Abstract
In contrast to other primary EGFR mutations in lung adenocarcinomas, insertions in exon 20 of EGFR have been generally associated with resistance to EGFR tyrosine kinase inhibitors. Their molecular spectrum, clinicopathologic characteristics and prevalence are not well established. Tumors harboring EGFR exon 20 insertions were identified through an algorithmic screen of 1500 lung adenocarcinomas. Cases were first tested for common mutations in EGFR (exons 19 and 21) and KRAS (exon 2) and, if negative, further analyzed for EGFR exon 20 insertions. All samples underwent extended genotyping for other driver mutations in EGFR, KRAS, BRAF, NRAS, PIK3CA, MEK1 and AKT by mass spectrometry; a subset was evaluated for ALK rearrangements. We identified 33 EGFR exon 20 insertion cases (2.2%, 95% CI 1.6 to 3.1%), all mutually exclusive with mutations in the other genes tested (except PIK3CA). They were more common among never-smokers (p<0.0001). There was no association with age, sex, race, or stage. Morphologically, tumors were similar to those with common EGFR mutations, but with frequent solid histology. Insertions were highly variable in position and size, ranging from 3 to 12bp, resulting in 13 different insertions which, by molecular modeling, are predicted to have potentially different effects on erlotinib binding. EGFR exon 20 insertion testing identifies a distinct subset of lung adenocarcinomas, accounting for at least 9% of all EGFR mutated cases, representing the third most common type of EGFR mutation after exon 19 deletions and L858R. Insertions are structurally heterogeneous with potential implications for response to EGFR inhibitors.
Keywords: EGFR exon 20, EGFR, epidermal growth factor receptor, lung adenocarcinoma, driver oncogenes
INTRODUCTION
The identification of activating mutations within the tyrosine kinase (TK) domain of EGFR has transformed the management of patients with non-small cell lung cancers. Starting with the initial studies, two mutation types have been recognized as the most prevalent and clinically significant: in-frame deletions in exon 19 and the point mutation L858R (1-3). Together, these represent approximately 90% of all EGFR mutations and their association with response to tyrosine kinase inhibitors (TKIs) is well characterized. Mutations involving codons G719 and L861 are also associated with sensitivity but their incidence is much lower.
Insertions in exon 20 are included among the rarer activating mutations in the TK domain of EGFR.(4-9) They represent a combination of in-frame insertions and/or duplications of 3 to 21 base pairs, predominantly clustered between codons 767 and 774. Importantly, in contrast to the more classic activating EGFR mutations, these insertions have been associated with de-novo resistance to approved EGFR TKIs (erlotinib and gefitinib) (10-14) and to irreversible inhibitors that have recently entered clinical trials (neratinib, afatinib and dacomitinib)(10-16). In vitro studies show that cells harboring some of the most prevalent insertions require an average of 100-fold higher concentrations of these agents for inhibition, well beyond clinically achievable plasma levels. Clinical studies, although limited, confirm the pre-clinical findings (6, 8, 9, 12, 15-20) but rare cases with better clinical responses have been reported (8, 18, 20). Importantly, many of the insertions identified in patient samples have not been tested against these inhibitors. Further understanding of the biology, prognostic and predictive implications of these mutations is needed but has remained limited by the small number of patients included in clinical trials and the lack of preclinical models, such as patient derived cell lines or genetically engineered mouse models.
Despite the importance of EGFR exon 20 insertions as potentially targetable driver mutations, to date only a few reports have been dedicated to these tumors and most have been confined to East Asian populations. In this setting, with the exception of EGFR TKI sensitivity, the clinical and pathologic characteristics seem to closely match those of the classic EGFR mutations, including predilection for females, never smokers and adenocarcinoma histology. While the true incidence of these mutations is not yet well defined, with reports ranging from 0-13% (4, 6-8, 21, 22), reviews have suggested that insertions in exon 20 may represent up to 4% of all EGFR mutations (23). The incidence, clinicopathologic characteristics and molecular spectrum of these mutant tumors remain to be explored in the US population.
The aim of the current study was 1) to determine the frequency and molecular spectrum of EGFR exon 20 insertions in a large cohort of patients with lung adenocarcinomas, 2) to assess the clinical and histopathologic characteristics and 3) to confirm their mutually exclusive nature with mutations in EGFR, KRAS, BRAF, ERBB2/HER2, NRAS, PIK3CA, MAP2K1/MEK1 and AKT as well as ALK rearrangements.
