Abstract
During the last decade, microsatellites (short tandem repeats or STRs) have been successfully used for animal genetic identification, traceability and paternity, although in recent year single nucleotide polymorphisms (SNPs) have been increasingly used for this purpose. An efficient SNP identification system requires a marker set with enough power to identify individuals and their parents. Genetic diagnostics generally include the analysis of related animals. In this work, the degree of information provided by SNPs for a consanguineous herd of cattle was compared with that provided by STRs. Thirty-six closely related Angus cattle were genotyped for 18 STRs and 116 SNPs. Cumulative SNPs exclusion power values (Q) for paternity and sample matching probability (MP) yielded values greater than 0.9998 and 4.32E−42, respectively. Generally 2–3 SNPs per STR were needed to obtain an equivalent Q value. The MP showed that 24 SNPs were equivalent to the ISAG (International Society for Animal Genetics) minimal recommended set of 12 STRs (MP ∼ 10−11). These results provide valuable genetic data that support the consensus SNP panel for bovine genetic identification developed by the Parentage Recording Working Group of ICAR (International Committee for Animal Recording).
Keywords: microsatellite, single nucleotide polymorphism, exclusion probability, genetic identification, bovine
Introduction
DNA markers are becoming increasingly important in animal breeding and have been successfully used in bovine identification, in parentage testing and to establish relationships between two or more individuals (Glowatzki-Mullis et al., 1995; Heyen et al., 1997; Williams et al., 1997; Heaton et al., 2002). These markers have also been used to trace meat through the entire food chain (Arana et al., 2002) because of the reliable and accurate traceability they provide based on matching genetic marker profiles (Dalvit et al., 2007); the use of such markers has the potential to improve the rate of genetic progress (Van Eenennaam et al., 2007).
Microsatellites or short tandem repeats (STRs) have been the genetic markers of choice for more than two decades. Despite being highly polymorphic, informative and interspersed throughout the entire genome (Baumung et al., 2004; Tian et al., 2007), the results obtained with STRs by different laboratories are not always comparable because of inconsistencies in allele size calling and errors in size determination. Furthermore, STRs are time consuming for trained personnel to analyze, even with the use of appropriate software or other automated methods for allele analysis (Vignal et al., 2002). Recent advances in high-throughput DNA sequencing, computer software and bioinformatics have made the use of SNPs more popular (Heaton et al., 2002). Although in terms of genetic information a biallelic marker may be considered as a step backwards, SNPs have some promising advantages, including greater abundance (Heaton et al., 2005), genetic stability in mammals (Markovtsova et al., 2000; Nielsen, 2000; Thomson et al., 2000), simpler nomenclature and suitability to automated analysis and data interpretation (Wang et al., 1998; Lindblad-Toh et al., 2000). Furthermore, SNPs have been successfully used in the discovery of quantitative trait loci (QTL) and the association of genes with specific productive traits (Chen and Abecasis 2007; Wollstein et al., 2007) and in the identification of individuals and breeds (Negrini et al., 2008).
A prerequisite for the development of efficient SNP-based identification systems is the description of a minimal set with sufficient power to uniquely identify individuals and their parents in a variety of popular breeds and cross-bred populations (Heaton et al., 2002), even though the information content in a SNP set may vary significantly between populations (Krawczak, 1999). Previous studies designed strategies to sample the entire genetic diversity in beef cattle or purebred populations and simulated populations of purebred gene frequencies have been used to estimate the resolution and sensitivity of these methods in identifying individuals and in parental analysis (Table S7) (Heaton et al., 2002; Werner et al., 2004; López Herráez et al., 2005; Van Eenennaam et al., 2007; Baruch and Weller, 2008; Karniol et al., 2009; Allen et al., 2010; Hara et al., 2010a,b).
Most of the routine work done in livestock genetic laboratories includes the analysis of closely related animals (herdbook registry, half-sibs, etc.). Since high consanguinity is common in commercial ranches, additional markers are required to maintain the accuracy of the analysis (Pollak, 2005). In dealing with this problem, Anderson and Garza (2005) calculated the discriminatory power of SNPs in large scale parentage studies by considering the occurrence of related individuals among the members of putative mother-father-offspring trios. More recently, Fisher et al. (2009) used simulated and empirical data to evaluate the effectiveness of SNPs and STRs for parentage matching based on different degrees of relatedness.
Recently, the Parentage Recording Working Group of the ICAR (International Committee for Animal Recording) developed a cattle consensus panel of 99 SNPs, and a final ring test to certify laboratories around the world is underway. Considering this scenario, and the fact that there is considerably more experience in the use of microsatellites than SNPs (in terms of laboratory and statistical methods for analysis), the aim of this work was to compare the amount of information provided by microsatellites and SNPs within a consanguineous Angus herd.
