Abstract
Regulation of gene expression at the level of transcription controls many crucial biological processes. Transcription factors (TFs) play a great role in controlling cellular processes and MYB TF family is large and involved in controlling various processes like responses to biotic and abiotic stresses, development, differentiation, metabolism, defense etc. Here, we review MYB TFs with particular emphasis on their role in controlling different biological processes. This will provide valuable insights in understanding regulatory networks and associated functions to develop strategies for crop improvement.
Keywords: Abiotic stress, Biotic stress, Gene expression, MYB, Transcription factor
Introduction
Transcription factors (TFs) naturally act as master regulators of cellular processes, so they are expected to be excellent candidates for modifying complex traits in crop plants and TF-based technologies are likely to be prominent part of the next generation of successful biotechnology crops. Traditional breeding is limited by the amount of genetic diversity in the germplasm of a particular crop; transgenic technologies bypass genetic barriers and allow modification of regulatory pathways in one plant using TFs from another plant (Century et al. 2008). Genes for a variety of transcription factors that contain typical DNA binding motifs, such as bZIP, MYB, MYC, ERF/AP2 and Zinc fingers have been demonstrated to be signal inducible (Bray 1997; Shinozaki and Yamaguchi-Shinozaki 2000). These transcription factors function in further regulation of various functional genes under particular development and stress conditions. Identification of novel transcription factor genes and their role in regulating the expression of important genes will help in understanding signaling pathways leading to development of novel transgenic crops. Several transcription factor genes i.e. MYB, CBF/DREB1, HSF, TGA6, BOS1, bZIP, AP2/EREBP etc. are indicated for their role in plant development and stress tolerance (Meshi and Iwabuchi 1995; Xiong et al. 1997; Kizis et al. 2001; Narasuka et al. 2003; Mengiste et al. 2003).
MYB transcription factor
The MYB family of proteins is large, functionally diverse and represented in all eukaryotes. Most MYB proteins function as transcription factors with varying numbers of MYB domain repeats conferring their ability to bind DNA. They are widely distributed in plants and have been implicated in the ABA-response and also interact with other transcription factors. Members of this family function in a variety of plant-specific processes, as evidenced by their extensive functional characterization in Arabidopsis (Arabidopsis thaliana). The ‘classical’ MYB factors, which are related to c-Myb, seem to be involved in the control of the cell cycle in animals, plants and other higher eukaryotes.
The first MYB gene was identified as the ‘oncogene’ v-MYB from the avian myeloblastosis virus. This gene appears to have originated from a vertebrate gene and has three members- c-MYB, A-MYB and B-MYB. The first plant MYB gene identified was C1 from Zea mays (Paz-Ares et al. 1987). The MYB gene family is represented by only five 3R-MYB genes, compared with up to 190 R2R3-MYB genes in Arabidopsis (Stracke et al. 2001; Yanhui et al. 2006). Recently, Du et al. (2012a) performed a genome-wide survey of the R2R3-MYB gene family in maize and identified a putative full set of R2R3-MYBs in the maize genome, comprising a total of 157 typical R2R3-MYB encoding genes. The Populus genome contains five 3R-MYB genes and 192 R2R3-MYB genes (Wilkins et al. 2009). A total of 252 MYBs, including 244 R2R-MYB (2R-MYB) genes, six R1R2R3-MYB (3R-MYB) genes, and two R0R1R2R3-MYB (4R-MYB) genes were identified in soybean (Du et al. 2012b). Supriya et al. (2006) and Sharma et al. (2010) reported MYB TF in Brassica species. AtMYB2 gene has been reported to have role in drought tolerance. A list of some MYB proteins is given in Table 1.
Table 1.
Some MYB proteins and their function in plants
| Name of MYB protein | Species | Function | References |
|---|---|---|---|
| Role in plant development | |||
| AtMYB021/ AtMYB033/ AtMYB065/ | Arabidopsis thaliana | Stamen development/Anther development (tapetum), Filament length, GA– and JA–mediated | Mandaokar et al. 2006 |
| AtMYB037/ AtMYB038 | Arabidopsis thaliana | Axillary meristem regulation/Lateral organ formation (shoot branching, GA–mediated) | Lee et al. 2009 |
| AtMYB068/ AtMYB084 | Arabidopsis thaliana | Root elongation, Axillary meristem regulation/ Lateral organ formation | Feng et al. 2004 |
| OsMYB2P-1 | Oryza sativa | Root system architecture | Dai et al. 2012 |
| GmMYB-G20-1 | Glycine max | Flower color | Takahashi et al. 2013 |
| AtMYB115/ AtMYB118 | Arabidopsis thaliana | Embryogenesis | Wang et al. 2008 |
| Cell shape and petal morphogenesis | |||
| AmMYBMx | Antirrhinum majus | Cell shape | Noda et al. 1994 |
| PhMYB1 | Petunia hybrida | Petal development | Noda et al. 1994 |
| AtMYB016 | Arabidopsis thaliana | Petal development | Baumann et al. 2007 |
| Cellular proliferation and differentiation | |||
| AtMYB005 | Arabidopsis thaliana | Seed coat differentiation | Gonzalez et al. 2009 |
| AtMYB017 | Arabidopsis thaliana | Early inflorescence development and seed germination | Zhang et al. 2009 |
| AtMYB046/ AtMYB057 | Arabidopsis thaliana | Expression in siliques/ flower buds | Kranz et al. 1998 |
| GhMYB109 | Gossypium hirsutum | Fibre elongation | Suo et al. 2003 |
| Trichome development | |||
| AtMYB011 | Arabidopsis thaliana | Trichome formation | Oppenheimer et al. 1991 |
| AtMYB017/ AtMYB023 | Arabidopsis thaliana | Cell fate/Trichome initiation and branching, Root hair patterning | Kang et al. 2009 |
| AtMYB066 | Arabidopsis thaliana | Root hair development | Kranz et al. 1998 |
| Phenylpropanoid metabolism | |||
| AmMYB305/ AmMYB340 | Antirrhinum majus | Phenylpropanoid metabolism | Jackson et al. 1991 |
| AtMYB003/AtMYB007/ AtMYB032 | Arabidopsis thaliana | Phenylpropanoid metabolism | Dubos et al. 2008 |
| AtMYB011/AtMYB012/ AtMYB111 | Arabidopsis thaliana | Phenylpropanoid pathway/ Flavonol biosynthesis | Stracke et al. 2007 |
| AtMYB075/AtMYB090/ AtMYB113/ AtMYB114 | Arabidopsis thaliana | Phenylpropanoid pathway/Anthocyanin biosynthesis | Gonzalez et al. 2008 |
| EsMYB | E. sagittatum | Flavonoid biosynthesis | Huang et al. 2013 |
| PhMYB3 | Petunia hybrida | Anthocyanin synthesis | Solano et al. 1995 |
| PhMYBAn2 | Petunia hybrida | Anthocyanin synthesis | Quattrocchio et al. 1993 |
| ZmMYBC1 | Zea mays | Anthocyanin synthesis | Paz-Ares et al. 1987 |
| ZmMYBP1 | Zea mays | Anthocyanin synthesis | Grotewold et al. 1994; Du et al. 2012b |
| Hormone responses | |||
| AtMYB33/ AtMYB65 | Arabidopsis thaliana | Hormone response (GA signaling) | Kranz et al. 1998 |
| AtMYB101 | Arabidopsis thaliana | Hormone response (GA signaling) | Quaedvlieg et al. 1996 |
| HvMYBGa | Hordeum vulgare | Hormone response (GA signaling) | Gubler et al. 1995 |
| OsGAMYB | Oryza sativa | Hormone response (GA signaling) | Gubler et al. 1997 |
| Abiotic stress | |||
| AtMYB002 | Arabidopsis thaliana | Drought response | Urao et al. 1993 |
| AtMYB015 | Arabidopsis thaliana | Cold stress tolerance | Agarwal et al. 2006 |
| AtMYB030 | Arabidopsis thaliana | Abiotic stress response, SA–mediated pathway | Li et al. 2009 |
| AtMYB060/ AtMYB094 | Arabidopsis thaliana | Drought, ABA–mediated (stomatal closure) | Cominelli et al. 2005 |
| AtMYB070/ AtMYB073/ AtMYB077 | Arabidopsis thaliana | Abiotic stress response/ Drought, Light, Wounding | Jung et al. 2008 |
| AtMYB096 | Arabidopsis thaliana | Drought tolerance (ABA and JA–mediated) | Seo et al. 2009 |
| BcMYB1 | Boea crassifolia | Drought tolerance | Chen et al. 2005a |
