Abstract
We performed whole-exome sequencing of a family with autosomal dominant Dandy-Walker malformation and occipital cephaloceles (ADDWOC) and detected a mutation in the extracellular matrix protein encoding gene NID1. In a second family, protein interaction network analysis identified a mutation in LAMC1, which encodes a NID1 binding partner. Structural modeling the NID1-LAMC1 complex demonstrated that each mutation disrupts the interaction. These findings implicate the extracellular matrix in the pathogenesis of Dandy-Walker spectrum disorders.
Keywords: ADDWOC, Dandy-Walker, NID1, LAMC1, Extracellular Matrix
The Dandy-Walker spectrum of disorders including ADDWOC are characterized by variable cerebellar hypoplasia, meningeal anomalies, and occipital skull defects. (Barkovich, et al., 2009) We have previously reported deletions that encompass FOXC1 (Aldinger, et al., 2009) or ZIC genes (Grinberg, et al., 2004) in rare familial and sporadic cases of Dandy-Walker spectrum disorders. These mutations likely affect the extracellular matrix (ECM) of bone, meninges, and brain. (Aldinger, et al., 2009; Grinberg, et al., 2004; Inoue, et al., 2008)
We previously ascertained the phenotype of a family with ADDWOC and normal eye examination of Vietnamese origin (Fig. 1a, family; previously described by Bassuk et al. (Bassuk, et al., 2004), and Jalali et al. (Jalali, et al., 2008)). Genome-wide linkage analysis in the seven affected family members with obvious cephaloceles followed by resequencing failed to identify a mutation in the only significant LOD peak (Jalali, et al., 2008). To directly determine if a protein-coding mutation segregated with the phenotype in this pedigree, we re-examined this family by whole-exome capture and massively parallel sequencing.
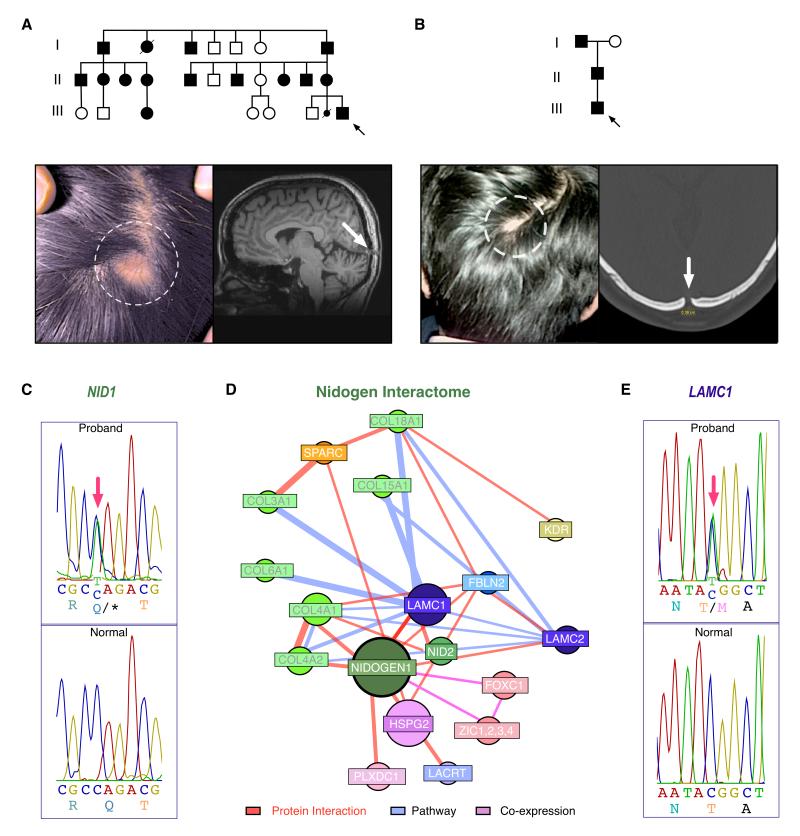
Figure 1. Pedigrees and Phenotyptes of Affected ADDWOC Families, NID1 and LAMC1 mutations, and NID1 interactome.