Materials and Methods
Sample and DNA extraction
The study was done using 36 consanguineous Angus calves from a herd in Buenos Aires Province. This herd belongs to a typical commercial farm that produces, selects and sells bulls to breeding farms. The samples analyzed included half-sibs from six bulls that shared a grandfather and were obtained from the nucleus herd (consanguinity ∼0.2). Figure S1 provides a schematic diagram of the breeding system used. DNA was extracted from blood using NucleoSpin Blood purification kits (Macherey-Nagel, Düren, Germany), according to the manufacturer’s instructions.
Genotyping
DNA genotyping was done with microsatellites and SNPs. The microsatellite markers used were BM1818, BM1824, BM2113, BRR, CSRM60, CSSM66, ETH3, ETH10, ETH225, HAUT27, HEL1, INRA023, RM067, SPS115, TGLA53, TGLA122, TGLA126, and TGLA227. These 18 STRs belong to the standard FAO panel (Van de Goor et al., 2009) and/or to the standardized recommended list of the International Society for Animal Genetics (ISAG). A self-developed kit was used for PCR and the fragments were identified in an automatic MegaBACE 1000 DNA sequencer (GE Healthcare, USA). Allele sizes were standardized to the ISAG nomenclature. For SNP genotyping, 116 parentage SNPs from the Illumina BovineHD BeadChip were used (the list of SNPs is detailed in the Supplementary Material). This set comprised all SNPs included in the consensus panel for cattle identification developed by the Parentage Recording Working Group of ICAR (International Committee for Animal Recording). Genotypes with auto-calling < 85% were excluded from the analyses despite the fact that they were highly curated; 30 duplicates were included in the chip used. SNP genotyping was done using the genotyping services of GeneSeek Inc. (Lincoln, NE, USA).
Statistical analysis
Allele frequencies were determined by direct counting. ARLEQUIN 3.5 software (Schneider et al., 2000) was used to estimate the levels of genetic variability through allelic diversity (na; total number of alleles, average number of alleles and number of alleles per locus) and the unbiased expected (he) and observed heterozygosity (ho) for each locus and all loci. Hardy-Weinberg equilibrium (HWE) was estimated by FIS using the exact test implemented in GENEPOP 4 (Rousset and Raymond, 1997; Rousset, 2007). The FIS index was also used to estimate the degree of molecular consanguinity instead of pedigree consanguinity or kinship because the entire matrilineage was unavailable.
The match probability (MP) and exclusion power (Q) were estimated for cases involving two known parents, one known parent, missing parents and individual identification based on one (Q1) and two (Q2) marker exclusion criteria. These parameters were calculated for each marker and for the whole set as described by Weir (1996), using algorithms programmed with Visual Basic and implemented in Excel software (available upon request from the corresponding author).
Results
Thirty-six related animals were studied for 18 STRs and 116 SNPs. The animals belonged to a farm that uses artificial insemination (AI) and a natural multi-sire mating system. The exclusion of data with an auto-calling < 85% resulted in 4144 genotypes (32 missing data), with an average of 35.72 successful genotype (range: 34–36) per locus. All of the SNPs analyzed were polymorphic (na = 2) while an STR na of 5.22 ±1.35 (mean ±SD; range: 3–8) (Table 1). The minimum allele frequency (MAF) for SNPs was > 0.05 in 114 of the 116 SNP markers, the exceptions being the SNPs ARS-USMARC-Parent-EF034087-no-rs and ARS-USMARC-Parent-AY842472-rs29001941. The SNP he values ranged from 0.028 to 0.507, with an average value of 0.417 (Table 1). For STRs, the he values ranged from 0.255 to 0.816, with an average of 0.640 (Tables S1 and S2). In total, 133 HWE tests were done (115 for SNPs and 18 for STRs), nine of which (five for SNPs and four for STRs) showed significant deviations (p < 0.05) from theoretical proportions (Tables S1 and S2). The allele frequencies for SNPs and STRs are available from the corresponding author upon request.
Table 1.
Average number of alleles (na), unbiased expected (he), standard deviation of na and he, range of na and he among loci and FIS estimated for the SNP and STR sets of markers in Angus inbred cattle.
| Marker type | na (range) | He (range) | FIS p value |
|---|---|---|---|
| SNP | 2 ± 0* (2) | 0.417 ± 0.0098 (0.028–0.507) | < 0.001 |
| STR | 5.22 ± 1.35 (3–8) | 0.640 ±0.015 (0.255–0.816) | < 0.001 |
Mean ± SD.