METHODS
Patients and mutation analysis
Clinical cases of lung adenocarcinomas received for routine EGFR and KRAS testing at Memorial Sloan-Kettering Cancer Center between January 2009 and January 2011 were selected for the study, under an IRB-approved waiver. The study period was chosen to allow a minimum of 1 year of potential follow-up time.
Clinical testing for the detection of major mutations in EGFR (exon 19 deletions and L858R) and KRAS (exon 2) was carried out by fragment analysis and Sanger sequencing, respectively, using previously described methods (24, 25). Extended mutation analysis for other recurrent point mutations in EGFR, KRAS, BRAF, ERBB2/HER2, NRAS, AKT, MAP2K1 and PIK3CA was performed in all cases by mass spectrometry genotyping (Sequenom) as previously described (26). Briefly, samples were subjected to a series of multiplexed assays designed to interrogate a total of 92 non-synonymous mutations in 6 multiplex reactions (see Supplementary table S1 for complete list of tested mutations). Amplification and single base pair extension primers were designed with the Sequenom Assay Designer v3.1 software. Allele-specific single base extension products were quantitatively analyzed using matrix-assisted laser desorption/ionization-time of flight/mass spectrometry (MALDI-TOF/MS) on the Sequenom MassArray Spectrometer. All automated system mutation calls were confirmed by manual review of the spectra. All testing was carried out in duplicate.
When sufficient tissue was available, samples that were EGFR/KRAS wild type were also tested for ALK rearrangements by fluorescent in-situ hybridization (Vysis ALK Break Apart FISH Probe Kit) using standard protocols (Supplementary Figure S1 outlines the sequential genotyping algorithm).
Testing for EGFR exon 20 insertions
Assessment for insertions in exon 20 of EGFR primarily targeted cases known to be negative for major EGFR (exon 19 del, L858R) and KRAS mutations, given previous reports of their mutual exclusivity and further based on DNA availability. Initial screening was performed by a sizing assay (9, 24) using primers FW1:5′-TCTTCACCTGGAAGGGGTCCA-3′ and REV1:5′-Fam-TGCCACCTCCACTCCGTCTA-3′). Positive cases were characterized by Sanger sequencing using primers FW1:5′-CATTCATGCGTCTTCACCTG-3′ and REV1:5′-GTATAGGGGTACCGTTTGAG-3′ following previously described protocols.
To confirm the mutually exclusivity of EGFR exon 20 insertions with major EGFR and KRAS mutations, as well as other rarer mutations not well represented in our cohort, we tested a separate set of adenocarcinomas with a known positive mutation profile and sufficient DNA. Also, to confirm that mutations were confined to adenocarcinomas, we tested sets of squamous and small cell carcinomas following similar protocols.
Histopathology
Morphologic analysis was performed by semi-quantitatively recording 6 patterns – lepidic (bronchioloalveolar), acinar, papillary, micropapillary, solid and mucinous. The distribution of morphologic patterns in adenocarcinomas with exon 20 insertions was compared to a control group of adenocarcinomas with canonical EGFR mutations. The groups were compared by two-tailed Fisher’s exact test.
Statistical Analysis
The association between EGFR mutation status and clinical and biological characteristics was analyzed by Fisher’s exact test. Age differences were compared using the t test for independent samples. The two-sided significance level was set at p < 0.05.
Prediction of functional impact of exon 20 insertions
To examine the likelihood that previously unreported mutations identified in our series would have similar impact on the function of the protein, we used a computational biology approach utilizing the publicly available Mutation Assessor software as described in detail elsewhere (27).
RESULTS
Initial screening
A total of 1500 adenocarcinomas were reviewed for the study and screened under the clinical and extended mass spectrometry genotyping assays. Of these, 901 were positive for mutations (60%, 901/1500), 36 had ALK rearrangements (437 tested) and 563 had no alterations detected. The latter group of tumors were termed “pan-negative”. The detailed distribution of mutations is outlined in supplementary table 2.
