SUMMARY
Reactive oxygen species (ROS) are proposed to play a major role in telomere length alterations during aging. The mechanisms by which ROS disrupt telomeres remain unclear. In Saccharomyces cerevisiae, telomere DNA consists of TG(1-3) repeats, which are maintained primarily by telomerase. Telomere length maintenance can be modulated by the expression level of telomerase subunits and telomerase activity. Additionally, telomerase-mediated telomere repeat addition is negatively modulated by the levels of telomere-bound Rap1-Rif1-Rif2 protein complex. Using a yeast strain defective in the major peroxiredoxin Tsa1 that is involved in ROS neutralization, we have investigated the effect of defective ROS detoxification on telomere DNA, telomerase, telomere binding proteins, and telomere length. Surprisingly, the tsa1 mutant does not show significant increase in steady-state levels of oxidative DNA lesions at telomeres. The tsa1 mutant displays abnormal telomere lengthening, and reduction in oxidative exposure alleviates this phenotype. The telomere lengthening in the tsa1 cells was abolished by disruption of Est2, subtelomeric DNA, Rap1 C-terminus, or Rif2, but not by Rif1 deletion. Although telomerase expression and activity are not altered, telomere-bound Est2 is increased, while telomere-bound Rap1 is reduced in the tsa1 mutant. We propose that defective ROS scavenging can interfere with pathways that are critical in controlling telomere length homeostasis.
Keywords: reactive oxygen species, peroxiredoxin, telomere length, telomerase
INTRODUCTION
It has been proposed that ROS play an important role in age-related cellular dysfunction and disease. ROS are generated as byproducts of normal cellular respiration or from exposure to exogenous physical and chemical agents and can result in a variety of oxidative damage to cellular components. To counteract the deleterious effects of ROS, most organisms have evolved multiple defense pathways to scavenge ROS and repair oxidized cellular components. Peroxiredoxins (Prxs) are a family of small peroxidases which catalyze peroxide reduction of hydrogen peroxide (H2O2) and other peroxide substrates. All Prxs contain a conserved peroxidatic cysteine residue (CP) that reduces peroxide, though some Prxs carry additional resolving cysteine (CR) to react with oxidized CP to form a disulfide bond that is reduced by thioredoxin (Trx)(Hall et al. 2009). There are five peroxiredoxins isoforms in the budding yeast S. cerevisiae; Tsa1 (cTPx1), Tsa2 (cTPxII), Prx1 (mTPx), Dot5 (nTPx), and Ahp1 (cTPxIII)(Park et al. 2000). These Prxs are localized to different cellular compartments with varying abundance, and Tsa1 is the most highly-expressed Prx (Ghaemmaghami et al. 2003), consistent with its role as the major scavenger for ROS (Iraqui et al. 2009). The tsa1 mutant displays reduced ability to scavenge ROS (Wong et al. 2002). In addition, the tsa1 mutant accumulates a variety of mutations (Huang et al. 2003; Smith et al. 2004), supporting its role in maintaining genome stability.
Telomeres are the protective caps of chromosomes that prevent linear chromosomes from being recognized as double-strand DNA breaks. Telomere dysfunction activates a DNA damage response and subsequently leads to genome instability, cellular senescence, and/or cell death (de Lange 2011). In S. cerevisiae, telomere DNA is composed of ~300±75 bp of double-stranded tandem repeats (TG1-3)n and a short, single-stranded 3′ overhang. All chromosome ends contain subtelomeric repeats; X elements are present in all chromosome ends, while Y′ elements can be found in about 2/3 of telomeres (Teng & Zakian 1999). Although many factors influence telomere length maintenance (Askree et al. 2004; Ungar et al. 2009), telomerase plays an essential role in telomere repeat addition. Telomerase consists of two core components: a reverse-transcriptase protein subunit (encoded by EST2) and a template RNA (encoded by TLC1) (Singer & Gottschling 1994; Lendvay et al. 1996; Counter et al. 1997). Additionally, telomerase-associated proteins, such as Est1, play a vital role in telomerase recruitment to telomeres (Lendvay et al. 1996; Evans & Lundblad 1999). Telomerase core components and telomerase-associated proteins are essential in telomere repeat addition and defects in these genes cause gradual telomere shortening and eventually cell death (Singer & Gottschling 1994; Lendvay et al. 1996). Telomere-associated protein complex Rap1/Rif1/Rif2 also regulates telomerase-mediated telomere repeat addition. Rap1 binds to double-stranded telomere DNA and recruits its interacting proteins, Rif1 and Rif2 via its C-terminus. It has been suggested that these proteins negatively regulate telomerase function and thus telomere length through a protein-counting mechanism (Bianchi & Shore 2008). Proteins binding to subtelomeric DNA may also regulate telomerase (Berthiau et al. 2006; Arneric & Lingner 2007).
Oxidative stress has been proposed to be the major factor causing telomere shortening in human cells in culture (von Zglinicki 2000). To further understand the mechanisms through which oxidative damage disrupts telomere integrity, we deleted peroxiredoxins in S. cerevisiae and investigated the effects of defective ROS neutralization on telomere DNA and telomere length maintenance. Ablation of Tsa1 function leads to an increase in ROS levels, but does not cause telomere attrition. Unexpectedly, the tsa1 mutant displays telomerase-, Rap1-, or subtelomeric DNA-dependent telomere over-elongation. Our data suggest that physiological level of oxidative stress interferes with telomerase regulation.