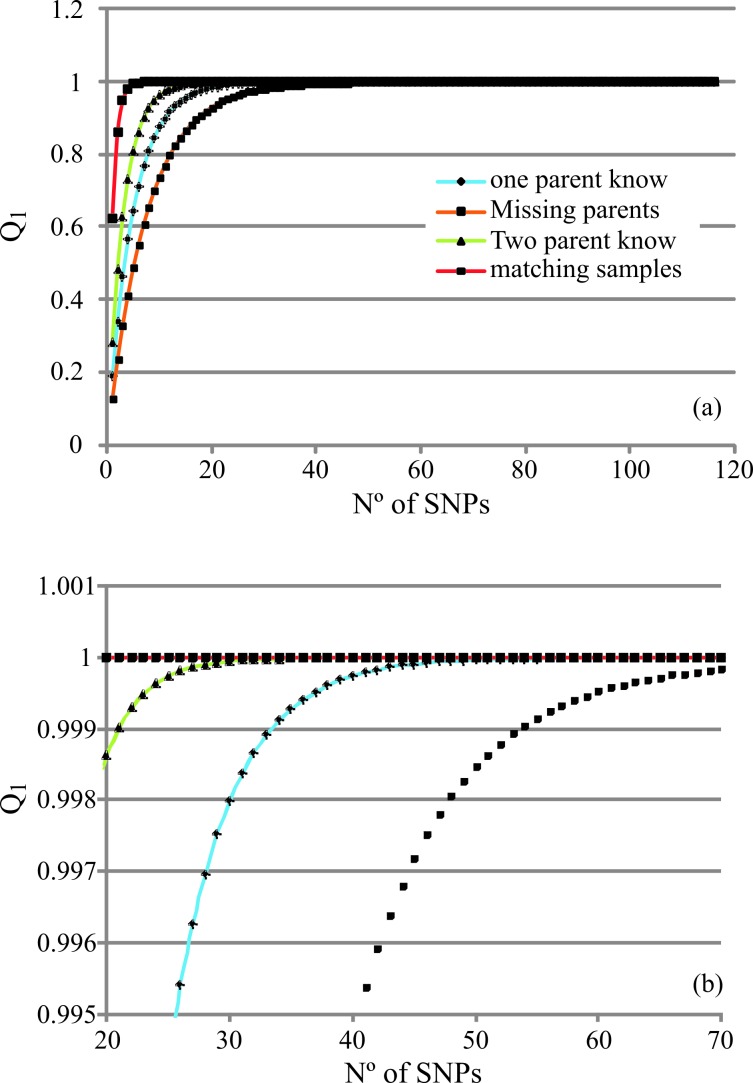
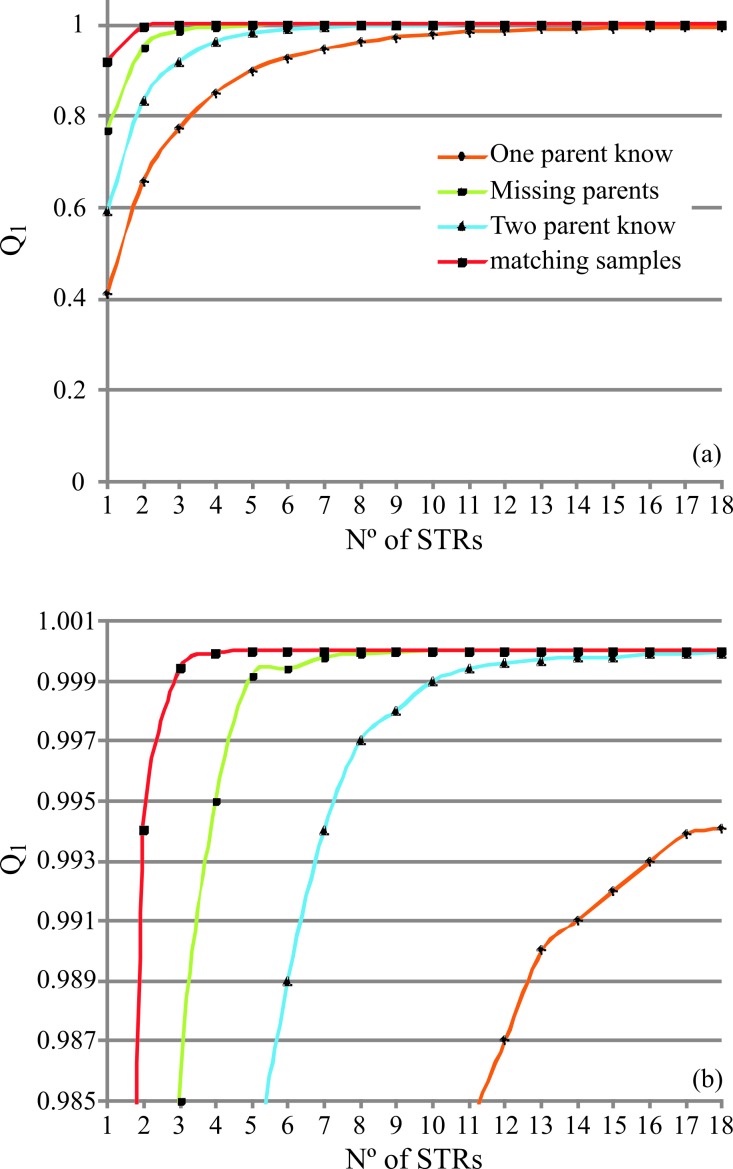
Q was estimated for each SNP marker for the most common cases of genetic identification (two known parents, one known parent, missing parents and matching samples), while MP was calculated only for matching samples (Tables S3 and S4). As shown in Figure S2, the distribution of the number of SNPs based on their individual Q values yielded a logarithmic curve. In the case of matching samples, more than 50% of the SNPs had Q values > 0.60. When the genotypes of both parents were known, more than 50% of the SNPs had a Q value ≥ 0.17, while in the worst scenario (one known parent) this value was ≥ 0.10. In addition, Q was estimated for each whole set of markers by considering one and two mismatch criteria. The corresponding Q1 and Q2 values were > 0.999991 and > 0.9998 for SNPs and > 0.994 and > 0.957 for STRs, respectively; the MP values were 2.45E−42 and 3.0E−12 for SNPs and STRs, respectively (Table 2). Figures 1 and 2 and Tables S5 and S6 show the cumulative Q1, Q2 and MP values for all of the cases studied. These results show that it is necessary to analyze between eight (matching samples scenario) and 55 (one known parent) SNPs to achieve a Q1 ≥ 0.999 [or cumulative non-exclusion power (1 - Q) = 1.0E−4]. On the other hand, for STRs, three and more than 18 markers, respectively, are necessary. When using the Q2 ≥ 0.999 criterion, 10 (matching samples) and 79 (one known parent) SNPs are needed, whereas for STRs five and > 18, respectively, are required. Finally, in the population studied here, 24 SNPs or 11 STRs were necessary to obtain an MP ≥ 10−11.