| OsMYB55 | Oryza sativa | Heat stress tolerance | El-kereamy et al. 2012 |
| ScMYBAS1 | Saccharum officinarum | Drought and salt tolerance | Prabu and Theertha 2011, Prabu and Prasad 2012 |
| Biotic stress | |||
| AtMYB030 | Arabidopsis thaliana | Hypocotyl elongation, brassinosteroid pathway | Li et al. 2009; Segarra et al. 2009 |
| AtMYB44 | Arabidopsis thaliana | Plant defense response against aphid | Liu et al. 2010 |
| AtMYB060/ AtMYB094/ AtMYB096 | Arabidopsis thaliana | Biotic stress response | Cominelli et al. 2005; Seo and Park 2010 |
| Light response | |||
| AmMYB305 | Antirrhinum majus | UV light response | Jackson et al. 1991 |
| AtMYB004 | Arabidopsis thaliana | UV light response | Kranz et al. 1998 |
| PcMYB1 | Petroselinum crispum | Light response | Feldbrugge et al. 1997 |
| Nutrient deficiency | |||
| AtMYB28 | Arabidopsis thaliana | Sulfur-starvation response | Hirai et al. 2007 |
| AtMYB29 | Arabidopsis thaliana | Sulfur-starvation response | Hirai et al. 2007 |
| MYB as negative regulators | |||
| AtMYB60 | Arabidopsis thaliana | Inhibits anthocyanin biosynthesis in the lettuce plant. | Park et al. 2008 |
| MYB MIXTA | M. guttatus | Negative regulator of trichome development | Scoville et al. 2011 |
| ZmMYB31 | Zea mays | Inhibits sinapoylmalate and phenylpropanoid biosynthesis | Fornale et al. 2010 |
| Regulation of primary and secondary metabolism | |||
| AtMYB058/ AtMYB063 | Arabidopsis thaliana | Lignin biosynthesis (fibers and vessels) | Zhou et al. 2009 |
| AtMYB123 | Arabidopsis thaliana | Proanthocyanidins (PAs) biosynthesis | Lepiniec et al. 2006 |
| AtMYB028/AtMYB034/ AtMYB122 | Arabidopsis thaliana | Glucosinolate biosynthesis | Gigolashvili et al. 2007 |
| AtMYB052/AtMYB054/ AtMYB069 | Arabidopsis thaliana | Cell wall thickening (fibers) | Zhong et al. 2008 |
Structure of MYB
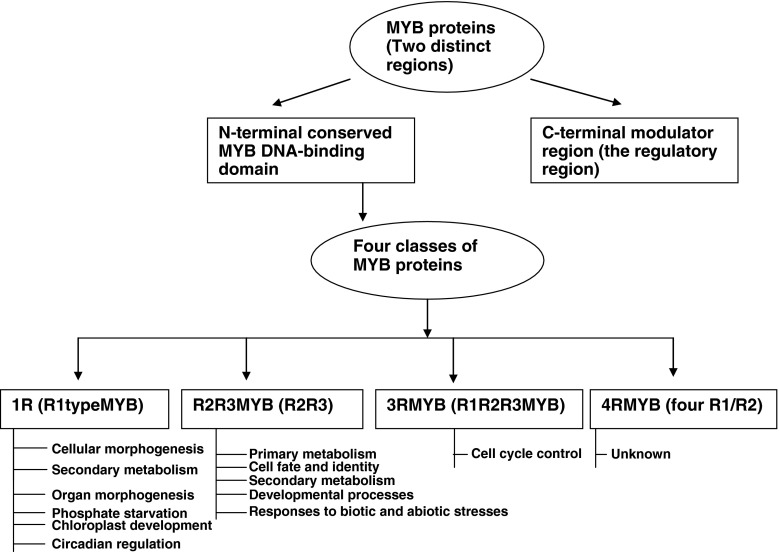
MYB factor represents a family of proteins that include a conserved domain, the MYB DNA-binding domain. In contrast to animals, plants contain mainly MYB protein subfamily which is characterized by the R2R3-type MYB domain. MYB proteins have two distinct regions, an N-terminal conserved MYB DNA-binding domain and a diverse C-terminal modulator region responsible for the regulatory activity of the protein. MYB domain generally consists of up to four imperfect amino acid sequence repeats (R) of about 52 amino acids, each forming three α–helices (Fig. 1). The second and third helices of each repeat build a helix-turn-helix (HTH) structure with three regularly spaced tryptophan (or hydrophobic) residues, forming a hydrophobic core in the 3D HTH structure (Ogata et al. 1996). In plants, the first tryptophan of R3 is substituted by phenylalanine or isoleucine.
Fig. 1.
Structure of MYB Protein and schematic illustration of different MYB protein classes and their functions depending on the number of adjacent MYB repeats (R)
Based on the number of MYB domain, the MYB family can be divided into four classes, 1R-, R2R3-, 3R- and 4R-MYB proteins (Dubos et al. 2010; Stracke et al. 2001). R2R3-MYB proteins are specific to plants and are also the most abundant type in plants, with more than 100 R2R3-MYB members in the genomes of dicots and monocots (Jiang et al. 2004a; Wilkins et al. 2009). The three repeats of the prototypic MYB protein c-Myb are referred to as R1, R2 and R3, and repeats from other MYB proteins are named according to their similarity to R1, R2or R3 of c-Myb. All four classes are found in plants, representing the taxon with the highest diversity of MYB proteins. The smallest class is the 4R-MYB group, whose members contain four R1/R2-like repeats. A single 4R-MYB protein is encoded in several plant genomes. The second class contains R1R2R3-type MYB (3R-MYB) proteins, typically encoded by five genes in higher plant genomes. In contrast to the highly conserved MYB domain, the other regions of R2R3-MYB proteins are highly variable. Based on the conservation of the DNA binding domain and of amino acid motifs in the C terminal domains, R2R3-MYB proteins have been divided into subgroups (Stracke et al. 2001). Therefore, the plant R2R3-MYB family is categorized into three major subdivisions on the basis of the sequence of the DNA binding domain: subgroup A whose members are most similar to c-MYB and other animal MYB proteins; subgroup B, which is a relatively small group (four members in Arabidopsis); and subgroup C, which encompasses 70 members in Arabidopsis.
Evolution of MYB
Evolutionary studies based on the sequences of MYB domains from several organisms indicate that plant MYB ancestors may have had three MYB repeats and that the first repeat was lost. A model for evolution of MYB proteins has been presented by Lipsick (1996). According to this model, R1R2R3MYBs were generated by successive intragenic duplications or triplications in the primitive eukaryotes, and these evolved into today’s two repeat (R2R3-MYB) and three repeat (R1R2R3-MYB) genes in plants and animals. It is considered that, upon loss of R1, several subgroups of genes encoding R2R3-MYB proteins were formed through selective amplification and subgroup expansion during plant evolution (Jin and Martin 1999). MYB genes were generated by successive gain of repeat units. Du et al. (2009) and Dubos et al. (2010) have reviewed the structure, characteristic, classification, multi-functionality, mechanism of combinational control, the “gain” model for evolution and function redundancy of MYB genes in detail.
The third heterogeneous class comprises proteins with a single or a partial MYB repeat, collectively designated “MYB-related” that fall into several subclasses (Rosinski and Atchley 1998). Most plant MYB genes encode proteins of the R2R3MYB class, which are thought to have evolved from an R1R2R3-MYB gene ancestor, by the loss of the sequences encoding the R1 repeat and subsequent expansion of the gene family (Rosinski and Atchley 1998). However, the evolution of 3R-MYB genes from R2R3-MYB genes by the gain of the sequences encoding the R1 repeat through an ancient intragenic duplication has also been proposed (Jiang et al. 2004b). Two R2R3-MYB genes, Arabidopsis AtMYB59 and AtMYB48, and their rice homologues (OsMYBAS1 and OsMYBAS2) undergo a similar pattern of alternative splicing, producing four differently spliced transcripts in Arabidopsis and three in rice. Thus, elucidation of the mechanism behind the alternative splicing of MYB genes will not only provide information on gene evolution in monocots and dicots, but also facilitate our understanding of the regulation of MYB transcription factor genes in development (Jigang et al. 2006).
Tissue-specific expression
Some MYB genes were expressed in most tissues and after most treatments (AtMYB1, AtMYB3, AtMYB38 and AtMYB44), while others were only expressed under very specific conditions. For example, AtMYB19 transcripts were detected only after infection with Pseudomonas syringae. Similar observations were made in terms of organ-specific expression patterns: AtMYB7, AtMYB44 and AtMYB73 were expressed in all plant organs studied while AtMYB46 was only detected in siliques and AtMYB21 only in flower buds (Shin et al. 2002). Several MYB-related genes expressed in anthers have been identified, namely AtMYB26 and AtMYB103 in Arabidopsis (Steiner-Lange et al. 2003; Higginson et al. 2003) and NtMYBAS1 in tobacco (Yang et al. 2001). In Arabidopsis, AtMYB103 gene expression was restricted to the tapetum of developing anthers and trichomes, down-regulation results in early tapetal degeneration and aberrant pollen. Similarly, AtMYB32 was expressed in most tissues, but strongly expressed in the anther tapetum, stigma papillae, and lateral root primordia.
