LETTER
A 35-year-old patient with a remote history of Hodgkin's lymphoma and a postradiation and postchemotherapy status presented to our institution in January 2013 with a 4-day history of dyspnea, productive cough, fever, and chills that began while he was traveling abroad. Upon admission, the patient demonstrated persistent hypoxemia, requiring emergent intubation. Due to acute respiratory distress and septic shock, empirical treatment with vancomycin, cefepime, levofloxacin, and oseltamivir was initiated. Laboratory testing included Legionella and Streptococcus pneumoniae urine antigen testing, PCR studies for Pneumocystis jirovecii, adenovirus, and Legionella species, viral cultures of bronchoalveolar lavage (BAL) fluid, and bacterial cultures of his blood, urine, sputum, and stool. All laboratory results were negative. In addition, real-time PCR for influenza A/B and respiratory syncytial virus (RSV) (Prodesse ProFlu+; Hologic/Gen-Probe, Waukesha, WI) was performed on a nasopharyngeal (NP) swab and BAL fluid, and results were negative.
Four days following admission, viral cell culture of BAL fluid was reported as positive for influenza A virus, which was determined by rapid antigen testing (BinaxNOW Influenza A&B; Alere, Waltham, MA) of the culture supernatant. The BAL fluid was subsequently tested by four additional influenza real-time PCR assays, including the FilmArray respiratory panel (BioFire, Salt Lake City, UT), Focus Simplexa Flu A/B and RSV kit (Quest Diagnostics, Cypress, CA), Prodesse ProFAST+ kit (Hologic/Gen-Probe), and a laboratory-developed test (LDT) (1, 5), all of which were positive for influenza A virus. Furthermore, the FilmArray assay, ProFAST+ assay, and LDT typed the virus as influenza A 2009/H1N1. The strain type was confirmed by the Minnesota Department of Health (MDH), and pyrosequencing analysis for the detection of the H275Y oseltamivir resistance mutation at the MDH showed results consistent with susceptibility to oseltamivir. Repeat testing of the NP swab and BAL fluid and testing of the culture isolate by the Prodesse ProFlu+ assay yielded negative results.
Due to these findings, we speculated that the virus may have acquired mutations in the matrix (M) gene, impacting the ability of the ProFlu+ assay to detect this strain. Therefore, sequencing studies of the culture isolate were performed by the manufacturer (Hologic/Gen-Probe), which identified 2 point mutations in the region of the M gene targeted by the ProFlu+ probe; however, the sequence analysis demonstrated that both primer regions were 100% conserved (see the supplemental material; primer/probe regions were not identified). These findings were supported by an analysis of the PCR amplicon, which showed an amplified product of the expected size by agarose gel electrophoresis but no fluorescent signal in a real-time PCR (Fig. 1).
Fig 1.
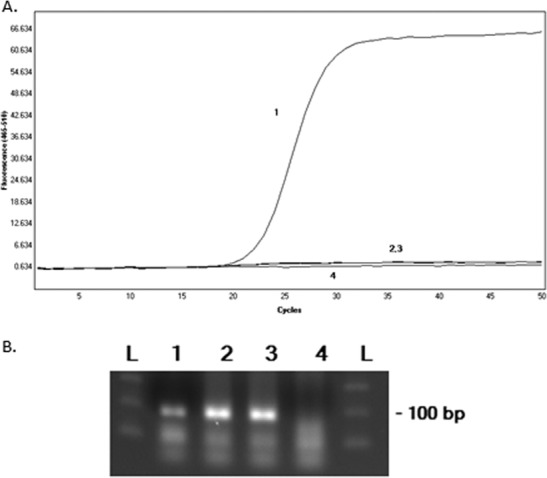
Analysis of an influenza A virus 2009/H1N1 strain with point mutations in the matrix gene by real-time PCR (A) and agarose gel electrophoresis (B). The positive control (line and lane 1), patient culture isolate (lines and lanes 2 and 3), and negative control (line and lane 4) were tested by the Prodesse ProFlu+ real-time PCR assay. The PCR product was analyzed in real time for fluorescent output (A) and by agarose gel electrophoresis with ethidium bromide staining (B). Lanes L, molecular weight ladders.