Table 2.
Non-exclusion power (1 - Q) estimated for the whole set of SNPs and STRs considering one (Q1) and two (Q2) mismatch criteria for the cases of two known parents, one known parent, missing parents and matching samples. MP - match probability calculated for matching samples.
| Locus type | N | Both parents | One parent | Missing parent | Matching samples | MP | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| 1 - Q1 | 1 - Q2 | 1 - Q1 | 1 - Q2 | 1 - Q1 | 1 - Q2 | 1 - Q1 | 1 - Q2 | |||
| SNPs | 116 | 1.4E-09 | 3.2E-08 | 1.6E-05 | 2.1E-04 | 4.0E-15 | 1.6E-13 | < 4.1E-15 | < 4.1E-15 | 2.4E-42 |
| STRs | 18 | 6.0E-05 | 9.0E-04 | 5.9E-03 | 4.2E-02 | 1.0E-08 | 3.0E-06 | 3.0E-14 | 3.0E-12 | 2.6E-14 |
Figure 1.
Cumulative exclusion power (Q) calculated for SNPs considering (A) one (Q1) mismatch criterion and (B) two mismatch criteria (Q2) for cases of two known parents, one known parent, missing parents and matching samples. Markers are listed based on decreasing expected heterozygosity (he).
Figure 2.
Cumulative exclusion power (Q) calculated for STRs considering (A) one (Q1) mismatch criterion and (B) two mismatch criteria (Q2) for cases of two known parents, one known parent, missing parents and matching samples. Markers are listed based on decreasing expected heterozygosity (he).
