. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- Correnti JM, Brindley PJ, Pearce EJ. Long-term suppression of cathepsin B levels by RNA interference retards schistosome growth. Molecular and Biochemical Parasitology. 2005;143:209–215. doi: 10.1016/j.molbiopara.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Correnti JM, Jung E, Freitas TC, Pearce EJ. Transfection of Schistosoma mansoni by electroporation and the description of a new promoter sequence for transgene expression. International Journal for Parasitology. 2007;37:1107–1115. doi: 10.1016/j.ijpara.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Correnti JM, Pearce EJ. Transgene expression in Schistosoma mansoni: introduction of RNA into schistosomula by electroporation. Molecular and Biochemical Parasitology. 2004;137:75–79. doi: 10.1016/j.molbiopara.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Coustau C, Yoshino TP. Flukes without snails: advances in the in vitro cultivation of intramolluscan stages of trematodes. Experimental Parasitology. 2000;94:62–66. doi: 10.1006/expr.1999.4462. [DOI] [PubMed] [Google Scholar]
- Couto LB, High KA. Viral vector-mediated RNA interference. Current Opinion in Pharmacology. 2010;10:534–542. doi: 10.1016/j.coph.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Damasceno JD, Beverley SM, Tosi LR. A transposon toolkit for gene transfer and mutagenesis in protozoan parasites. Genetica. 2010;138:301–311. doi: 10.1007/s10709-009-9406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Parra A, LoVerde PT, Ribeiro E, Glorioso G, Hodgson S. Transient expression of DNA and RNA in parasitic helminths by using particle bombardment. Proceedings of the National Academy of Sciences, USA. 1999;96:8687–8692. doi: 10.1073/pnas.96.15.8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks C, Miller AD. Many nonmammalian cells exhibit postentry blocks to transduction by gammaretroviruses pseudotyped with various viral envelopes, including vesicular stomatitis virus G glycoprotein. Journal of Virology. 2001;75:6375–6383. doi: 10.1128/JVI.75.14.6375-6383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, Beckmann S, Lim KC, Engel JC, Grevelding CG, McKerrow JH, Caffrey CR. Biolistic transformation of Schistosoma mansoni: Studies with modified reporter-gene constructs containing regulatory regions of protease genes. Molecular and Biochemical Parasitology. 2010;170:37–40. doi: 10.1016/j.molbiopara.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Elick TA, Bauser CA, Fraser MJ. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica. 1996;98:33–41. doi: 10.1007/BF00120216. [DOI] [PubMed] [Google Scholar]
- Emi N, Friedmann T, Yee JK. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. Journal of Virology. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghiri Z, Skelly PJ. The role of tegumental aquaporin from the human parasitic worm, Schistosoma mansoni, in osmoregulation and drug uptake. FASEB Journal. 2009;23:2780–2789. doi: 10.1096/fj.09-130757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser MJ, Brusca JS, Smith GE, Summers MD. Transposon-mediated mutagenesis of a baculovirus. Virology. 1985;145:356–361. doi: 10.1016/0042-6822(85)90172-2. [DOI] [PubMed] [Google Scholar]
- Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Molecular Biology. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Freitas TC, Jung E, Pearce EJ. TGF-beta signaling controls embryo development in the parasitic flatworm Schistosoma mansoni. PLoS pathogens. 2007;3:e52. doi: 10.1371/journal.ppat.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Schaack S, Pace JK, 2nd, Brindley PJ, Feschotte C. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature. 2010;464:1347–1350. doi: 10.1038/nature08939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano-Santini R, Dupuy D. Selectable genetic markers for nematode transgenesis. Cellular and Molecular Life Sciences. 