a. Pedigree of the Vietnamese family (Family 1) with photo of cephalocele (dotted circle) and skull defect (white arrow) on sagittal brain MRI (T1 weighted image). Black arrow=proband.
b. Pedigree of the Indian family (Family 2) with examples photos of cephalocele and skull defect on x-ray. Black arrow=proband.
c. NID1 mutant sequence from Family 1 (top) compared with the normal NID1 sequence (bottom). Red arrow indicates site of mutant nucleotide.
d. Construction of a NID1 interactome reveals putative candidates for the ADDWOC phenotype that include LAMC1.
e. LAMC1 mutant sequence from Family 2 (top) compared to the normal LAMC1 sequence (bottom).
c and e. Nucleotide numbering reflects cDNA with +1 as the A of the ATG translation initiation codon in the reference sequence (see text). Initiation codon is codon 1.
Whole-exome capture and massively parallel sequencing were performed for seven affected family members with cephalocele. Re-sequencing of putative segregating variants by Sanger sequencing in the entire pedigree revealed only a single mutation that was present in all fourteen affected family members (all with Dandy-Walker variant/cerebellar vermal hypoplasia ± cephalocele (Jalali, et al., 2008)) and absent from 384 Vietnamese control chromosomes, dbSNP135, the 1000 Genomes Project, or from the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP). This segregating variant at position 236201527 of chromosome 1 (hg19) corresponded to a nonsense mutation in the gene NID1 [MIM# 131390; www.lovd.nl/NID1; NM_002508.2 (NID1_v001): c.1162C>T, p.(Gln388*)] (Fig. 1c). The NID1 protein coordinates extracellular matrix protein interactions. (Mayer, et al., 1998; Mayer, et al., 1993; Takagi, et al., 2003)
A second ADDWOC family from India with normal eye examinations (Fig. 1b; previously described by Ghonge et al(Ghonge, et al., 2011)) was examined for NID1 mutations by Sanger resequencing. No NID1 mutations were discovered. Since the functional consequence of the NID1 stop mutation found in Family 1 was the deletion of several domains that interact with other ECM proteins (Figure 2a), (Mayer, et al., 1998; Mayer, et al., 1993; Takagi, et al., 2003) we investigated binding partner interactions that might be disrupted. A NID1 interactome was generated, and the NID1 interacting proteins were considered candidate genes for Family 2 (Figure 1d, Supp. Tables S1 and S2). The coding sequence for each of these genes was then interrogated following whole-exome capture and massively parallel sequencing of Family 2. Only one single mutation from the NID1-interactome segregated with the phenotype in the Indian ADDWOC family. This segregating variant at position 183091222 of chromosome 1 (hg19) corresponded to a missense mutation in the gene LAMC1 [MIM# 150290; NM_002293.3 (LAMC1_v001): c.2237C>T, p.(Thr746Met)] (Fig. 1e). The LAMC1 gene encodes the laminin gamma chain. This mutation was not found in dbSNP135, the 1000 Genomes or data from the NHLBI Exome Sequencing Project. The variant alters a threonine residue conserved throughout evolution (Fig. 2c). Note that there was insufficient DNA samples available to analyze an English and Brazilian pedigree described previously (Jalali, et al., 2008).