RESULTS
Tsa1, the major peroxiredoxin, is required for telomere length maintenance
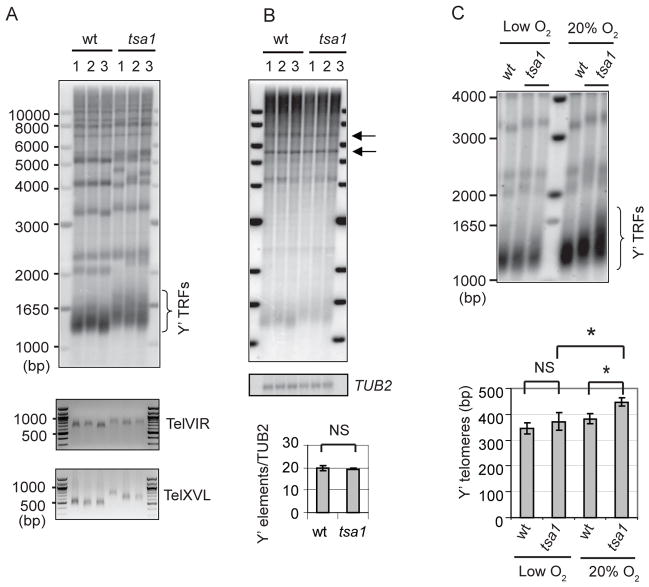
ROS include superoxide anions (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH·). H2O2 is a relatively long-lived molecule that can readily penetrate cell membranes, causing oxidative damage at a distant site of its formation. Peroxiredoxins (Prxs) are the major ROS detoxification enzymes for H2O2 and are ubiquitously expressed in many organisms including S. cerevisiae (Park et al. 2000). To investigate the consequence of H2O2 and/or peroxiredoxin deficiency in telomere length maintenance, we individually disrupted five peroxiredoxin isoforms, Tsa1, Tsa2, Prx1, Dot5, and Ahp1 in S. cerevisiae. Among the five Prx isoform mutants, only the tsa1 deletion mutant displayed abnormal telomere length, which is consistent with the fact that Tsa1 is the major scavenger of H2O2 (Park et al. 2000). It is noteworthy that Tsa1 deficiency does not affect cell proliferation or cell cycle progress (Park et al. 2000; Wong et al. 2002). Surprisingly, the tsa1 mutant showed longer Y′-containing telomere restriction fragments (Y′-containing TRFs) than the wild-type cells (Fig. 1A), suggesting its telomeres were over-lengthened.
Figure 1. Loss of Tsa1 causes abnormal telomere lengthening.
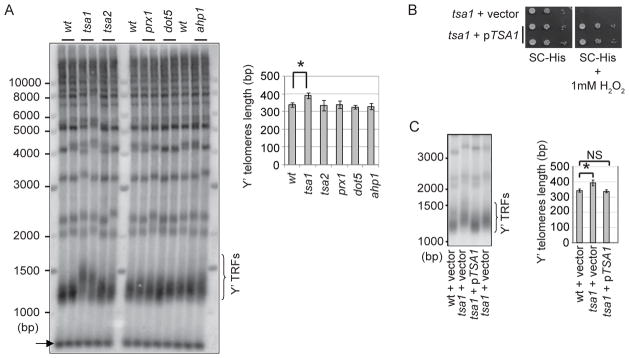
(A) Genomic DNA from Prx mutants was cut with XhoI and analyzed by Southern blot using a telomere repeat-specific probe. Y′ TRFs are bracketed. A DNA fragment (~700bp) containing telomere repeats was included as the migrating control (arrow). (B) The wild type S. cerevisiae TSA1 gene (S.c. TSA1) was cloned into a S. cerevisiae vector and transformed into a tsa1 strain. Transformants were spotted on plates containing H2O2 for testing H2O2 sensitivity (upper panel). (C) The indicated strains (~90 generations after spore germination) were analyzed for telomere length as in (A). Error bars indicate SD of the length of TRFs from at least three independent clones. Student’s t test was used to compare means between two groups. NS, not significant; *, p<0.05.
To test if telomere lengthening is specifically caused by Tsa1 deficiency, we cloned the wild-type TSA1 gene into an expressing plasmid and transformed the recombinant DNA into a heterozygous tsa1/TSA1 diploid strain. This strain was sporulated to obtain the wild-type and tsa1 spores carrying the TSA1 plasmid. H2O2-sensitivity of the tsa1 mutant was complemented by a plasmid harboring the wild-type TSA1 gene, but not by the vector alone, suggesting that the TSA1 gene in the plasmid is functional (Fig. 1B). The tsa1 mutant carrying the TSA1 plasmid displayed a telomere length similar to that of the wild type (Fig. 1C). Thus, telomere lengthening in the tsa1 mutant is caused by loss of Tsa1 function.
We further characterized telomere lengthening phenotype among individual telomeres and clones in the tsa1 mutant by Southern blot analysis. Several wild-type and tsa1 clones were examined, and the tsa1 mutant clones displayed variably lengthened Y′-telomeres (Fig. 2A, top panel). In addition, telomere PCR revealed that individual telomeres, Tel VIR and Tel XVL, were lengthened to different extents among the tsa1 clones (Fig 2A, bottom panel). Thus, ablation of the Tsa1 function leads to disparate lengthening of telomere repeats in individual clones and telomeres.
Figure 2. Characterization of telomere lengthening in the tsa1 mutant.
(A) The representative wild-type and tsa1 clones were analyzed for telomere length using Southern blot as in Figure 1A. Tel VIR and Tel XVL were analyzed by telomere PCR (lower panel). (B) The same blot was stripped sequentially and probed with the probes specific for the Y′ elements (top panel) and the TUB2 gene (bottom panel). The positions of Y′ elements are indicated by arrows. The intensity ratio between the Y′ elements and TUB2 was compared between the wild-type and tsa1 clones using Student’s t test. NS, not significant. (C) The wild-type and tsa1 spores were dissected from a TSA1/tsa1 heterozygous strain. Immediately after dissection, one plate was allowed to germinate in a closed jar in which oxygen was depleted by GasPak EZ Gas Generating Sachets (low O2). Another plate was incubated in an ambient oxygen condition (20% O2). Genomic DNA was isolated after 5 days and analyzed for telomere length using Southern blot as in Figure 1A. Representative clones are shown. The length of Y′ TRFs was compared between two groups using Student’s t test. Error bars indicate SD from at least three independent clones. NS, not significant; *, p<0.05.
Given that tandem amplification of Y′ elements can attribute to telomere maintenance (Teng & Zakian 1999), we investigated if Y′ elements are amplified in the tsa1 mutant. Arrays of amplified Y′ elements generate 5.2 and 6.7 kb fragments after digestion with XhoI, which reflect the short or long isoforms of Y′ elements, each of which contains a single XhoI site. As shown in Fig. 2B, Southern blot analysis using a Y′-specific probe of Xho-digested DNA revealed no amplification in the tsa1 mutant.