EGFR exon 20 insertion testing
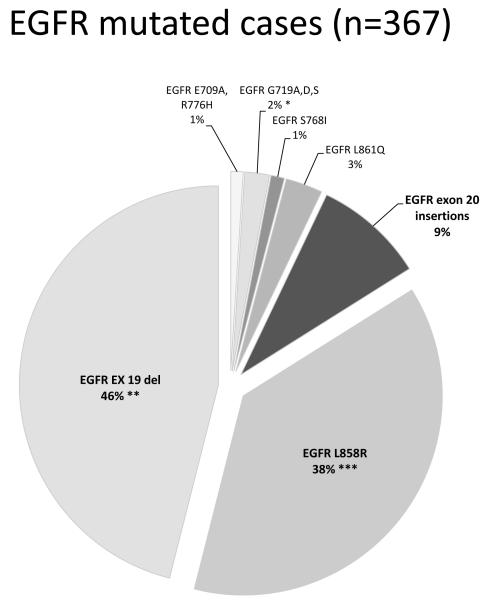
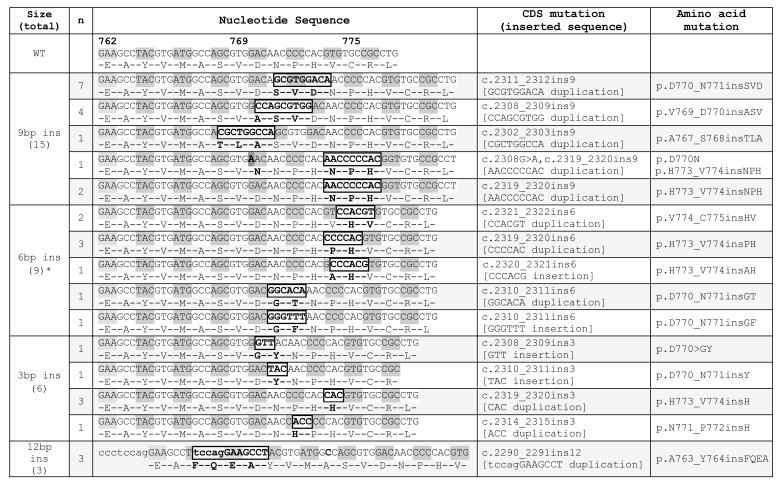
A total of 600 cases were tested. This included 464 pan-negative tumors (out of the 563 above cases negative for all other mutations and ALK rearrangements) and 136 mutation positive tumors as specified in supplementary table 3. We could not test the remaining 99 of the 563 pan-negative cases due to unavailable material. Among the tested group, we detected 33 insertion mutations (6% of tested, 7% of the pan-negative set), all mutually exclusive with other genetic alterations except for 2 with concurrently mutated PIK3CA (both H1047R). Insertions of 9 base pairs (bp) were the most common mutation type (48%, 16/32). Sanger sequencing of 32 positive cases showed all mutations were confined to the 5′ end of the exon, between codons A763 and C775 comprising duplications and insertions as outlined in Table 1. At the amino acid level, mutations were highly variable with 13 different types identified. One sample harbored a concurrent D770N point mutation. The specific insertion sequence could not be ascertained in one tumor (6bp insertion) withvery low mutant peaks due to very low tumor content. EGFR exon 20 insertions represented 9% (33/367) of all EGFR mutated tumors (figure 1).
Table 1.
EGFR exon 20 insertions identified in the study
|
One 6bp insertion could not be characterized by Sanger sequencing due to very low mutant peaks
Figure 1.
Distribution of all primary EGFR mutations identified in the current study. Although, based on our analysis, insertions in exon 20 corresponded to 9% of all EGFR mutated samples, we estimate that the true incidence may be even higher, closer to 11%, factoring in the expected positive cases which would have been detected if the entire negative group had been tested. Figure includes 5 cases with double mutations as follows *2 cases with double mutations G719A/S768I and G719S/E709A, **2 cases with concurrent T790M at baseline, *** 1 case with concurrent T790M at baseline.
Testing of the separate set of adenocarcinomas with a known positive mutation profile (n=311, 70 EGFR ex 19 del, 70 L858R, 120 KRAS G12&G13, 7 NRAS, 3 MAP2K1, 2 AKT, 30 BRAF) identified no EGFR exon 20 insertions, confirming their mutually exclusive relationship with these other driver mutations. No EGFR exon 20 insertions were found among 105 squamous cell carcinomas and 8 small cell carcinomas tested.
Morphologic features
Morphologically, all tumors were highly heterogeneous with a mixture of various patterns. Predominant patterns included acinar/papillary/micropapillary (n=21; 70%), lepidic (n=5; 17%) and solid (n=4; 13%); all tumors were entirely non-mucinous. This distribution of morphologic patterns was similar to the control group of 36 adenocarcinomas with classic sensitizing EGFR mutations in exon 19 and 21, although tumors with exon 20 insertions showed a trend for a greater proportion of solid component, but this was not statistically significant (supplementary table 4, supplementary figure 2).
