Summary
Caloric restriction (CR) and down-regulation of the insulin/IGF pathway are the most robust interventions known to increase longevity in lower organisms. However, little is known about the molecular adaptations induced by CR in humans. Here we report that long-term CR in humans inhibits the IGF-1/insulin pathway in skeletal muscle, a key metabolic tissue. We also demonstrate that CR-induced dramatic changes of the skeletal muscle transcriptional profile that resemble those of younger individuals. Finally, in both rats and humans CR evoked similar responses in the transcriptional profiles of skeletal muscle. This common signature consisted of three key pathways typically associated with longevity: IGF-1/insulin signaling, mitochondrial biogenesis and inflammation. Furthermore, our data identifies promising pathways for therapeutic targets to combat age-related diseases and promote health in humans.
Keywords: human, caloric restriction, skeletal muscle, insulin/IGF-1 signaling
CR without malnutrition and inhibition of the insulin/IGF-1 signaling are the most robust and reproducible interventions for extending lifespan and preventing or delaying age-related disease in a variety of species (Anderson & Weindruch 2007; Kennedy et al. 2007; Piper & Bartke 2008). Aging was believed to be the consequence of the inevitable wear and tear process, but in 1993 Kenyon and her associates published the first paper indicating that the inhibition of the insulin/IGF-1/FOXO pathway dramatically extends lifespan in worms (Kenyon et al. 1993). Since then, accumulating data have shown that this pathway is evolutionarily conserved, and that dietary and genetic manipulations of the insulin/IGF-1/FOXO pathway extend lifespan in rodents as well (Kenyon et al. 1993; van Heemst et al. 2005; Kennedy et al. 2007; Piper & Bartke 2008). Moreover, evidence derived from exceptionally long-lived people also supports a role for the IGF signaling pathway in human longevity (van Heemst et al. 2005; Suh et al. 2008). However, nothing is known on the molecular adaptations induced by long-term CR in humans, and in particular on the effects of CR on the regulation of the insulin/IGF-1/FOXO pathway.
As one of the primary metabolic tissues, skeletal muscle is extremely vulnerable to the effects of aging, and decreases in muscle mass and strength have been associated with increased mortality (Cesari et al. 2009). Interestingly, CR prevents or delays this age-related loss of muscle function and mass, thereby contributing to increases in both the quantity and quality of life (Aspnes et al. 1997; McKiernan et al. 2010). Although research on CR in humans is still at an early stage, information from population and physiologic studies suggest that many of the same beneficial metabolic adaptations that occur in experimental animal models are also observed in humans (Fontana & Klein 2007; Fontana et al. 2010b). We hypothesized that the impressive metabolic changes previously described in these individuals on long-term CR would result in several molecular adaptations, including a down-regulation of the insulin/IGF-1/FOXO pathway, a key nutrient sensing pathway, shown to slow aging in several experimental animals (Kenyon et al. 1993; van Heemst et al. 2005; Kennedy et al. 2007; Piper & Bartke 2008). We also hypothesized that the relative changes of the skeletal muscle gene expression profiles would be similar between CR rats and humans. In this study, we studied the molecular adaptations induced by long-term CR in healthy lean men and women, and compared the skeletal muscle transcriptional profiles induced by CR in rats and humans. Our results show that chronic moderate (~30%) CR in lean and weight-stable adult individuals, despite their genetic heterogeneity, results in a uniform dramatic transcriptional reprogramming of molecular pathways in skeletal muscle, which shifts cellular metabolism from growth to maintenance/repair activities. We also found that CR induces a significant down-regulation of the IGF-1/insulin/FOXO pathway both at the transcriptional and post-transcriptional level. In addition, we found that humans and long-lived CR rats, despite significant genetic, physiologic and anatomical differences, exhibit a number of key common transcriptional adaptations in a major metabolic organ (i.e. skeletal muscle), suggesting that the beneficial gains of CR on healthy longevity would be observed in humans.