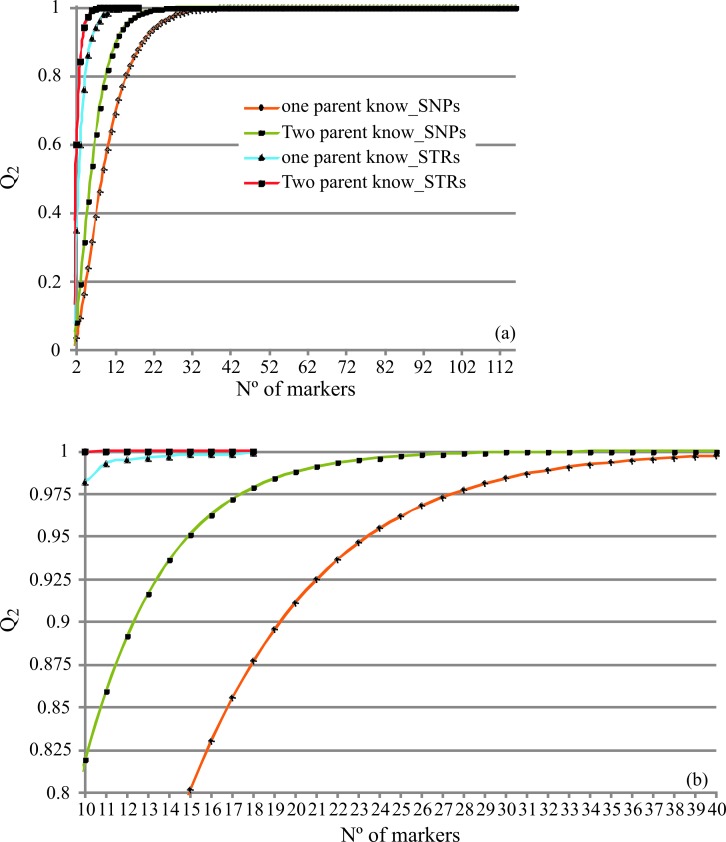
The minimum number of markers recommended by the ISAG for bovine genetic identification is 12 STRs. In our work, around 24 SNPs were necessary to achieve an MP (1.78E−11) equivalent to the standard marker set, and 31 SNPs (MP = 1.87E−14) were equivalent to the 18 STR set (Tables S5 and S6). For paternity testing, and when the two parents were known, 37 SNPs were needed for a Q value similar to the standard marker set. The resolution of more complex cases requires the use of additional markers. In these situations, such as one known parent or missing parents, around 39 and 49 SNPs are required, respectively, to obtain the same Q values as the 18 STRs (Figure 3).
Figure 3.
Comparison of the cumulative exclusion power (Q) curves calculated for SNPs and STRs considering two mismatch criteria (Q2) for cases of two known parents and matching samples. Markers are listed based on decreasing expected heterozygosity (he).
Discussion
Unrelated animal sampling has been successfully used to determine breed genetic profiles in phylogeographic studies and to estimate general theoretical Q and MP values for DNA identification (traceability, parentage analysis, etc.). Several studies have evaluated and compared the Q and/or MP values obtained for STR and SNP sets (Table S7). Most of them used only representative (unrelated) purebred samples to determine the entire genetic diversity. For example, Heaton et al. (2002) analyzed three composite bovine beef groups to identify SNPs useful for animal identification and paternity testing. Werner et al. (2004) selected unrelated bulls belonging to three dairy or dual-purpose pure breeds to identify SNPs and estimate their respective allelic frequencies. López Herráez et al. (2005) genotyped Galloway animals from different farms and used STRs and SNPs to compare the Q values in the identification of individuals and parental analysis. More recently, Karniol et al. (2009) evaluated the statistical power of the 25-plex assay in traceability (identity control) and parentage testing by genotyping unrelated animals from six cattle breeds.
These common approaches do not take into account population structure and consanguinity. Furthermore, most of the routine genotyping of livestock done in genetic laboratories consists of the analysis of highly related pedigree animals rather than unrelated animals from beef breeding or dairy farms. In this framework, a marker set should have enough exclusion power to resolve any possible situation, including cases of paternity with multi-putative consanguineous sires. In view of this scenario, and considering that there is generally much more experience in the use of STRs compared with SNPs, in this work we examined the amount of information obtained with SNP and STR markers for paternity testing and genetic identification within a consanguineous commercial Angus herd.
Almost all of the SNPs examined were polymorphic, with a mean MAF of 0.328, while more than 50% of the SNPs had a high Q value because both alleles had balanced gene frequencies. These findings were not unexpected given that SNPs from the Illumina BovineHD BeadChip were validated in Angus breeds and showed a high rate of polymorphic loci (573,437 out of 770,000). Comparison of the mean MAF values for the parentage subset of 116 SNPs showed that our inbred population gave a similar result in the Illumina test to that of the Red Angus (MAF = 0.327) and Angus (MAF = 0.346) samples used to validate the chips (ftp.illumina.com). These values ranked in the upper third distribution among 29 breeds (MAF = 0.135 to 0.395), as reported by the manufacturer. The average MAF of the parentage subset was greater than those reported for the entire SNP panel (0.13–0.27), perhaps because this subset had been carefully selected and highly curated for this purpose.
