Abstract
An 85-kDa Group VI phospholipase A2 enzyme (iPLA2) that does not require Ca2+ for catalysis has recently been cloned from three rodent species. A homologous 88-kDa enzyme has been cloned from human B-lymphocyte lines that contains a 54-amino acid insert not present in the rodent enzymes, but human cells have not previously been observed to express catalytically active iPLA2 isoforms other than the 88-kDa protein. We have cloned cDNA species that encode two distinct iPLA2 isoforms from human pancreatic islet RNA and a human insulinoma cDNA library. One isoform is an 85-kDa protein (short isoform of human iPLA2 (SH-iPLA2)) and the other an 88-kDa protein (long isoform of human iPLA2 (LH-iPLA2)). Transcripts encoding both isoforms are also observed in human promonocytic U937 cells. Recombinant SH-iPLA2 and LH-iPLA2 are both catalytically active in the absence of Ca2+ and inhibited by a bromoenol lactone suicide substrate, but LH-iPLA2 is activated by ATP, whereas SH-iPLA2 is not. The human iPLA2 gene has been found to reside on chromosome 22 in region q13.1 and to contain 16 exons represented in the LH-iPLA2 transcript. Exon 8 is not represented in the SH-iPLA2 transcript, indicating that it arises by an exon-skipping mechanism of alternative splicing. The amino acid sequence encoded by exon 8 of the human iPLA2 gene is proline-rich and shares a consensus motif of PX5PX8HHPX12NX4Q with the proline-rich middle linker domains of the Smad proteins DAF-3 and Smad4. Expression of mRNA species encoding two active iPLA2 isoforms with distinguishable catalytic properties in two different types of human cells demonstrated here may have regulatory or functional implications about the roles of products of the iPLA2 gene in cell biologic processes.
Phospholipases A2 (PLA2)1 catalyze hydrolysis of sn-2 fatty acid substituents from glycerophospholipid substrates to yield a free fatty acid and a 2-lysophospholipid (1–7). PLA2 is a diverse group of enzymes, and the first well characterized members have low molecular masses (approximately 14 kDa), require millimolar [Ca2+] for catalytic activity, and function as extracellular secreted enzymes (sPLA2) (3, 6). The first cloned PLA2 that is active at [Ca2+] achieved in the cytosol of living cells is an 85-kDa protein classified as a Group IV PLA2 and designated cPLA2 (3, 5). This enzyme is induced to associate with its substrates in membranes by rises in cytosolic [Ca2+] within the range achieved in cells stimulated by extracellular signals that induce Ca2+ release from intracellular sites or Ca2+ entry from the extracellular space, is also regulated by phosphorylation, and prefers substrates with sn-2 arachidonoyl residues (5).
Recently, a second PLA2 that is active at [Ca2+] that can be achieved in cytosol has been cloned (8–10). This enzyme does not require Ca2+ for catalysis, is classified as a Group VI PLA2, and is designated iPLA2 (3, 4). The iPLA2 enzymes cloned from hamster (8), mouse (9), and rat (10) cells represent species homologs and all are 85-kDa proteins containing 752 amino acid residues with highly homologous (approximately 95% identity) sequences. Each contains a GXSXG lipase consensus motif and eight stretches of a repeating motif homologous to a repetitive motif in the integral membrane protein-binding domain of ankyrin (8–10). The substrate preference of these iPLA2 enzymes varies with the mode of presentation (8), but each is inhibited (8–10) by a bromoenol lactone (BEL) suicide substrate (11, 12) that is not an effective inhibitor of sPLA2 or cPLA2 enzymes at comparable concentrations (4, 11–14).
Proposed functions for iPLA2 include a housekeeping role in phospholipid remodeling that involves generation of lysophospholipid acceptors for arachidonic acid incorporation into P388D1 macrophage-like cell phospholipids (4, 15, 16). Signaling roles for iPLA2 in generating substrate for leukotriene biosynthesis (17) and lipid messengers that regulate ion channel activity (10, 18, 19) and apoptosis (20) have also been suggested. Recent observations with human iPLA2 suggest that the enzyme might serve distinct functions in different cells that involve regulatory interactions among splice variants (17, 21). Human iPLA2 cloned from B-lymphocyte lines and testis differs from iPLA2 cloned from cells of rodent species in that it is an 88-kDa rather than an 85-kDa protein and contains a 54-amino acid insert interrupting the eighth ankyrin repeat (21). The human B-lymphocyte iPLA2 sequence is otherwise highly homologous to hamster, mouse, and rat sequences and includes the seven other ankyrin-like repeats and a GXSXG lipase sequence (21). Catalytically active iPLA2 other than the 88-kDa isoform have not yet been observed in human cells (21).
Human B-lymphocyte lines do express truncated, inactive iPLA2 sequences that contain the ankyrin repeat domain but lack the catalytic domain and are thought to arise from alternative splicing of the transcript (21). Co-expression of the truncated sequences with full-length human iPLA2 attenuates catalytic activity (21). Because the active form of iPLA2 is an oligomeric complex (8, 22) that may result from subunit associations through ankyrin repeat domains (8), this suggests that formation of hetero-oligomeric complexes represents a means to regulate iPLA2 activity (21). That mechanisms of iPLA2 regulation differ among human cell types is suggested by the fact that stimuli that induce iPLA2-catalyzed arachidonate release and leukotriene production in human granulocytes fail to induce these events in human lymphocyte lines, even though both classes of cells express iPLA2 and leukotriene biosynthetic enzymes (17, 21).
One human cell type in which iPLA2 may be biomedically important is the pancreatic islet beta cell. Impaired beta cell survival and signaling functions underlie development of types I and II diabetes mellitus, respectively; these are the most prevalent human endocrine diseases. In rodent islets, iPLA2 has been proposed to play a signaling role in glucose-induced insulin secretion (8, 23–25) and in experimentally induced beta cell apoptosis (26). Human islets express a BEL-sensitive PLA2 activity that does not require Ca2+ (27, 28), but iPLA2 mRNA has not been demonstrated in human islets. We have cloned human beta cell iPLA2 cDNA here and find that human islets express mRNA species encoding two iPLA2 isoforms with different sizes (85 and 88 kDa) and catalytic properties. We have also determined the human iPLA2 gene structure and its chromosomal location and find that the transcript encoding the short isoform arises from an exon-skipping mechanism of alternative splicing.
EXPERIMENTAL PROCEDURES
Materials
The compounds [32P]dCTP (3000 Ci/mmol), [35S]dATPS (1000 Ci/mmol), and l-α-1-palmitoyl-2-[14C]arachidonoyl-phosphatidylethanolamine (50 mCi/mmol) and ECL detection reagents were obtained from Amersham Pharmacia Biotech, and the BEL ((E)-6-(bromomethylene)tetrahydro-3-(1-naphthalenyl)-2H-pyran-2-one) iPLA2 suicide substrate was obtained from BIOMOL (Plymouth Meeting, PA). A human placental genomic DNA library in lambda FIX II was obtained from Stratagene (La Jolla, CA). Human promonocytic U937 cells (30) were obtained from American Type Culture Collection (Manassas, VA) and cultured as described (20, 31). Sources of other common materials are identified elsewhere (10, 14, 23–25, 28).
Cloning cDNA Species Containing iPLA2 Sequence from a Human Insulinoma Cell cDNA Library
Rat islet iPLA2 cDNA was isolated (10), labeled with 32P, and used to screen a human insulinoma cDNA library (32) provided by Dr. Alan Permutt of Washington University. Insert sizes in clones that hybridized with the probe were determined by digestion with restriction endonucleases, and their sequences were determined from the double strand (33). Two cDNA species were obtained that contained about 1.80 and 1.59 kb, respectively, of the 3′-sequence of human iPLA2 cDNA, including the poly(A) tail. Neither contained the 5′-end of the full coding sequence, and RT-PCR was therefore performed with human islet RNA.
Isolation of RNA from Human Islets and Human Promonocytic U937 Cells, Reverse Transcription, and Polymerase Chain Reactions
Islets were isolated from human pancreata in the Washington University Diabetes Research and Training Center (34) and cultured as described (35). Total RNA was isolated from human islets and promonocytic U937 cells and first strand cDNA prepared by reverse transcription (RT) using standard procedures (36). PCRs were performed under described conditions (10), and products were analyzed by agarose gel electrophoresis (36). Primers used to generate the 5′ portion of human iPLA2 cDNA were sense (5′-GATGCAGTTCTTTGGACGCCTGG-3′), anti-sense (5′-T-CAGCATCACCTTGGGT-TTCC-3′), and nested antisense (5-AATGGCCAGGGCCAGGATG-C-3′). Two distinct cDNA fragments were obtained, subcloned, sequenced, and found to extend from the 5′-initiator codon through about 1.59 and 1.79 kb of DNA, respectively.