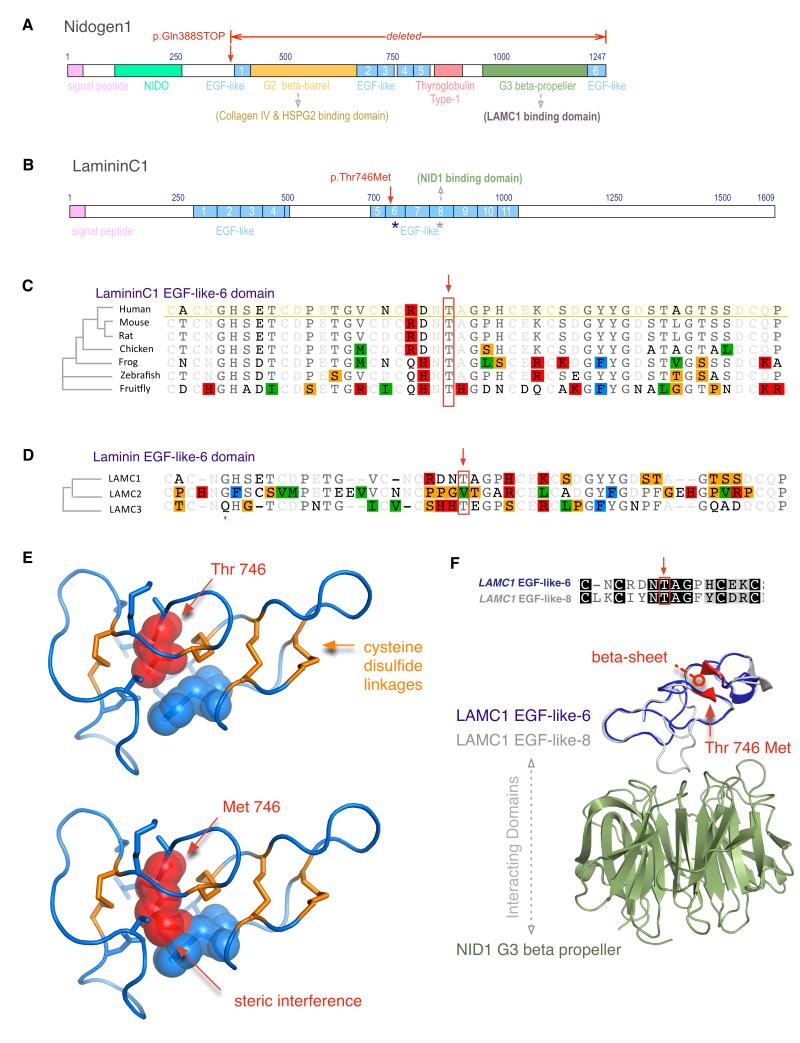
Figure 2. Protein Domain and Structure analysis.
a. Nidogen1 domains. A stop codon leads to deletion of the Nidogen1 G2 beta barrel domain (green), several EGF-like domains (blue), and the thyroglobulin Type-1 domain.
b. LamininC1 domains. The family-2 mutation was found in EGF-6-like (blue asterisk). There are 11 EGF-like domains (blue), and a previously known interaction with Nidogen-1 occurs through EGF-8-like domain (grey asterisk).
c. Ortholog alignment shows high evolutionary conservation of the Threonine-746 residue (red box).
d. Paralog alignment of three human EGF-6-like domains from LAMC1, C2, and C3 shows size conservation of the 746-residue, Threonine (M.W. 119 Da) and Valine (M.W. 117 Da, red box).
e. The normal threonine-746 (red) occupies a small space between rigid cysteine disulfide bridges (orange), and a mutation to methionine leads to steric interference with surrounding residues (blue).
f. EGF-like-6 domain (homology model in blue) shows 33% sequence identity to EGF-like-8 domain (grey), which interacts with the Nidogen1 G2 beta barrel domain (green). The mutation is expected to disrupt a beta sheet (red).