We first sought to determine if CR modifies the gene expression profile in human skeletal muscle. To this end, we recruited and studied 15 middle-aged (58.7±7.4 yrs), weight-stable very lean (BMI=19.2±1.1 kg/m2) members of the Calorie Restriction Society who have been practicing ~30% CR with adequate nutrition (at least 100% of RDI for each nutrient) for an average of 9.6 years and a control group of 10 non-obese (BMI=25.3±2.3 kg/m2) age-matched controls eating a typical Western diet (Table 1). Principle Component Analysis (PCA) revealed a very distinct separation of groups based on dietary manipulation (Fig. 1A). Remarkably, we found that CR in humans induces dramatic and uniform changes to the gene expression profile within skeletal muscle that are strong enough to overcome the genetic, nutritional and geographical heterogeneity of this human population. Furthermore, comparison of our study population expression profiles with a younger (31.2±2.0 yrs) non-obese (BMI=25.3±2.3 kg/m2) human cohort showed that our CR was grouped closer to the younger profile by PCA (Table 1 and Fig. 1B). In order to further investigate the role of CR in modifying skeletal muscle gene expression in humans, we next performed parametric analysis of gene set enrichment (PAGE), a computational method that determines differences between genes and pathways using a priori defined gene sets (Kim & Volsky 2005; Subramanian et al. 2005). PAGE analysis indicated that aging caused a significant down-regulation of pathways associated with muscle contraction, electron transport and oxidative phosphorylation (Fig. 1D, for a complete list see Table S1). Notably, CR opposes the majority of the transcriptional changes observed in skeletal muscle of middle-aged individuals following a WD. These results indicate that CR in humans can slow down the transcriptional changes associated with aging in skeletal muscle.
Table 1.
Characteristics of the study subjects
| HUMANS | RATS | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| WD-Y (n=5) | WD-M (n=10) | CR (n=15) | P value | AL (n=27) | CR (n=27) | P value | |
| Age (years) | 31.2±2.0 | 58.0±7.4 | 58.7±7.4 | 0.810 | 1.3 | 1.3 | - |
| Sex (M/F) | 5/0 | 8/2 | 11/4 | - | 27/0 | 27/0 | |
| BODY COMPOSITION | |||||||
| Height (m) | 1.73±8.8 | 1.76±0.0 | 1.73±0.0 | 0.409 | - | - | - |
| Body weight (kg) | 78.4±16.3 | 78.1±9.7 | 57.5±6.4 | <0.0001 | 0.40±0.7 | 0.24±0.2 | 0.001 |
| Body mass index (kg/m2) | 25.0±5.0 | 25.3±2.3 | 19.2±1.1 | <0.0001 | - | - | - |
| DEXA - NMR | |||||||
| Fat mass (%) | - | 24.0±7.4 | 13.1±4.8 | <0.0001 | 30.1±0.2 | 27.2±0.2 | 0.01 |
| Trunk fat (%) | - | 25.6±6.8 | 11.0±3.5 | <0.0001 | - | - | - |
| Lean mass (%) | - | 73.1±6.0 | 81.1±4.8 | 0.001 | 60.1±0.2 | 63.1±0.1 | 0.03 |
Values are mean ± SD. WD-Y=younger individuals eating Western diets (WD); WD-M=middle-aged individuals on WD; AL=ad libitum laboratory diet. For humans, p-value comparison CR – WD-M.
Fig. 1. Principal component analysis (PCA) and parametric analysis of gene-set enrichment (PAGE) were performed on microarray data from humans and rats.
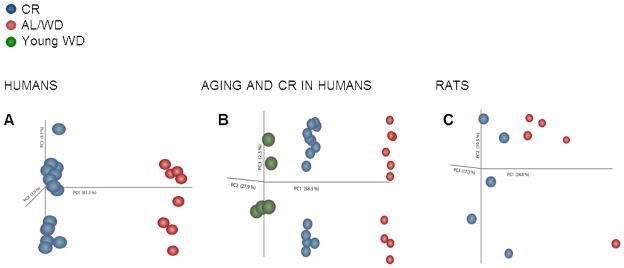
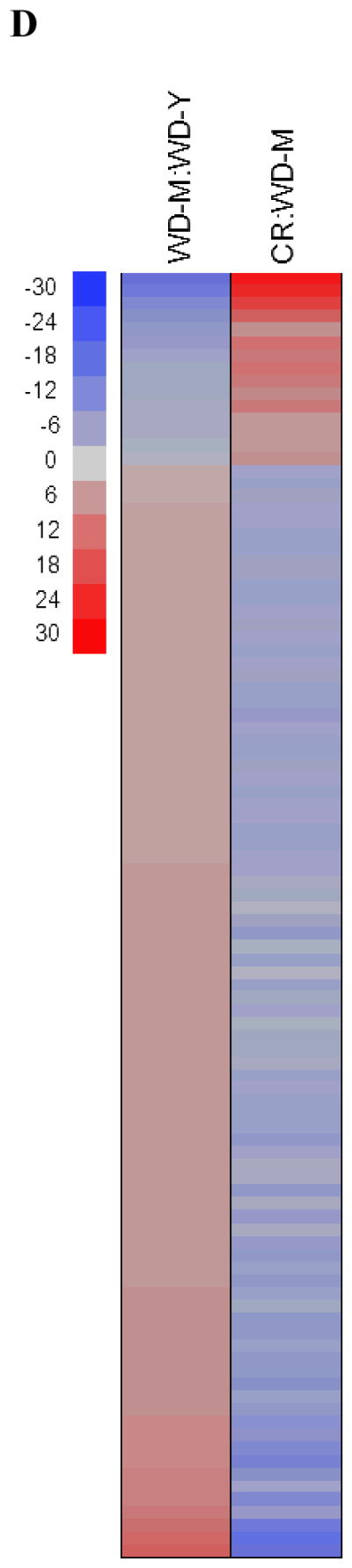
PCA of (A) humans on caloric restriction (CR) (blue) or ad libitum (AL)/Western diet (WD) (red), (B) those of younger subjects on a WD (green), and (C) rats. n=5 for both AL and CR-fed rats; n=10 and n=15 for humans on WD and CR, respectively; n=5 for younger individuals on WD. (D) PAGE comparing the top 100 significantly pathways up-regulated (red) or down-regulated (blue) by either the WD or CR (WD-Y=younger individuals eating Western diets (WD); WD-M=middle-aged individuals on WD).