2011;68:1917–1927. doi: 10.1007/s00018-011-0670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Estevez C, Momose T, Gehring WJ, Salo E. Transgenic planarian lines obtained by electroporation using transposon-derived vectors and an eye-specific GFP marker. Proceedings of the National Academy of Sciences, USA. 2003;100:14046–14051. doi: 10.1073/pnas.2335980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevelding CG. Transgenic flatworms. In: Maule AG, Marks NJ, editors. Parasitic Flatworms. Molecular Biology, Biochemisrty, Immunolgy and Physiology. CABI; Wallingford, UK: 2006. pp. 149–173. [Google Scholar]
- Han ZG, Brindley PJ, Wang SY, Chen Z. Schistosoma genomics: new perspectives on schistosome biology and host-parasite interaction. Annual Review of Genomics and Human Genetics. 2009;10:211–240. doi: 10.1146/annurev-genom-082908-150036. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Heyers O, Walduck AK, Brindley PJ, Bleiss W, Lucius R, Dorbic T, Wittig B, Kalinna BH. Schistosoma mansoni miracidia transformed by particle bombardment infect Biomphalaria glabrata snails and develop into transgenic sporocysts. Experimental Parasitology. 2003;105:174–178. doi: 10.1016/j.exppara.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Hoffmann KF, Johnston DA, Dunne DW. Identification of Schistosoma mansoni-gender associated transcripts by cDNA microarray profiling. Genome Biology. 2002;3:research0041–research0041.12. doi: 10.1186/gb-2002-3-8-research0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genetics. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, Jacobson J. Helminth infections: the great neglected tropical diseases. Journal of Clinical Investigation. 2008;118:1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, Izsvak Z. Transposon-mediated genome manipulation in vertebrates. Nature Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurberg AD, Goncalves T, Costa TA, de Mattos AC, Pascarelli BM, de Manso PP, Ribeiro-Alves M, Pelajo-Machado M, Peralta JM, Coelho PM, Lenzi HL. The embryonic development of Schistosoma mansoni eggs: proposal for a new staging system. Development Genes and Evolution. 2009;219:219–234. doi: 10.1007/s00427-009-0285-9. [DOI] [PubMed] [Google Scholar]
- Kalinna BH, Brindley PJ. Manipulating the manipulators: advances in parasitic helminth transgenesis and RNAi. Trends in Parasitology. 2007;23:197–204. doi: 10.1016/j.pt.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Kapp K, Coustau C, Wippersteg V, Jourdane J, Kunz W, Grevelding CG. Transplantation of in vitro-generated Schistosoma mansoni mother sporocysts into Biomphalaria glabrata. Parasitology Research. 2003;91:482–485. doi: 10.1007/s00436-003-0951-1. [DOI] [PubMed] [Google Scholar]
- Kines KJ, Mann VH, Morales ME, Shelby BD, Kalinna BH, Gobert GN, Chirgwin SR, Brindley PJ. Transduction of Schistosoma mansoni by vesicular stomatitis virus glycoprotein-pseudotyped Moloney murine leukemia retrovirus. Experimental Parasitology. 2006;112:209–220. doi: 10.1016/j.exppara.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Kines KJ, Morales ME, Mann VH, Gobert GN, Brindley PJ. Integration of reporter transgenes into Schistosoma mansoni chromosomes mediated by pseudotyped murine leukemia virus. FASEB Journal. 2008;22:2936–2948. doi: 10.1096/fj.08-108308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kines KJ, Rinaldi G, Okatcha TI, Morales ME, Mann VH, Tort JF, Brindley PJ. Electroporation facilitates introduction of reporter transgenes and virions into schistosome eggs. PLoS Neglected Tropical Diseases. 2010;4:e593. doi: 10.1371/journal.pntd.0000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautz-Peterson G, Bhardwaj R, Faghiri Z, Tararam CA, Skelly PJ. RNA interference in schistosomes: machinery and methodology. Parasitology. 