Predicted functional impact
Computational prediction of the functional impact of exon 20 insertions showed functional scores ranging from 2 to 3.6 corresponding to the medium to high functional impact categories. Insertions in codons 762 and 766 affect residues conserved in the entire family of tyrosine kinases; insertions in codons 768 to 769 affect residues conserved in the large specific subfamily of EGFR homologs; insertions in codons 774 and 775 affect residues which are conserved both across all tyrosine kinase homologs and within the specific EGFR subfamily. The point mutation D770N, concurrently found with one H773_V774insNPH, had a low impact score of 1.2 and not likely to be comparable to the impact of the associated insertion.
Clinical characteristics
The clinical characteristics of patients with tumors harboring EGFR exon 20 insertions are summarized in Table 2. Sixty seven percent of patients were female, 48% were never smokers and 12% were of Asian descent. When compared to patients whose tumors lacked them, EGFR exon 20 insertions were more common among never smokers (p<0.0001) but there was no significant difference in age, sex, ethnic origin or stage at diagnosis. No significant differences were noted in comparison to patients with classic sensitizing EGFR mutations (including EGFR exon 19 del, L858R, L861Q, and G719 mutations). The proportion of EGFR exon 20 insertions among all EGFR mutations was the same (9%) for both the Caucasian and the Asian patient subsets.
Table 2.
Clinical characteristics of patients with EGFR exon 20 insertions and comparison to all other patients and to patients with other EGFR mutations
|
EGFR exon 20 insertions (n=33) |
No EGFR exon 20 ins (n=1368) |
Other EGFR
mutations (n=291)** |
||
|---|---|---|---|---|
| Gender | ||||
| (female/male) | 22/11 | 867/501 | 212/79 | |
| Median age (range) | 66 (38-85) | 66 (20-96) | 66 (32-90) | |
| Smoking Status (never/former or current) | 16/17* | 347/1021* | 138/153 | |
| Stage | I-II/III-IV | 18/15 | 629/739 | 121/170 |
| I/II-IV | 17/16 | 521/847 | 106/185 | |
| Ethnicity | Asian/Caucasian | 4/28 | 74/1272 | 32/249 |
| other | 1 | 22 | 10 | |
|
|
||||
p=0.0045 (no other comparisons were statistically significant)
patients with classic sensitizing EGFR mutations only (EGFR exon 19 deletions, L858R, L861Q, and G719 mutations). EGFR T790M mutations associated with clinical resistance are excluded.
Response to treatment and survival analysis
Of the 33 patients with exon 20 insertions, 5 received erlotinib for advanced disease: 1 patient with A763_Y764insFQEA was treated with erlotinib in combination with chemotherapy and had partial response; 1 patient with V774_C775insHV was treated with a combination of chemotherapy and gefitinib with partial response followed by 1 year of erlotinib maintenance prior to disease progression, at which time he was switched to neratinib without benefit; 2 patients with D770_N771insGT and V774_C775insHV were treated with erlotinib as single agent with no response. Finally, one patient with V769_D770insASV was lost to follow up.
For the 15 patients who presented with advanced disease, the median overall survival was >4 years. These patients received combinations of standard chemotherapies including cisplatin or carboplatin with a taxane or pemetrexed. Two patients with remarkable survival had multimodality therapies, 1 with resection of multifocal lung lesions and another with unilateral surgery and contralateral radiation therapy. Both patients had prolonged disease control with these interventions. Other patients received standard chemotherapy agents with typical or less than average duration of benefit.
DISCUSSION
Insertions in exon 20 are a subset of activating EGFR mutations primarily known for their reported association with de novo resistance to TKIs. To date, however, few studies have focused on this subset, each confined to a limited number of mutation positive East Asian patients (4-9). Many of the mutations appear in the literature a single time, some anecdotally associated with response to EGFR inhibitors. It is therefore difficult, even with the combination of these studies, to draw conclusions as to the true prevalence of these mutations, their molecular spectrum, clinicopathologic characteristics or their pattern of resistance. Table 3 summarizes the largest studies and the mutations identified.
Table 3.