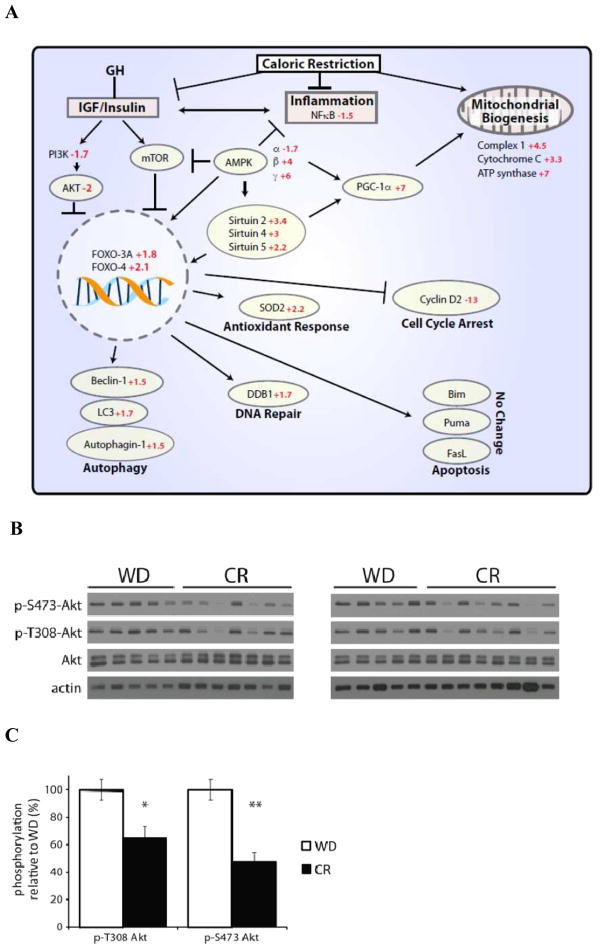
As the IGF-1/insulin/FOXO pathway is implicated in improved healthspan, we then sought to determine if CR modifies this pathway in human skeletal muscle. Importantly, we found that a highly significant number of transcripts along the IGF-1/insulin/FOXO pathway were altered by CR. The PI3K and AKT transcripts were significantly down-regulated 1.7 and 2 folds, respectively (Fig. 2A). Consistently with the gene expression changes, skeletal muscle of humans on long-term CR showed a 35–50% reduction in AKT phosphorylation of the serine and threonine residues (Fig. 2B and C). To our knowledge, this is the first set of data showing that long-term CR in humans down-regulates the activity of the insulin/IGF pathway, which has been shown by Kenyon and others to play a key role in promoting health and longevity in several experimental model organisms (Kenyon et al. 1993; van Heemst et al. 2005; Kennedy et al. 2007; Piper & Bartke 2008). Importantly, we also found that other transcription factors downstream of AKT, such as FOXO-3A and FOXO-4, were up-regulated (Fig. 2A). It is well known that the inhibition of AKT activates FOXO, which modifies several “longevity genes” (i.e. stress resistance, antioxidants, DNA repair, protein turnover and cell death genes) (Lanvin et al. 2007). Consistently, we found that in human skeletal muscle CR alters several transcripts downstream of FOXO. For example, SOD2, an important antioxidant enzyme, and DDB1, a key DNA repair molecule were both up-regulated, while cyclin D2, a master regulator of cell cycle progression, was powerfully down-regulated. Several transcripts downstream of FOXO that induce macroautophagy (i.e. beclin-1, autophagin-1 and MAP1LC3B) were also significantly up-regulated (Fig. 2A). FOXO is also regulated by two central energy-sensing pathways, SIRT1 and AMPK, which are both activated by decreased nutrient availability (Brunet et al. 2004; Greer et al. 2007). In our study, SIRT2, SIRT4, SIRT5, AMPKβ and AMPKγ transcripts were significantly up-regulated in the human skeletal muscle of the CR participants (Fig. 2A). It is well established that SIRT1 and AMPK control the activity of PGC-1α, a key transcriptional regulator of mitochondrial metabolism (Wu et al. 1999). Consistently, our findings demonstrate that in human skeletal muscle, CR powerfully increases PGC-1α gene expression (Fig. 2A), which leads to an up-regulation of several genes that induce mitochondrial biogenesis (Wu et al. 1999), thus opposing the decline in mitochondrial function with age (Anderson & Weindruch 2007). Transcriptional profiling also indicates a CR-induced decrease in the inflammatory response, which is implicated in a range of age-related diseases. The beneficial effects of CR may involve an elaborate crosstalk between these highly conserved inflammatory, nutrient- and energy-sensing pathways.