The comparison of the two types of markers showed that, in the case of matching samples, two SNPs were necessary to provide the same statistical power as one STR (five STRs and 10 SNPs for a Q2 ≥ 0.999). In the parentage analysis, 2.55 SNPs had a Q value equivalent to one STR when both parents were known and the two exclusion (Q2) criteria were used. In this case, 18 STRs and 46 SNPs were required to reach a Q2 ≥ 0.999. The SNP/STR ratios obtained here were similar to those reported by others using unrelated animals. For example, Werner et al. (2004) observed that 37 SNPs provided the same power as a typical, commonly used microsatellite set, whereas Weller et al. (2006) reported a ratio of 2–2.25 (25 SNPs were equivalent to 11 microsatellites with five alleles) using simulated data. More recently, Fisher et al. (2009), based on an analysis of simulated data and data from a test Jersey herd, indicated that 40 SNPs (with a mean MAF of 0.35, similar to that observed here) would be at least as effective for parentage matching as the 14 STR panel currently used for parentage testing in New Zealand dairy animals.
With regard to the MP, our results agreed with previously published data in that 25 SNPs were equivalent to 11–12 STRs (MP ∼10−11) (Table S7), sufficient to resolve simple cases of genetic identification. However, in routine work, more markers (17–18) are usually needed to resolve complicated cases such as parentage analysis with one known parent and multiple, closely related putative sires. As shown in Table S7, an MP value of 10−13 to 10−15 can be obtained by analyzing 17–18 STRs in a purebred breed, whereas 29–34 SNPs were required to reach an equivalent MP in our inbreeding Angus population. Interestingly, by using 12 and 18 STRs we achieved MP values of 10−11 and 10−14, similar to that obtained with 24 and 31 SNPs, respectively.
Recently, Baldo et al. (2010) showed that in beef traceability ∼25% more microsatellite markers were needed to identify consanguineous animals vs. unrelated animals. In contrast, our results show that, in this same context, the number of SNPs needed to provide the same Q in consanguineous samples and in the Illumina reference samples would be similar. The difference between these two studies can be explained by the fact that biallelic SNP markers are less affected by consanguinity than multiallelic STRs. In this sense, consanguinity affects the number of alleles first and then gene diversity, thereby easily purging rare STR alleles.
In conclusion, our results show that approximately twice as many SNP markers were needed to provide the same effectiveness as STRs for genetic identification and parentage analysis in a consanguineous Angus herd. This ratio is similar to previously reported values and provides evidence that biallelic SNPs are apparently less affected by consanguinity and population structure than STRs. International collaborations by the ISAG and ICAR have sought to select and validate SNPs that can be used in a standard panel for genetic identification in cattle. The results described here provide genetic information that supports the consensus SNP panel developed by the Parentage Recording Working Group of ICAR.
Acknowledgments
We thank Flores Chicas de Areco for help during the experiments described here and Dr. Patricia M. Mirol for reviewing the manuscript. M.E.F. is a Fellow of Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) and D.E.G. and A.R.M. are Fellows of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). This work was supported by ANPCyT, CONICET and Universidad Nacional de La Plata (UNLP).
Footnotes
Associate Editor: Alexandre Rodrigues Caetano
Supplementary Material
The following online material is available for this article:
- Figure S1 - Schematic diagram of the breeding system used.
- Figure S2 - Distribution of the SNP exclusion power.
- Table S1 - Alleles observed per SNP.
- Table S2 - Alleles observed per STR.
- Table S3 - Exclusion power (Q) estimated for each SNP.
- Table S4 - Exclusion power (Q) estimated for each STR.
- Table S5 - Cumulative non-exclusion power (1 - Q) calculated for SNPs.
- Table S6 - Cumulative non-exclusion power (1 - Q) calculated for STRs.
- Table S7 - Match probability values obtained in recent studies.
This material is available as part of the online article from http://www.scielo.br/gmb.