Largest studies reporting lung cancers harboring EGFR exon 20 insertions
| Study | Year | % TOTAL | % of EGFR mutant |
Reported mutations | Cosmic database nomenclature |
|---|---|---|---|---|---|
| (EGFR ex20/Total) | (EGFR ex20/total) | ||||
| Huang et al | 2004 | 2% (2/101) | 5.1% (2/39) | D761_E762insEAFQ(1) | D763_E764insFQEA |
| S768_D770dup(1) | D770_N771insSVD | ||||
| Kosaka et al | 2004 | 1.4% (4/277) | 3.6% (4/111) | diagram** | p.A761_Y762insEAFQ? |
| p.A767_S768insTLA | |||||
| p.V769_D770insASV | |||||
| p.D770>GY | |||||
| Shigematsu et al | 2005 | 1.9% (12/617) | 8.9% (12/134) | ASV770-772ins (4) | p.V769_D770insASV |
| H774ins(2) | p.V773_C774insH | ||||
| G771ins (1) | p.D770_N771insG | ||||
| CV770-771ins (1) | p.V769_D770insC | ||||
| NP773-774ins, H775Y(1) | p.P772_H773insNP | ||||
| PH774-775ins(1) | p.H773_V774insPH | ||||
| NPH774-776ins(1) | p.H773_V774insNPH | ||||
| HV775-776ins(1) | p.V774_C775insHV | ||||
| Mitsudomi et al | 2005 | 0% (0/59) | 0% (0/33) | ||
| Chou et al | 2005 | 0% (0/54) | 0% (0/33) | ||
| Sasaki et al* | 2007 | 2.1 (7/322) | 13% (7/54) | 774_776insNPH(2) | p.H773_V774insNPH |
| 770_772insASV(1) | p.V769_D770insASV | ||||
| 771_773insSVD(1) | p.D770_N771insSVD | ||||
| 772_773insV(1) | p.P772_H773insV | ||||
| 772_773insV(1) | p.P772_H773insN | ||||
| Sequist et al | 2007 | 1.8% (5/278) | 7% (5/68) | D770_N771 ins SVD(1) | D770_N771 ins SVD |
| D770 del ins GI(1) | D770>GI | ||||
| N771 del ins TH(1) | N771>TH | ||||
| P772_H773 dup(1) | H773_V774insPH | ||||
| H773_V774 dup(1) | V774_C775insHV | ||||
| Wu et al* | 2008 | 2.5% (13/515) | 5% (13/253) | S768_D770dupSVD(3) | D770_N771insSVD |
| A767_V769dupASV(3) | p.V769_D770insASV | ||||
| D770_N771insD 1(1) | p.D770_N771insD | ||||
| P772_H773insYNP, H773Y | p.P772_H773insYNP + H773Y | ||||
| N771_H773dupNPH(2) | p.H773_V774insNPH | ||||
| D770_N771insG 2 | p.D770_N771insG |
Studies specifically dedicated to exon 20 insertions
mutations diagrammed but specific sequence not reported
To our knowledge, our study represents the largest assessment for EGFR exon 20 insertions and the most comprehensive analysis for other mutations in the same cohort. We used an algorithmic approach for our initial screening, focusing on the group negative for major mutations in EGFR and KRAS, given the previous reports of their mutually exclusive relationship (7). Based on this analysis, we identified 33 patients with insertions, corresponding to 9% of all EGFR mutated samples. We estimate, however, that the true incidence may be even higher, closer to 11%, factoring in the expected positive cases which would have been detected if our entire driver mutation negative group had been tested (99 “mutation negative” samples were not tested due to unavailable DNA). This rate has been further validated by our subsequent clinical testing data for the year 2011, following the inclusion of EGFR exon 20 insertion analysis as part of our standard reflex testing of clinical samples. During an 8 month period, 19 additional EGFR exon 20 insertion cases were detected among 179 EGFR mutant samples (19/179 or 11%, 95% CI 7 to 16%). This rate is consistent with the highest rates previously reported in smaller studies (6, 7), confirming that exon 20 insertions are the third most common EGFR mutation after exon 19 deletions and L858R. The overall underestimation of EGFR exon 20 insertions in the literature may reflect the fact that many studies have focused on the two major mutations and that indels, especially in the setting of low tumor content, may occasionally be mistaken for “high background” on Sanger sequencing traces, a pitfall that is avoided by the simple and more sensitive PCR product sizing assay used in the present study. We estimate that the overall incidence of EGFR exon 20 insertions among all adenocarcinomas is approximately 3%. Through concurrent extended mass spectrometry genotyping and additional testing of known positive EGFR and KRAScases, we also confirmed the mutually exclusive nature of these mutations with all other tested oncogenes, with the exception of PIK3CA. The coexistence of PIK3CA mutations with other oncogene mutations is a frequent event in lung adenocarcinomas (28).