Fig. 2. Transcriptional and post-transcriptional modifications of the PI3K/AKT/FOXO pathway in human skeletal muscle by CR.
(A) Transcriptional down-regulation of the PI3K/AKT/FOXO signaling pathway by CR. GH: growth hormone; IGF: insulin-like growth factor; PI3K: phosphatidylinositol 3-kinase; AKT: protein kinase B (PKB); FOXO-3A: forkhead box O3; FOXO-4: forkhead box O4; mTOR: mammalian target of rapamycin; LC3: microtubule-associated protein 1 light chain 3; AMPK: adenosine monophosphate-activated protein kinase; NFκB: nuclear factor-kappaB; SOD2: superoxide dismutase 2; DDB1: damage-specific DNA binding protein 1; FASL: fas ligand.
Western blot of human skeletal muscle from individuals on a Western diet (WD) or caloric restricted (CR) diet. Immunoblot (B) and quantification (C) of western blots in panel a for p-T308 AKT and p-S473 AKT was performed using NIH ImageJ, and normalized to total AKT expression. (*p<0.01, **P<0.00003 (n=10 WD, 15 CR samples)). Bars indicate mean ± SEM.
We then wanted to determine if the overall physiological and molecular response of long-term CR was similar in humans and rats. Rats were subjected to either 40% CR or ad libitum (AL) diets early in life. As expected, our rats on a CR-fed diet had increased mean and maximum lifespan compared to those on an ad libitum-fed diet (Fig. S1). In humans, nevertheless, such early initiation of CR would be detrimental to growth and development. Therefore, human subjects included in this study were middle-aged men and women. However, it is well known that in rodents CR initiated at adulthood (12–19 months of age) is as effective as CR begun early in life at decelerating mortality rate, extending remaining lifespan, and inducing dramatic changes in the gene expression profile (Dhahbi et al. 2004). As expected, CR reduced body weight, reduced fat mass and increased the lean to fat mass ratio in both middle-aged humans and rats compared to those on Western or AL diets, respectively (Table 1). Interestingly, both species exhibited similar responses to CR in several physiological, hormonal and biochemical markers. Previous studies on these individuals indicate that long-term CR in humans results in improved cardiometabolic profile, markedly lower blood pressure and inflammation, improved left ventricular diastolic function and heart rate variability, and lower serum concentration of insulin, leptin, sex hormones, triiodothyronine and core body temperature (Fontana & Klein 2007; Cangemi et al. 2010; Fontana et al. 2010a; Fontana et al. 2010b; Soare et al. 2011), all of which are hallmark adaptations also seen in rats in response to CR (Weindruch & Walford 1988; de Cabo et al. 2003). Interestingly, the effect of CR on circulating IGF-1 concentrations was not significantly reduced in humans unless protein intake is also reduced (Fontana et al. 2008).