Preparation of cDNA Species Containing the Complete Human Islet iPLA2 Coding Sequence
The cDNA fragments obtained from screening the human insulinoma cell cDNA library overlapped at their 5′-ends with 3′-ends of cDNA fragments from RT-PCR of human islet RNA, and the overlapping region contained an NcoI site. The fragments were subcloned into pBluescript SK. Fragments from RT-PCR of human islet RNA contained the iPLA2 5′-coding sequence and were released from plasmids with EcoRI and NcoI. Products were isolated by agarose gel electrophoresis and ligated with a plasmid containing the 3′-end of human iPLA2 cDNA that had been treated with NcoI. Ligation product plasmids were used to transform bacterial host cells and sequenced. The resultant cDNA species contained complete coding sequences of human iPLA2 isoforms and were inserted into appropriate vectors for expression and used to prepare 32P-labeled human iPLA2 cDNA for genomic screening.
Bacterial Expression of Recombinant Human Islet iPLA2 Isoforms
The cDNA species encoding full-length human islet iPLA2 isoforms were subcloned in-frame into the EcoRI and XhoI sites of pET-28c (Novagen). The constructs were analyzed by restriction endonuclease digestion, sequenced, and transformed into bacterial expression host BL21(DE3) (Novagen). Cells transformed with pET28c without insert were negative controls. Protein expression was induced by treating cells with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside (IPTG) and assessed by SDS-polyacrylamide gel electrophoresis analyses with Coomassie Blue staining and by immunoblotting under described conditions (10) with a rabbit polyclonal antibody against recombinant rat islet iPLA2.
Eukaryotic Expression of Recombinant Human Islet iPLA2 Isoforms
The Spodoptera frugiperda (Sf9) insect cell-baculovirus system used to express other PLA2 enzymes in catalytically active forms (37, 38) was used to express human iPLA2 isoforms. The iPLA2 cDNA inserts were released from pBluescript SK plasmids by digestion with EcoRI and XhoI and subcloned into Eco RI and Xho I sites of pBAC-1 baculovirus transfer plasmid (Novagen). Sf9 cells were co-transfected with this transfer plasmid and linearized baculovirus DNA (BacVector-2000, Novagen) to construct recombinant baculovirus with human islet iPLA2 isoform cDNA inserts. Infection and culture were performed under described conditions (47). At 48 h after infection, Sf9 cells were collected by centrifugation, washed, resuspended in buffer (250 mm sucrose, 25 mm imidazole, pH 8.0), and disrupted by sonication. Cytosolic and membranous fractions were prepared by sequential centrifugations (10,000 × g for 10 min and 100,000 × g for 60 min) and used for PLA2 activity assays.
Phospholipase A2 Activity Assays
The protein content of Sf9 cell cytosolic and membranous fractions was determined by Bio-Rad assay, and iPLA2 activity was measured in aliquots (approximately 20 μg of protein) added to assay buffer (200 mm Tris-HCl, pH 7.0; total assay volume, 200 μl) containing 5 mm EGTA with or without 1 mm ATP. Some aliquots were pretreated (2 min) with BEL (10 mm) before the assay. Reactions were initiated by injecting substrate (l-α-1-palmitoyl-2-[14C]arachidonoyl-phosphatidylethanolamine; specific activity, 50 Ci/mol; final concentration, 5 μm) in ethanol (5 μl). Assay mixtures were incubated (3 min at 37 °C), and reactions were terminated by adding butanol (0.1 ml) and vortexing. After centrifugation (2000 × g for 4 min), products in the butanol layer were analyzed by silica gel G TLC in hexane/ethyl ether/acetic acid (80:20:1). The TLC region containing free arachidonic acid (RF, 0.58) was scraped into vials, and its 14C content was determined.
Cloning Human iPLA2 Genomic DNA Fragments, Determination of Intron-Exon Boundaries, and Estimation of Intron Size
A 32P-labeled human islet iPLA2 cDNA was used to screen a human placental Lambda FIX II genomic DNA library (Stratagene). Clones that hybridized with the probe were isolated and plaque-purified, and the lambda DNA fragments containing genomic DNA inserts were purified by standard procedures (36). Inserts were excised with NotI and subcloned into a pBluescript SK plasmid for restriction site mapping. Sequences of intron-exon boundaries were determined by comparing sequences of genomic DNA and cDNA. Intron sizes were estimated from lengths of PCR products from reactions using genomic DNA as template and primers that hybridize to sequences in adjacent exons.
Chromosomal Mapping of Human iPLA2 Gene by Fluorescence in Situ Hybridization
A human iPLA2 genomic DNA clone was biotinylated with dATP (40, 41) and used as a probe to map the chromosomal location of the human iPLA2 gene. Fluorescence in situ hybridization (FISH) detection of the locus of hybridization of the fluorescent probe with chromosomal DNA was performed by See DNA Biotech Inc. (Downsview, Ontario, Canada) using described methods (40, 41). Human blood lymphocytes were cultured in α-minimal essential medium supplemented with 10% fatal calf serum and phytohemagglutinin at 37 °C for 68–72 h. Bromodeoxyuridine (0.18 mg/ml, Sigma) was used to synchronize the cell populations, which were then washed three times with serum-free medium to release the block and recultured (37 °C, 6 h) in α-minimal essential medium with thymine (2.5 μg/ml, Sigma). Cells were harvested and slides were prepared by standard procedures, including hypotonic treatment, fixation, and air-drying (40, 41), and the slides were baked (55 °C, 1 h). After RNase treatment, slides were denatured in 70% formamide and dehydrated with ethanol. Probes were denatured (75 °C, 5 min) in a hybridization mixture (50% formamide, 10% dextran sulfate, and human cot I DNA). After incubation (15 min, 37 °C) to suppress repetitive sequences, probes were loaded on denatured chromosomal slides, which were incubated overnight, washed, and subjected to detection and amplification procedures. FISH signals and DAPI banding patterns were recorded in separate photographs. Assignment of FISH mapping data with chromosomal bands was achieved by superimposing FISH signals with DAPI banding pattern on the chromosomes (40, 41).
RESULTS
Characterization of iPLA2 cDNA from Human Islets
To determine whether human pancreatic islet beta cells express mRNA species encoding iPLA2, a human insulinoma cell cDNA library (32) was screened with a 32P-labeled rat iPLA2 cDNA (10) probe. Two clones (INS-C1 and INS-C2) of about 1.59 and 1.80 kb in length, respectively, hybridized to the probe and were sequenced. Both clones contained identical 3′-sequences that included a presumptive polyadenylation sequence and a poly(A) tail, and their sequences were identical except for additional 5′-sequence in the longer clone not contained in the shorter clone. Alignment with the rat iPLA2 cDNA sequence indicated that the clones contained the 3′-end of human iPLA2 cDNA, but neither contained the full 5′-coding sequence (Fig. 1). RNA from human islets was therefore used as template in RT-PCRs with primers designed from the 5′-sequence of rat iPLA2 cDNA and from sequence in INS-C1 and INS-C2 clones. The primers were designed to amplify cDNA from the initiator methionine codon at the 5′-end through the region of sequence at the 3′-end recognized by primers designed from INS-C1 and INS-C2 sequences. A nested primer approach was employed in 3′-end primers to verify specificity of amplification products. When used with the same 5′-primer, one of the 3′-primers was expected to yield a longer product than the other.
Fig. 1. Summary of cDNA fragments used to construct cDNA species containing the complete coding sequences of human islet iPLA2 isoforms.
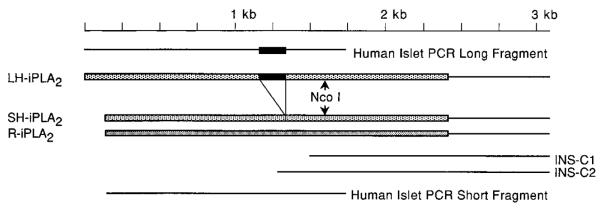
The two cDNA clones obtained by screening the human insulinoma cell cDNA library that contain the 3′-sequence of the human islet iPLA2 cDNA are designated INS-C1 and INS-C2. The RT-PCR products obtained using human islet RNA as template that contain the 5′-end of the human islet iPLA2 coding sequence (see Fig. 2) are designated human islet PCR long fragment and human islet PCR short fragment. Arrows indicate the location of the recognition site for the restriction endonuclease NcoI that is contained in the region of overlap between the insulinoma cell cDNA fragments and the human islet RNA RT-PCR products. The cDNA species containing the complete coding sequence of the long and short isoforms of human iPLA2 are designated LH-iPLA2 and SH-iPLA2, respectively. These cDNA species were prepared by NcoI digestion and ligation of the insulinoma cell cDNA fragment and one of the two RT-PCR products derived from human islet RNA. R-iPLA2, rat islet iPLA2 cDNA. The region of the 162-bp insert that distinguishes LH-iPLA2 from SH-iPLA2 is indicated by the black bar. The remainder of the coding sequence is indicated by shaded bars. The lighter shading represents human iPLA2 coding sequence; the darker shading represents rat iPLA2 coding sequence.
