Abstract
This study compared three sample preparation methods (direct transfer, the direct transfer-formic acid method with on-target formic acid treatment, and ethanol-formic acid extraction) for the identification of Gram-positive cocci with matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). A total of 156 Gram-positive cocci representing the clinically most important genera, Aerococcus, Enterococcus, Staphylococcus, and Streptococcus, as well as more rare genera, such as Gemella and Granulicatella, were analyzed using a Bruker MALDI Biotyper. The rate of correct genus-level identifications was approximately 99% for all three sample preparation methods. The species identification rate was significantly higher for the direct transfer-formic acid method and ethanol-formic acid extraction (both 77.6%) than for direct transfer (64.1%). Using direct transfer-formic acid compared to direct transfer, the total time to result was increased by 22.6%, 16.4%, and 8.5% analyzing 12, 48, and 96 samples per run, respectively. In a subsequent prospective study, 1,619 clinical isolates of Gram-positive cocci were analyzed under routine conditions by MALDI-TOF MS, using the direct transfer-formic acid preparation, and by conventional biochemical methods. For 95.6% of the isolates, a congruence between conventional and MALDI-TOF MS identification was observed. Two major limitations were found using MALDI-TOF MS: the differentiation of members of the Streptococcus mitis group and the identification of Streptococcus dysgalactiae. The Bruker MALDI Biotyper system using the direct transfer-formic acid sample preparation method was shown to be a highly reliable tool for the identification of Gram-positive cocci. We here suggest a practical algorithm for the clinical laboratory combining MALDI-TOF MS with phenotypic and molecular methods.
INTRODUCTION
Gram-positive cocci are among the most frequently isolated bacterial species in clinical diagnostic laboratories. Their identification is currently based on conventional, i.e., morphological and biochemical, methods (1). In addition, molecular methods, such as sequence analysis of the 16S rRNA gene and other housekeeping genes, are applied for identification (1, 2). This variety of methods allows a reliable identification of Gram-positive cocci. However, biochemical and molecular methods are time-consuming and cost- and labor-intensive, and molecular techniques are not routinely available in many diagnostic laboratories.
Recently, several studies evaluated the performance of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for the identification of bacteria in the diagnostic laboratory (3–8). MALDI-TOF MS promises to be a fast, reliable, and cost-effective alternative to currently used methods (4, 6, 8). Apart from the species coverage of the reference database and the identification algorithm of the software (9, 10), the sample preparation procedure seems to be critical for the performance of the system (11–13). Most studies evaluating the Bruker MALDI-TOF MS system used a direct transfer protocol, measuring whole bacterial cells prepared directly from freshly grown colonies. When no identification was achieved with this method, a chemical extraction method was performed, where the cells were suspended in water and ethanol, and the bacterial proteins were subsequently extracted with formic acid and acetonitrile (14). Sample preparation using the extraction protocol significantly increases the yield of successful identifications compared to the direct transfer method (3, 5, 6, 11, 15). However, the extraction method significantly increases the processing time (14) and seems not suitable for high-throughput applications as required in clinical diagnostic laboratories. An adapted direct transfer method with on-target formic acid treatment leading to in situ cell lysis promises an increased identification rate without the time-consuming extraction method (10, 12). To date, few studies have systematically compared different sample preparation methods for the identification of bacteria with MALDI-TOF MS (11, 15).
This study was organized in two parts. In a retrospective study (part one), we compared three sample preparation methods, namely, direct transfer, the direct transfer-formic acid method, and ethanol-formic acid extraction, for the identification of Gram-positive cocci with the Bruker MALDI Biotyper system (Bruker Daltonik) using the clinical strain collection of our institution (n = 156). We focused on evaluating rates of identification and preparation time. In a prospective study (part two), we selected the direct transfer-formic acid method based on data from part one for comparison with the conventional routine identification algorithms of our laboratory for clinically relevant isolates of Gram-positive cocci (n = 1,619) (1). From this comparison, we derived a practical algorithm to integrate MALDI-TOF MS in the routine diagnostic laboratory. We summarize reliable species and genus identifications for Gram-positive cocci and suggest options to resolve problematic identifications.
MATERIALS AND METHODS
Bacterial isolates and culture conditions.