References
- Allen AR, Taylo M, McKeown B, Curry AI, Lavery JF, Mitchell A, Hartshorne D, Fries R, Skuce RA. Compilation of a panel of informative single nucleotide polymorphisms for bovine identification in the Northern Irish cattle population. BMC Genet. 2010;11:e5. doi: 10.1186/1471-2156-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EC, Garza JC. The power of single nucleotide polymorphisms for large-scale parentage analysis. Genetics. 2005;172:2567–2582. doi: 10.1534/genetics.105.048074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana A, Soret B, Lasa I, Alfonso L. Meat traceability using DNA markers: Application to the beef industry. Meat Sci. 2002;61:367–373. doi: 10.1016/s0309-1740(01)00206-6. [DOI] [PubMed] [Google Scholar]
- Baldo A, Rogberg-Muñoz A, Prando A, Mello Cesar AS, Lirón JP, Sorarrain N, Ramelli P, Posik DM, Pofcher E, Ripoli MV, et al. Effect of consanguinity on Argentinean Angus beef DNA traceability. Meat Sci. 2010;85:671–675. doi: 10.1016/j.meatsci.2010.03.023. [DOI] [PubMed] [Google Scholar]
- Baruch E, Weller JI. Estimation of the number of SNP genetic markers required for parentage verification. Anim Genet. 2008;39:474–479. doi: 10.1111/j.1365-2052.2008.01754.x. [DOI] [PubMed] [Google Scholar]
- Baumung R, Simianer H, Hoffmann I. Genetic diversity studies in farm animals - A survey. J Anim Breed Genet. 2004;121:361–373. [Google Scholar]
- Chen WM, Abecasis GR. Family-based association tests for genome-wide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvit C, De Marchi M, Cassandro M. Genetic traceability of livestock products: A review. Meat Sci. 2007;77:437–449. doi: 10.1016/j.meatsci.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Fisher PJ, Malthus B, Walker MC, Corbett G, Spelman RJ. The number of single nucleotide polymorphisms and on-farm data required for whole-herd parentage testing in dairy cattle herds. J Dairy Sci. 2009;92:369–374. doi: 10.3168/jds.2008-1086. [DOI] [PubMed] [Google Scholar]
- Glowatzki-Mullis ML, Gaillard C, Wigger G, Fries R. Microsatellite-based parentage control in cattle. Anim Genet. 1995;26:7–12. doi: 10.1111/j.1365-2052.1995.tb02612.x. [DOI] [PubMed] [Google Scholar]
- Hara K, Kon Y, Sasazaki S, Mukai F, Mannen H. Development of novel SNP system for individual and pedigree control in a Japanese Black cattle population using whole-genome genotyping assay. J Anim Sci. 2010a;81:6–12. doi: 10.1111/j.1740-0929.2010.00766.x. [DOI] [PubMed] [Google Scholar]
- Hara K, Watabe H, Sasazaki S, Mukai F, Mannen H. Development of SNP markers for individual identification and parentage test in a Japanese black cattle population. J Anim Sci. 2010b;81:152–157. doi: 10.1111/j.1740-0929.2009.00720.x. [DOI] [PubMed] [Google Scholar]
- Heaton MP, Harhay GP, Bennett GL, Stone RT, Grosse WM, Casas E, Keele JW, Smith TP, Chitko-McKown CG, Laegreid WW. Selection and use of SNP markers for animal identification and paternity analysis in U.S. beef cattle. Mamm Genome. 2002;13:272–281. doi: 10.1007/s00335-001-2146-3. [DOI] [PubMed] [Google Scholar]
- Heaton MP, Keen JE, Clawson ML, Harhay GP, Bauer N, Schultz C, Green BT, Durso L, Chitko-McKown CG, Laegreid WW. Use of bovine single nucleotide polymorphism markers to verify sample tracking in beef processing. J Am Vet Med Assoc. 2005;226:1311–1314. doi: 10.2460/javma.2005.226.1311. [DOI] [PubMed] [Google Scholar]
- Heyen DW, Beever JE, Da Y, Evert RE, Green C, Bates SR, Ziegle JS, Lewin HA. Exclusion probabilities of 22 bovine microsatellite markers in fluorescent multiplexes for semiautomated parentage testing. Anim Genet. 1997;28:21–27. doi: 10.1111/j.1365-2052.1997.t01-1-00057.x. [DOI] [PubMed] [Google Scholar]
- Karniol B, Shirak A, Baruch E, Singrün C, Tal A, Cahana A, Kam M, Skalski Y, Brem G, Weller JI, et al. Development of a 25-plex SNP assay for traceability in cattle. Anim Genet. 2009;40:353–356. doi: 10.1111/j.1365-2052.2008.01846.x. [DOI] [PubMed] [Google Scholar]
- Krawczak M. Informativity assessment for biallelic single nucleotide polymorphisms. Electrophoresis. 1999;20:1676–1681. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1676::AID-ELPS1676>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Winchester E, Daly MJ, Wang DG, Hirschhorn JN, Laviolette JP, Ardlie K, Reich DE, Robinson E, Sklar P, et al. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat Genet. 2000;24:381–386. doi: 10.1038/74215. [DOI] [PubMed] [Google Scholar]
- López Herráez D, Schafer H, Mosner J, Fries HR, Wink M. Comparison of microsatellite and single nucleotide polymorphism markers for the genetic analysis of a Galloway cattle population. Z Naturforsch C. 2005;60:637–643. doi: 10.1515/znc-2005-7-821. [DOI] [PubMed] [Google Scholar]