Enhanced ROS correlates with telomere length abnormalities in the tsa1 mutant
Because the tsa1 mutant displays reduced ability to scavenge ROS (Wong et al. 2002), we hypothesize that telomere lengthening in the tsa1 mutant is caused by oxidative stress; a reduction in oxidative stress should therefore alleviate telomere lengthening. We thus investigated telomere length of both wild-type and tsa1 cells grown under the oxygen-limiting condition. Freshly dissected wild-type and tsa1 cells were grown in ambient condition (20% oxygen) or a sealed jar in which oxygen was depleted (Ragu et al. 2007). First, we measured intracellular ROS level in the wild-type and tsa1 cells by dihydroethidium (DHE) staining. When grown in 20% oxygen, the tsa1 cells displayed a higher DHE fluorescence signal intensity than the wild-type cells. However, when cells were grown in an oxygen-limiting condition, there was no significant difference in DHE signals between the wild-type and tsa1 cells (Supplementary Fig. 1). Concurrently, when grown in 20% oxygen, the tsa1 cells had longer telomeres than the wild-type cells. However, the wild type and tsa1 mutant showed similar telomere length when grown in an oxygen-limiting condition (Fig. 2C). These results confirm the oxygen-dependent nature of telomere lengthening and support that oxidative stress is responsible for telomere lengthening in the tsa1 mutant.
Telomerase is required for telomere lengthening in the tsa1 mutant
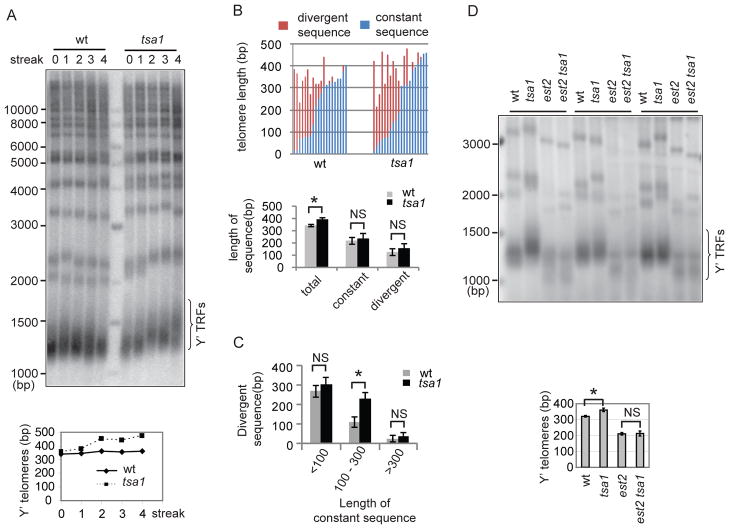
Abnormal telomere lengthening in the tsa1 mutant suggests that telomere length homeostasis is disrupted in this mutant. Many mutations that result in telomere elongation through telomerase-dependent mechanisms cause telomeres to gradually reach a longer length. To explore the dynamic change in telomere length, a heterozygous tsa1/TSA1 diploid strain was sporulated to obtain the wild-type and tsa1 spores. After successive re-streaking, telomere length was checked by Southern blot analysis at different generations. While the wild type had relatively stable telomere length over time (Y′-containing telomeres ranging from 338 to 363 bp), the tsa1 mutant increased its telomere length with progressive re-streaking (Fig. 3A).
Figure 3. Telomerase is required for telomere lengthening in the tsa1 mutant.
(A) The wild-type and tsa1 spores from a TSA1/tsa1 heterozygous strain were re-streaked successively on YEPD plates. Genomic DNA was isolated at the indicated time and analyzed for telomere length using Southern blot as in Figure 1A. (B) Telomere XVL from either wild-type or tsa1 cells was amplified using a high fidelity DNA polymerase, cloned, and sequenced. Telomere sequences were aligned and constant and divergent sequences were indicated by blue and red color. The average length of constant and divergent sequences is shown below the graph. Error bars indicate standard errors. NS, not significant; *, p<0.05. (C) Telomeres were categorized according to the length of constant sequences and the length of divergent sequences and compared between the wild-type and tsa1 cells. Error bars indicate standard errors. NS, not significant; *, p<0.05. (D) Spores with the indicated genotype generated from a TSA1/tsa1 EST2/est2 heterozygous strain were re-streaked successively on YEPD plates for ~ 90 generations. Genomic DNA was analyzed for telomere length using Southern blot as in Figure 1A. Representative clones are shown. Calculated Y′-telomere lengths (mean±SD) from three clones for each genotype are shown at the bottom.
Telomeres consist of stable sequences at the centromere-proximal region and degenerate repeats at the distal region; and sequence divergence is the result of telomere shortening followed by telomerase-mediated extension (Forstemann et al. 2000). To further characterize the involvement of telomerase in telomere lengthening, we verified telomere sequences in individual telomeres in the wild type and tsa1 mutant. Telomere XV-L (TEL15L) was PCR amplified, cloned, and sequenced from an early passage (~50 generations after spore germination) of the tsa1 mutant. It is noteworthy that the sequencing data present here may reflect multiple telomere repeats addition events and are distinct from data generated during a single-round of telomerase action (Teixeira et al. 2004). Consistent with the overall telomere length changes determined by Southern blot analysis, the PCR-amplified TEL15L showed an increase in average telomere length in the tsa1 cells (393±16 and 344±10 bps for the tsa1 and wild-type cells, respectively). The sequences of telomere repeats (TG1-3) were aligned and constant and divergent sequences were identified (Supplementary Fig. 2). The number of sequenced telomeres with divergent sequences was the same (15/20) in the wild-type and tsa1 mutant, which is not in favor of the idea that telomerase elongates a greater number of telomeres in the tsa1 mutant. The tsa1 and wild-type cells contain 236±36 and 219±28 bps of constant sequences and 157±32 and 125±27 bps of divergent sequences, respectively (Fig. 3B). Thus, the average lengths of constant and divergent sequences were not significantly different in the wild-type and tsa1 cells. Because maintenance of very short or long telomeres can involve different mechanisms, e.g. very short telomeres are prone to recombination and very long telomeres are rarely elongated by telomerase (Teixeira et al. 2004), we therefore categorized telomeres into three groups according to the length of constant telomere sequences (<100, 100–300, >300 bp) and compared the length of divergent sequences between the wild-type and tsa1 cells. Telomeres with <100 or >300 bp constant telomere sequences did not show more divergent sequences, but telomeres with 100–300 bp constant telomere sequences had more divergent sequences in the tsa1 mutant (Fig. 3C). Collectively, these data suggest that telomerase elongation is stimulated in the tsa1 mutant.