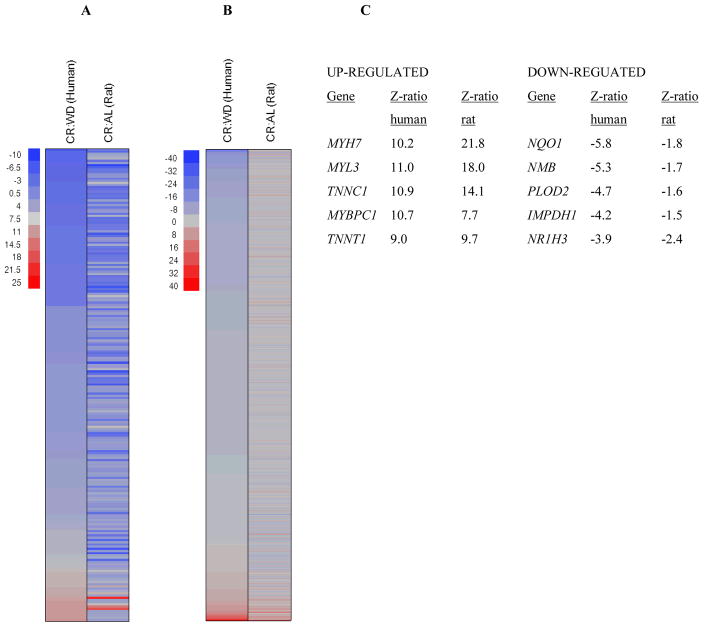
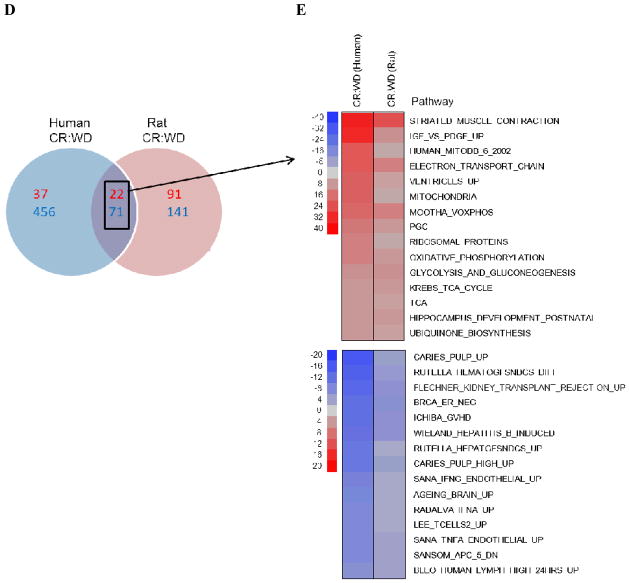
Based on the broad phenotypical similarities observed in both species following long-term CR, we next examined if similarities between humans and rats occurred in gene expression profiles of skeletal muscle as well. Principle Component Analysis (PCA) revealed a very distinct separation of groups based on dietary manipulation regardless of species (Fig. 1C). PAGE analysis indicated that out of the shared changes CR evoked a number of common genes and pathways in both species (83% genes and 93% pathways), with the majority of genes being down-regulated by CR (Fig. 3A and B). A subset of gene expression changes was verified by quantitative real-time PCR or QuantiGene based assays (Table S2). Notably, the 5 most highly elevated transcripts were contractile proteins (Fig. 3C). The list of the 5 most highly down-regulated transcripts include proteins involved in antioxidant responses (NQO1), cholesterol metabolism (NR1H3), fibrosis (PLOD2) and cell growth (IMPDH1). The complete dataset is available at http://www.ncbi.nlm.nih.gov/geo/. We identified 93 pathways (22 up-regulated, 71 down-regulated) that were common among rats and humans in response to CR. These included an up-regulation of genes involved in energy metabolism (e.g. TCA cycle, mitochondrial ETC, and glycolysis), as well as a decrease of those in insulin/IGF-1 signaling, and in both the classical and alternative inflammatory pathways (Fig. 3D and E); for a complete list see Table S3, S4). Interestingly, the effects observed on these pathways by CR oppose those normally occurring during the aging process (Park & Prolla 2005). These results indicate that humans share with other species, such as rats, similar transcriptional responses to CR that are typically associated with improved health and survival. However, this association does not imply causation for increased longevity. In summary, transcriptional patterns of CR humans suggest that CR may retard the aging process by shifting cellular metabolism from growth to maintenance and repair activities. A molecular trigger for this shift may be the down-regulation of the IGF/insulin/FOXO signaling pathway, a result that connects our findings to those obtained through genetic manipulation of aging in worms, fruit flies and rodents (Kenyon et al. 1993; van Heemst et al. 2005; Kennedy et al. 2007; Piper & Bartke 2008).
Fig. 3. Parametric analysis of gene-set enrichment (PAGE) of microarray data from humans and rats.