In other plants evidence of tissue-specific regulation was also reported, HbMYB1 being expressed in leaves, bark, and latex of rubber trees, but its expression was significantly decreased in bark of TPD (tapping panel dryness) trees (Chen et al. 2003). AtMYB33 and AtMYB65 were co-expressed in many tissues and AtMYB101 expression was only restricted to subapical plant cells and the hypocotyls hook. In Arabidopsis, AtMYB102 was upregulated in root, leaf and young flowers and down regulated in stem when treated with ABA indicating that each R2R3-MYB gene had a unique expression pattern (Kranz et al. 1998).
In cotton, GhMYB109 was specifically expressed in cotton fiber initial cells as well as elongating fibers (Suo et al. 2003). GhMYB7 and GhMYB9 were expressed in flowers and fibers, and their expression in fibers is developmentally regulated (Chuan et al. 2005). Furthermore, some recent studies have suggested that GAMYB may be involved in floral initiation, stem elongation, anther development and seed development (Woodger et al. 2003), GAMYB was expressed at a high level in the floral meristem at the double-ridge stage and in stamen primordia of the grass L. temulentum. The two soybean R2R3-MYB genes, GmMYBJ6 (DQ902863) and GmMYBJ7 (DQ902864) were expressed only in leaf and stem indicating that it may be a common characteristic of the MYB TFs showing different expression patterns in higher plants.
Functions of MYB
The large size of the MYB family in plants indicates their importance in the control of plant specific processes. A tremendous amount of data is available on the roles of MYB transcription factors in plants (Du et al. 2009; Dubos et al. 2010). The functions of MYB proteins have been investigated in numerous plant species such as Arabidopsis, maize, cotton, rice (Oryza sativa), petunia (Petunia hybrida), snapdragon (Antirrhinum majus), grapevine (Vitis vinifera L.), poplar (Populus tremuloides) and apple (Malus domestica), using both genetic and molecular analyses. In the past decade, the R2R3-MYB genes have been extensively studied and members of the MYB family have been found to be involved in a variety of biological functions like phenylpropanoid metabolism (Grotewold et al. 1994; Hichri et al. 2011), biotic and abiotic stress (Segarra et al. 2009; Lippold et al. 2009), cell shape such as Am MIXTA (Noda et al. 1994), differentiation (Oppenheimer et al. 1991; Kang et al. 2009; Xie et al. 2010), hormone responses i.e. AtMYB2 (Urao et al. 1993), GAMYB and CpMYB (Gubler et al. 1995 ; Iturriaga et al. 1996), formation of B-type cyclin (Ito et al. 2001) or during plant defense reactions i.e. NtMYB1(Yang and Klessig 1996; Liu et al. 2008). Myb1R is involved in regulation of circadian clock (Schaffer et al. 1998, 2001) and telomeric DNA-binding protein (Yu et al. 2000).
Arabidopsis thaliana dedicates over 5 % of its genome to code for more than 1,500 transcription factors, about 45 % of which are from families specific to plants. The three largest families of transcription factors in Arabidopsis AP2/EREBP (Apetala/ethylene responsive element binding protein, MYB(R1) R2R3 and bHLH (basic helix-loop-helix) each represent only approximately 9 % of the total families of transcription factors and there are several other families with comparable numbers of genes (Riechmann et al. 2000). The R3-type, TRIPTYCHON (TRY), CAPRICE (CPC) and MYBL2, are likely to have evolved from R2R3-MYB genes and involved in the control of cellular morphogenesis (Pesch and Hulskamp 2009) and in secondary metabolism control (Dubos et al. 2008). Those MYB-related genes of the evolutionarily older R1/R2-type, including CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), encode core components of the central circadian oscillator (Lu et al. 2009).
The various functions of MYB are enlisted in Table 1 and discussed below-
Role in plant development
Several R2R3-MYB genes control anther development and/ or its function including AtMYB21, AtMYB24, AtMYB57, AtMYB108/BOS1, AtMYB35/TDF1, AtMYB80 and AtMYB99 (Cheng et al. 2009; Mandaokar and Browse 2009). AtMYB125/ DUO1 is a pollen-specific factor controlling male germ cell division and differentiation (Brownfield et al. 2009). AtMYB33 and AtMYB65 redundantly facilitate both anther and pollen development (Millar and Gubler 2005). (AtMYB105/LOF2 and AtMYB117/LOF1) control lateral organ separation and axillary meristem formation upstream of AtMYB37 (Lee et al. 2009). AtMYB115 and AtMYB118/PGA37 have been proposed to play roles in embryogenesis (Wang et al. 2008).
Cell shape and petal morphogenesis
Another role for plant MYB genes is in the control of cell shape where the MIXTA gene of Antirrhinum and the orthologous PhMYB1 gene from Petunia have been shown to be essential for developing the conical form of petal epidermal cells and the GL1 gene of Arabidopsis has been shown to be essential for the differentiation of hair cells (trichomes) in some parts of the leaf and stem (Noda et al. 1994; Oppenheimer et al. 1991). WEREWOLF is a MYB-related protein in Arabidopsis and it is a position dependent regulator of epidermal cell patterning (Lee and Schiefelbein 1999).
Baumann et al. (2007) reported that AmMYBML2, encodes an R2R3 MYB factor very closely related to MIXTA, is also expressed in flowers of A. majus. They analysed the roles of AmMYBML2 and two MIXTA-related genes, PhMYB1 from Petunia hybrida and AtMYB16 from Arabidopsis thaliana, in petal development. The structural similarity between these genes, their comparable expression patterns and the similarity of the phenotypes they induce when ectopically expressed in tobacco, suggest they share homologous functions closely related to, but distinct from, that of MIXTA. Detailed phenotypic analysis of a phMYB1 mutant confirmed the role of PhMYB1 in the control of cell morphogenesis in the petal epidermis.
Cellular proliferation and differentiation
c- MYB has been shown to activate transcription factor from the CDC2 kinase gene promoter in animal cells and control the G1-S phase transition. Thus, it has role in cellular proliferation. But no such role has been demonstrated for a plant MYB gene product although the CDC2 α gene from Arabidopsis has been shown to contain MYB recognition motifs within its promoter (Martin and Paz-Ares 1997). AtMYB5 regulated the outer seed coat differentiation (Gonzalez et al. 2009) while AtMYB66 controlled root hair patterning. AtMYB23 regulated trichome extension and branching in combination with AtMYB5 (Li et al. 2009). AtMYB16/MIXTA, is proposed to control the shape of petal epidermal cells (Baumann et al. 2007), and AtMYB17 is reported to be a putative regulator of early inflorescence development and seed germination (Zhang et al. 2009). AtMYB98 regulates synergid cell differentiation during female gametophyte development, pollen tube guidance and the formation of the filiform apparatus (Punwani et al. 2008). Similarly, the male germline-specific R2R3 MYB transcription factor DUO1 POLLEN1 (DUO1) is reported to have an essential role in sperm cell differentiation in Arabidopsis (Borg et al. 2011).
Role of MYBs in root development and differentiation has been reported by various workers (Feng et al. 2004; Shin et al. 2007; Mu et al. 2009). AtMYB59 regulated root development through the control of cell cycle progression at the root tips (Mu et al. 2009) and the expression of auxin-inducible genes was modulated by AtMYB77 regulating lateral root formation (Shin et al. 2007). Feng et al. (2004) found that AtMYB68 is expressed specifically in root pericycle cells and was involved in some steps in root development. They further pointed out that the closest MYB68 homolog MYB84, exhibited an overlapping expression pattern in pericycle cells, suggesting that their functions may be partly redundant. Dai et al. (2012) suggested that R2R3 MYB transcriptional factor, OsMYB2P-1 was associated with the regulation of root system architecture in rice. Takahashi et al. (2013) revealed that a MYB transcription factor gene GmMYB-G20-1 controlled flower color in soybean.
Trichome development
AtMYB0 and AtMYB23 control trichome initiation in shoots Arabidopsis thaliana. TRANSPARENT TESTA GLABRA2 (TTG2) encodes a WRKY transcription factor and is expressed in young leaves, trichomes, seed coats, and root hairless cells. An examination of several trichome and root hair mutants indicated that MYB and bHLH genes regulated TTG2 expression. Two MYB binding sites in the TTG2 5′ regulatory region act as cis regulatory elements and as direct targets of R2R3 MYB transcription factors such as WEREWOLF, GLABRA1, and TRANSPARENT TESTA2. Mutations in TTG2 cause phenotypic defects in trichome development and seed color pigmentation. Transgenic plants expressing a chimeric repressor version of the TTG2 protein (TTG2: SRDX) showed defects in trichome formation, anthocyanin accumulation, seed color pigmentation and differentiation of root hairless cells. GLABRA2 (GL2) expression was markedly reduced in roots of ProTTG2:TTG2:SRDX transgenic plants, suggesting that TTG2 is involved in the regulation of GL2 expression, although GL2 expression in the ttg2 mutant was similar to that in the wild type and this suggests a new step in a regulatory cascade of epidermal differentiation (Ishida et al. 2007).