2010;137:485–495. doi: 10.1017/S0031182009991168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T, Osada Y, Ohta N, Kanazawa T. Peroxiredoxin-1 from Schistosoma japonicum functions as a scavenger against hydrogen peroxide but not nitric oxide. Molecular and Biochemical Parasitology. 2009;164:26–31. doi: 10.1016/j.molbiopara.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Lambeth LS, Moore RJ, Muralitharan M, Dalrymple BP, McWilliam S, Doran TJ. Characterisation and application of a bovine U6 promoter for expression of short hairpin RNAs. BMC Biotechnology. 2005;5:13. doi: 10.1186/1472-6750-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Research. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis FA. Schistosomiasis. Current Protocols in Immunology. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, editors. Animal Models for Infectious Diseases. Suppl. 28. Wiley; New York: 1998. [Google Scholar]
- Linford AS, Moreno H, Good KR, Zhang H, Singh U, Petri WA., Jr. Short hairpin RNA-mediated knockdown of protein expression in Entamoeba histolytica. BMC Microbiology. 2009;9:38. doi: 10.1186/1471-2180-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath N, Dykxhoorn DM. Advances in synthetic siRNA delivery. Discovery Medicine. 2010;9:418–430. [PubMed] [Google Scholar]
- Mann VH, Morales ME, Kines KJ, Brindley PJ. Transgenesis of schistosomes: approaches employing mobile genetic elements. Parasitology. 2008;135:141–153. doi: 10.1017/S0031182007003824. [DOI] [PubMed] [Google Scholar]
- Mann VH, Morales ME, Rinaldi G, Brindley PJ. Culture for genetic manipulation of developmental stages of Schistosoma mansoni. Parasitology. 2010;137:451–462. doi: 10.1017/S0031182009991211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann VH, Suttiprapa S, Rinaldi G, Brindley PJ. Establishing transgenic schistosomes. PLoS Neglected Tropical Diseases. 2011 doi: 10.1371/journal.pntd.0001230. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastromarino P, Conti C, Goldoni P, Hauttecoeur B, Orsi N. Characterization of membrane components of the erythrocyte involved in vesicular stomatitis virus attachment and fusion at acidic pH. Journal of General Virology. 1987;68:2359–2369. doi: 10.1099/0022-1317-68-9-2359. [DOI] [PubMed] [Google Scholar]
- Mates L, Izsvak Z, Ivics Z. Technology transfer from worms and flies to vertebrates: transposition-based genome manipulations and their future perspectives. Genome Biology. 2007;8(Suppl 1):S1. doi: 10.1186/gb-2007-8-s1-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AD. Human gene therapy comes of age. Nature. 1992;357:455–460. doi: 10.1038/357455a0. [DOI] [PubMed] [Google Scholar]
- Morales ME, Mann VH, Kines KJ, Gobert GN, Fraser MJ, Jr., Kalinna BH, Correnti JM, Pearce EJ, Brindley PJ. piggyBac transposon mediated transgenesis of the human blood fluke, Schistosoma mansoni. FASEB Journal. 2007;21:3479–3489. doi: 10.1096/fj.07-8726com. [DOI] [PubMed] [Google Scholar]
- Morales ME, Rinaldi G, Gobert GN, Kines KJ, Tort JF, Brindley PJ. RNA interference of Schistosoma mansoni cathepsin D, the apical enzyme of the hemoglobin proteolysis cascade. Molecular and Biochemical Parasitology. 2008;157:160–168. doi: 10.1016/j.molbiopara.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourão MM, Dinguirard N, Franco GR, Yoshino TP. Phenotypic screen of early-developing larvae of the blood fluke, Schistosoma mansoni, using RNA interference. PLoS Neglected Tropical Diseases. 2009;3:e502. doi: 10.1371/journal.pntd.0000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MT, Phuong T, Healer J, Richard D, Cowman AF. Gene deletion from Plasmodium falciparum using FLP and Cre recombinases: implications for applied site-specific recombination. International Journal for Parasitology. 2011;41:117–123. doi: 10.1016/j.ijpara.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Petrus I, Chuah M, VandenDriessche T. Gene therapy strategies for hemophilia: benefits versus risks. Journal of Gene Medicine. 2010;12:797–809. doi: 10.1002/jgm.1500. [DOI] [PubMed] [Google Scholar]