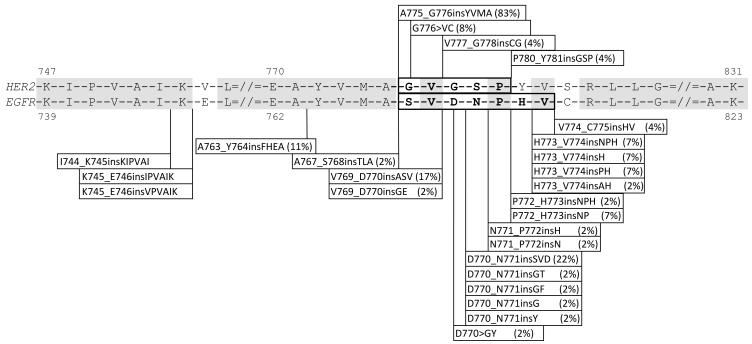
At the molecular level, in agreement with prior studies (7, 8, 11), we found that insertions in exon 20 are all in-frame and confined to the proximal region of the exon, between codons 763 and 775. Compared to other studies, we identified greater heterogeneity with 13 different mutation types within the hotspot region, varying significantly in size and position. Insertions such as V769_D770insASV, previously considered among the most prevalent insertions (7, 8, 29), represented only 12% (4/33) of the mutations in our series. In contrast, the rarely reported mutation A763_Y764insFQEA, a duplication spanning the intron-exon junction (Figure 2), represented 9% of all cases and several other insertions had not been previously reported. Based on our experience over a 3 year period, we have identified 20 different insertions, confirming their wide variability. This degree of heterogeneity is unlike other insertions such as those in exon 19 of EGFR (30) or in exon 20 of HER2 (31), where the vast majority of inserted sequences share the same length and amino acid content. Similarly, contrasting features can even be noted in comparison with EGFR deletions which involve a common defining region within E746 to A750 (Figure 3). How this structural heterogeneity may impact on biologic behavior and response to targeted therapy is not yet known. Computational analysis of these mutations predicts that all insertions, regardless of length and amino acid composition, would confer a significant functional impact by affecting the evolutionary conserved protein regions.
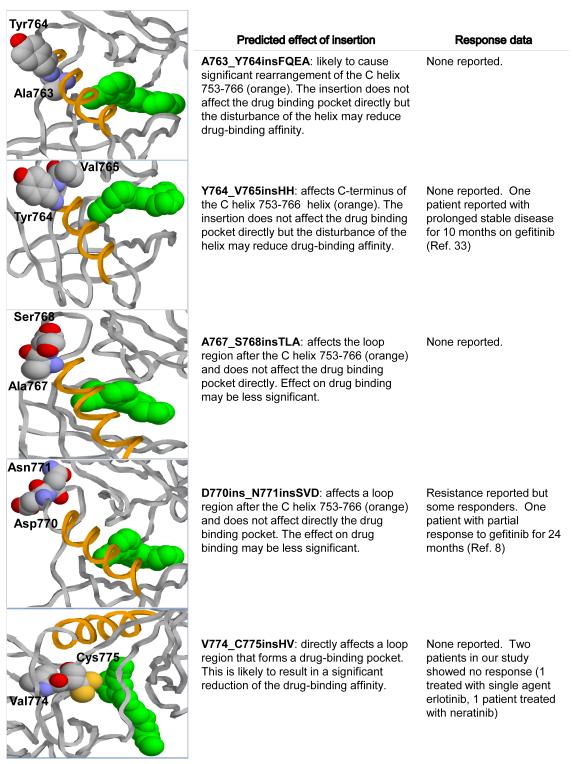
Figure 2.
Modeling of EGFR exon 20 insertions using the 3-dimensional structure of the EGFR kinase domain predicts different interactions with the erlotinib binding region. The X-ray structure at 2.6 Å resolution (PDB code 1M17) is used to show the drug and the positions of mutations. Yellow – C helix; green – erlotinib; labeled residues identify the region of the insertion.
Figure 3.
Positions of the EGFR exon 20 insertions identified over a 3 year period and comparison with the spectrum of EGFR exon 19 and HER2 insertion mutations detected within the same time frame. Insertions in exon 20 of EGFR show higher heterogeneity compared to both HER2 and EGFR exon 19. Most insertions in HER2 are represented by the A775_G776insYVMA while insertions in exon 19 of EGFR all share the inserted sequence PVAI and are located in the same region I744-E746.