To genetically confirm the involvement of telomerase in telomere over-elongation, we deleted the EST2 gene (which encodes the catalytic subunit of telomerase complex) in the tsa1 mutant. The heterozygous tsa1/TSA1 est2/EST2 diploid strains were sporulated to obtain single and double mutants. Senescence rate of the est2 mutant is similar to that of the tsa1 est2 double mutant (Supplementary Fig. 3A). Telomere length of single and double mutants was analyzed by Southern blot at ~90 generations after spore germination prior to senescence. Consistent with earlier observations (Lendvay et al. 1996; Counter et al. 1997; Lingner et al. 1997), telomere length in the est2 mutant became shorter after successive subcultures (Supplementary Fig. 3B and C). Telomere length of the tsa1 est2 double mutant was significantly shorter than that of the tsa1 mutant but similar to that of the est2 mutant (Fig. 3D and Supplementary Fig. 3B and C), suggesting that Tsa1 deficiency is unable to cause telomere lengthening in the est2 mutant. The requirement of Est2 for telomere lengthening supports the idea that telomerase participates in telomere lengthening in the tsa1 mutant.
Telomerase-dependent telomere lengthening in the tsa1 mutant could be due to increased telomerase expression/activity or enhanced telomerase recruitment. We therefore checked the expression of telomerase core components (TLC1 and Est2) and a telomerase-associated protein Est1 (which is involved in the recruitment of telomerase to its substrate) in the tsa1 mutant. Quantitative real-time PCR was utilized to measure the transcripts of these genes. The transcripts of TLC1, EST2, and EST1 genes were not significantly altered in the tsa1 mutant (Supplementary Fig. 4A). To compare telomerase activity between the wild type and tsa1 mutant, the TSA1 gene was deleted in a strain carrying a Myc-tagged EST2 gene (Osterhage et al. 2006). The expression of Myc-Est2 was confirmed by Western blot analysis (Supplementary Fig. 4B). Myc-Est2 was immunoprecipitated by α-Myc antibody (Supplementary Fig. 4C) and its capacity to extend an oligonucleotide complementary to the telomerase template was measured. As shown in Supplementary Fig. 4D, the intensity of primer extension products was similar in the wild type and tsa1 mutant, suggesting that telomerase activity was not affected in the tsa1 mutant.
The involvement of telomere protein complex in telomere lengthening in the tsa1 mutant
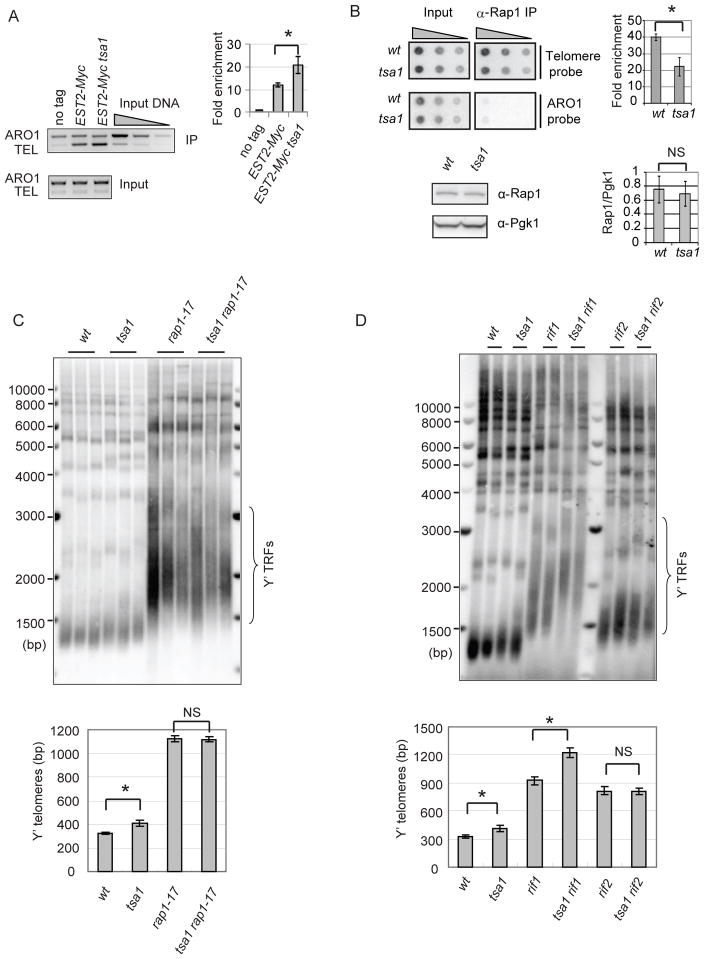
Telomerase occupancy on telomeres can influence telomere repeat addition (Bianchi & Shore 2008). We therefore used chromatin immunoprecipitation (ChIP) to analyze the binding of Est2 protein to telomere DNA. Antibody against Myc was used to immunoprecipitate Myc-tagged Est2 and quantitative multiplex PCR was used to monitor the level of telomere VII-L (TEL) and nontelomeric control (ARO1) fragments. As shown in Fig. 4A, the tsa1 mutant had approximately two-fold increase of Est2 binding to telomeres.
Figure 4. Altered binding of telomerase and Rap1p to telomere DNA in the tsa1 mutant.
(A) The association of Myc-tagged Est2 with telomere DNA was determined by ChIP using an antibody to Myc. A strain carrying untagged EST2 allele (no tag) was used as a control. Multiplex PCR results from a representative ChIP experiment are shown. Quantitative analysis was derived from four independent experiments and fold enrichment of TEL over background (no tag) was calculated as described (Taggart et al. 2002) and shown on the right. (B) Upper panel: binding of Rap1 protein to telomere DNA was analyzed by ChIP using an antibody to Rap1. The representative dot-blot results are shown. Fold enrichment of telomere signal from three independent experiments was calculated as described (Ji et al. 2008) and shown on the right. Error bars represent SD values from multiple independent experiments. Student’s t t-test was used to determine significance. *, p<0.05. Lower panel: Rap1 protein level in the wild-type and tsa1 cells were determined by Western blot using the antibody against Rap1. Pgk1 level was measured as the loading control. The signals of a band corresponding to Rap1 were normalized to those of Pgk1p for each sample. Error bars indicate SD from three independent clones. Student’s t test was used to compare means between groups. NS, not significant. (C and D) Genomic DNA from the indicated strains was analyzed for telomere length using Southern blot as in Figure 1A. Representative clones are shown.