RT-PCRs with a given set of these primers using human islet RNA as template yielded two products (Fig. 2A). The experiments shown in lanes 1–4 of the agarose gel electrophoretic analysis of the RT-PCR products (Fig. 2A) were performed with a primer set expected to yield a product of 1.65 kb in length based on the rat iPLA2 cDNA sequence. Lanes 1 and 3 represent PCRs performed without an RT step to exclude contamination with genomic DNA, and no amplification products were observed. Lanes 2 and 4 represent RT-PCRs performed with two different preparations of human islet RNA, which both yielded two products. The more intensely staining product exhibited the 1.65-kb length expected from the rat iPLA2 cDNA sequence. There was also a less intensely staining band at 1.85 kb. Lanes 5 and 6 represent RT-PCRs performed with human RNA as template, the same 5′-end primer used in lanes 1–4, and a nested 3′-end primer expected to yield a shorter product than that obtained with the 3′-end primer used in the experiments shown in lanes 1–4. Two products were again observed. The more intensely staining band exhibited the length expected based on the rat iPLA2 cDNA sequence, and it was accompanied by another band about 0.2 kb longer.
Fig. 2. Agarose gel electrophoretic analyses of products of RT-PCRs performed with human islet RNA or U937 cell RNA as template and primers designed to amplify the 5′-end of human iPLA2 cDNA.
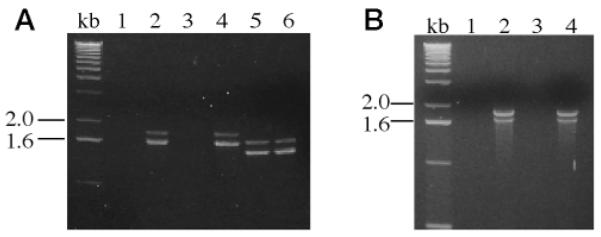
RT-PCRs were performed with RNA isolated from human islets from two different donors (A) or from two different preparations of human promonocytic U937 cell RNA (B). In A, experiments shown in lanes 1, 2, and 5 were performed with RNA from islets from donor 1, and experiments shown in lanes 3, 4, and 6 were performed with RNA from islets from donor 2. Reverse transcriptase was omitted from the reactions analyzed in lanes 1 and 3 to exclude contamination from genomic DNA in the human islet RNA preparations. In reactions analyzed in lanes 1–4 (A), a set of PCR primers was used that was expected to yield a single 1.65-kb product, based on the rat islet iPLA2 cDNA sequence. In reactions analyzed in lanes 5 and 6 (A), the same 5′-primer was used as in the reactions analyzed in lanes 1–4, but a different 3′-primer was used that was expected to yield a shorter product, based on the rat iPLA2 cDNA sequence. The sequences of the 5′-primer and of the two 3′-primers used in these reactions are specified under “Experimental Procedures.” In B, experiments shown in lanes 1 and 2 were performed with RNA from U936 cell preparation 1, and experiments shown in lanes 3 and 4 were performed with RNA from U937 cell preparation 2. Reverse transcriptase was omitted from the reactions analyzed in lanes 1 and 3 (B). In reactions analyzed in lanes 1–4 (B), the set of PCR primers was the same as that in lanes 1–4 of A. Both of the RT-PCR products visualized in lanes 2 and 4 (B) were subcloned and sequenced, and the results were identical to those for the products in lanes 2 and 4 of A.
The 1.65- and 1.85-kb human islet RT-PCR products in Fig. 2A, lanes 2 and 4, were subcloned and sequenced. Each contained a 5′-coding sequence that specified an amino acid sequence highly homologous to the N-terminal portion of rat iPLA2. The nucleotide sequences of the two human islet RT-PCR products were identical except for a 162-bp insert in the longer product that was not contained in the shorter product. This insert did not interrupt the reading frame and encoded a 54-amino acid insert in the eighth ankyrin repeat of the iPLA2 amino acid sequence. Similar RT-PCRs using RNA from human U937 promonocytic cells as template and the primer set employed in Fig. 2A, lanes 1–4, indicated that U937 cells also express two distinct iPLA2 mRNA species (Fig. 2B), and the lengths of the two RT-PCR products corresponded exactly to those from human islet RNA. The relative intensities of the two products differed, however, and staining of the band for the longer product was more intense than that for the shorter product when U937 cell RNA was used as template. The converse was true with human islet RNA. The U937 cell RT-PCR products were subcloned and sequenced and were identical to the products from human islet RNA. RT-PCRs in Fig. 2 are analogous to competitive PCR (10, 66, 67) and involve amplification of two distinct cDNA species from the same primer set in the same reaction mixture. As with competitive RT-PCR, relative abundances of reaction products in Fig. 2 may reflect the relative abundances of different cDNA species in the original reaction mixture.
These findings indicate that some human cells express mRNA species that encode two distinct isoforms of iPLA2. While these experiments were in progress, cloning of human iPLA2 from lymphocyte lines and testis was reported (21). That report identified only one human mRNA species that encoded a full-length iPLA2 sequence, and it corresponded to the longer isoform predicted from our results. No mRNA species corresponding to the shorter human iPLA2 isoform predicted from our results was observed in human B-lymphocyte lines or human testis, but two mRNA species, thought to represent alternative splicing products, were observed that encoded truncated iPLA2 variants that contained the ankyrin repeat region but lacked the catalytic domain (21). We sought evidence for expression mRNA encoding these truncated iPLA2 variants in human islets and human U937 promonocytic cells and observed them in U937 cells but not in islets (not shown), suggesting that there is heterogeneity among human cells in expression of products of the iPLA2 gene.
Fig. 3 illustrates nucleotide and deduced amino acid sequences predicted from our results for cDNAs encoding the long and short isoforms of human iPLA2. The predicted amino acid sequence for the long isoform differs from that for the short isoform only by the presence of a 54-amino acid insert interrupting the eighth ankyrin repeat. The short isoform is highly homologous to the hamster, mouse, and rat iPLA2 sequences, all of which also lack the 54-amino acid insert (8–10). This insert is proline-rich, and a BLAST search revealed similarities to the proline-rich middle linker domain of the DAF-3 Smad protein from Caenorhabditis elegans (42), which is most closely related (42) to mammalian Smad4 (43). The proline-rich middle linker region of Smad4 shares a PX5PX8HHPX12NX4Q motif with the corresponding region of DAF-3 and the proline-rich insert in the long human iPLA2 isoform. In Fig. 4, residues that are identical among the three sequences are indicated by dark boxes, and residues with chemically similar side chains are indicated by light boxes. The Smad4 middle linker domain mediates protein interactions with signaling partners (43), is located near the center of the protein, and separates an N-terminal MH1 domain with DNA binding activity from a C-terminal MH2 domain with transcriptional activity (54). The proline-rich insert in the long iPLA2 isoform is also located near the center of the protein and separates an N-terminal domain with protein binding activity from a C-terminal catalytic domain.
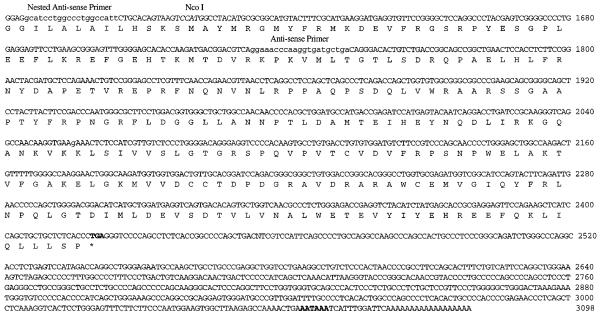
Fig. 3. Nucleotide and deduced amino acid sequences of cDNA species encoding the long and short isoforms of human iPLA2.
Species of cDNA containing the full coding region and the 3′-untranslated region for transcripts encoding LH-iPLA2 and SH-iPLA2 were constructed with cDNA fragments from the INS-C1 and INS-C2 clones and from the long and short fragments obtained from RT-PCRs with human islet RNA, as illustrated schematically in Fig. 1. The resultant cDNA species were subcloned and sequenced. The figure displays the nucleotide (top row) and deduced amino acid sequences (bottom row) for the LH-iPLA2 cDNA. The sequences of the forward primer, the reverse primer, and the nested reverse primer used in the RT-PCRs with human islet RNA are indicated, as is the location of the NcoI restriction endonuclease site. The 162-bp insert that is present in LH-iPLA2 cDNA but absent from SH-iPLA2 cDNA is underlined. The stop codon TGA is indicated by an asterisk. The presumptive polyadenylation signal sequence is indicated in boldface type.