To further explore the possible functional differences between the different EGFR exon 20 insertions, we examined their effect on the 3-dimensional structure of the EGFR kinase domain. These in-silico molecular modeling studies showed that the various insertions are predicted to interact differently with the erlotinib binding region. Those involving amino acids 764 to 770 showed the least interaction with the drug binding pocket. In contrast, insertions between A763 and 764 are predicted to cause significant rearrangement of the C helix, which could markedly reduce drug affinity. Insertions in the more distal region of the hotspot, particularly those affecting the 773 to 775 region would affect the drug binding pocket directly, predicting the most significant obstructive effect on erlotinib binding. These predictions suggest a basis for the observed variability of response to erlotinib in patients with different EGFR exon 20 insertions. A better understanding of the biology of these mutations is therefore needed and will require further research into the structure of a wide variety of insertions and the development of additional preclinical models. Based on available literature and our present findings, it appears that insertions between codons 769 and 775, despite variable length or amino acid composition, are associated with resistance to currently approved EGFR TKIs (6, 8, 9, 12) and to irreversible inhibitors entering clinical trials (15, 16). It should be mentioned that mutations such as the A767_V769dupASV and S768_D770dupAVD, commonly associated with resistance, do not involve codons 767 or 768 but represent a duplication of the indicated wild-type sequence inserted distally between 769 to 770 and 770 to 771, respectively. In contrast, we found no specific literature to support a resistance pattern for mutations in the region between codons 762 to 768 which would encompass mutations A763_Y764insFQEA, A767_S768insTLA, V765insHH and M766_A767insAI. In fact, 2 patients with tumors harboring the latter 2 mutations have been reported to show prolonged periods of disease control with reversible EGFR TKIs (18, 32). Of interest, the point mutation S768I in this region, also associated with de-novo resistance, has been reported to show different responses to TKIs depending on the presence of other mutations; specifically S768I has been associated with resistance when found in conjunction with G719A (8) or V769L (33) but not with L858R.(8) In our series, we identified 5 patients with the S768I mutation, one in conjunction with G719A but none were treated with TKI’s. Also, we note the incidental finding of a D770N mutation concurrently with an H773_V774insNPH. While this mutation has been previously reported (34), its association with resistance is not established. In our computational assessment, this mutation had a low score, predicting no functional impact.
In terms of clinical characteristics, patients with EGFR exon 20 insertions were more often never-smokers, but there was no clear association with age, sex, or race. The subset of patients with advanced disease had variable clinical outcomes following either chemotherapy or multi-modality interventions. This treatment heterogeneity precluded a rigorous analysis of the prognostic significance of these mutations. However, among the 3 patients who received single-agent EGFR TKIs, harboring mutations D770_N771insGT and V774_C775insHV, there were no objective responses to therapy.
In conclusion, we find that EGFR exon 20 insertions are a highly heterogeneous family of activating mutations with an incidence that is notably higher than previously reported, placing them as the third most common EGFR mutation after EGFR exon 19 deletions and the L858R point mutation. The high variability identified in these mutations may confer diversity in biologic behavior and response to targeted therapies, arguing against their blanket designation as “non-responsive” mutations. Given the high incidence of lung adenocarcinomas, we estimate that testing could identify over 5000 patients with these mutations every year in the U.S.A alone. Preclinical research and drug development represent unmet needs in this underestimated subgroup of lung cancer patients.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Laetitia Borsu, Angela Marchetti, Talia Mitchell, Justyna Sadowska, Jacklyn Casanova for technical assistance with molecular testing assays; Edyta Brzostowski for help with the MSKCC Lung Cancer Mutation Analysis Project database.
GRANT SUPPORT:
NIH P01 CA129243 (to M. Ladanyi, M.G. Kris.)
Financial support: NIH P01 CA129243 (to M. Ladanyi, M.G. Kris.)
Footnotes
Disclosures: Mark Kris has served as a consultant to Pfizer, Boehringer, Ingelheim and Genentech.
A portion of this material was presented at the 2012 ASCO Annual Meeting, Chicago, June 4, 2012.