In yeast, the recruitment of telomerase to telomeres is negatively regulated by the levels of telomere-bound Rap1-Rif1-Rif2 protein complexes (Bianchi & Shore 2008). To determine the binding of Rap1 to telomere DNA, ChIP was performed using a Rap1 antibody and the amount of pulled-down telomere DNA was determined by dot-blot. Rap1-bound telomere DNA in the tsa1 mutant was about 50% of that in the wild type (Fig. 4B, upper panel). This mild reduction is consistent with a mild increase in telomere length in the tsa1 mutant. This reduction cannot be explained by the change of Rap1 expression, since Rap1 expression was not altered in the tsa1 mutant (Fig. 4B, lower panel). Thus, Tsa1p deficiency alters telomere-bound telomerase and Rap1, implying that loss of Tsa1 impairs Rap1’s negative regulation on telomerase recruitment to telomeres.
To further confirm the role of Rap1-Rif1-Rif2 in telomere lengthening, we carried out genetic epistasis studies with rap1, rif1, rif2, and tsa1 mutation. Double mutants were created and telomere length was examined by Southern blot analysis. Because deletion of
Rap1 leads to cellular lethality, we utilized the rap1-17 allele that encodes a Rap1 protein with C-terminal truncation (missing amino acids 663-827) (Kyrion et al. 1992). The rap1-17 mutant is incapable of interacting Rif1 and Rif2, but efficiently binds to telomere DNA (Kyrion et al. 1992). As shown in Fig. 4C, the tsa1 rap1-17 double mutant had a similar telomere length as the rap1-17 single mutant, supporting that Rap1 is required for telomere lengthening in the tsa1 mutant. Similarly, the tsa1 rif2 double mutant displayed a similar telomere length as the rif2 single mutant (Fig. 4D), implying the importance of Rif2 in telomere lengthening in the tsa1 mutant. Interestingly, telomere length of the tsa1 rif1 double mutant is significantly longer than that of the rif1 single mutant, suggesting that tsa1 and rif1 mutations causes telomere lengthening via distinct pathways.
Telomerase regulation has been shown to be influenced by the proteins that bind to subtelomeric DNA (Berthiau et al. 2006; Arneric & Lingner 2007). We thus tested if removal of subtelomeric DNA would affect telomere lengthening in the tsa1 mutant. Subtelomeric DNA adjacent to the telomere at chromosome VII-L was replaced by the URA3 gene in UMY2584 strain (Lu & Liu 2010). Deletion of the TSA1 gene caused general lengthening of Y′ telomeres in this strain (361 and 429 for the wild type and tsa1 mutant, respectively) (Supplementary Fig. 5A). In the tsa1 mutant the telomere at the URA3-Tel VII-L locus showed a length close to wild-type length (304 and 322 for the wild type and tsa1 mutant, respectively). Thus, proteins binding to subtelomeric DNA might contribute to telomere length alteration in the tsa1 mutant. However, over-lengthening of URA3-Tel was found in some of the tsa1 mutant clones (Supplementary Fig. 5B). Hence, subtelomeric proteins cannot be the solo factors causing telomere over-lengthening in the tsa1 mutant.
Ablation of Tsa1 does not significantly enhance steady-state level of oxidative DNA lesions at telomeres
Oxidation can result in oxidative guanine lesions in telomeric repeats in vitro (Kawanishi et al. 1999; Rhee et al. 2011), and an exposure to a high oxidative environment causes more single-strand breaks (SSBs) and oxidative guanine lesions in telomeric DNA compared to non-telomeric DNA in human and murine cells (Petersen et al. 1998; Rhee et al. 2011). Furthermore, oxidative DNA lesions impair the binding of telomere binding proteins to telomere DNA (Opresko et al. 2005; Lu & Liu 2010). We reasoned that enhanced ROS might cause oxidative DNA damage and consequently perturb the association of Rap1 and telomere DNA in the tsa1 mutant. We therefore measured the levels of oxidative base modifications, abasic sites, or DNA strand breaks in telomere DNA in the tsa1 mutant. Restricted genomic DNAs from the wild type and tsa1 mutant were treated with E. coli DNA glycosylases, either Fpg alone or EndoIII plus EndoVIII, which release oxidized purines and/or pyrimidines and incise at abasic sites via N-glycosylase and lyase activity (Lu & Liu 2010; Wang et al. 2010; Rhee et al. 2011). The extent of telomere DNA fragmentation caused by DNA glycosylase treatment reflects the amount of base modifications and abasic lesions at telomeres, which are revealed as a decreased medium telomere length (MTL) on an alkaline gel (Supplementary Fig. 6A). DNA glycosylase treatment resulted in a slight decrease of MTL in the wild type and tsa1 mutant, indicating that normal cellular respiration can cause low levels of oxidative base damage at telomeres. Surprisingly, there was no difference in DNA glycosylase-sensitive lesions between the wild type and tsa1 mutant (Supplementary Fig. 6B).
Single-strand breaks (SSBs) can lead to faster migrating DNA fragments in alkaline electrophoresis, yet no significant increase in faster migrating telomere restriction fragments was observed in the tsa1 mutant (Supplementary Fig. 6B, lanes without DNA glycosylase). Furthermore, SSBs as well as other DNA replication blocking lesions (e.g. abasic sites) in telomere DNA should block DNA polymerases, resulting in a decrease in telomere PCR products. We therefore checked the presence of these DNA lesions at telomeres by telomere PCR in the wild type and tsa1 mutant. As shown in Supplementary Fig. 7, both strains yielded similar levels of telomere PCR products.
S. cerevisiae telomere DNA consists of TG(1-3) repeats. G or tandem Gs are relatively sensitive to oxidative damage in comparison to other bases or base combinations, causing G to T transversion if oxidative guanine damage is left unrepaired. Because G to T transversion in one or a series of guanine bases tandemly adjacent to thymidine can alter telomere TG(1-3) repeats, e. g. TG to TT, TGG to TTT, or TGGG to TTTT, we counted the occurrence of tandem Ts in the telomere repeats in the wild type and tsa1 mutant. Telomere VIR (Tel 06R) and telomere XVL (Tel 15L) were amplified by PCR using a high-fidelity DNA polymerase. The PCR product was cloned and sequenced. Tandem Ts were not found in the wild-type telomeres, but were found in two of the tsa1 telomeres (t21 and t25, Supplementary Fig. 2). Because of the degenerate nature of S. cerevisiae telomeres repeats, telomere sequence analysis cannot determine G to T transversions that occur in guanine bases not adjacent to thymidine (e.g, TGGG to TGTG). Thus, more G to T transversions might exist in the telomere DNA. Lastly, strings of five or four G’s were also found in the tsa1 telomeres at low frequency. The origin of these abnormal sequences remains unknown.