Fig. 4. Comparison of the amino acid sequence of the proline-rich insert in the long form of human iPLA2 to sequences in the proline-rich middle linker domains of the Smad proteins DAF-3 and Smad4.
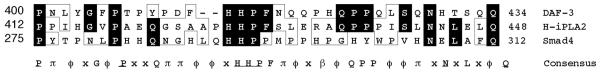
The deduced amino acid sequence between residues 412–448 for the long isoform of human iPLA2 is designated H-iPLA2 in the figure. A BLAST search indicated similarities between this sequence and that of residues 400–434 of the C. elegans DAF-3 Smad protein, which falls within the proline-rich middle linker domain of that protein. DAF-3 is most closely related to the mammalian protein Smad4, and residues 275–312 within the proline-rich middle linker domain of the Smad4 sequence are illustrated in the figure. Amino acid residues that are contained in the iPLA2 sequence and one or both of the other sequences are illustrated by dark boxes, and residues with chemically similar side chains are illustrated by light boxes. In the consensus sequence, residues that are identical among the three sequences are indicated by underlined, capitalized Roman letters. Residues that are common to the iPLA2 sequence and to one but not both of the other sequences are indicated by capitalized Roman letters that are not underlined. Positions at which residues with chemically similar side chains are observed in two or three of the sequences are designated with Greek letters. Acidic residues (Asp and Glu) are denoted by α; basic residues (His, Lys, and Arg) by b; neutral, nonpolar residues (Ala, Phe, Ile, Leu, Met, Pro, Val, and His) by ϕ; and neutral, polar residues (Gly, Asn, Gln, Ser, Thr, and Tyr) by p. Positions at which there is no similarity among the three sequences are denoted with an x.
Fig. 1 summarizes relationships among the human beta cell iPLA2 cDNA fragments obtained from screening the insulinoma cell cDNA library and from RT-PCRs with human islet RNA relative to the predicted sequences for the two full-length iPLA2 isoforms. The 5′-fragments obtained from RT-PCR overlap the 3′-fragments obtained from library screening, and within the region of overlap is a NcoI restriction endonuclease site. There are no other NcoI sites in the sequences. To obtain cDNA species with the full coding sequences for the long (LH-iPLA2) and the short (SH-iPLA2) human islet iPLA2 isoforms, appropriate 5′- and 3′-fragments were digested with NcoI, and ligation reactions were performed. The resultant plasmids were used to transform bacterial host cells and sequenced. The cDNA species so obtained contained the full coding sequences of human iPLA2 isoforms; they were inserted into vectors for expression in bacteria and in Sf9 insect cells and used to prepare 32P-labeled human iPLA2 cDNA to generate a probe for genomic screening.
Bacterial Expression of Recombinant Human Islet iPLA2 Isoforms
To demonstrate that the human islet iPLA2 isoform cDNA species encoded proteins of expected sizes, they were subcloned into expression vector pET-28c, and the resultant constructs were used to transform bacterial host BL21(DE3). Expression of proteins encoded by cDNA inserts was induced by IPTG treatment, and proteins were analyzed by SDS-polyacrylamide gel electrophoresis (Fig. 5). In IPTG-treated cells, proteins of the expected sizes, 85 (lane 2) or 88 (lane 4) kDa, were produced from cDNA for SH-iPLA2 or LH-iPLA2, respectively, in much greater abundance than in non-IPTG-treated cells (lanes 1 and 3). Both proteins were recognized by a polyclonal antibody against rat iPLA2 (not shown).
Fig. 5. Bacterial expression of the long isoform and short isoform human islet iPLA2 proteins.
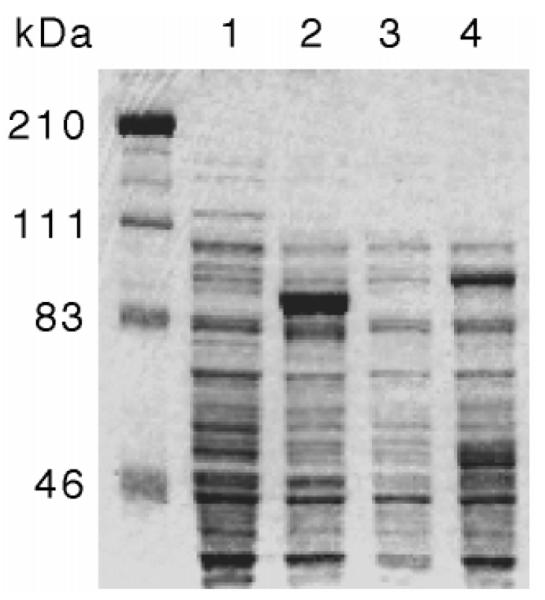
The cDNA species containing the complete coding regions of the short isoform (lanes 1 and 2) or the long isoform (lanes 3 and 4) of human islet iPLA2 were subcloned into the bacterial expression vector pET-28c (Novagen). The pET28-iPLA2 constructs were then used to transform the bacterial expression host BL21(DE3) (Novagen). Expression of proteins encoded by the cDNA inserts was induced by treating the cells with IPTG, and proteins expressed by induced (lanes 2 and 4) and noninduced (lanes 1 and 3) cells were compared by SDS-polyacrylamide gel electrophoresis analyses with Coomassie Blue staining. The expected molecular mass of the short isoform of human islet iPLA2 is 85 kDa (lane 2), and that of the long isoform of human islet iPLA2 is 88 kDa (lane 4).
Eukaryotic Expression of Recombinant Human Islet iPLA2 Isoforms
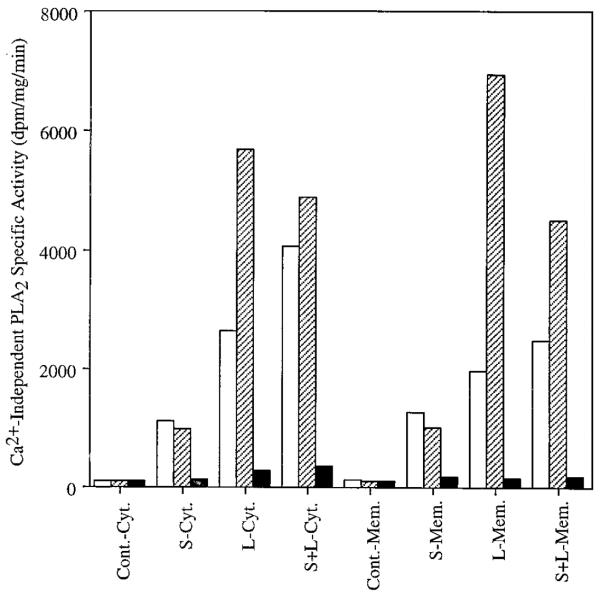
To determine whether human islet iPLA2 isoform cDNA species encoded catalytically active enzymes, a baculovirus vector-Sf9 cell system was used in which other PLA2 enzymes have been expressed (38, 39). Recombinant baculovirus that contained inserts encoding LH-iPLA2 or SH-iPLA2 were used for infection, and subcellular fractions from infected cells were assayed for iPLA2 activity. Uninfected Sf9 cells exhibited no detectable iPLA2 activity, but such activity was observed in cytosolic and membranous fractions of cells infected with baculovirus that contained cDNA inserts encoding LH-iPLA2 or SH-iPLA2 (Fig. 6). The iPLA2 activities expressed in cells infected with baculovirus that contained cDNA encoding either human islet iPLA2 isoform were inhibited by the iPLA2 suicide substrate (4, 8–12) BEL (Fig. 6). We believe this to be the first demonstration that recombinant human iPLA2 is inhibited by BEL, as this issue was not examined in a recent report on human iPLA2 cloned from lymphocyte lines and testis (21). Activities of recombinant LH-iPLA2 and SH-iPLA2 were affected differently by 1 mM ATP (Fig. 6). ATP exerted a stimulatory effect on LH-iPLA2 activities in cytosol or membranes but did not affect SH-iPLA2 activities. ATP has been reported to stimulate iPLA2 activities from rat islets (10) and murine P388D1 cells (9) but not to affect the iPLA2 activity of Chinese hamster ovary cells (8). These findings indicate that cDNA species for both LH-iPLA2 and SH-iPLA2 encode catalytically active enzymes and that catalytic properties of the two human iPLA2 isoforms differ. The experiments also suggest that the ratio of membranous to cytosolic activity may differ somewhat for LH-iPLA2 and SH-iPLA2 under these assay conditions (Fig. 6).