Columns show every (A) gene or (B) pathway significantly up-regulated (red) or down-regulated (blue) by CR. (C) The five most highly up-regulated and down-regulated genes, based on Z-ratio in CR compared to WD and AL respectively. (D) Venn diagram showing the pathway interactions between humans and rats (up-regulated and down-regulated pathways are shown in red and blue respectively). (E) Cell plot showing the most highly significant up-regulated (red) and down-regulated (blue) common pathways between humans and rats.
Experimental procedures
Study subjects
Rats
Male Fisher 344 rats (n=54) were randomly assigned to two groups at 2 months of age. One group was kept ad libitum (AL) fed throughout their lifespan while the calorie restriction (CR) group was progressively brought down to a 40% CR. All animals were fed a NIH-31 standard chow (Harlan Teklad, Indianapolis, IN, USA). Rats were singly housed in an environmentally controlled vivarium with unlimited access to water and a controlled photoperiod (12 hr light;12 hr dark). Body weights and food intake were recorded biweekly. All rats were maintained between 68–72°F according to animal protocols and NIH guidelines. All animal procedures for this study were reviewed and approved by the Animal Care and Use Committee (ACUC) at the Biomedical Research Center (NIA/NIH).
Humans
Fifteen individuals had been on CR for an average of 9.6 years (4–20 yrs). Subjects were instructed by an experienced research dietician to record all food and beverages consumed, preparation methods, and approximate portion sizes for 7 consecutive days. Food records were analyzed by using the NDS-R program (version 4.03_31), which is the Nutrition Data System for research from the Nutrition Coordinating Center at the University of Minnesota. CR subjects consumed a variety of nutrient-dense unprocessed foods (i.e. vegetables, fruits, nuts, egg whites, fish, poultry, low-fat dairy products, whole grains and beans) which supplied > 100% of the Recommended Daily Intake for all essential nutrients. Refined foods rich in empty calories, and trans-fatty acids were avoided. Energy intake was 30% lower in the CR group (1773±239 kcal/d) than in the Western diet (WD) group (n=10) (2483±378 kcal/d) (p≤0.0001). The percentage of total energy intake derived from protein, carbohydrate, fat and alcohol was 22%, 50%, 28% and 0.2%, respectively in the CR group and 17%, 47%, 35% and 1% in the WD group. The human study was approved by the Human Studies Committee of Washington University School of Medicine, and all participants gave informed consent before their participation. Moreover, a younger cohort of humans (n=5) who underwent surgery for hip dysplasia was included in this study. Written informed consent was obtained from those participants, and the study was approved by the medical ethics committee of the Rizzoli Hospital.
Body composition
Rats
Measurements of lean and fat mass in whole, live rats were acquired by nuclear magnetic resonance (NMR) using the Minispec LF90 (Bruker Optics, Billerica, MA).
Humans
Total body fat mass and fat free mass were determined by dual-energy X-ray absorptiometry (DXA) (QDR 1000/w, Hologic, Waltham, MA).
Micro-array analysis
Five rats per group at 27 months of age were sacrificed and vastus lateralis muscle were flash frozen. Human percutaneous biopsy specimens of vastus lateralis muscle of middle-aged individuals following CR (n=15), and young (n=5) and middle-aged (n=10) participants eating a standard Western diet (WD) were obtained. RNA was extracted from skeletal muscle samples from both species using Trizol Reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions and hybridized to RatRef-12 v1 Expression or Human6 Expression v3 and v4 beadchips (Illumina, San Diego, CA) following protocols listed on the Gene Expression and Genomics Unit of the NIA (http://www.grc.nia.nih.gov/branches/rrb/dna/index/protocols.htm). Microarray florescent signals were extracted using an Illumina BeadArray 500GX reader. The signals on each sample are normalized by log z-transformation to obtained z-scores and tests for distributions as previously described (Cheadle et al. 2003). Correlation analysis, sample clustering analysis and principal component analysis including all probes were performed to identify/exclude any possible outliners. The resulting dataset was next analyzed with DIANE 6.0, a spreadsheet based microarray analysis program. Gene set enrichment analysis used gene expression values or gene expression change values for all of the genes on the microarray. Parametric analysis of gene set enrichment (PAGE) was used [pubmed 20682848] for gene set analysis. Gene Sets include the MSIG database [Link], Gene Ontology Database [Link], GAD human disease and mouse phenotype gene sets [pubmed: 20092628] were used to explore functional level changes.