GhMYB109, encoding a R2R3 MYB transcription factor of 234 amino acids, was found to be structurally related to AtMYBGL1 and AtWER controlling the trichome initiation in Arabidopsis thaliana. GhMYB109 is present as a unique-copy gene in cotton genome and RNA expression analysis showed that it was specifically expressed in cotton fibre initial cells as well as elongating fibres (Suo et al. 2003). Wang et al. (2010) revealed distinct relationships between GL2 and single MYBs in the regulation of trichome vs. root hair development which could provide new insights into the molecular mechanism of epidermal patterning.
Phenylpropanoid metabolism
MYB proteins are known to play an important role in the control of phenylpropanoid metabolism. The C1 protein activates transcription of genes encoding enzymes involved in the anthocyanin biosynthesis in the outer layer of cells (aleurone) of the maize seed endosperm (Paz-Ares et al. 1987; Cone et al. 1986). While C1 is active in aleurone, a very similar MYB protein, P1, is functional in controlling anthocyanin biosynthesis in the maize plant where it interacts with other members of the R-protein family to activate anthocyanin biosynthetic gene expression (Cone et al. 1993). In maize, another MYB protein, ZmMYB1 activated the structural genes required for anthocyanin biosynthesis while ZmMYB38 inhibited C1-mediated activation of the same promoter (Du et al. 2012a).
In other plant species, MYB proteins served similar roles in control of phenylpropanoid metabolism, for example, in Petunia flowers where the AN2 gene product is required for anthocyanin production, it has been shown to encode a MYB-related product (Quattrocchio et al. 1993). Gene encoding Chalcone synthase (CHS) was activated by another MYB protein from Petunia, PhMYB3, which was expressed specifically in petal epidermis where anthocyanin pigment was made (Solano et al. 1995). MYB protein also served to regulate other branches of phenylpropanoid metabolism. In Antirrhinum majus and tobacco, AmMYB305 (or its orthologue in tobacco) activated the gene encoding the first enzyme of phenylpropanoid metabolism, phenylalanine ammonia lyase (PAL) (Urao et al. 1993). Morita et al. (2006) reported that spatial and temporal expression of the structural genes encoding the enzymes for anthocyanin biosynthesis was determined by combinations of the R2R3-MYB, bHLH and WDR factors and their interaction. Lin-Wang et al. (2010) described the association of MYBA or MYB1 and MYB10 with regulation of the anthocyanin biosynthetic pathway in Rosaceae.
Maize expresses at least 82 genes encoding R2R3 Myb proteins and the five genes encoding R2R3 Myb-domain proteins have been identified in maize corresponding to P, C1, Pl, ZmI and ZM 38 (Paz-Ares et al. 1987; Cone et al. 1993). All of these genes products have been implicated directly or indirectly in the control of flavonoid biosynthesis. Rabinowicz et al. (1999) found that sequence analysis of maize R2R3 revealed a novel line of evidence for the amplification of the R2R3 MYB gene family in the early history of land plants. It suggested that maize provided a possible model system to examine the hypothesis that the expansion of MYB gene family was associated with the regulation of novel plant cellular functions. MYB340 (Moyano et al. 1996), MYB308 and MYB330 (Tamagnone et al. 1998) were reported to be other MYB genes involved in phenylpropanoid metabolism. The Arabidopsis TRANSPARENT TESTA (TT2) gene encoded an R2R3 MYB domain protein that acted as a key determinant for proanthocyanidin accumulation in developing seed (Nesi et al. 2001). Huang et al. (2013) isolated and characterized 13 full-length cDNA clones of R2R3-MYB TFs from E. sagittatum (EsMYB) and found to regulate the flavonoid biosynthetic pathway.
Yang et al. (2001) isolated two cDNA clones (NtMyb AS1 and NtMyb AS2) from tobacco encoding MYB-related proteins with strong sequence similarity to petunia PhMYB3 and pointed out that NtMYBAS1 was a functional anther-specific transcription factor, which is likely to be a positive regulator of PAL1 expression and phenylpropanoid synthesis in sporophytic tissues but not in gametophytic tissues of the anther. Similarly, GmMYB176 has been found to play key role in flavoniod biosynthesis in soybean (Yi et al. 2010). Moyano et al. (1996) reported that AtMYB21 and 57 were specifically expressed in flower buds, reminiscent of the expression pattern of AmMYB305 and 340 from A. majus. The inhibition of this branch of phenylpropanoid metabolism was found to be specific to AmMYB308 and AmMYB330, suggesting that they recognized their normal target genes in these transgenic plants. Experiments with yeast indicate that AmMYB308 can act as a very weak transcriptional activator and overexpression competitively inhibited the activity of stronger activators recognizing the same target motifs (Tamagnone et al. 1998).
Hormone responses
Plant MYB proteins have also been reported to have role in hormonal responses during seed development and germination. Some MYB genes are expressed in response to GA treatment in Petunia petals. Another plant hormone, Abscisic acid (ABA), induces expression of AtMYB2 in Arabidopsis, a MYB gene that is also induced in response to dehydration or salt stress (Shinozaki et al. 1992). Gocal et al. (2001) identified three Arabidopsis genes AtMYB33, AtMYB65 and AtMYB101 with GAMYB like activity. GAMYB is involved in transactivating the barley α- amylase promoter. These AtMYB genes may also play a role in the root tip and later in stem tissue during germination. They further observed increase in growth rate and erectness of petiole with accumulation of GAs in the petioles (GA3 by 11-fold and GA4 by 3-fold), and an increase in expression of AtMYB33 at the shoot apex. They indicated that GAMYB-like genes mediate GA signaling in growth and flowering responses.
HvGAMYB from barley and OsGAMYB from rice, which are required for the expression of the α-amylase in aleurone, are both regulated by GA signal (Gubler et al. 1995, 1997) and over-expression of HvGAMYB caused abnormal anther phenotype (Murray et al. 2003). It was first identified in barley aleurone cells and was shown to be upregulated and strongly expressed in barley anthers by gibberellin (GA) (Murray et al. 2003). They further found that with the increase in HvGAMYB levels, there was a progressive decrease in anther size, particularly a decrease in anther length. Anthers also became increasingly lighter in color. Anthers with fourfold or more HvGAMYB protein than non-transgenic controls failed to dehisce and were male-sterile, while anthers with approximately three to fourfold endogenous GAMYB protein levels were smaller and paler but still shed normally.
Chen et al. (2005b) isolated 23 MYB gene fragments and 6 nearly complete ORF encoding putative wheat MYB TFs (TaMYB1 to TaMYB6). Sequence analysis indicated that these putative wheat MYB TFs represent typical R2R3 MYBs. Expression analysis of the six TaMYB genes indicated that they were expressed in root, sheath and leaf, but at different abundance. Another study by Song et al. (2011) demonstrated that the R2R3-MYB transcription factors MYB21 and MYB24 function as direct targets of JAZs to regulate male fertility specifically. They speculated that JAZs interact with MYB21 and MYB24 to attenuate their transcriptional function; upon perception of JA signal, COI1 recruits JAZs to the SCFCOI1 complex for ubiquitination and degradation through the 26S proteasome. MYB21 and MYB24 are then released to activate expression of various genes essential for JA-regulated anther development and filament elongation.
Abiotic stress
AtMYB2, AtMYB74 and AtMYB102 were up-regulated by drought stress (Denekamp and Smeekens 2003; Abe et al. 2003; Urao et al. 1993). AtMYB2 was induced by dehydration and salt stress but not by cold and heat stress and thus AtMYB2 is responsive to dehydration at the transcriptional level. The putative protein (AtMyb2) encoded by AtMYB2 has 274 amino acids, a molecular mass of 32 kDa and a putative DNA binding domain that shows considerable homology to plant MYB-related proteins, such as maize C1. In addition to this, Zhang et al. (2011) reported that AtMYBL functions in the leaf senescence process, and thus modulates an abiotic stress response in Arabidopsis.
AtMYB60 and AtMYB96 acted through the ABA signaling cascade to regulate stomatal movement (Cominelli et al. 2005), and drought stress and disease resistance (Seo et al. 2009; Seo and Park 2010) respectively. The transcriptional activation of cuticular wax biosynthesis by MYB96 contributed to drought resistance in Arabidopsis thaliana (Seo et al. 2011).