DISCUSSION
High oxidative stress associates with telomere shortening in human cells in culture (von Zglinicki 2000). However, it is not known if mild oxidative stress, such as ROS would generate a similar telomere phenotype in vivo. Peroxiredoxin proteins reduce H2O2 and other hydroperoxide substrates. These proteins are ubiquitously expressed in many organisms and play important roles in protecting cells against ROS-induced damage. Here, we demonstrate that deletion of the major peroxiredoxin gene TSA1 can disrupt telomere length regulation in S. cerevisiae. Surprisingly, elevated ROS is associated with telomerase-dependent telomere over-lengthening in the tsa1 mutant. This observation is in agreement with a recent study that oxidative stress leads to telomere lengthening in humans (Shlush et al. 2011). Thus, oxidative stress can alter telomere length homeostasis, and its level may influence the outcome of telomere length changes.
Oxidative stress induces SSBs at 5′ of GGG bases and 8-oxo-7,8-dihydroguanine (8-oxoG) base damage in telomere DNA in vitro (Kawanishi et al. 1999). These observations lead to the hypothesis that oxidative stress damages telomere DNA, especially at or close to guanine bases in vivo. Unexpectedly, the tsa1 mutant does not display a significant increase in oxidative DNA damage, e.g. SSBs, 8-oxoG and its derived G to T transversion at telomeres. The same method has been successful in detecting oxidative guanine lesions in a budding yeast mutant deficient in a DNA glycosylase, Ogg1 (Lu & Liu 2010). Because the tsa1 mutant has intact repair pathways, it is possible that oxidative DNA lesions are efficiently removed in the mutant and could not therefore accumulate and reach a significant steady-state level at telomeres for easy detection. However, we cannot rule out the possibility that the tsa1 mutant suffers slightly enhanced oxidative DNA damage that is not readily detectable by the current method.
Several lines of evidence support that telomere over-lengthening is dependent on telomerase in the tsa1 mutant. First, Southern blot and sequence analysis reveals that telomere lengthening results from telomere repeat expansion, while the length of the subtelomeric region is not affected. Second, the progressive telomere lengthening suggests a mechanism that involves telomerase. Consistently, the elongated portion of a telomere contains the irregular TG repeats, a pattern that is characteristic of telomerase action. Third, the requirement of Est2 for telomere lengthening further supports telomerase’s participation. Because Rad52 is essential in the tsa1 mutant ((Huang & Kolodner 2005) and data not shown), it is not feasible to determine its involvement in telomere lengthening. However, all the clones of the tsa1 est2 strains display similar telomere length to the est2 strain, suggesting that telomerase is the major (if not the sole) pathway involved in telomere lengthening.
Telomerase-mediated telomere repeat addition can be influenced by the expression level of telomerase subunits. ROS have been proposed to activate or increase telomerase activity, resulting in telomere lengthening in individuals with oxidative exposure (Shlush et al. 2011). We thus tested if increased ROS levels would alter telomerase expression or activity in the tsa1 mutant. Over-expression of Est1 or Est2 can lead to increased telomerase activity, while over-expression of TLC1 decreases telomerase activity in S. cerevisiae (Singer & Gottschling 1994; Larose et al. 2007). However, expression of EST1, EST2, and TLC1 is not altered in the tsa1 mutant. Furthermore, the wild type and tsa1 mutant have comparable telomere primer extension activity, implying that telomerase activity is not enhanced in the tsa1 mutant.
Telomerase recruitment to telomeres is highly regulated, and it preferentially acts on short telomeres over long telomeres (Teixeira et al. 2004). The tsa1 mutant has increased telomerase occupancy at telomeres, and the longer divergent telomere sequences observed in the tsa1 mutant could be therefore caused by telomerase. These findings suggest that telomerase elongation is dysregulated in the tsa1 mutant. Rap1p directly binds telomere DNA, where it negatively regulates telomerase recruitment and elongation via a protein counting mechanism (Bianchi & Shore 2008). The tsa1 mutant has decreased telomere-bound Rap1, which may contribute to increased telomerase occupancy at telomeres in the tsa1 mutant. Previous studies have shown that oxidative DNA lesions in the telomere DNA affect the binding of telomere binding proteins to telomere DNA (Opresko et al. 2005; Lu & Liu 2010). However, oxidative DNA lesions are not significantly elevated in the tsa1 mutant. We cannot rule out the possibility that the tsa1 mutant harbors a moderate increase of telomeric oxidative DNA damage that is undetectable by the current method, nevertheless contributes to reduced Rap1 binding to telomere DNA. Alternatively, DNA binding properties of Rap1 might be sensitive to ROS levels, so the amount of active Rap1 could be reduced in the tsa1 mutant. Although Rap1-Rif1-Rif2 complexes have been proposed to regulate the recruitment of telomerase to telomeres (Bianchi & Shore 2008), telomere lengthening in the tsa1 cells can be abolished by disruption of Rif2, but not by Rif1 deletion, suggesting a functional separation of Rif1 and Rif2 in telomere length regulation in the tsa1 mutant. In support of these observations, it has been shown that Rif1 and Rif2 are differentially required for Tel1-mediated telomere length regulation (Hirano et al. 2009) and accumulation of TERRA at telomeres (Iglesias et al. 2012). Rif1 is also involved in Rap1-independent anti-checkpoint function (Xue et al. 2012) and CST complex-modulated telomere capping (Anbalagan et al. 2012).
In addition to Rap1/Rif1/Rif2, other proteins binding to subtelomeric DNA have been suggested to regulate telomerase (Berthiau et al. 2006; Arneric & Lingner 2007). Like Rap1/Rif1/Rif2 complex, the binding of subtelomeric proteins to subtelomeric DNA might also be sensitive to ROS levels and therefore become less functional in the tsa1 mutant. When subtelomeric DNA adjacent to Tel VII-L is replaced by the URA3 gene, URA3-Tel VII-L does not show significant lengthening in the tsa1 mutant. However, over-lengthening of URA3-Tel VII-L is indeed found in particular tsa1 mutant clones, and it appears to be less prominent than over-lengthening of bulk telomeres. Collectively, these results suggest a possible role of subtelomeric proteins in telomere lengthening in some, but not all the tsa1 mutant clones. An alternative explanation is that both Rap1/Rif2 and one or more subtelomeric proteins play a role in telomere lengthening in the tsa1 mutant, and the over-lengthening seen in some URA3-telomere tsa1 clones is due a stochastic effect on Rap1/Rif2 in that cell lineage.