Fig. 6. Catalytic activities of recombinant long and short isoforms of human islet iPLA2 expressed in Sf9 cells.
Recombinant baculovirus that contained cDNA inserts encoding either the long or short isoforms of human islet iPLA2 were prepared and used to infect Sf9 cells. At 48 h after infection, subcellular fractions were prepared from the Sf9 cells and assayed for iPLA2 activity with a radiolabeled phospholipid substrate. Uninfected Sf9 cells exhibited no detectable iPLA2 activity in either cytosol (Cont.-Cyt.) or membranes (Cont.-Mem.). Activity was observed in both cytosolic (S-Cyt. and L-Cyt.) and membranous (S-Mem. and L-Mem.) fractions of cells infected with baculovirus that contained cDNA inserts encoding LH-iPLA2 or SH-iPLA2, and activity was also observed in both subcellular fractions when cells were co-infected with baculovirus mixtures that contained both the LH-iPLA2 and SH-iPLA2 cDNA inserts (S+L-Cyt. and S+L-Mem.). The iPLA2 activities expressed in cells infected with baculovirus that contained cDNA encoding either human islet iPLA2 isoform were susceptible to inhibition by the iPLA2 suicide substrate BEL (filled bars). ATP (1 mM) was included in some assay solutions (hatched bars). Open bars, no ATP.
Characterization of the Human iPLA2 Gene
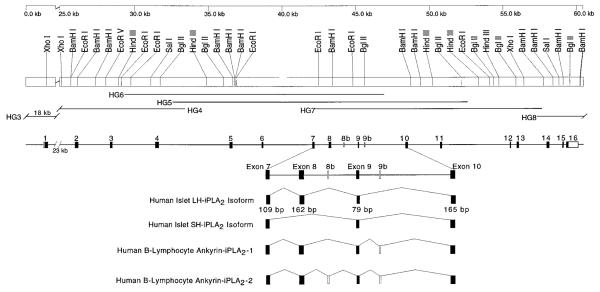
To explore the basis for producing human islet mRNA species that encode the two distinct iPLA2 isoforms, the structure of the human iPLA2 gene was determined. A 32P-labeled LH-iPLA2 cDNA was used as probe to screen a human Lambda FIX II genomic DNA library. Eight genomic clones with overlapping regions of sequence were isolated and analyzed by Southern blotting and restriction endonuclease digestion. Fig. 7 is a schematic representation of the human iPLA2 gene structure. The cloned sequence spans over 60 kb of DNA and includes 16 exons representing 5′-untranslated region, the entire coding sequence, and 3′-untranslated region of the LH-iPLA2 transcript. Intron sizes were estimated from the length of PCR fragments produced by using genomic DNA as template and primers that hybridize with sequences in adjacent exons. The sequences of intron-exon boundaries were determined by comparing the sequences of genomic DNA and cDNA. Table I summarizes the sequences at the 39-acceptor sites and the 5′-donor sites at these boundaries. In each case, the intron sequence at the 5′-boundary of the exon ended in the dinucleotide AG and that at the 3′-boundary of the exon began with the dinucleotide GT, conforming to recognized rules for sequences at such junctions (29).
Fig. 7. Schematic representation of the structure of the human iPLA2 gene and its restriction endonuclease map.
The line at the top of the diagram indicates the scale in kb. There is an interruption in the scale between 0 and 25 kb because of the long length of the first intron. The locations of cleavage sites for restriction endonucleases are illustrated beneath the scale line. Below the summary of endonuclease sites, the lines designated HG6, HG5, HG4, HG7, HG3, and HG8 represent the regions of sequence contained in separate genomic clones (HGn) obtained from screening a human genomic library with 32P-labeled cDNA containing the full coding sequence of the long isoform of human islet iPLA2 and the 3′-untranslated region of its transcript. The genomic clones span over 60 kb of DNA and contain 16 exons represented in the cDNA for the long isoform of human iPLA2 that include 5′-untranslated region, the entire coding region, and 3′-untranslated region. The line below the genomic clone lines represents the locations of exons, which are identified by black rectangles, and the approximate lengths of the intervening introns are indicated by the lengths of the lines between the exons. The dark portions of the rectangles representing the exons indicate coding regions, and the unshaded portions of the rectangles representing exons 1 and 16 indicate untranslated regions. In the lower portion of the diagram, the region of the gene that includes exons 7–10 and the intervening introns is represented on an expanded scale, and the number of base pairs in each exon in this region is indicated. The regions of the gene that are included in four recognized splice-variant products iPLA2 gene are illustrated schematically in the bottom four lines. The human islet LH-iPLA2 isoform transcript contains exons 1–16 but not the alternate exons 8b and 9b. The human islet SH-iPLA2 isoform transcript contains exons 1–7 and 9–16 but not exon 8 or alternate exons 8b or 9b. Two iPLA2 splice variants have been reported by others in human B-lymphocyte lines (21). The transcripts for these variants contain intron sequences that result in premature stop codons and encode truncated forms of iPLA2 that contain the ankyrin repeat domain but lack the catalytic domain. These variants are designated human B-lymphocyte ankyrin-iPLA2-1 and human B-lymphocyte ankyrin-iPLA2-2. The open rectangles reflect the sites of the intron sequences that are contained in these truncation variants, and the location of these intron sequences in the iPLA2 gene are designated by sites 8b and 9b.
Table I.
Exon-intron boundary sequences of the human iPLA2 gene
| 3′ Acceptor site | Exon | 5′ Donor site | |
|---|---|---|---|
| (c/t)11n(c/t)ag: | :gt(a/g)agt | ||
| ..................Exon | 1..................GTGGATTCCG | gtgggtgatg | |
| ggctctacag | ACTCTTCCAG..................Exon | 2..................GTATCATCAG | gtgagcaagg |
| tctcctgcag | CTGTCCAATT..................Exon | 3..................GGTGCTGCAG | gtgagcaggg |
| ctttcatcag | CTCCTTGGAA..................Exon | 4..................CTCAGAAGGG | gtaagacctc |
| ctctcaccag | GTGTGCGGAA..................Exon | 5..................GAACGCAGAG | gtgagtggat |
| tctcccgcag | ATGGCCCGCA..................Exon | 6..................GGCCATGTCG | gtgagcccag |
| tatccaacag | AAAGACAACG..................Exon | 7..................ATCGGCAGAC | gtatgtgctc |
| ccatgacaag | TTGTCACCAG..................Exon | 8..................AACAACCTAG | gtaggcctcg |
| aattttgcag | GCAGTCACCC..................Exon | 8b.................CGTCAGATGG | gtaacgccct |
| tgcctcacag | AACTACAGGA..................Exon | 9..................AGAAGCGGAC | gtaagtggat |
| tgttttacag | ATGCAGAACC..................Exon | 9b.................GTCACACGGA | gtgagtgtca |
| cccactgcag | CCACGACCAC..................Exon | 10.................ATTCTGCACA | gtgagggcgg |
| ttgttcgtag | GTAAGTCCAT..................Exon | 11.................GGAAACCCAA | gtaagccctg |
| ttctctccag | GGTGATGCTG..................Exon | 12.................CAGCCCTCAG | gtttaaacca |
| tggtttacag | ACCAGCTGGT..................Exon | 13.................GATCCGCAAG | gtgagtgccg |
| ttcccaacag | GGTCAGGCCA..................Exon | 14.................GGTGGACTGT | gtgagtgtgg |
| ttctcaccag | TGCACGGATC..................Exon | 15.................AGTACTTCAG | gtgagggctc |
| gcccgaacag | ATTGAACCCC..................Exon | 16.................ATTTGGATTC | |
The sequences of exon-intron boundaries were determined by comparing the sequences of genomic DNA and of human islet LH-iPLA2 cDNA. The table displays the 10 nucleotides at the 5′-end and at the 3′-end of each identified exon in the human iPLA2 gene and that portion of the nucleotide sequences of the immediately adjacent introns that form junctions with the exons. Alternate exons 8b and 9b are not observed in the transcripts for human islet LH-iPLA2 or SH-iPLA2 but are observed in transcripts encoding catalytically inactive, truncated iPLA2 variants in human lymphoma cell lines (21). Exon sequence is represented by upper case and intron sequence by lower case letters.