- Plasterk RH, Izsvak Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends in Genetics. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- Rinaldi G, Morales ME, Alrefaei YN, Cancela M, Castillo E, Dalton JP, Tort JF, Brindley PJ. RNA interference targeting leucine aminopeptidase blocks hatching of Schistosoma mansoni eggs. Molecular and Biochemical Parasitology. 2009;167:118–126. doi: 10.1016/j.molbiopara.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi G, Morales ME, Cancela M, Castillo E, Brindley PJ, Tort JF. Development of functional genomic tools in trematodes: RNA interference and luciferase reporter gene activity in Fasciola hepatica. PLoS Neglected Tropical Diseases. 2008;2:e260. doi: 10.1371/journal.pntd.0000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi G, Suttiprapa S, Brindley PJ. Quantitative retrotransposon anchored PCR confirms transduction efficiency of transgenes in adult Schistosoma mansoni. Molecular and Biochemical Parasitology. 2011;177:70–76. doi: 10.1016/j.molbiopara.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Wippersteg V, Klinkert MQ, Grevelding CG. Cloning of 5′ and 3′ flanking regions of the Schistosoma mansoni calcineurin A gene and their characterization in transiently transformed parasites. Molecular and Biochemical Parasitology. 2003;130:133–138. doi: 10.1016/s0166-6851(03)00158-0. [DOI] [PubMed] [Google Scholar]
- Sayed AA, Cook SK, Williams DL. Redox balance mechanisms in Schistosoma mansoni rely on peroxiredoxins and albumin and implicate peroxiredoxins as novel drug targets. Journal of Biological Chemistry. 2006;281:17001–17010. doi: 10.1074/jbc.M512601200. [DOI] [PubMed] [Google Scholar]
- Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple JI, Garcia-Verdugo R, Lehner B. Rapid selection of transgenic C. elegans using antibiotic resistance. Nature Methods. 2010;7:725–727. doi: 10.1038/nmeth.1495. [DOI] [PubMed] [Google Scholar]
- Shibata N, Rouhana L, Agata K. Cellular and molecular dissection of pluripotent adult somatic stem cells in planarians. Development, Growth and Differentiation. 2010;52:27–41. doi: 10.1111/j.1440-169X.2009.01155.x. [DOI] [PubMed] [Google Scholar]
- Skelly PJ, Da'dara A, Harn DA. Suppression of cathepsin B expression in Schistosoma mansoni by RNA interference. International Journal for Parasitology. 2003;33:363–369. doi: 10.1016/s0020-7519(03)00030-4. [DOI] [PubMed] [Google Scholar]
- Sliva K, Schnierle BS. Selective gene silencing by viral delivery of short hairpin RNA. Virology Journal. 2010;7:248. doi: 10.1186/1743-422X-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis M, Mizukami C, Oku Y, Kiss F, Brehm K, Gottstein B. Echinococcus multilocularis primary cells: improved isolation, small-scale cultivation and RNA interference. Molecular and Biochemical Parasitology. 2010;174:83–87. doi: 10.1016/j.molbiopara.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Stefanic S, Dvorak J, Horn M, Braschi S, Sojka D, Ruelas DS, Suzuki B, Lim KC, Hopkins SD, McKerrow JH, Caffrey CR. RNA interference in Schistosoma mansoni schistosomula: selectivity, sensitivity and operation for larger-scale screening. PLoS Neglected Tropical Diseases. 2010;4:e850. doi: 10.1371/journal.pntd.0000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K, Luban J, Jeang KT. Human cellular restriction factors that target HIV-1 replication. BMC Medicine. 2009;7:48. doi: 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Matano T. Host factors involved in resistance to retroviral infection. Microbiology and Immunology. 2008;52:318–325. doi: 10.1111/j.1348-0421.2008.00040.x. [DOI] [PubMed] [Google Scholar]