RT-qPCR
Rats
Vastus lateralis total RNA from rats was isolated as described above and first strand cDNA was synthesized using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions. Real-time quantitative PCR was performed with SYBR® Green PCR master mix on an ABI Prism 7300 sequence detection system (Applied Biosystems). Each sample was analyzed in triplicate and relative gene expression was normalized to GAPDH. The data were analyzed using the 2−ΔΔCT method. Primer sequences are listed in Table S5.
Humans
Total RNA was extracted from human muscle biopsies using Trizol as described above. Gene expression levels were quantitated using the Quantigene QuantiGene Plex 2.0 assay according to the manufacturer’s protocols. (Panomics/Affymetrix, Fremont, CA) (probe set regions are provided in Table S5). In order to determine the fold-change of the genes of interest, we used Partek Genomic Suite to conduct an ANOVA analysis on log2 transformed background-subtracted Quantigene data, generating a fold-change of the CR RNA samples over the WD RNA samples.
Immunoblotting
Vastus lateralis muscles were lysed in 1mL of cold RIPA buffer (50 mM HEPES ph 7.4, 40 mM NaCl, 2 mM EDTA, 1.5 mM sodium Orthovanadate (Na3VO4), 50 mM NaF, 10 mM Sodium Pyrophosphate, 10 mM Sodium beta glycerophosphate, 0.1% SDS, 1% Sodium Deoxycholate, 1% Triton) supplemented with phosphatase inhibitor and protease inhibitor cocktail tablets, using a FastPrep 24 (MP Biomedical) instrument. Protein concentration was determined by Bradford Assay (Bio-Rad). 20ug protein was separated by sodium dodecylsulpahte-polyacrylamide gel electrophoresis (SDS-PAGE) on 8%, 10% or 16% Tris-Glycine gels (Invitrogen), and transferred to Immobilon P 0.45uM PVDF membrane (Millipore). Membranes were blocked in 5% milk/TBST, and antibodies were incubated overnight at 4°C in 5% BSA/TBST (phospho-antibodies) or 5% milk/TBST (all other antibodies). Quantification was performed by densitometry using ImageJ software, and loading was verified by blotting for actin. Antibodies to p-T308 AKT (CST 2965), p-S473 AKT (CST 4060), and total AKT (CST 4691) were from Cell Signaling Technology. The actin (sc-7210) antibody was from Santa Cruz Biotechnology.
Statistical analysis
Subject characteristics were analyzed using Student’s t-test (two-sample equal variance; two-tailed distribution). Data are expressed as mean ± SD unless indicated otherwise. Survival curves were plotted using the Kaplan-Meier method, which included 27 rats for both CR and AL. Statistical analysis were performed using Sigma-Stat Program for Windows, version 3.5 (Systat Software, Inc). A difference with p-values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
Supported by grants from AFAR (American Federation for Aging Research), the National Center for Research Resources (UL1 RR024992), the National Institute of Diabetes And Digestive And Kidney Diseases (P30DK056341). This research was supported in part by the National Institute on Aging-Intramural Research Program, the Longer Life Foundation (an RGA/Washington University Partnership), the Bakewell Foundation, and a donation from the Calorie Restriction Society and the Scott and Annie Appleby Charitable Trust. The funding agencies had no role in the analysis or interpretation of the data or in the decision to submit the report for publication. We thank William Wood and Elin Lehrmann for help with microarray processing and Kevin Becker for microarray data analysis. We also thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1RR024992 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. DWL is a Charles A. King Trust Postdoctoral Fellow; DMS is an Investigator of the Howard Hughes Medical Institute. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. Data have been deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession code GSE38063.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
Author Contributions
E.M.M., S.D.C, R.de C. and L.F. designed the experiments. Protein assays were performed by D.W.L. E.M.M., S.D.C., D.V., D.M.S., R.de C. and L.F. contributed to data analysis and interpretation. Computational methods for analysis of microarray data were developed and applied by L.S., Y.Z., and K.G.B. M.C. and C.F. provided young human muscle biopsies. E.M.M., S.D.C, S.K.-W., R.de C. and L.F. wrote the manuscript.
Contributor Information
Evi M. Mercken, Email: merckenem@mail.nih.gov.
Seth D. Crosby, Email: scrosby@watson.wustl.edu.
Dudley W. Lamming, Email: lamming@wi.mit.edu.
Lellean JeBailey, Email: Lellean@genego.com.
Susan Krzysik-Walker, Email: susan.walker@nih.gov.
Dennis Villareal, Email: dennis.villareal@va.gov.
Miriam Capri, Email: miriam.capri@unibo.it.
Claudio Franceschi, Email: claudio.franceschi@unibo.it.
Yongqing Zhang, Email: zhangyon@grc.nia.nih.gov.