Alternatively Spliced mRNA Species Encoding Long and Short Isoforms of Human iPLA2
The 54-amino acid insert interrupting the last ankyrin repeat in the LH-iPLA2 isoform corresponds exactly to the amino acid sequence encoded by exon 8 of the human iPLA2 gene. This indicates that mRNA encoding the SH-iPLA2 isoform arises from an exon-skipping mechanism of alternative splicing (29) in transcription of the iPLA2 gene. Different mechanisms of alternative splicing are involved in producing iPLA2 truncation variants observed in human B-lymphocyte lines (21), as illustrated in Fig. 7. The variants contain additional sequence arising from introns that results in premature stop codons, and they encode truncated proteins that contain the ankyrin repeat domain but lack the iPLA2 catalytic domain. The locations within the human iPLA2 gene of intron sequences in the transcripts for the truncation variants were determined from PCR experiments using primers designed from the identified exon sequences and from the published (21) sequences of cDNA species encoding the truncation variants. The truncation variant human B-lymphocyte ankyrin-iPLA2-1 contains sequence from the intron between exons 9 and 10. The truncation variant human B-lymphocyte ankryin-iPLA2-2 contains sequence from two intron regions. The first resides between exons 8 and 9 and the second between exons 9 and 10. The second region of intron sequence occurs in transcripts encoding each of the truncation variants (Fig. 7). Table I indicates the sequences at the intron-exon junctions for these alternate exons.
Chromosomal Localization of Human iPLA2 Gene
To determine the location of the iPLA2 gene on human chromosomes, a clone identified in screening the human genomic DNA library with LH-iPLA2 cDNA was biotinylated to generate a probe for FISH experiments with human lymphocyte chromosomes (40, 41). Using this probe, 91 of 100 examined mitotic figures exhibited fluorescent signals on one pair of chromosomes (Fig. 8), indicating a hybridization efficiency of 91%. The human chromosomes were identified by their DAPI banding patterns (40, 41), and these patterns were correlated with the site of fluorescent signal from biotinylated probe. Such comparisons indicated that the iPLA2 gene resides on human chromosome 22. A detailed positional assignment achieved from analyses of 10 photographs indicated that the iPLA2 gene resides in region q13.1 of chromosome 22 (Fig. 8). No other loci of hybridization of the probe were observed.
Fig. 8. Localization of the human iPLA2 gene to chromosome 22q13.1 by fluorescence in situ hybridization.
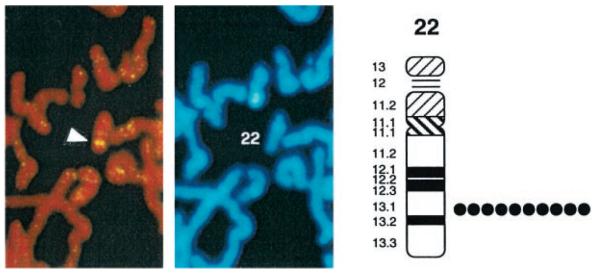
A genomic clone identified in screening the human genomic DNA library with the iPLA2 cDNA was biotinylated to generate a probe for FISH experiments with human lymphocyte chromosomes. Using this probe, 91 of 100 examined mitotic figures exhibited fluorescent signals on one pair of chromosomes. The white arrowhead in the left panel indicates the location of the two intense fluorescent spots reflecting hybridization of the probe with each member of the chromosome pair. The center panel illustrates the DAPI staining pattern of the same mitotic figure and identifies the chromosome as number 22. The diagram in the right panel illustrates regions of human chromosome 22 determined by DAPI banding patterns. These patterns were correlated with the site of fluorescent signal from the biotinylated iPLA2 genomic clone. A detailed positional assignment was achieved from analyses of 10 photographs from different preparations. The black circles indicate the location of the probe observed in each of the 10 experiments. In each case, the probe localized to region q13.1 of human chromosome 22. No other loci of hybridization of the biotinylated probe were observed.
DISCUSSION
Our results indicate that human pancreatic islets express mRNA species encoding two distinct, catalytically active isoforms of iPLA2 that are distinguishable by size and by their susceptibility to activation by ATP. These two mRNA species are also observed in human U937 promonocytic cells. These two human iPLA2 isoforms differ only by the presence of a 54-amino acid insert in the longer isoform that is absent from the shorter isoform, and the deduced amino acid sequences of the two isoforms are otherwise identical. This insert is encoded in its entirety by exon 8 of the human iPLA2 gene, which resides in region q13.1 of human chromosome 22, indicating that the mRNA species encoding the short iPLA2 isoform arises from an exon-skipping mechanism of alternative splicing. The mRNA encoding the short iPLA2 isoform has not previously been identified in human cells, and we believe our studies are the first to demonstrate expression of iPLA2 mRNA by human islets or U937 cells and to examine effects of BEL and ATP on activities of recombinant human iPLA2 enzymes.
The demonstration that both human islets and U937 cells express iPLA2 mRNA species that encode BEL-sensitive enzymes is of interest in the context of recent reports suggesting that iPLA2 may participate in apoptosis in both U937 cells (20) and in islet beta cells (26). U937 cells express the protein Fas on their surfaces (20), and ligation of Fas molecules with the protein Fas ligand or with agonistic anti-Fas antibodies induces a cell death program that involves activation of caspase intracellular proteases (44–46). U937 cells also express a PLA2 activity distinct from cPLA2 or sPLA2 that does not require Ca2+ and exhibits a profile of sensitivity to inhibitors such as BEL and methyl arachidonylfluorophosphate that is similar to that of iPLA2 (20). Our demonstration that U937 cells express iPLA2 mRNA indicates that this PLA2 activity may reside in the iPLA2 protein. Anti-Fas antibodies induce U937 cell apoptosis and hydrolysis of arachidonic acid from cell membranes (20). During this process, cPLA2 is proteolytically inactivated by caspases, but iPLA2 activity is preserved (20). Inhibitors of iPLA2 both suppress Fas antibody-induced arachidonate release from U937 cells and retard cell death (20), suggesting that iPLA2 may participate in apoptosis.
Similarly, stimuli that induce Ca2+ store depletion in islet beta cells induce apoptosis by a mechanism that does not require a rise in cytosolic [Ca2+] but that does require hydrolysis of arachidonic acid from membrane phospholipids and its conversion to 12-lipoxygenase metabolites (26). Ca2+ store depletion-induced hydrolysis of arachidonic acid from islet phospholipids also does not require a rise in cytosolic [Ca2+] and is mediated by a BEL-sensitive phospholipase (47), such as iPLA2. Although phosphatidate phosphohydrolase is also inhibited by BEL (48), the phosphatidate phosphohydrolase inhibitor propranolol (49) does not block Ca2+ store depletion-induced release of arachidonate from islet phospholipids (47), suggesting that iPLA2 may mediate this phenomenon. Inter-leukin-1 also induces accumulation of non-esterified arachidonate and its 12-lipoxygenase products in islets by a BEL-sensitive mechanism (50), and interleukin-1 induces apoptosis of human islet beta cells through Fas-mediated events (51). In the context of these observations, our findings that human islets express iPLA2 mRNA raise the possibility that iPLA2 might participate in Fas-mediated apoptosis in human beta cells in a manner similar to that in U937 cells (20). Beta cell apoptosis is thought to contribute to development of diabetes mellitus (52).
The amino acid sequence of the insert that distinguishes the long and short isoforms of human iPLA2 is of interest in the context of the potential involvement of iPLA2 in apoptosis. This insert shares a PX5PX8HHPX12NX4Q consensus motif with the proline-rich middle linker domains of the C. elegans Smad protein DAF-3 (42) and the mammalian protein Smad4 (43). Smad proteins participate in controlling cell proliferation and apoptosis and form hetero-oligomers with signaling partners (54), via the proline-rich middle linker domain in the case of Smad 4 (43). Smad4 and Smad2 are products of tumor-suppressor genes that are deleted or mutated in some human carcinomas (54–58). Studies of allelic losses in human breast and head and neck carcinomas indicate that a tumor suppressor gene(s) resides on human chromosome 22q13.1 (59, 60), which is the chromosomal location of the human iPLA2 gene. If iPLA2 does participate in apoptosis (20, 26), it might exert tumor suppressor activity.
The active form of iPLA2 appears to be an oligomer of interacting protein subunits. Radiation inactivation studies of iPLA2 activity in crude cytosol indicate a size of 337 kDa for the active complex (22).The iPLA2 activity in crude cytosol also migrates with an apparent molecular mass of 250–350 kDa on gel filtration chromatography (8, 22, 61), and this is also the case for the iPLA2 activity of the purified 85-kDa iPLA2 protein (8). This has been taken to suggest that the active form of iPLA2 is an oligomer of 85-kDa subunits and that the subunits may associate with each other via their ankyrin repeat regions (8) because such ankyrin repeats participate in many other protein-protein interactions (53). Consistent with this possibility is the observation that iPLA2 deletion mutants lacking the ankyrin repeat domain but retaining the catalytic domain are catalytically inactive (8). In the long isoform of human iPLA2, the last iPLA2 ankyrin repeat is interrupted by a proline-rich insert with some similarities to the Smad4 domain that mediates interactions with signaling partners (43). This raises the possibility that the proline-rich insert in human iPLA2 might allow it to interact with proteins not recognized by the short isoform of iPLA2.