For the retrospective part of the study, we selected 156 Gram-positive cocci of the clinically most relevant genera from the strain collection of our institute, i.e., aerococci (n = 17 isolates), enterococci (n = 16 isolates), staphylococci (n = 32 isolates), and streptococci (n = 75 isolates), as well as more rarely encountered genera (n = 16 isolates), including Abiotrophia, Gemella, Granulicatella, Kocuria, Lactococcus, Leuconostoc, and Rothia (see Table S1 in the supplemental material). The isolates were characterized by conventional methods and 16S rRNA gene analysis. Pneumococci were, in addition, identified by recA gene analysis. During the 4-month prospective study period from November 2011 to February 2012, 1,619 clinical Gram-positive cocci isolates were identified in parallel by conventional methods and MALDI-TOF MS. Bacteria were routinely cultivated on Columbia agar containing 5% sheep blood (bioMérieux, Marcy l'Etoile, France) at 37°C with 7.5% CO2 for 24 to 48 h for MALDI-TOF MS measurement. Granulicatella adiacens was grown on chocolate agar with PolyViteX (bioMérieux) under anaerobic conditions.
Phenotypic identification.
Bacterial isolates were characterized by a combination of conventional assays (e.g., Gram staining, testing of catalase, coagulase or hemolytic activity) (1) and the commercially available identification systems Vitek 2 (GP ID card) and API 20 Strep (bioMérieux) according to the manufacturer's instructions. When identification to the species level is not required as per clinical relevance, identification was limited to the genus or group level, e.g., coagulase-negative staphylococci (CoNS; including all staphylococcal species except Staphylococcus aureus and Staphylococcus lugdunensis), viridans streptococci (alpha-hemolytic streptococcal species except Streptococcus pneumoniae), the Streptococcus anginosus group (Streptococcus anginosus/constellatus/intermedius), or Enterococcus species originating from normally nonsterile body sites.
Molecular identification.
Molecular characterization of the isolates by 16S rRNA gene and recA gene sequence analysis was done as previously described (1, 2).
Sample preparation for MALDI-TOF MS.
Preparation of bacterial isolates for MALDI-TOF MS measurement was done as previously described (12, 14). Briefly, for the direct transfer method, fresh colony material was smeared on a polished steel MSP 96 target (Bruker Daltonik) using a toothpick, overlaid with 1 μl of a saturated α-cyano-4-hydroxy-cinnamic acid (HCCA) matrix solution in 50% acetonitrile-2.5% trifluoroacetic acid (Bruker Daltonik), and air dried at room temperature. For the direct transfer-formic acid method, 1 μl of 70% formic acid was added to the bacterial spot and allowed to air dry, before the matrix solution was added. For the protein extraction procedure, a loopful of bacterial material was suspended in 300 μl of distilled water and 900 μl ethanol was added. The cell suspension was centrifuged at 17,000 × g for 2 min, and the supernatant was discarded. The centrifugation was repeated, and the residual ethanol was discarded. The pellet was air dried and thoroughly resuspended in 5 to 50 μl formic acid-water (70:30 [vol/vol]) depending on the size, and finally an equal volume of acetonitrile was added. After centrifugation at 17,000 × g for 2 min, 1 μl of the supernatant was transferred to the MALDI target plate and allowed to dry at room temperature before being overlaid with 1 μl of matrix solution.
MALDI-TOF MS analysis.
The acquisition and analysis of mass spectra was performed by a Microflex LT mass spectrometer (Bruker Daltonik) using the MALDI Biotyper software package (version 3.0) with the reference database version 3.1.2.0 (3,995 database entries; Bruker Daltonik) and default parameter settings (positive linear mode; laser frequency, 60 Hz; ion source 1 voltage, 20 kV; ion source 2 voltage, 16.7 kV; lens voltage, 7.0 kV; mass range, 2,000 to 20,000 Da). For each spectrum, 240 laser shots in 40-shot steps from different positions of the sample spot were accumulated and analyzed (automatic mode, default settings). The Bruker bacterial test standard (Bruker Daltonik) was used for daily calibration according to the instructions of the manufacturer. For each isolate, two similar applications of colony/sample material were measured.
MALDI-TOF MS data interpretation.
The Biotyper software compares each sample mass spectrum to the reference mass spectra in the database, calculates an arbitrary unit score value between 0 and 3 reflecting the similarity between sample and reference spectrum, and displays the top 10 matching database records. As specified by the manufacturer, identification scores of ≥2.0 were accepted for a reliable identification to the species level (green), and scores of ≥1.7 but <2.0 were accepted for identification to the genus level (yellow). Scores below 1.7 were considered unreliable (red). In addition, the consistency categories A, B, and C, assigned to the identifications by the Biotyper software based on analysis of the top 10 matching identifications, were considered. The following category descriptions were applied. For category A, species consistency, the best match was classified as green (species identification) by MALDI. Further green identifications are of the same species. Further yellow (genus identification) identifications are at least the same genus. For category B, genus consistency, the best match was classified as green or yellow by MALDI. Further green or yellow identifications have at least the same genus. The conditions of species consistency are not fulfilled. For category C, no consistency, neither species nor genus consistency exists. Evaluation of cutoff adaption was done by reducing the species cutoff to 1.9, 1.8, and 1.7 and by then reinterpreting the top 10 matching database records as described above for a cutoff 2.0.