- Tchoubrieva EB, Ong PC, Pike RN, Brindley PJ, Kalinna BH. Vector-based RNA interference of cathepsin B1 in Schistosoma mansoni. Cellular and Molecular Life Sciences. 2010;67:3739–3748. doi: 10.1007/s00018-010-0345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, Zehe C, Bode J. Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. Journal of Molecular Biology. 2011;407:193–221. doi: 10.1016/j.jmb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- van Ooij C, Tamez P, Bhattacharjee S, Hiller NL, Harrison T, Liolios K, Kooij T, Ramesar J, Balu B, Adams J, Waters AP, Janse CJ, Haldar K. The malaria secretome: from algorithms to essential function in blood stage infection. PLoS Pathogens. 2008;4:e1000084. doi: 10.1371/journal.ppat.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Matsumoto T, Yokoyama S. Drosophila U6 promoter-driven short hairpin RNAs effectively induce RNA interference in Schneider 2 cells. Biochemical and Biophysical Research Communications. 2005;331:1163–1170. doi: 10.1016/j.bbrc.2005.03.240. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Coates CJ, George AL., Jr. PiggyBac transposon-mediated gene transfer in human cells. Molecular Therapy. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- Wippersteg V, Kapp K, Kunz W, Grevelding CG. Characterisation of the cysteine protease ER60 in transgenic Schistosoma mansoni larvae. International Journal for Parasitology. 2002a;32:1219–1224. doi: 10.1016/s0020-7519(02)00092-9. [DOI] [PubMed] [Google Scholar]
- Wippersteg V, Kapp K, Kunz W, Jackstadt WP, Zahner H, Grevelding CG. HSP70-controlled GFP expression in transiently transformed schistosomes. Molecular and Biochemical Parasitology. 2002b;120:141–150. doi: 10.1016/s0166-6851(01)00446-7. [DOI] [PubMed] [Google Scholar]
- Wippersteg V, Ribeiro F, Liedtke S, Kusel JR, Grevelding CG. The uptake of Texas Red-BSA in the excretory system of schistosomes and its colocalisation with ER60 promoter-induced GFP in transiently transformed adult males. International Journal for Parasitology. 2003;33:1139–1143. doi: 10.1016/s0020-7519(03)00168-1. [DOI] [PubMed] [Google Scholar]
- Wippersteg V, Sajid M, Walshe D, Khiem D, Salter JP, McKerrow JH, Grevelding CG, Caffrey CR. Biolistic transformation of Schistosoma mansoni with 5′ flanking regions of two peptidase genes promotes tissue-specific expression. International Journal for Parasitology. 2005;35:583–589. doi: 10.1016/j.ijpara.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Wise TG, Schafer DJ, Lambeth LS, Tyack SG, Bruce MP, Moore RJ, Doran TJ. Characterization and comparison of chicken U6 promoters for the expression of short hairpin RNAs. Animal Biotechnology. 2007;18:153–162. doi: 10.1080/10495390600867515. [DOI] [PubMed] [Google Scholar]
- Yang S, Brindley PJ, Zeng Q, Li Y, Zhou J, Liu Y, Liu B, Cai L, Zeng T, Wei Q, Lan L, McManus DP. Transduction of Schistosoma japonicum schistosomules with vesicular stomatitis virus glycoprotein pseudotyped murine leukemia retrovirus and expression of reporter human telomerase reverse transcriptase in the transgenic schistosomes. Molecular and Biochemical Parasitology. 2010;174:109–116. doi: 10.1016/j.molbiopara.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan XS, Shen JL, Wang XL, Wu XS, Liu DP, Dong HF, Jiang MS. Schistosoma japonicum: a method for transformation by electroporation. Experimental Parasitology. 2005;111:244–249. doi: 10.1016/j.exppara.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Yuan YW, Wessler SR. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proceedings of the National Academy of Sciences, USA. 2011;108:7884–7889. doi: 10.1073/pnas.1104208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZR, Lei L, Liu M, Zhu SC, Ren CP, Wang XN, Shen JJ. Schistosoma japonicum: inhibition of Mago nashi gene expression by shRNA-mediated RNA interference. Experimental Parasitology. 2008;119:379–384. doi: 10.1016/j.exppara.2008.03.015. [DOI] [PubMed] [Google Scholar]