Since LAMC1 was selected as a candidate gene based on its physical interaction with NID1, we investigated how the mutations might disrupt this interaction by computational three-dimensional structural homology modeling of the involved domains in the context of known NID1-LAMC1 interaction (pdb ID: 1NPE, Fig. 2).(Takagi, et al., 2003) The NID1 stop mutation (c.1162C>T, p.(Gln388*)) results in a loss of the entire G2 and G3 regions of NID1, including the β-propeller domain that directly interacts with LAMC1 (Fig. 2a). The LAMC1 mutation (c.2237C>T, p.(Thr746Met)) occurs in the epidermal growth factor-like (EGF-like)-6 domain (Fig. 2b, 2d), a highly rigid structure with 4 disulfide linkages compared to 3 disulfide linkages of a typical EGF domain. This mutation maps to a four-residue beta sheet, one of only 3 short stretches in this domain with secondary structure. Normally the buried threonine-746 (M.W. 117 Da) is constrained by rigid disulfide linkages on either side, and mutation to a 25% larger residue such as methionine (M.W. 149 Da) is predicted to significantly decrease the stability of the protein (△△G of 7.7 kcal/mol for Thr→Met), primarily due to steric clashes with neighboring residues (Fig. 2e) (Schymkowitz, et al., 2005). While there are no reports of EGF-like-6 domain directly interacting with NID1, unraveling of this domain due to a highly destabilizing mutation could indirectly lead to loss of the well-documented binding of neighboring EGF-like-7-8-9 stretch to the NID1 β-propeller domain.(Mayer, et al., 1998; Mayer, et al., 1993; Takagi, et al., 2003) Interestingly, of the eleven EGF-like domains in LAMC1, the EGF-like-6 domain aligns best to the NID1 binding EGF-like-8 domain (Fig 2f, see Supp. Methods).
While a genetic etiology for the Dandy-Walker spectrum of disorders has long been appreciated, to date, causative mutations have been identified only in a few cases. Loss of Foxc1 and Zic cause ECM abnormalities(Grinberg, et al., 2004; Inoue, et al., 2008),(Zarbalis, et al., 2007) and analysis of these Dandy-Walker mouse models has lead to the hypothesis that disruptions of mesenchymal development adjacent to the developing cerebellum underlie the developmental pathogenesis of Dandy-Walker spectrum phenotypes(Aldinger, et al., 2009; Blank, et al., 2011). This mesenchyme directly gives rise to the meninges and skull, directly accounting for structural abnormalities of the posterior fossa. Additionally, since signaling from the mesenchyme is essential for normal cerebellar development, the hindbrain itself is secondarily affected (Aldinger, et al., 2009). Notably, in mice, both Nid1 (http://www.informatics.jax.org/image/MGI:4534594) and Lamc1 (various figures in http://www.informatics.jax.org/searches/expression.cgi?60953) are highly expressed in the mesenchyme and the ECM surrounding the developing cerebellum in the posterior fossa. Our current finding of Nid1 and Lamc1 mutations in ADDWOC patients indicates that the ECM is required for the normal structural integrity of the developing posterior fossa. Interestingly, mutations in the COL18A1 gene (encoding type XVIII collagen), another member of the Nid1 interactome cause Knobloch syndrome, which is characterized by occipital cephaloceles (Mahajan, et al., 2010),(Sertie, et al., 2000), and type XVIII collagen and Nid1 interact physically to support the BM even in species as distant from human as Caenorhabditis elegans.(Ackley, et al., 2003) Mice harboring missense or stop mutations in Nid1 and Lamc1 have not been described, but mice with deletions in these genes do display BM abnormalities and neurological deficits. Homozygous deletion of Nid1 leads to BM defects in brain capillaries, and behavioral abnormalities(Dong, et al., 2002). Mice null for Lamc1 in Shwann cells display disrupted basal lamina and peripheral nerve function,(Chen and Strickland, 2003) while mice with specific homozygous deletion of the Lamc1 binding site of Nid1 die in utero with multiple BM defects.(Willem, et al., 2002) It remains to be determined if the hindbrain defects our ADDWOC patients are a direct result of loss of normal NID1 and LAMC1 function within the developing hindbrain or if their loss in the ECM surrounding the developing brain is sufficient to impair normal mesenchymal to brain signaling.
In addition to furthering our understanding of the genetics of Dandy-Walker spectrum disorders, our study demonstrates several important aspects of combining next-generation sequencing technologies, proteomics, and tertiary structural modeling. First, we show that implementation of whole-exome capture and sequencing allowed us to rapidly identify a mutation in a family where genome-wide linkage analysis failed. Second, proteomic network analysis followed by whole-exome capture and massively parallel sequencing allowed us to rapidly identify a disease causing variant in a second pedigree too small to perform linkage analysis. Third, combining the known high-resolution atomic structure of the complex of the two interacting proteins with structural bioinformatics tools allowed us to model the pathogenic interaction of the two mutations. It seems likely that as more mutations are identified by next-generation sequencing technologies and as more tertiary structures are deposited in public repositories, genotype-structural solutions will become increasingly common. In many cases, this improved facility will greatly enhance our ability to design novel pharmaceuticals.