In summary, our data support the notion that defective ROS neutralization can alter pathways that regulate telomerase elongation and thus telomere length homeostasis. Human stem cells, highly proliferating tissues and majority human cancers express telomerase (Shay & Wright 2005). High levels of ROS are commonly observed in cancer cells and can also result from chronic inflammation or decreased scavenging capacity in aging (Trachootham et al. 2009). Enhanced ROS may alter telomerase-mediated telomere repeat addition and the pathways that regulate telomerase in highly proliferating tissues and cancer cells, which could be one of the contributing factors influencing aging and cancer.
EXPERIMENTAL PROCEDURES
Yeast strains and genetic procedures
Yeast transformation, media, and genetic procedures have been described previously (Lu & Liu 2010). Except the rap1 mutant, the rest yeast mutants used in this study are null mutants in which the corresponding open reading frame (ORF) was replaced with a deletion marker. The details of the yeast strains were described in supplementary material.
Oxygen-limiting growth condition and ROS detection
The heterozygous TSA1/tsa1 diploid strains were sporulated, and tetrads were picked under normal aeration condition. After tetrads were dissected and spores were placed on YEPD plates, the dissecting plates were immediately placed in an airtight jar containing GasPak EZ Gas Generating Sachets (BD Diagnostics)(Ragu et al. 2007). Anaerobic Indicator Strips (BD Diagnostics) were used to monitor the anaerobic conditions. The jar was incubated at 30 °C for 5 days before Southern blot analysis and stained for cytoplasm and chromatin-bound dihydroethidium bromide (DHE) for detecting intracellular ROS (Polysciences, Inc, Catalog No. 17084-50). 30,000 cells were measured for DHE fluorescence intensity by Accuri C6 flow cytometer (Accuri Cytometers) using FlowJo software (Tree Star).
RNA preparation, quantitative PCR, and telomerase activity
Total RNA was prepared using Trizol (Gibco BRL). cDNA was reverse-transcribed by SuperScript II reverse transcriptase (Invitrogen) using random primers according to the manufacturer’s instruction. Quantitative PCR of the EST1, EST2, TLC1, TUB1, and ACT1 cDNA was conducted using iTaq Fast SYBR Green Supermix (BioRad) and primers listed in Table S1. A strain carrying the Myc-tagged EST2 allele (Osterhage et al. 2006) was used for determining telomerase activity as described (Ji et al. 2005).
Southern blot analysis
DNA was isolated from exponentially growing cultures and digested with XhoI. Southern blot analysis was performed as previously described (Lu & Liu 2010) using radio-labeled polyTG1-3 fragments (the EcoRI/KpnI fragments from pCT300 containing 256 bps of TG1-3) (Ray & Runge 1998). The length of TRFs containing Y′-telomeres was estimated by plotting the peak of signal intensity of TRFs against the position of the molecular weight standards (Askree et al. 2004). The length of TRFs was subtracted by that of Y′ element fragment (~875 bp) to yield Y′-containing telomere length. For URA3-TEL analysis, genomic DNA was digested with ApaI and a 300 bp URA3 fragment (PCR amplified, corresponding to 400 – 700 nt of URA3 gene) was used as probe.
ChIP and western blot
The binding of Est2 and Rap1 protein to the telomere DNA was measured by ChIP as described (Ji et al. 2008; Lu & Liu 2010). Myc-tagged Est2 was pulled down by antibody to Myc. Quantitative multiplex PCR was used to measure two DNA fragments associated with Est2: TEL, a 252-bp fragment that is 7 bp from telomere repeats at the left telomere of chromosome VII; ARO1, a control non-telomere fragment (372-bp). The PCR products were quantified by ethidium bromide staining using Image Lab (Biorad). Dot blot was used to measure telomere DNA pulled down by the Rap1 antibody. Threefold serial dilutions of DNA were dot blotted onto positively charged nylon membrane. The radioautography results were quantified by Image Quant (GE Healthcare). For each sample, the fold enrichment was calculated as (TELIP / ARO1IP) / (TELinput / ARO1input).
Western blot was employed to measure Rap1 and Myc-tagged Est2 protein levels using an anti-Rap1 antibody and anti-Mcy antibody (Ji et al. 2008; Lu & Liu 2010). Pgk1 was probed in the same blot as a loading control.
Telomere PCR and sequencing
Telomere XVL (Tel 15L) was amplified from genomic DNA using PCR (Teixeira et al. 2004; Hector et al. 2007). Briefly, polyC was added to a telomere using terminal transferase. Tel 15L telomere was then amplified using a polyG primer and a primer specific for Tel 15L using Phusion high fidelity DNA polymerase (New England Biolabs). PCR product was gel purified and cloned into pCR2.1 vector (Invitrogen). 20–30 colonies were picked from one transformation and plasmids isolated from these clones were sequenced using M13R or M13F primer. Sequencing service was performed by Genewiz Corporation using the difficult template protocols (http://www.genewiz.com/public/rnai.aspx).
Detection of oxidative DNA lesions in telomeres
DNA isolation and identification of oxidative base lesions in telomeres were performed as previously described with minor modifications (Lu & Liu 2010; Wang et al. 2010). In brief, DNA was treated with AluI, HaeI, HinfI, and MspI and then with mock buffer or Fpg- or EndoIII/EndoVIII. The resulting single-stranded DNA fragments were detected by Southern blot using 32P-labeled polyTG1-3 fragments (Ray & Runge 1998). The frequencies of DNA glycosylase-sensitive sites were calculated as described previously (Lu & Liu 2010; Wang et al. 2010).
To quantify single strand breaks and other DNA blocking lesions, telomere VI-R or XV-L ((Tel 6R or Tel 15L) was amplified from genomic DNA of four clones of each genotype using the similar method (see telomere PCR and sequencing), except that polyG primer and a primer specific for these telomeres were used and Phusion DNA polymerase was replaced by Taq (New England Biolabs) (Hector et al. 2007). PCR was controlled so that amplification was in an exponentially increasing phase.
Supplementary Material
Acknowledgments
We sincerely thank Drs. Anders Byström, Kurt W. Runge, Katherine L. Friedman, Stéphane Marcand, and Virginia Zakian for providing us with yeast strains and plasmids; Drs. Vilhelm Bohr, David Wilson, Sarah I. West and Kent Horvath for critical reading of the manuscript. This work was supported by funds from the Intramural Research Program of the NIA, National Institutes of Health.