Chen et al. (2005a) isolated a drought-inducible MYB gene, designated as BcMYB1, from drought tolerant Boea crassifolia which encoded a typical R2R3-MYB transcription factor belonging to the subgroup 11 of the R2R3-MYB protein family. It was strongly induced by drought stress and also responded to PEG, high salinity and low temperature to some extent. BcMYB1 might be involved in the regulation of gene expression in response to dehydration stress through an ABA-independent pathway, whereas it seemed not to be a regulatory component in wounding signaling. It shared high similarity with AtMYB102 from Arabidopsis and both genes responded to water stress. However, their expression patterns were quite different. AtMYB102 could be induced by exogenous ABA, while BcMYB1 was insensitive to ABA treatment. In addition, AtMYB102 expression was dependent on and integrated signals from both wounding and water stress while BcMYB1 could hardly be induced by wound signaling.
Seo and Park (2009) have recently reported that an Arabidopsis R2R3-type MYB transcription factor, MYB96, regulated lateral root meristem activation under drought conditions via an ABA-auxin signaling crosstalk. In this signaling scheme, the MYB96-mediated ABA signals were incorporated into an auxin signaling pathway that involved a subset of GH3 gene encoding auxin-conjugating enzymes. The MYB96-overexpressing, activation tagging mutant, which was featured by dwarfed growth and reduced lateral root formation, exhibited an enhanced drought resistance. The sugarcane (Saccharum officinarum) stress-related MYB transcription factor gene, ScMYBAS1-3 was also induced in response to water-deficit and salt stress. Prabu and Theertha (2011) elucidated its sequence-to-structure-to-function paradigm, the putative three-dimensional structure of ScMYBAS1. Prabu and Theertha (2011) further isolated and characterized the promoter (PScMYBAS1) which they found to be helpful in understanding the regulation of ScMYBAS1 expression and providing a new stress-inducible promoter system in transgenic plants.
The expression of Arabidopsis R2R3 AtMYB102 transcription factor gene was dependent on signals derived from both wounding and osmotic stress (Denekamp and Smeekens2003). AtMYB102/AtM4 and AtMYB41 (subgroup 11) contributed to plant resistance against insects and probably affected dehydration after wounding (De Vos et al. 2006) and osmotic stress responses (Lippold et al. 2009), respectively. Akagi et al. (2010) characterized DkMYB2, a Myb-like transcription factors (MYB-TF), which was placed in a subclade including a PA regulator of Arabidopsis (Arabidopsis thaliana), TRANSPARENT TESTA2 (TT2), and was co-induced with PA pathway genes after wound stress. AtMYB44/AtMYBR1 regulated ABA-mediated stomatal closure in response to abiotic stresses and three other members of this subgroup (AtMYB70, AtMYB73 and AtMYB77/AtMYBR2) are likely to be associated with stress responses (Jung et al. 2008). AtMYB13, AtMYB15, AtMYB33 and AtMYB101 (Reyes and Chua 2007) were involved in ABA-mediated responses to environmental signals. AtMYB15 was also involved in cold stress tolerance (Agarwal et al. 2006).
OsMYB3R-2 in rice transgenic plants enhanced tolerance to freezing, dehydration and salt stress and decreased sensitivity to ABA (Dai et al. 2007). Similarly, El-kereamy et al. (2012) studied the rice R2R3-MYB transcription factor OsMYB55 and concluded that overexpression of OsMYB55 improved rice plant tolerance to high temperature which was associated with enhanced amino acid metabolism through transcription activation. Liao et al. (2008) identified 156 GmMYB genes in soyabean (Glycine max) of which the expression of 43 genes changed on treatment with ABA, salt, drought and/or cold stress. Three MYB proteins have been reported to be involved in response to abiotic stress in rice. For instance, overexpression of OsMYB4 significantly enhanced tolerance to chilling and freezing stress in transgenic Arabidopsis (Vannini et al. 2004; Pasquali et al. 2008). Ma et al. (2009) reported that OsMYB3R-2 participated in cold signalling pathway by targeting the cell cycle and a putative DREB/CBF. Moreover, a recent study revealed that OsMYBS3 was essential for conferring tolerance to cold stress in rice plants (Su et al. 2010) while R2R3MYB gene, OsMYB2, was involved in salt, cold, and dehydration tolerance in rice (Yang et al. 2012).
Biotic stress
AtMYB30 encodes an activator of the hypersensitive cell death program in response to pathogen attack, acting through the regulation of very-long-chain fatty acids synthesis. In seedlings, AtMYB30 has also been shown to act in the brassinosteroid pathway controlling hypocotyls cell elongation (Li et al. 2009). AtMYB62 is reported to be induced in response to phosphate starvation (Devaiah et al. 2009) while AtMYB108 in both biotic and abiotic stress responses (Mengiste et al. 2003). The family of R2R3-MYB-like transcription factors has repeatedly been implicated in JA-dependent defense responses. For instance, the OsLTR1 gene from rice regulated JA-dependent defense whereas AtMYB15, AtMYB34, AtMYB51 and AtMYB75 were associated with the wound response or resistance against insect herbivores (Cheong et al. 2002; Johnson and Dowd 2004). The BOTRYTIS SUSCEPTIBLE 1(BOS1) gene encoded an R2R3 MYB transcription factor protein which was found to be involved in biotic as well as abiotic stress response. It interacted with jasmonate signaling pathway and mediated response to signals by reactive oxygen intermediates from biotic as well as abiotic stress (Mengiste et al. 2003).
AtMYB44 has been found to play role in the plant defense response against aphid (Liu et al. 2010). Similarly, AtMYB102 has been reported to be effective in defense against the insect herbivore Pieris rapae (De Vos et al. 2006). Raffaele et al. (2008) proposed that AtMYB30 modulated hypersensitive response via very-long-chain fatty acids (VLCFAs) by themselves or VLCFA derivatives and thus playing a role in programmed cell death. Segarra et al. (2009) and Van der Ent et al. (2008) demonstrated that the defence pathways triggered by beneficial Trichoderma, Rhizobacteria and Pseudomonas spp. strains were highly similar and root-specific transcription factor MYB72 functioned as an early node of convergence in the signalling pathways that were induced by these different beneficial microorganisms playing role in defence response in Arabidopsis. AtMYB96-mediated ABA signals enhanced pathogen resistance response by inducing salicylic acid biosynthesis and thus MYB96 acted as a molecular link in ABA-SA crosstalks (Seo and Park 2010).
Light response
MRECH (a MYB recognition element for chalcone synthase) is known to have a functional core that is essential for light responsiveness and is specifically recognized by two distantly related MYB-like proteins: MYB305 and the novel factor MYB1 from Petroselinum crispum. PcMYB1 was identified by both its specific binding to MRECHSin vitro and recognition of MRECHS in vivo. The deduced amino acid sequence revealed that PcMYB1 contained only one MYB-like repeat (Feldbrugge et al. 1997). Stracke et al. (2010) reported that the bZIP transcriptional regulator ELONGATED HYPOCOTYL5 (HY5) was required for the transcriptional activation of the PFG1/MYB12 and PFG3/MYB111 genes under UV-B and visible light respectively.
Nutrient deficiency
Rubio et al. (2001) found PHR1 (Phosphate Starvation Response1), a mutant which was related to the Phosphorus Starvation Response1 (PSR1) gene from Chlamydomonas reinhardtii. They further reported that PHR1 is expressed in Pi sufficient conditions and in contrast to PSR1, is only weakly responsive to Pi starvation. PHR1, PSR1 and other members of the protein family shared a MYB domain and a predicted coiled-coil (CC) domain, defining a subtype within the MYB superfamily, the MYB-CC family and was involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Similarly, Rouached et al. (2011) reported that PHR1 played an important role in sulfate inter-organ transport, in particular in the regulation of the SULTR1;3 gene and its impact on shoot-to-root sulfate transport in phosphate-deficient plants. MYB28 and MYB29 transcription factors and dozens of downstream enzymes were involved in the production of glucosinolate (GSL) in the sulfur-starvation stress response synthesis in Arabidopsis (Hirai et al. 2007). Dai et al. (2012) reported a novel R2R3 MYB transcriptional factor, OsMYB2P-1 which was associated with Pi starvation signaling in rice.