Kevin Becker, Email: beckerk@grc.nia.nih.gov.
David M. Sabatini, Email: Sabatini@wi.mit.edu.
Rafael de Cabo, Email: decabora@grc.nia.nih.gov.
Luigi Fontana, Email: lfontana@dom.wustl.edu.
References
- Anderson RM, Weindruch R. Metabolic reprogramming in dietary restriction. Interdisciplinary topics in gerontology. 2007;35:18–38. doi: 10.1159/000096554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspnes LE, Lee CM, Weindruch R, Chung SS, Roecker EB, Aiken JM. Caloric restriction reduces fiber loss and mitochondrial abnormalities in aged rat muscle. Faseb J. 1997;11:573–581. doi: 10.1096/fasebj.11.7.9212081. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science (New York, NY. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Cangemi R, Friedmann AJ, Holloszy JO, Fontana L. Long-term effects of calorie restriction on serum sex-hormone concentrations in men. Aging cell. 2010;9:236–242. doi: 10.1111/j.1474-9726.2010.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari M, Pahor M, Lauretani F, Zamboni V, Bandinelli S, Bernabei R, Guralnik JM, Ferrucci L. Skeletal muscle and mortality results from the InCHIANTI Study. The journals of gerontology. 2009;64:377–384. doi: 10.1093/gerona/gln031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Cho-Chung YS, Becker KG, Vawter MP. Application of z-score transformation to Affymetrix data. Applied bioinformatics. 2003;2:209–217. [PubMed] [Google Scholar]
- de Cabo R, Furer-Galban S, Anson RM, Gilman C, Gorospe M, Lane MA. An in vitro model of caloric restriction. Experimental gerontology. 2003;38:631–639. doi: 10.1016/s0531-5565(03)00055-x. [DOI] [PubMed] [Google Scholar]
- Dhahbi JM, Kim HJ, Mote PL, Beaver RJ, Spindler SR. Temporal linkage between the phenotypic and genomic responses to caloric restriction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5524–5529. doi: 10.1073/pnas.0305300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Klein S. Aging, adiposity, and calorie restriction. Jama. 2007;297:986–994. doi: 10.1001/jama.297.9.986. [DOI] [PubMed] [Google Scholar]
- Fontana L, Klein S, Holloszy JO. Effects of long-term calorie restriction and endurance exercise on glucose tolerance, insulin action, and adipokine production. Age (Dordrecht, Netherlands) 2010a;32:97–108. doi: 10.1007/s11357-009-9118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science (New York, NY. 2010b;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Weiss EP, Villareal DT, Klein S, Holloszy JO. Long-term effects of calorie or protein restriction on serum IGF-1 and IGFBP-3 concentration in humans. Aging cell. 2008;7:681–687. doi: 10.1111/j.1474-9726.2008.00417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. The Journal of biological chemistry. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Steffen KK, Kaeberlein M. Ruminations on dietary restriction and aging. Cell Mol Life Sci. 2007;64:1323–1328. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanvin O, Bianco S, Kersual N, Chalbos D, Vanacker JM. Potentiation of ICI182,780 (Fulvestrant)-induced estrogen receptor-alpha degradation by the estrogen receptor-related receptor-alpha inverse agonist XCT790. The Journal of biological chemistry. 2007;282:28328–28334. doi: 10.1074/jbc.M704295200. [DOI] [PubMed] [Google Scholar]
- McKiernan SH, Colman RJ, Lopez M, Beasley TM, Aiken JM, Anderson RM, Weindruch R. Caloric restriction delays aging-induced cellular phenotypes in rhesus monkey skeletal muscle. Experimental gerontology. 2010;46:23–29. doi: 10.1016/j.exger.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Prolla TA. Lessons learned from gene expression profile studies of aging and caloric restriction. Ageing research reviews. 2005;4:55–65. doi: 10.1016/j.arr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Piper MD, Bartke A. Diet and aging. Cell metabolism. 2008;8:99–104. doi: 10.1016/j.cmet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Soare A, Cangemi R, Omodei D, Holloszy JO, Fontana L. Long-term calorie restriction, but not endurance exercise, lowers core body temperature in humans. Aging. 2011;3:374–379. doi: 10.18632/aging.100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst D, Beekman M, Mooijaart SP, Heijmans BT, Brandt BW, Zwaan BJ, Slagboom PE, Westendorp RG. Reduced insulin/IGF-1 signalling and human longevity. Aging cell. 2005;4:79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Walford RL. The Retardation of Aging and Disease by Dietary Restriction. Springfield, IL: Thomas; 1988. [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.