Supplementary Material
Acknowledgments
Author contribution statement: BWD performed most of the genotype bioinformatics analysis with contributions from EC, JC, and AS. VM, LG, and JMS performed the structural and network modeling. SW, XB, KM, WBD, JK, AJ, JRM, JM, SS, PN, NG, PF, NN, MO, HDH, LTV, SL, EMV, LB, KAA, and AGB provided genotype-phenotype correlations. AGB, BWD, VM, and LG wrote the paper with contributions from all authors. AGB oversaw all aspects of the work. This work was supported by JSPS KAKENHI Grant Number 24249092 (to SS), European Research Council (ERC Starting Grant 260888 to EMV), Italian Ministry of Health (Ricerca Corrente 2013), and NIH 1R01 NS064159-01A1 (AGB).
Grant Sponsor: This work was supported by JSPS KAKENHI Grant Number 24249092 (to SS), European Research Council (ERC Starting Grant 260888 to EMV), Italian Ministry of Health (Ricerca Corrente 2013), and NIH 1R01 NS064159-01A1 (AGB).
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- Ackley BD, Kang SH, Crew JR, Suh C, Jin Y, Kramer JM. The basement membrane components nidogen and type XVIII collagen regulate organization of neuromuscular junctions in Caenorhabditis elegans. J Neurosci. 2003;23(9):3577–87. doi: 10.1523/JNEUROSCI.23-09-03577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldinger KA, Lehmann OJ, Hudgins L, Chizhikov VV, Bassuk AG, Ades LC, Krantz ID, Dobyns WB, Millen KJ. FOXC1 is required for normal cerebellar development and is a major contributor to chromosome 6p25.3 Dandy-Walker malformation. Nat Genet. 2009;41(9):1037–42. doi: 10.1038/ng.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asan, Xu Y, Jiang H, Tyler-Smith C, Xue Y, Jiang T, Wang J, Wu M, Liu X, Tian G. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol. 2011;12(9):R95. doi: 10.1186/gb-2011-12-9-r95. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovich AJ, Millen KJ, Dobyns WB. A developmental and genetic classification for midbrain-hindbrain malformations. Brain. 2009;132(Pt 12):3199–230. doi: 10.1093/brain/awp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuk AG, McLone D, Bowman R, Kessler JA. Autosomal dominant occipital cephalocele. Neurology. 2004;62(10):1888–90. doi: 10.1212/01.wnl.0000125255.90915.5c. [DOI] [PubMed] [Google Scholar]
- Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5(15165179):433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- Blank MC, Grinberg I, Aryee E, Laliberte C, Chizhikov VV, Henkelman RM, Millen KJ. Multiple developmental programs are altered by loss of Zic1 and Zic4 to cause Dandy-Walker malformation cerebellar pathogenesis. Development. 2011;138(6):1207–16. doi: 10.1242/dev.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZL, Strickland S. Laminin gamma1 is critical for Schwann cell differentiation, axon myelination, and regeneration in the peripheral nerve. J Cell Biol. 2003;163(4):889–99. doi: 10.1083/jcb.200307068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MJ, Chen R, Lam HYK, Karczewski KJ, Chen R, Euskirchen G, Butte AJ, Snyder M. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29(21947028):908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Chen Y, Lewis M, Hsieh JC, Reing J, Chaillet JR, Howell CY, Melhem M, Inoue S, Kuszak JR. Neurologic defects and selective disruption of basement membranes in mice lacking entactin-1/nidogen-1. Lab Invest. 2002;82(12):1617–30. doi: 10.1097/01.lab.0000042240.52093.0f. others. [DOI] [PubMed] [Google Scholar]