MYB as negative regulators
Most MYB genes were positive regulators of transcription, for example, ZmC1 positively regulated flavonoid biosynthesis by controlling chalcone syntheses (CHS) gene expression (Paz-Ares et al. 1987) and WER was a positive regulator of GL2 expression (Lee and Schiefelbein 1999). However, R2R3 Myb gene also acted as negative regulator. For example, Antirrhinum AmMyb305 and its Arabidopsis orthologue AtMyb4 regulated the accumulation of UV protective napoylmalate by repressing the expression of cinnamate hydroxylase (C4H) gene (Jin et al. 2000). AtMYB4 expression was down regulated by exposure to UV-B light, indicating that the de-repression was an important mechanism for acclimation of UV-B in Arabidopsis thaliana. AtMYB4 worked as a repressor of target gene expression and included a repression domain. It belonged to a novel group of plant R2R3 MYB proteins involved in transcriptional silencing (Jin et al. 2000). Park et al. (2008) reported that AtMYB60 inhibited anthocyanin biosynthesis in lettuce. The correlation between the overexpression of AtMYB60 and the inhibition of anthocyanin accumulation suggested that the transcription factor AtMYB60 controlled anthocyanin biosynthesis. Kazan (2006) suggested that transcriptional repression of gene expression by EAR-motif-containing repressor proteins played a key role in modulating plant defense and stress response. Scoville et al. (2011) identified M. guttatus MYB MIXTA-like 8 as a possible negative regulator of trichome development and found that parental leaf damage induced down-regulation of MYB MIXTA-like 8 in progeny, which was associated with epigenetically inherited increased trichome density. ZmMYB31 downregulated several genes involved in the synthesis of monolignols and transgenic plants were dwarf and showed a significantly reduced lignin content with unaltered polymer composition. In addition, ZmMYB31 repressed the synthesis of sinapoylmalate and phenylpropanoid resulting in plants that were more sensitive to UV irradiation, and induced several stress-related proteins (Fornale et al. 2010).
Low oxygen induction of the Arabidopsis ADH1 gene
The transcription factor AtMYB2, induced by hypoxia, acted as a key regulatory factor in the induction of the ADH1 promoter by low oxygen (Hoeren et al. 1998). Like ADH1, AtMYB2 had root-limited expression. When driven by a constitutive promoter, AtMYB2 was able to transactivate ADH1 expression in transient assays in both Arabidopsis and Nicotiana plumbaginifolia protoplasts, and leaves of Pisum sativum.
Regulation of primary and secondary metabolism
The MYB transcription factors play important roles in the regulation of many secondary metabolites at the transcriptional level other than anthocyanin synthesis. Stracke et al. (2007) found that AtMYB11/PFG1, AtMYB12/PFG1 and AtMYB111/PFG3 controled flavones biosynthesis in all tissues while AtMYB75/ PAP1, AtMYB90/PAP2, AtMYB113 and AtMYB114 regulated anthocyanin biosynthesis in vegetative tissues (Gonzalez et al. 2008) while AtMYB123/TT2 controlled the biosynthesis of proanthocyanidins (PAs) in the seed coat of Arabidopsis (Lepiniec et al. 2006). Verdier et al. (2012). Found that MtPAR (Medicago truncatula proanthocyanidin regulator) was a MYB family transcription factor that functioned as a key regulator of proanthocyanidin (PA) biosynthesis in the model legume Medicago truncatula and could be of great potential for biotechnological strategies to increase PAs in forage legumes for reduction of pasture bloat in ruminant animals.
LoAtMYB5 was recently proposed to be partially redundant with AtMYB123 in regulating tannin biosynthesis (Gonzalez et al. 2009). AtMYB58, AtMYB63 and AtMYB85 activated lignin biosynthesis in fibers and/or vessels (Zhou et al. 2009). AtMYB46 is found to be a positive regulator of lignin biosynthesis in fibers and vessels and also regulated cellulose and xylan deposition (Zhong et al. 2007). AtMYB26/ MS35 controlled secondary wall deposition in anthers (Yang et al. 2007). AtMYB52, AtMYB54 and AtMYB69 and AtMYB103 were positive regulators dedicated to cell wall thickening in fiber cells. Zhong et al. (2008) proposed that AtMYB52, AtMYB54 and AtMYB69 regulated lignin, xylan and cellulose biosynthesis, and cellulose biosynthesis. AtMYB61 plays a pleiotropic role, influencing lignin deposition (Newman et al. 2004), mucilage production (Penfield et al. 2001) and stomatal aperture (Liang et al. 2005), suggesting that it might act upstream of the different pathways by regulating carbon allocation. The R2R3-MYB proteins of subgroup 12 i.e. AtMYB28/HAG1/PMG1, AtMYB29/HAG3/PMG2 and AtMYB76/HAG2 regulate the biosynthesis of aliphatic glucosinolates in aerial issues (Gigolashvili et al. 2008). However, AtMYB34/ATR1, AtMYB51/HIG1 and AtMYB122 regulate the production of indolic glucosinolates in roots and late stage rosette leaves (Gigolashvili et al. 2007).
Signal transduction pathways
R2R3-MYB genes are involved in the signal transduction pathways of salicylic acid (Raffaele et al. 2006), abscisic acid (Abe et al. 2003), gibberellic acid (Murray et al.2003) and jasmonic acid (Lee et al. 2001) as well. The phytohormone ABA, produced under water deficit conditions, caused stomatal closure and played an important role in the adaptation of vegetative tissues to abiotic environmental stresses, such as drought and high salinity (Vannini et al. 2004; Maeda et al. 2005).
Conclusion
MYB proteins play an important role in controlling various plant processes and new insights have been obtained into the mechanisms that control MYB protein activities and gene expression profiles and several target genes have been determined. The large family of plant-specific R2R3-MYB genes has contributed to the evolution of physiological or developmental processes specific to plants, especially those involved in responses to fluctuating biotic or abiotic environments, still a lot of work is required to fully characterize the roles of all MYB proteins in regulatory networks and inferring functions in more plant species. Once more data are available, it will be interesting to establish how the control of specific target genes relates to the biological functions that MYB factors control. It will also facilitate better understanding of gene regulation in plants by the MYB-type transcription factors and the development of new varieties and other commercially important plants with metabolic engineering approaches.
Acknowledgments
The authors (RCY and NRY) acknowledge the financial assistance provided by DBT, Govt. of India.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M, et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Akagi T, Ikegami A, Yonemori K (2010) DkMyb2 wound-induced transcription factor of persimmon (Diospyros kaki Thunb.), contributes to proanthocyanidin regulation. Planta 232:1045–1059 [DOI] [PubMed]
- Baumann K, Rodriguez MP, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development. 2007;134:1691–1701. doi: 10.1242/dev.02836. [DOI] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Khatab H, Sidorova, Lingaya M, Twell D. The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell. 2011;23:2534–2549. doi: 10.1105/tpc.110.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA. Plant responses to water deficit. Trends Plant Sci. 1997;2:48–54. doi: 10.1016/S1360-1385(97)82562-9. [DOI] [Google Scholar]
- Brownfield L, et al. A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 2009;5:e1000430. doi: 10.1371/journal.pgen.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century K, Reuber TL, Ratcliffe OJ. Regulating the regulators: the future prospects for transcription-factor based agricultural biotechnology products. Plant Physiol. 2008;147:20–29. doi: 10.1104/pp.108.117887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Peng S, Huang G, Wu K, Fu X, Chen Z. Association of decreased expression of a Myb transcription factor with the TPD (tapping panel dryness) syndrome in Hevea brasiliensis. Plant Mol Biol. 2003;51:51–58. doi: 10.1023/A:1020719420867. [DOI] [PubMed] [Google Scholar]
- Chen B-J, Wang Y, Hu YL, Wu Q, Lin ZP. Cloning and characterization of a drought inducible MYB gene from Boea crassifolia. Plant Sci. 2005;168:493–500. doi: 10.1016/j.plantsci.2004.09.013. [DOI] [Google Scholar]
- Chen R, Ni Z, Nie X, Qin Y, Dong G, Sun Q. Isolation and characterization of genes encoding Myb transcription factor in wheat (Triticum aestivum L.) Plant Sci. 2005;169:1146–1154. doi: 10.1016/j.plantsci.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Cheng H, et al. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009;5:e1000440. doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuan YH, Johnie N, Jenkins A, Sukumar SH, Din PM. Transcriptional regulation of the lipid transfer protein gene LTP3 in cotton fibers by a novel MYB protein. Plant Sci. 2005;168:167–181. doi: 10.1016/j.plantsci.2004.07.033. [DOI] [Google Scholar]
- Cominelli E, et al. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Cone KC, Burr FA, Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci U S A. 1986;83:9631–9635. doi: 10.1073/pnas.83.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone KC, Cocciolone SM, Burr FA, Burr B. Maize anthocyanin regulatory gene pl is a duplicate c1 that functions in the plant. Plant Cell. 1993;5:1795–1805. doi: 10.1105/tpc.5.12.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene OsMYB3R-2, increases tolerance to freezing, drought, salt stress in transgenic Arabidopsis. Plant Physiol 143:739–1751 [DOI] [PMC free article] [PubMed]
- Dai X, Wang Y, Yang A, Zhang WH. OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol. 2012;159:169–183. doi: 10.1104/pp.112.194217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M et al (2006) The Arabidopsis thaliana transcription factor AtMYB102 functions in defense against the insect herbivore Pieris rapae. Plant Signal Behav 1:305–311 [DOI] [PMC free article] [PubMed]