Abbreviation
- Prxs
Peroxiredoxins
- ROS
reactive oxygen species
- SSB
single strand break
Footnotes
AUTHOR CONTRIBUTIONS
J.L., H.V, and J.Y, performed the experiments. J.L. and Y.L. conceived and designed the experiments as well as wrote the paper.
References
- Anbalagan S, Bonetti D, Lucchini G, Longhese MP. Rif1 supports the function of the CST complex in yeast telomere capping. PLoS genetics. 2012;7:e1002024. doi: 10.1371/journal.pgen.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arneric M, Lingner J. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO reports. 2007;8:1080–1085. doi: 10.1038/sj.embor.7401082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiau AS, Yankulov K, Bah A, Revardel E, Luciano P, Wellinger RJ, Geli V, Gilson E. Subtelomeric proteins negatively regulate telomere elongation in budding yeast. The EMBO journal. 2006;25:846–856. doi: 10.1038/sj.emboj.7600975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Shore D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Molecular cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harbor symposia on quantitative biology. 2011;75:167–177. doi: 10.1101/sqb.2010.75.017. [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science (New York, NY. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Hoss M, Lingner J. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic acids research. 2000;28:2690–2694. doi: 10.1093/nar/28.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins--structures, mechanisms and functions. Febs J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Molecular cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Fukunaga K, Sugimoto K. Rif1 and rif2 inhibit localization of tel1 to DNA ends. Molecular cell. 2009;33:312–322. doi: 10.1016/j.molcel.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ME, Kolodner RD. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Molecular cell. 2005;17:709–720. doi: 10.1016/j.molcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Huang ME, Rio AG, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Redon S, Pfeiffer V, Dees M, Lingner J, Luke B. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO reports. 2012;12:587–593. doi: 10.1038/embor.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui I, Kienda G, Soeur J, Faye G, Baldacci G, Kolodner RD, Huang ME. Peroxiredoxin Tsa1 is the key peroxidase suppressing genome instability and protecting against cell death in Saccharomyces cerevisiae. PLoS genetics. 2009;5:e1000524. doi: 10.1371/journal.pgen.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Adkins CJ, Cartwright BR, Friedman KL. Yeast Est2p affects telomere length by influencing association of Rap1p with telomeric chromatin. Molecular and cellular biology. 2008;28:2380–2390. doi: 10.1128/MCB.01648-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Platts MH, Dharamsi LM, Friedman KL. Regulation of telomere length by an N-terminal region of the yeast telomerase reverse transcriptase. Molecular and cellular biology. 2005;25:9103–9114. doi: 10.1128/MCB.25.20.9103-9114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanishi S, Oikawa S, Murata M, Tsukitome H, Saito I. Site-specific oxidation at GG and GGG sequences in double-stranded DNA by benzoyl peroxide as a tumor promoter. Biochemistry. 1999;38:16733–16739. doi: 10.1021/bi990890z. [DOI] [PubMed] [Google Scholar]
- Kyrion G, Boakye KA, Lustig AJ. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Molecular and cellular biology. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larose S, Laterreur N, Ghazal G, Gagnon J, Wellinger RJ, Elela SA. RNase III-dependent regulation of yeast telomerase. The Journal of biological chemistry. 2007;282:4373–4381. doi: 10.1074/jbc.M607145200. [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Liu Y. Deletion of Ogg1 DNA glycosylase results in telomere base damage and length alteration in yeast. The EMBO journal. 2010;29:398–409. doi: 10.1038/emboj.2009.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL, Fan J, Danzy S, Wilson DM, 3rd, Bohr VA. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic acids research. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhage JL, Talley JM, Friedman KL. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nature structural & molecular biology. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- Park SG, Cha MK, Jeong W, Kim IH. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. The Journal of biological chemistry. 2000;275:5723–5732. doi: 10.1074/jbc.275.8.5723. [DOI] [PubMed] [Google Scholar]
- Petersen S, Saretzki G, von Zglinicki T. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Experimental cell research. 1998;239:152–160. doi: 10.1006/excr.1997.3893. [DOI] [PubMed] [Google Scholar]
- Ragu S, Faye G, Iraqui I, Masurel-Heneman A, Kolodner RD, Huang ME. Oxygen metabolism and reactive oxygen species cause chromosomal rearrangements and cell death. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9747–9752. doi: 10.1073/pnas.0703192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Runge KW. The C terminus of the major yeast telomere binding protein Rap1p enhances telomere formation. Molecular and cellular biology. 1998;18:1284–1295. doi: 10.1128/mcb.18.3.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee DB, Ghosh A, Lu J, Bohr VA, Liu Y. Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Repair (Amst) 2011;10:34–44. doi: 10.1016/j.dnarep.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Skorecki KL, Itzkovitz S, Yehezkel S, Segev Y, Shachar H, Berkovitz R, Adir Y, Vulto I, Lansdorp PM, Selig S. Telomere elongation followed by telomere length reduction, in leukocytes from divers exposed to intense oxidative stress--implications for tissue and organismal aging. Mechanisms of ageing and development. 2011;132:123–130. doi: 10.1016/j.mad.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science (New York, NY. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, Myung K. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science (New York, NY. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Molecular and cellular biology. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nature reviews. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- Ungar L, Yosef N, Sela Y, Sharan R, Ruppin E, Kupiec M. A genome-wide screen for essential yeast genes that affect telomere length maintenance. Nucleic acids research. 2009;37:3840–3849. doi: 10.1093/nar/gkp259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T. Role of oxidative stress in telomere length regulation and replicative senescence. Annals of the New York Academy of Sciences. 2000;908:99–110. doi: 10.1111/j.1749-6632.2000.tb06639.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Rhee DB, Lu J, Bohr CT, Zhou F, Vallabhaneni H, de Souza-Pinto NC, Liu Y. Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS genetics. 2010;6:e1000951. doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Zhou Y, Ng RW, Kung Hf HF, Jin DY. Cooperation of yeast peroxiredoxins Tsa1p and Tsa2p in the cellular defense against oxidative and nitrosative stress. The Journal of biological chemistry. 2002;277:5385–5394. doi: 10.1074/jbc.M106846200. [DOI] [PubMed] [Google Scholar]
- Xue Y, Rushton MD, Maringele L. A novel checkpoint and RPA inhibitory pathway regulated by Rif1. PLoS genetics. 2012;7:e1002417. doi: 10.1371/journal.pgen.1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.