REFERENCES
- 1.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang SF, Liu HP, Li LH, Ku YC, Fu YN, Tsai HY, et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res. 2004;10:8195–203. doi: 10.1158/1078-0432.CCR-04-1245. [DOI] [PubMed] [Google Scholar]
- 5.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–23. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki H, Endo K, Takada M, Kawahara M, Kitahara N, Tanaka H, et al. EGFR exon 20 insertion mutation in Japanese lung cancer. Lung Cancer. 2007;58:324–8. doi: 10.1016/j.lungcan.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 7.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 8.Wu JY, Wu SG, Yang CH, Gow CH, Chang YL, Yu CJ, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14:4877–82. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 9.Su Z, Dias-Santagata D, Duke M, Hutchinson K, Lin YL, Borger DR, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924–32. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 11.Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–33. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–11. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuza Y, Glatt KA, Jiang J, Greulich H, Minami Y, Woo MS, et al. Allele-dependent variation in the relative cellular potency of distinct EGFR inhibitors. Cancer biology & therapy. 2007;6:661–7. doi: 10.4161/cbt.6.5.4003. [DOI] [PubMed] [Google Scholar]
- 15.Gerecitano J, Gounder S, Teruya-Feldstein J, Arcila M, Ogilvie S, Gonzalez C, et al. Tissue microarray analysis reveals protein expression patterns and potential biomarkers of clinical benefit to bortezomib in relapsed/refractory non-Hodgkin lymphoma. British journal of haematology. 2012;158:290–2. doi: 10.1111/j.1365-2141.2012.09137.x. [DOI] [PubMed] [Google Scholar]
- 16.Oxnard GR, Miller VA, Robson ME, Azzoli CG, Pao W, Ladanyi M, et al. Screening for germline EGFR T790M mutations through lung cancer genotyping. J Thorac Oncol. 2012;7:1049–52. doi: 10.1097/JTO.0b013e318250ed9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cappuzzo F, Ligorio C, Janne PA, Toschi L, Rossi E, Trisolini R, et al. Prospective study of gefitinib in epidermal growth factor receptor fluorescence in situ hybridization-positive/phospho-Akt-positive or never smoker patients with advanced non-small-cell lung cancer: the ONCOBELL trial. J Clin Oncol. 2007;25:2248–55. doi: 10.1200/JCO.2006.09.4300. [DOI] [PubMed] [Google Scholar]
- 18.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 19.Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Janne PA, Riely GJ, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:5267–73. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Angelo SP, Park B, Azzoli CG, Kris MG, Rusch V, Ladanyi M, et al. Reflex testing of resected stage I through III lung adenocarcinomas for EGFR and KRAS mutation: report on initial experience and clinical utility at a single center. The Journal of thoracic and cardiovascular surgery. 2011;141:476–80. doi: 10.1016/j.jtcvs.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–20. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 22.Chou TY, Chiu CH, Li LH, Hsiao CY, Tzen CY, Chang KT, et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11:3750–7. doi: 10.1158/1078-0432.CCR-04-1981. [DOI] [PubMed] [Google Scholar]
- 23.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. The lancet oncology. 2012;13:e23–31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 24.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, Moreira AL, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18:1167–76. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic acids research. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza MC, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther. 2012;11:485–91. doi: 10.1158/1535-7163.MCT-11-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. The Lancet Oncology. 2012;13:e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 30.He M, Capelletti M, Nafa K, Yun CH, Arcila ME, Miller VA, et al. EGFR exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res. 2012;18:1790–7. doi: 10.1158/1078-0432.CCR-11-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arcila ME, Chaft JE, Nafa K, Roy-Chowdhuri S, Lau C, Zaidinski M, et al. Prevalence, Clinicopathologic Associations, and Molecular Spectrum of ERBB2 (HER2) Tyrosine Kinase Mutations in Lung Adenocarcinomas. Clin Cancer Res. 2012;18:4910–8. doi: 10.1158/1078-0432.CCR-12-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Janne PA, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 33.Asahina H, Yamazaki K, Kinoshita I, Yokouchi H, Dosaka-Akita H, Nishimura M. Non-responsiveness to gefitinib in a patient with lung adenocarcinoma having rare EGFR mutations S768I and V769L. Lung Cancer. 2006;54:419–22. doi: 10.1016/j.lungcan.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Penzel R, Sers C, Chen Y, Lehmann-Muhlenhoff U, Merkelbach-Bruse S, Jung A, et al. EGFR mutation detection in NSCLC--assessment of diagnostic application and recommendations of the German Panel for Mutation Testing in NSCLC. Virchows Archiv: an international journal of pathology. 2011;458:95–8. doi: 10.1007/s00428-010-1000-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.