- Denekamp M, Smeekens SC. Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol. 2003;132:1415–1423. doi: 10.1104/pp.102.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Athikkattuvalasu R, Karthikeyan S, Raghothama KG. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant. 2009;2:43–58. doi: 10.1093/mp/ssn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, Huang YB, Tang YX. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry. 2009;74:1–11. doi: 10.1134/s0006297909010015. [DOI] [PubMed] [Google Scholar]
- Du H, Feng BR, Yang SS, Huang YB, Tang YX. The R2R3-MYB transcription factor gene family in maize. PLoS One. 2012;7(6):e37463. doi: 10.1371/journal.pone.0037463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Gourrierec JL, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- El-kereamy A, Bi Y-M, Ranathunge K, Beatty PH, Good AG, et al. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS One. 2012;7(12):e52030. doi: 10.1371/journal.pone.0052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrugge M, Sprenger M, Hahlbrock K, Weisshaar B. PcMYB1 novel plant protein containing a DNA-binding domain with one Myb repeat, interacts in vivo with a light-regulatory promoter unit. Plant J. 1997;11:1079–1093. doi: 10.1046/j.1365-313X.1997.11051079.x. [DOI] [PubMed] [Google Scholar]
- Feng C, Andressson E, Maslak A, Mock HP, Mattsson O, Mundy L. Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci. 2004;167:1099–1107. doi: 10.1016/j.plantsci.2004.06.014. [DOI] [Google Scholar]
- Fornale S et al (2010) ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J 64(4):633–644 [DOI] [PubMed]
- Gigolashvili T, et al. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, et al. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008;177:627–642. doi: 10.1111/j.1469-8137.2007.02295.x. [DOI] [PubMed] [Google Scholar]
- Gocal GFW, Sheldon CC, Gubler F. GAMyb-like gene, flowering and gibberellins signaling in Arabidopsis. Plant Physiol. 2001;127:682–1691. doi: 10.1104/pp.010442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, et al. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol. 2009;325:412–421. doi: 10.1016/j.ydbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV. Gibberellin regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pl α amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Watts RJ, Kalla R, Matthews P, Keys M, Jacobsen JV. Cloning of rice cDNA encoding a transcription factor homologous to barley GAMyb. Plant Cell Physiol. 1997;38:362–365. doi: 10.1093/oxfordjournals.pcp.a029175. [DOI] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62(8):2465–2483 [DOI] [PubMed]
- Higginson T, Li SF, Parish RW. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 2003;35:177–192. doi: 10.1046/j.1365-313X.2003.01791.x. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci U S A. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics. 1998;149:479–490. doi: 10.1093/genetics/149.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sun W, Lv H, Xiao G, Zeng S, Wang Y. Isolation and molecular characterization of thirteen R2R3-MYB transcription factors from Epimedium sagittatum. Int J Mol Sci. 2013;14:594–610. doi: 10.3390/ijms14010594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, Okada K, Wada T (2007) Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19:2531–2543 [DOI] [PMC free article] [PubMed]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell. 2001;13:1891–1906. doi: 10.1105/tpc.13.8.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriaga G, Leyns L, Villeges A, Gharaibeh R, Salamini F, Bartels D (1996) A family of novel myb-related genes from the resurrection plant Craterostigma plantagineum are specifically expressed in callus and roots in response to ABA or dessication. Plant Mol Biol 32:707–716 [DOI] [PubMed]
- Jackson D, Culianez-Macia F, Prescott AG, Roberts K, Martin C. Expression patterns of myb genes from Antirrhinum flowers. Plant Cell. 1991;3:115–125. doi: 10.1105/tpc.3.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C et al (2004a) Ordered origin of the typical two- and three-repeat Myb genes. Gene 326:13–22 [DOI] [PubMed]
- Jiang C, Gu X, Peterson T. Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 2004;5:R46. doi: 10.1186/gb-2004-5-7-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41(5):577–585. doi: 10.1023/A:1006319732410. [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV- protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ET, Dowd PF. Differentially enhanced insect resistance, at a cost, in Arabidopsis thaliana constitutively expressing a transcription factor of defensive metabolites. J Agric Food Chem. 2004;52:5135–5138. doi: 10.1021/jf0308049. [DOI] [PubMed] [Google Scholar]
- Jung C, et al. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YH, Kirik V, Hulskamp M, Nam KH, Hagely K, Lee MM, Schiefelbein J. The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell. 2009;21:1080–1094. doi: 10.1105/tpc.108.063180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. Negative regulation of defense and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006;11:109–112. doi: 10.1016/j.tplants.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Kizis D, Lumbreras V, Pages M. Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Lett. 2001;498:187–189. doi: 10.1016/S0014-5793(01)02460-7. [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin HL, Leyva A, Meissner R, Petroni K, Urzainqui A, Bevan M, Martin C, Smeekens S, Tonelli C, Paz-Ares J, Weisshaar B. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB related protein in Arabidopsis, is a position dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/S0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Lee MW, Qi M, Yang Y. A novel jasmonic acid-inducible rice MYB gene associates with fungal infection and host cell death. Mol Plant Microbe Interact. 2001;14:527–535. doi: 10.1094/MPMI.2001.14.4.527. [DOI] [PubMed] [Google Scholar]
- Lee DK et al (2009) Lateral organ fusion1 and lateral organ fusion2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136:2423–2432 [DOI] [PubMed]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- Jigang L, et al. A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J Exp Bot. 2006;57:1263–1273. doi: 10.1093/jxb/erj094. [DOI] [PubMed] [Google Scholar]
- Li L, et al. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58:275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YK, et al. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Liao Y, Zou HF, Wang HW, Zhang WK, Ma B, Zhang JS. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 2008;18:1047–1060. doi: 10.1038/cr.2008.280. [DOI] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:50–66. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, Udvardi MK. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 2009;149:1761–1772. doi: 10.1104/pp.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- Liu L, Du H, Tang XF, Wu YM, Huang YB, Tang YX. The roles of MYB transcription factors on plant defense responses and its molecular mechanism. Hereditas. 2008;30:1265–1271. doi: 10.3724/SP.J.1005.2008.01265. [DOI] [PubMed] [Google Scholar]
- Liu R, Lü B, Wang X, Zhang C, Zhang S, Qian J, Chen L, Shi H, Dong H. Thirty-seven transcription factor genes differentially respond to a hairpin protein and affect resistance to the green peach aphid in Arabidopsis. J Biosci. 2010;35:435–450. doi: 10.1007/s12038-010-0049-8. [DOI] [PubMed] [Google Scholar]
- Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM. Circadian clock associated1 and late elongated hypocotyl function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009;150:834–843. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009;150:244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Kimura S, Demura T, Takeda J, Ozeki Y. DcMYB1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase gene (DcPAL1) in response to elicitor treatment, UV-B irradiation and the dilution effect. Plant Mol Biol. 2005;59:739–752. doi: 10.1007/s11103-005-0910-6. [DOI] [PubMed] [Google Scholar]
- Mandaokar A, Browse J (2009) MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol 149:851–862 [DOI] [PMC free article] [PubMed]
- Mandaokar A, et al. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006;46:984–1008. doi: 10.1111/j.1365-313X.2006.02756.x. [DOI] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/S0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. 2003;15:2551–2565. doi: 10.1105/tpc.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi T, Iwabuchi M. Plant transcription factors. Plant Cell Physiol. 1995;36:1405–1420. [PubMed] [Google Scholar]
- Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese Morning Glory. Plant Cell Physiol. 2006;47:457–470. doi: 10.1093/pcp/pcj012. [DOI] [PubMed] [Google Scholar]
- Moyano E, Martinez-Garcia JF, Martin C. Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers. Plant Cell. 1996;8:1519–1532. doi: 10.1105/tpc.8.9.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu RL et al (2009) An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res 19:1291–1304 [DOI] [PubMed]
- Murray F, Kalla R, Jacobsen J, Gubler F. A role for HvGMYB in anther development. Plant J. 2003;33:481–491. doi: 10.1046/j.1365-313X.2003.01641.x. [DOI] [PubMed] [Google Scholar]
- Narasuka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34:137–148 [DOI] [PubMed]
- Nesi N, Jond C, Debeaujon I, Caboche M. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2311–2322. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LJ, et al. Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 2004;37:239–250. doi: 10.1046/j.1365-313X.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- Noda KI, Glover BJ, Linstead P, Martin C. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature. 1994;369:661–664. doi: 10.1038/369661a0. [DOI] [PubMed] [Google Scholar]
- Ogata K, et al. The cavity in the hydrophobic core of Myb DNA binding domain is reserved for DNA recognition and trans-activation. Nat Struct Biol. 1996;3:178–818. doi: 10.1038/nsb0296-178. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Hertman PL, Sivakumaran S, Esch J (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67:483–493 [DOI] [PubMed]