- Markovtsova L, Marjoram P, Tavare S. The age of a unique event polymorphism. Genetics. 2000;156:401–409. doi: 10.1093/genetics/156.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini R, Nicoloso L, Crepaldi P, Milanesi E, Colli L, Chegdani F, Pariset L, Dunner S, Leveziel H, Williams JL, et al. Assessing for SNP markers for assigning individuals to cattle populations. Anim Genet. 2008;40:18–26. doi: 10.1111/j.1365-2052.2008.01800.x. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Estimation of population parameters and recombination rates from single nucleotide polymorphisms. Genetics. 2000;154:931–942. doi: 10.1093/genetics/154.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak EJ. Application and impact of new genetic technologies on beef cattle breeding: A ‘real world’ perspective. Aust J Exp Agric. 2005;45:739–748. [Google Scholar]
- Rousset F, Raymond M. Statistical analyses of population genetic data: Old tools, new concepts. Trends Ecol Evol. 1997;12:313–317. doi: 10.1016/S0169-5347(97)01104-X. [DOI] [PubMed] [Google Scholar]
- Rousset F. Inferences from spatial population genetics. In: Balding DJ, Bishop M, Cannings C, editors. Handbook of Statistical Genetics. 3rd edition. Wiley; Chichester: 2007. pp. 945–979. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin: A software for population genetics data analysis. 2000. Ver. 2.0. Genetics and Biometry Lab, Department of Anthropology, University of Geneva. [PMC free article] [PubMed]
- Thomson R, Pritchard JK, Shen P, Oefner PJ, Feldman MW. Recent common ancestry of human Y chromosomes: Evidence from DNA sequence data. Proc Natl Acad Sci USA. 2000;97:7360–7365. doi: 10.1073/pnas.97.13.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Sun D, Zhang Y. Establishment of paternity testing system using microsatellites markers in Chinese Holstein. J Genet Genomics. 2007;35:279–284. doi: 10.1016/S1673-8527(08)60040-5. [DOI] [PubMed] [Google Scholar]
- Van de Goor LH, Panneman H, van Haeringen WA. A proposal for standardization in forensic equine DNA typing: Allele nomenclature for 17 equine-specific STR loci. Anim Genet. 2009;41:122–127. doi: 10.1111/j.1365-2052.2009.01975.x. [DOI] [PubMed] [Google Scholar]
- Van Eenennaam AL, Weaber RL, Drake DJ, Penedo MC, Quaas RL, Garrick DJ, Pollak EJ. DNA-based paternity analysis and genetic evaluation in a large, commercial cattle ranch setting. J Anim Sci. 2007;85:3159–3169. doi: 10.2527/jas.2007-0284. [DOI] [PubMed] [Google Scholar]
- Vignal A, Milan D, San Cristobal M, Eggen A. A review on SNP and other molecular markers and their use in animal genetics. Genet Sel Evol. 2002;43:275–305. doi: 10.1186/1297-9686-34-3-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sinauer Associates; Sunderland: 1996. p. 437. [Google Scholar]
- Weller JI, Seroussi E, Ron M. Estimation of the number of genetic markers required for individual animal identification accounting for genotyping errors. Anim Genet. 2006;37:387–389. doi: 10.1111/j.1365-2052.2006.01455.x. [DOI] [PubMed] [Google Scholar]
- Werner FAO, Durstewitz G, Habermann FA, Thaller G, Krämer W, Kollers S, Buitkamp J, Georges M, Brem G, Mosner J, et al. Detection and characterization of SNPs useful for identity control and parentage testing in major European dairy breeds. Anim Genet. 2004;35:44–49. doi: 10.1046/j.1365-2052.2003.01071.x. [DOI] [PubMed] [Google Scholar]
- Williams JL, Usha AP, Urquhart BG, Kilroy M. Verification of the identity of bovine semen using DNA microsatellite markers. Vet Rec. 1997;140:446–449. doi: 10.1136/vr.140.17.446. [DOI] [PubMed] [Google Scholar]
- Wollstein A, Herrmann A, Wittig M, Nothnagel M, Franke A, Nürnberg P, Schreiber S, Krawczak M, Hampe J. Efficacy assessment of SNP sets for genome-wide disease association studies. Nucleic Acids Res. 2007;35:e113. doi: 10.1093/nar/gkm621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- International Society for Animal Genetics http://www.isag.org.uk (March 30, 2012).
- Illumina BovineHD BeadChip. http://www.illumina.com/Documents//products/datasheets/datasheet_bovineHD.pdf (March 30, 2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
- Figure S1 - Schematic diagram of the breeding system used.
- Figure S2 - Distribution of the SNP exclusion power.
- Table S1 - Alleles observed per SNP.
- Table S2 - Alleles observed per STR.
- Table S3 - Exclusion power (Q) estimated for each SNP.
- Table S4 - Exclusion power (Q) estimated for each STR.
- Table S5 - Cumulative non-exclusion power (1 - Q) calculated for SNPs.
- Table S6 - Cumulative non-exclusion power (1 - Q) calculated for STRs.
- Table S7 - Match probability values obtained in recent studies.