The Prodesse ProFlu+ assay is FDA approved for the detection of influenza A/B virus and RSV and, as indicated above, targets the M gene of influenza virus using TaqMan-based real-time PCR chemistry. Prior studies have demonstrated that ProFlu+ exhibits high overall percent agreement (>97%) with other commercially available influenza PCR methods and reliably detects the novel influenza A H1N1 virus that was predominant in 2009 (3, 4). Interestingly, the Prodesse ProFAST+ assay, used to subtype influenza A virus, targets the hemagglutinin (HA) gene, which explains the different detection capabilities of ProFlu+ and ProFAST+ in this case. Historically, it has been believed that the majority of the genetic diversity of influenza viruses is attributable largely to point mutations in the HA and neuraminidase (NA) genes (i.e., antigenic drift) or to genetic reassortment of the RNA genome (i.e., antigenic shift) (5). However, this case highlights the fact that point mutations also occur in the more highly conserved M gene, and this may impact the detection of certain strains of influenza virus.
In summary, this report underscores the potential for genetic variation among influenza viruses. While the prevalence of this particular strain of 2009/H1N1 may currently be rare, our findings corroborate those of a prior report showing missed detection of an influenza A 2009/H1N1 strain by the ProFlu+ assay (2). Together, these cases highlight the possibility of false-negative results due to mutations in targeted regions of molecular assays. Clinical laboratories should be aware that strains of influenza A virus with point mutations within the M gene do circulate among the population and that molecular assays, such as real-time PCR, may miss detecting them. In the future, molecular methods targeting multiple gene targets within the influenza virus genome (e.g., the matrix and HA/NA genes) may allow for more reliable detection of strains undergoing genetic variation. Finally, health care providers should interpret laboratory results for influenza in the context of a patient's clinical presentation, as the inherent variability of influenza virus strains makes the detection of this virus an ongoing challenge.
Nucleotide sequence accession number.
The sequence of our patient's virus was submitted to GenBank under accession no. KC809971.
Supplementary Material
ACKNOWLEDGMENT
Sequencing studies were performed by Hologic/Gen-Probe.
Footnotes
Published ahead of print 3 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00446-13.
REFERENCES
- 1. Dhiman N, Espy MJ, Irish C, Wright P, Smith TF, Pritt BS. 2010. Mutability in the matrix gene of novel influenza A H1N1 virus detected using a FRET probe-based real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 48:677–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng X, Todd KM, Yen-Lieberman B, Kaul K, Mangold K, Shulman ST. 2010. Unique finding of a 2009 H1N1 influenza virus-positive clinical sample suggests matrix gene sequence variation. J. Clin. Microbiol. 48:665–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Novak-Weekly SM, Marlowe EM, Poulter M, Dwyer D, Speers D, Rawlinson W, Baleriola C, Robinson CC. 2012. Evaluation of the Cepheid Xpert Flu assay for rapid identification and differentiation of influenza A, influenza A 2009 H1N1, and influenza B viruses. J. Clin. Microbiol. 50:1704–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tang Y, Lowery KS, Valsamakis A, Schaefer VC, Chappell JD, White-Abell J, Quinn CD, Li H, Washington CA, Cromwell J, Ciamanco CM, Forman M, Holden J, Rothman RE, Parker ML, Ortenberg EV, Zhang L, Lin Y, Gaydos CA. 2013. Clinical accuracy of a PLEX-ID flu device for simultaneous detection and identification of influenza viruses A and B. J. Clin. Microbiol. 51:40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zitterkopf NL, Leekha S, Espy MJ, Wood CM, Sampathkumar P, Smith TF. 2006. Relevance of influenza A virus detection by PCR, shell vial assay, and tube cell culture to rapid reporting procedures. J. Clin. Microbiol. 44:3366–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