That the long isoform of human iPLA2 can form heterooligomers with altered catalytic properties is suggested by the finding that activity of this protein is reduced when it is co-expressed with truncated iPLA2-like proteins that retain the ankyrin repeat domain but lack the catalytic domain (21). These truncated iPLA2 variants arise from alternatively spliced transcripts that contain intron sequences that result in a premature stop codon, and these transcripts are expressed in human lymphoma cell lines (21). These lymphoma cell lines do not express mRNA species encoding the catalytically active short isoform of iPLA2 observed in human islets that arises from another mechanism of alternative splicing of the transcript from the iPLA2 gene. Human islets do not express mRNA species encoding the truncated iPLA2 variants observed in human lymphoma cells. In contrast, human U937 promonocytic cells express mRNA species encoding both the long and short isoforms of catalytically active iPLA2 and also express mRNA species encoding an iPLA2 truncation variant. This indicates that there is heterogeneity among human cells in expression of iPLA2 gene products that arise from alternative splicing.
The presence of two distinct domains that might mediate protein-protein interactions in the long isoform of human iPLA2 could cause it to interact with a variety of other proteins. Various participants in hetero-oligomeric complexes with iPLA2 have been suggested to alter iPLA2 catalytic properties (21, 47, 61–64). These include calmodulin (63), which physically interacts with and negatively modulates the activity of recombinant 85-kDa iPLA2 cloned from Chinese hamster ovary cells (64) and rat islets (47). This has been offered as one explanation of why Ca2+ store depletion activates iPLA2 (65). Although the mechanism underlying this effect may be complex, the effect itself has occurs in vascular myocytes (64), pancreatic islet beta cells (47), and human granulocytes (17). Ca2+ store depletion also activates hydrolysis of arachidonate from phospholipids in differentiated human U937 promonocytic cells by a mechanism that does not require a rise in cytosolic [Ca2+] (31). Our demonstration that U937 cells express mRNA species encoding iPLA2 isoforms suggest that iPLA2 is one candidate for mediating Ca2+ store depletion-induced arachidonate release in those cells. The differences in products of the iPLA2 gene expressed in specific human cells suggest that iPLA2 regulation might be complex, as also indicated by the fact that stimuli that induce iPLA2-catalyzed arachidonate release and leukotriene production in granulocytes fail to induce these events in lymphocytes, even though both classes of cells express iPLA2 and leukotriene biosynthetic enzymes (17, 21).
Acknowledgments
We thank Alan Bohrer, Sheng Zhang, and Dr. Mary Wohltmann for excellent technical assistance, Dr. Alan Permutt for the human insulinoma cDNA library, and Dr. Simon Jones of Genetics Institute and the members of his laboratory group for advice and encouragement at the outset of these studies.
Footnotes
This research was supported by National Institutes of Health Grant R37-DK-34388. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This paper is available on line at http://www.jbc.org
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AF102988, AF102989, and AF116252 through AF116267.
The abbreviations used are: BEL, bromoenol lactone; bp, base pair(s); DAPI, 4′, 6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridization; IPTG, isopropyl-1-thio-β-d-galactopyranoside; kb, kilobase pair(s); PCR, polymerase chain reaction; RT, reverse transcription; Sf9, Spodoptera frugiperda, type 9; PLA2, phospholipase A2; cPLA2, Group IV PLA2; iPLA2, Group VI PLA2; sPLA2, secretory PLA2; LH-iPLA2, long isoform of human iPLA2; SH-iPLA2, short isoform of human iPLA2.
REFERENCES
- 1.Dennis EA. J. Biol. Chem. 1994;269:13057–13060. [PubMed] [Google Scholar]
- 2.Gijon MA, Leslie CC. Cell Dev. Biol. 1997;8:297–303. doi: 10.1006/scdb.1997.0151. [DOI] [PubMed] [Google Scholar]
- 3.Dennis EA. Trends Biochem. Sci. 1997;22:1–2. doi: 10.1016/s0968-0004(96)20031-3. [DOI] [PubMed] [Google Scholar]
- 4.Balsinde J, Dennis EA. J. Biol. Chem. 1997;272:16069–16072. doi: 10.1074/jbc.272.26.16069. [DOI] [PubMed] [Google Scholar]
- 5.Leslie CC. J. Biol. Chem. 1997;272:16709–16712. doi: 10.1074/jbc.272.27.16709. [DOI] [PubMed] [Google Scholar]
- 6.Tischfield JA. J. Biol. Chem. 1997;272:17247–17250. doi: 10.1074/jbc.272.28.17247. [DOI] [PubMed] [Google Scholar]
- 7.Stafforini DM, McIntyre TM, Zimmerman GA, Prescott SM. J. Biol. Chem. 1997;272:17895–17898. doi: 10.1074/jbc.272.29.17895. [DOI] [PubMed] [Google Scholar]
- 8.Tang J, Kriz RW, Wolfman N, Shaffer M, Seehra J, Jones S. J. Biol. Chem. 1997;272:8567–8575. doi: 10.1074/jbc.272.13.8567. [DOI] [PubMed] [Google Scholar]
- 9.Balboa MA, Balsinde J, Jones SS, Dennis EA. J. Biol. Chem. 1997;272:8576–8580. doi: 10.1074/jbc.272.13.8576. [DOI] [PubMed] [Google Scholar]
- 10.Ma Z, Ramanadham S, Kempe K, Chi XS, Ladenson J, Turk J. J. Biol. Chem. 1997;272:11118–11127. [PubMed] [Google Scholar]
- 11.Hazen SL, Zupan LA, Weiss RH, Getman DP, Gross RW. J. Biol. Chem. 1991;266:7227–7232. [PubMed] [Google Scholar]
- 12.Zupan LA, Weiss RH, Hazen S, Parnas BL, Aston KW, Lennon PJ, Getman DP, Gross RW. J. Med. Chem. 1993;36:95–100. doi: 10.1021/jm00053a012. [DOI] [PubMed] [Google Scholar]
- 13.Balsinde J, Dennis EA. J. Biol. Chem. 1996;271:6758–6765. doi: 10.1074/jbc.271.12.6758. [DOI] [PubMed] [Google Scholar]
- 14.Ma Z, Ramanadham S, Hu Z, Turk J. Biochim. Biophys. Acta. 1998;1391:384–400. doi: 10.1016/s0005-2760(98)00027-7. [DOI] [PubMed] [Google Scholar]
- 15.Balsinde J, Bianco ID, Ackerman EJ, Conde-Friebos K, Dennis EA. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8527–8531. doi: 10.1073/pnas.92.18.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balsinde J, Balboa MA, Dennis EA. J. Biol. Chem. 1997;272:29317–29321. doi: 10.1074/jbc.272.46.29317. [DOI] [PubMed] [Google Scholar]
- 17.Larsson Forsell PKA, Runarsson G, Ibrahim M, Bjorkholm M, Claesson H-E. FEBS Lett. 1998;434:295–299. doi: 10.1016/s0014-5793(98)00999-5. [DOI] [PubMed] [Google Scholar]
- 18.Gubitosi-Klug RA, Yu SP, Choi DW, Gross RW. J. Biol. Chem. 1995;270:2885–2888. doi: 10.1074/jbc.270.7.2885. [DOI] [PubMed] [Google Scholar]
- 19.Eddlestone GT. Am. J. Physiol. 1995;268:C181–C190. doi: 10.1152/ajpcell.1995.268.1.C181. [DOI] [PubMed] [Google Scholar]
- 20.Atsumi G, Tajima M, Hadano A, Nakatani Y, Murakami M, Kudo I. J. Biol. Chem. 1998;273:13870–13877. doi: 10.1074/jbc.273.22.13870. [DOI] [PubMed] [Google Scholar]