- Park JS, et al. Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa) Plant Cell Rep. 2008;27:985–994. doi: 10.1007/s00299-008-0521-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali G, et al. OsMYB4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 2008;27:1677–1686. doi: 10.1007/s00299-008-0587-9. [DOI] [PubMed] [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-ncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6:3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, et al. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell. 2001;13:2777–2791. doi: 10.1105/tpc.13.12.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Hulskamp M. One, two, three.models for trichome patterning in Arabidopsis. Curr Opin Plant Biol. 2009;12:587–592. doi: 10.1016/j.pbi.2009.07.015. [DOI] [PubMed] [Google Scholar]
- Prabu G, Prasad DT. Functional characterization of sugarcane MYB transcription factor gene promoter (PScMYBAS1) in response to abiotic stresses and hormones. Plant Cell Rep. 2012;4:661–669. doi: 10.1007/s00299-011-1183-y. [DOI] [PubMed] [Google Scholar]
- Prabu GR, Theertha PD. Structure of DNA binding Myb transcription factor protein (SCMYBAS1-3) from sugarcane-threading and AB initio modelling. J Phytol. 2011;3:77–82. [Google Scholar]
- Punwani JA, et al. The MYB98 subcircuit of the synergid gene regulatory network includes genes directly and indirectly regulated by MYB98. Plant J. 2008;55:406–414. doi: 10.1111/j.1365-313X.2008.03514.x. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg N, Dockx J, Keultjes G, Kock P, Wilmering J, Weisbeek P, Smeekens S. Identification of a light regulated MYB gene from an Arabidopsis transcription factor gene collection. Plant Mol Biol. 1996;32:987–993. doi: 10.1007/BF00020495. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. Regulatory genes controlling anthocyanin pigmentation are conserved among plant species and have distinct set of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Braun EL, Wolfe AD, Bowen B, Grotewold I (1999) Maize R2R3 Myb genes: sequence analysis reveals amplification in the higher plants. Genetics 153:427–444 [DOI] [PMC free article] [PubMed]
- Raffaele S, Rivas S, Roby D. An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett. 2006;580:3498–3504. doi: 10.1016/j.febslet.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Raffaele S, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of t he hypersensitive cell death Response in Arabidopsis. Plant Cell. 2008;20:752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samara RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Chandahari D, Sherman BK, Yu CL. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol. 1998;46:74–83. doi: 10.1007/PL00006285. [DOI] [PubMed] [Google Scholar]
- Rouached H, Secco D, Arpat B, Poirier Y. The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 2011;11:19. doi: 10.1186/1471-2229-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93:1219–1229 [DOI] [PubMed]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville AG, Barnett LL, Roels SB, Kelly JK, Hileman LC (2011) Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol 191(1):251–263 [DOI] [PMC free article] [PubMed]
- Segarra G, Van der Ent S, Trillas I, Pieterse CMJ. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial Microbe. Plant Biol. 2009;11:90–96. doi: 10.1111/j.1438-8677.2008.00162.x. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park CM. Auxin homeostasis during lateral root development under drought condition. Plant Signal Behav. 2009;4:1002–1004. doi: 10.4161/psb.4.10.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Park CM (2010) MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol 186:471–483 [DOI] [PubMed]
- Seo PJ, et al. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009;151:275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM. The MYB96 Transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell. 2011;23:1138–1152. doi: 10.1105/tpc.111.083485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Kumar H, Supriya, Kumar S, Yadav NR, Pradeep, Yadav RC, Singh D. Comparative myb gene expression in Brassica species under drought stress through semi-quantitative RT-PCR. Cruciferae Newslett. 2010;29:23–25. [Google Scholar]
- Shin B, Choi G, Yi H, Yang S, Cho I, Kim J, Lee S, Paek NC, Kim JH, Song PS, Choi G. AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J. 2002;30:23–32. doi: 10.1046/j.1365-313X.2002.01264.x. [DOI] [PubMed] [Google Scholar]
- Shin R, et al. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. doi: 10.1016/S1369-5266(00)00067-4. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Urao OT, Koizumi M (1992) Nucleotide sequence of a gene from Arabidopsis thaliana encoding a myb homologue. Plant Mol Biol 19:493–499 [DOI] [PubMed]
- Solano R, Nieto C, Avila J, Canas L, Diaz I, Paz-Ares J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYBPh3) from Petunia hybrida. EMBO J. 1995;14:1773–1784. doi: 10.1002/j.1460-2075.1995.tb07166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell. 2011;23:1000–1013. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Lange S, Unte US, Eckstein L, Yang C, Wilson ZA, Schmelzer E, Dekker K, Saedler H. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 2003;34:519–528. doi: 10.1046/j.1365-313X.2003.01745.x. [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3–MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456 [DOI] [PubMed]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010;33:88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- Su CF, et al. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010;153:145–158. doi: 10.1104/pp.110.153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo J, Liang X, Pu L, Zhang Y, Xue Y. Identification of GhMYB109 encoding a R2R3MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.) Biochem Biophys Acta. 2003;1630:25–34. doi: 10.1016/j.bbaexp.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Supriya YNR, Yadav RC, Singh D. Isolation of biologically active RNA for generating myb transcription factor cDNAs in Brassica species. Brassica. 2006;8:95–98. [Google Scholar]
- Takahashi R, Yamagishi N, Yoshika N (2013) A MYB transcription factor controls flower color in soybean. J Hered 104(1):149–153 [DOI] [PubMed]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent S, et al. MYB72 is required in early signaling steps of Rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 2008;146:1293–1304. doi: 10.1104/pp.107.113829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I. Overexpression of the rice OsMYB4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 2004;37:115–127. doi: 10.1046/j.1365-313X.2003.01938.x. [DOI] [PubMed] [Google Scholar]
- Verdier J et al (2012) MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. PNAS 109(5):1766–1771 [DOI] [PMC free article] [PubMed]
- Wang X, et al. Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 2008;19:224–235. doi: 10.1038/cr.2008.276. [DOI] [PubMed] [Google Scholar]
- Wang S, Barron C, Schiefelbein J, Chen JG. Distinct relationships between GLABRA2 and single-repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 2010;185:387–400. doi: 10.1111/j.1469-8137.2009.03067.x. [DOI] [PubMed] [Google Scholar]
- Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3–MYB family of transcription factors. Plant Physiol. 2009;149:981–993. doi: 10.1104/pp.108.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodger FJ, Gubler F, Pogson BJ, Jacobsen JV. A MAK-like kinase is a repressor of GAMYB in barley aleurone. Plant J. 2003;33:707–717. doi: 10.1046/j.1365-313X.2003.01663.x. [DOI] [PubMed] [Google Scholar]
- Xie Z, Lee EK, Lucas JR, Morohashi K, Li D, Murray JAH, Sack FD, Grotewold E. Plant Cell. 2010;22:2306–2321. doi: 10.1105/tpc.110.074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C, Zhonghe M, Eric L. DNA binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol Biol. 1997;34:403–415. doi: 10.1023/A:1005873500238. [DOI] [PubMed] [Google Scholar]
- Yang Y, Klessig DF. Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. Proc Natl Acad Sci. 1996;93:14972–14977. doi: 10.1073/pnas.93.25.14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Sweetman JP, Amirsadeghi S, Barghchi M, Huttly AK, Chung WI, Twell D. Novel anther-specific myb genes from tobacco as putative regulators of phenylalanine ammonia-lyase expression. Plant Physiol. 2001;126:1738–1753. doi: 10.1104/pp.126.4.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, et al. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell. 2007;19:534–548. doi: 10.1105/tpc.106.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Xiaoyan Dai X, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63(7):2541–2556 [DOI] [PMC free article] [PubMed]
- Yanhui C, et al. The MYB transcriptionfactor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Yi J, Derynck MR, Li X, Telmer P, Marsolais F, Dhaubhadel S. A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 2010;62:1019–1034. doi: 10.1111/j.1365-313X.2010.04214.x. [DOI] [PubMed] [Google Scholar]
- Yu EY, Kim SE, Kim JH, Ko JH, Cho MH, Chung IK. Sequence-specific DNA recognition by the Myb-like domain of plant telomeric protein RTBP1. J Biol Chem. 2000;275:24208–24214. doi: 10.1074/jbc.M003250200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. Characterization of Arabidopsis MYB transcription factor gene AtMYB17 and its possible regulation by LEAFY and AGL15. J Genet Genomics. 2009;36:99–107. doi: 10.1016/S1673-8527(08)60096-X. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ju HW, Chung MS, Huang P, Ahn SJ, Kim CS. The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 2011;52:138–148. doi: 10.1093/pcp/pcq180. [DOI] [PubMed] [Google Scholar]
- Zhong R, et al. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, et al. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, et al. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]