- Ghonge NP, Kanika SS, Poonam B. Familial occipital cephalocele in a fetus at 21 weeks’ gestation: imaging demonstration across 3 generations. J Ultrasound Med. 2011;30(12):1747–51. doi: 10.7863/jum.2011.30.12.1747. [DOI] [PubMed] [Google Scholar]
- Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet. 2004;36(10):1053–5. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- Inoue T, Ogawa M, Mikoshiba K, Aruga J. Zic deficiency in the cortical marginal zone and meninges results in cortical lamination defects resembling those in type II lissencephaly. J Neurosci. 2008;28(18):4712–25. doi: 10.1523/JNEUROSCI.5735-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali A, Aldinger KA, Chary A, McLone DG, Bowman RM, Le LC, Jardine P, Newbury-Ecob R, Mallick A, Jafari N. Linkage to chromosome 2q36.1 in autosomal dominant Dandy-Walker malformation with occipital cephalocele and evidence for genetic heterogeneity. Hum Genet. 2008;123(3):237–45. doi: 10.1007/s00439-008-0467-y. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan VB, Olney AH, Garrett P, Chary A, Dragan E, Lerner G, Murray J, Bassuk AG. Collagen XVIII mutation in Knobloch syndrome with acute lymphoblastic leukemia. Am J Med Genet A. 2010;152A(11):2875–9. doi: 10.1002/ajmg.a.33621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Kohfeldt E, Timpl R. Structural and genetic analysis of laminin-nidogen interaction. Ann N Y Acad Sci. 1998;857:130–42. doi: 10.1111/j.1749-6632.1998.tb10113.x. [DOI] [PubMed] [Google Scholar]
- Mayer U, Nischt R, Poschl E, Mann K, Fukuda K, Gerl M, Yamada Y, Timpl R. A single EGF-like motif of laminin is responsible for high affinity nidogen binding. EMBO J. 1993;12(5):1879–85. doi: 10.1002/j.1460-2075.1993.tb05836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(20644199):1297–1303. doi: 10.1101/gr.107524.110. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parla JS, Iossifov I, Grabill I, Spector MS, Kramer M, McCombie WR. A comparative analysis of exome capture. Genome Biol. 2011;12(21958622) doi: 10.1186/gb-2011-12-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382–8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertie AL, Sossi V, Camargo AA, Zatz M, Brahe C, Passos-Bueno MR. Collagen XVIII, containing an endogenous inhibitor of angiogenesis and tumor growth, plays a critical role in the maintenance of retinal structure and in neural tube closure (Knobloch syndrome) Hum Mol Genet. 2000;9(13):2051–8. doi: 10.1093/hmg/9.13.2051. [DOI] [PubMed] [Google Scholar]
- Sulonen A-M, Ellonen P, Almusa H, Lepisto M, Eldfors S, Hannula S, Miettinen T, Tyynismaa H, Salo P, Heckman C. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12(21955854) doi: 10.1186/gb-2011-12-9-r94. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J, Yang Y, Liu JH, Wang JH, Springer TA. Complex between nidogen and laminin fragments reveals a paradigmatic beta-propeller interface. Nature. 2003;424(6951):969–74. doi: 10.1038/nature01873. [DOI] [PubMed] [Google Scholar]
- Willem M, Miosge N, Halfter W, Smyth N, Jannetti I, Burghart E, Timpl R, Mayer U. Specific ablation of the nidogen-binding site in the laminin gamma1 chain interferes with kidney and lung development. Development. 2002;129(11):2711–22. doi: 10.1242/dev.129.11.2711. [DOI] [PubMed] [Google Scholar]
- Zarbalis K, Siegenthaler JA, Choe Y, May SR, Peterson AS, Pleasure SJ. Cortical dysplasia and skull defects in mice with a Foxc1 allele reveal the role of meningeal differentiation in regulating cortical development. Proc Natl Acad Sci U S A. 2007;104(35):14002–7. doi: 10.1073/pnas.0702618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.