- 21.Larsson PKA, Claesson H-E, Kennedy BP. J. Biol. Chem. 1998;273:207–214. doi: 10.1074/jbc.273.1.207. [DOI] [PubMed] [Google Scholar]
- 22.Ackerman EJ, Kempner ES, Dennis EA. J. Biol. Chem. 1994;269:9227–9233. [PubMed] [Google Scholar]
- 23.Ramanadham S, Gross RW, Han X, Turk J. Biochemistry. 1993;32:337–346. doi: 10.1021/bi00052a042. [DOI] [PubMed] [Google Scholar]
- 24.Ramanadham S, Bohrer A, Mueller M, Jett P, Gross R, Turk J. Biochemistry. 1993;32:5339–5351. doi: 10.1021/bi00071a009. [DOI] [PubMed] [Google Scholar]
- 25.Ramanadham S, Wolf MJ, Jett PA, Gross RW, Turk J. Biochemistry. 1994;33:7442–7452. doi: 10.1021/bi00189a052. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y-P, Teng D, Drayluk F, Ostrega D, Roe MW, Philipson L, Polonsky KS. J. Clin. Invest. 1998;101:1623–1632. doi: 10.1172/JCI1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross RW, Ramanadham S, Kruszka K, Han X, Turk J. Biochemistry. 1993;32:327–336. doi: 10.1021/bi00052a041. [DOI] [PubMed] [Google Scholar]
- 28.Ramanadham S, Bohrer A, Gross RW, Turk J. Biochemistry. 1993;32:13499–13509. doi: 10.1021/bi00212a015. [DOI] [PubMed] [Google Scholar]
- 29.McKeown M. Annu. Rev. Cell Biol. 1992;8:133–155. doi: 10.1146/annurev.cb.08.110192.001025. [DOI] [PubMed] [Google Scholar]
- 30.Sundstrom C, Nilsson K. Int. J. Cancer. 1976;17:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 31.Rzigalinsky BA, Blackmore PR, Rosenthal MD. Biochim. Biophys. Acta. 1996;1299:342–352. doi: 10.1016/0005-2760(95)00224-3. [DOI] [PubMed] [Google Scholar]
- 32.Ferrer J, Wasson J, Permutt A. Diabetologia. 1996;38:891–898. doi: 10.1007/BF00403907. [DOI] [PubMed] [Google Scholar]
- 33.Sanger F, Nickeln S, Coulson AR. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricordi C, Lacy PE, Finke EH, Olack BJ, Scharp DW. Diabetes. 1988;37:413–420. doi: 10.2337/diab.37.4.413. [DOI] [PubMed] [Google Scholar]
- 35.Ramanadham S, Hsu F-F, Bohrer A, Nowatzke W, Ma Z, Turk J. Biochemistry. 1998;37:4553–4567. doi: 10.1021/bi9722507. [DOI] [PubMed] [Google Scholar]
- 36.Davis LG, Kuehl WM, Battery JF. In: Basic Methods in Molecular Biology. 2nd Ed. Wonsiewicz M, Greenfield S, editors. Appleton and Lange; East Norwalk, CT: 1994. pp. 335–338. [Google Scholar]
- 37.Becker GW, Miller JR, Kovacevic S, Ellis R, Louis AI, Small JS, Stark DH, Roberts EF, Wyrick TK, Hoskins J, Chious G, Sharp JD, McClure DB, Rioggin RM, Kramer RM. Bio/Technology. 1994;12:69–74. doi: 10.1038/nbt0194-69. [DOI] [PubMed] [Google Scholar]
- 38.de Carvalho MS, McCormack AL, Olsen E, Ghomaschchi F, Gelb MH, Yates JR, III, Leslie CC. J. Biol. Chem. 1996;271:6987–6997. doi: 10.1074/jbc.271.12.6987. [DOI] [PubMed] [Google Scholar]
- 39.O'Reilly DR, Miller LK, Luckow VA. Baculovirus Expression Vectors: A Laboratory Manual. W. H. Freeman and Co.; New York: 1992. [Google Scholar]
- 40.Heng HHQ, Squire J, Tsui L-C. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9509–9513. doi: 10.1073/pnas.89.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heng HHQ, Tsui L-C. Chromosoma. 1993;102:325–332. doi: 10.1007/BF00661275. [DOI] [PubMed] [Google Scholar]
- 42.Patterson GI, Koweek A, Wong A, Liu Y, Parkin G. Genes Dev. 1997;11:2679–2690. doi: 10.1101/gad.11.20.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Caestecker MP, Hemmati P, Larisch-Bloch S, Ajmera R, Roberts AB, Lechleider RJ. J. Biol. Chem. 1998;272:13690–13696. doi: 10.1074/jbc.272.21.13690. [DOI] [PubMed] [Google Scholar]
- 44.Nagata S, Golstein P. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 45.Cleveland JL, Ihle JN. Cell. 1995;81:479–482. doi: 10.1016/0092-8674(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 46.Longthorne VL, Williams GT. EMBO J. 1997;16:3805–3812. doi: 10.1093/emboj/16.13.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowatzke W, Ramanadham S, Ma Z, Hsu F-F, Bohrer A, Turk J. Endocrinology. 1998;139:4073–4085. doi: 10.1210/endo.139.10.6225. [DOI] [PubMed] [Google Scholar]
- 48.Balsinde J, Dennis EA. J. Biol. Chem. 1998;271:31937–31941. doi: 10.1074/jbc.271.50.31937. [DOI] [PubMed] [Google Scholar]
- 49.Balboa MA, Balsinde J, Dennis EA. J. Biol. Chem. 1998;273:7684–7690. doi: 10.1074/jbc.273.13.7684. [DOI] [PubMed] [Google Scholar]
- 50.Ma Z, Ramanadham S, Corbett JA, Bohrer A, Gross RW, McDaniel ML, Turk J. J. Biol. Chem. 1996;271:1029–1042. doi: 10.1074/jbc.271.2.1029. [DOI] [PubMed] [Google Scholar]
- 51.Loweth AC, Williams GT, James RFL, Scarpello JHB, Morgan NG. Diabetes. 1998;47:727–732. doi: 10.2337/diabetes.47.5.727. [DOI] [PubMed] [Google Scholar]
- 52.Pick A, Clark J, Kubstrap C, Levisetti M, Pugh W, Bonner-Weir S, Polonsky KS. Diabetes. 1998;47:358–364. doi: 10.2337/diabetes.47.3.358. [DOI] [PubMed] [Google Scholar]
- 53.Bennett V. J. Biol. Chem. 1992;267:8703–8706. [PubMed] [Google Scholar]
- 54.Kretzschmer M, Massague J. Curr. Opin. Genet. Dev. 1998;8:103–111. doi: 10.1016/s0959-437x(98)80069-5. [DOI] [PubMed] [Google Scholar]
- 55.de Winter JP, Roelen BAJ, ten Dijke P, van der Burg B, van den Eijnden-van Aaij AJM. Oncogene. 1997;14:1891–1899. doi: 10.1038/sj.onc.1201017. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Musci T, Derynck R. Curr. Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 57.Attisano L, Wrana JL. Curr. Opin. Cell Biol. 1998;10:188–194. doi: 10.1016/s0955-0674(98)80141-5. [DOI] [PubMed] [Google Scholar]
- 58.Padgett RW, Das P, Krishna S. BioEssays. 1998;20:382–391. doi: 10.1002/(SICI)1521-1878(199805)20:5<382::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 59.Iida A, Kurose K, Isobe R, Akiyama F, Sakamoto G, Yoshimoto M, Kasumi F, Nakamura Y, Emi M. Genes Chromosomes Cancer. 1998;21:108–112. [PubMed] [Google Scholar]
- 60.Miyakawa A, Wang X-L, Nakanishi H, Imai FL, Shiiba M, Miya T, Tanzawa H. Int. J. Oncol. 1998;13:705–709. doi: 10.3892/ijo.13.4.705. [DOI] [PubMed] [Google Scholar]
- 61.Ramanadham S, Wolf MJ, Ma Z, Li B, Wang J, Gross RW, Turk J. Biochemistry. 1996;35:5464–5471. doi: 10.1021/bi952652j. [DOI] [PubMed] [Google Scholar]
- 62.Hazen SL, Gross RW. J. Biol. Chem. 1993;268:9892–9900. [PubMed] [Google Scholar]
- 63.Wolf MJ, Gross RW. J. Biol. Chem. 1996;271:20989–20992. doi: 10.1074/jbc.271.35.20989. [DOI] [PubMed] [Google Scholar]
- 64.Wolf MJ, Gross RW. J. Biol. Chem. 1996;271:30879–30885. doi: 10.1074/jbc.271.48.30879. [DOI] [PubMed] [Google Scholar]
- 65.Wolf MJ, Wang J, Turk J, Gross RW. J. Biol. Chem. 1997;272:1522–1526. doi: 10.1074/jbc.272.3.1522. [DOI] [PubMed] [Google Scholar]
- 66.Gilliland G, Perrin S, Blanchard K, Bunn HF. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siebert PD, Larrick JW. Nature. 1992;359:557–558. doi: 10.1038/359557a0. [DOI] [PubMed] [Google Scholar]