Generation of in-house reference database.
For 48 isolates of the retrospective study that did not yield a score of ≥2.0 using the direct transfer method, reference spectra were created and added to the Bruker database version 3.1.2.0. For each isolate, a set of 24 spectra was measured and checked manually for flat-line, outlier, and single spectra with peaks differing from the other spectra. Such questionable spectra were removed, and a total of 20 to 24 spectra were used to calculate a reference spectrum, using the automated function of the Biotyper software.
Preparation time determination.
The preparation time included smearing 12, 48, or 96 samples on an MSP 96 target following the direct transfer or the direct transfer-formic acid protocol and using a toothpick for each spot. Twelve strains (four S. aureus strains, three Staphylococcus epidermidis strains, two Enterococcus faecalis strains, one Enterococcus faecium strain, one Streptococcus agalactiae strain, and one Streptococcus salivarius strain) were selected, reflecting the relative frequency of genus and species as found in the prospective study. The total time to result included administrative time comprising the recording of a 12-digit strain number for each spot and programming the run. Furthermore, total time to result included measuring the spectra and analyzing the data with the Biotyper software.
Statistical analysis.
Statistical calculations were done using IBM SPSS Statistics software, version 20 (SPSS Inc., Chicago, IL). Agreements between two methods were analyzed using a two-tailed McNemar test or a Wilcoxon rank sum test for paired samples. A P value of <0.05 was considered statistically significant.
RESULTS
Retrospective study. (i) Identification rates of Gram-positive cocci using direct transfer, direct transfer-formic acid, and extraction preparation methods for MALDI-TOF MS.
We evaluated the Bruker MALDI Biotyper system analyzing 156 Gram-positive cocci from the clinical strain collection of our institute. With all three methods, the direct transfer, the direct-transfer-formic acid, and the extraction methods, the MALDI system correctly identified around 99% of the isolates to the genus level. A correct identification to the species level was achieved for 100 (64.1%) of the isolates using the direct transfer method. This identification rate was significantly higher (121; 77.6%) when using the direct transfer-formic acid and the extraction methods. These species identification rates varied in taxonomic groups (Table 1). All enterococci were correctly identified to the species level independently of the preparation method. In contrast, correct species identification rates for streptococci ranged from 54.7% (direct transfer) to 69.3% (extraction) and 74.7% (direct transfer-formic acid). Identification rates to the species level for rare genera, including Abiotrophia, Gemella, Granulicatella, Kocuria, Lactococcus, Leuconostoc, and Rothia, were lower, reaching 37.5%, 50%, and 56.3% for the direct transfer, direct transfer-formic acid, and extraction methods, respectively. The formic acid overlay and extraction protocol increased average MALDI score values by 0.10 and 0.15 score units, respectively, compared to the direct transfer method. No peaks were detected in approximately 1% of measurements. This rate did not differ significantly between the direct transfer and direct transfer-formic acid methods.
Table 1.
Retrospective analysis of Gram-positive coccus clinical isolates with MALDI-TOF MS: comparison of three sample preparation methods
| Genus | Cutoff | No. of isolates (%) correctly identified at the species level by:a |
||
|---|---|---|---|---|
| Direct transfer | Direct transfer-formic acid | Extraction | ||
| Aerococcus (n = 17 isolates) | 2.0 | 10 (58.8)r | 15 (88.2) | 15 (88.2) |
| 1.9 | 13 (76.5) | 16 (94.1)* | 16 (94.1)* | |
| 1.8 | 15 (88.2) | 17 (100)* | 17 (100)* | |
| 1.7 | 17 (100)* | 17 (100)* | 17 (100)* | |
| Enterococcus (n = 16 isolates) | 2.0 | 16 (100)r | 16 (100) | 16 (100) |
| 1.9 | 16 (100) | 15 (93.8) | 16 (100) | |
| 1.8 | 16 (100) | 10 (62.5)* | 15 (93.8) | |
| 1.7 | 15 (93.8) | 5 (31.3)* | 9 (56.3)* | |
| Staphylococcus (n = 32 isolates) | 2.0 | 26 (81.3)r | 26 (81.3) | 29 (90.6) |
| 1.9 | 28 (87.5) | 27 (84.4) | 32 (100)* | |
| 1.8 | 30 (93.8) | 29 (90.6) | 31 (96.9) | |
| 1.7 | 28 (87.5) | 28 (87.5) | 30 (93.8) | |
| Streptococcus (n = 75 isolates) | 2.0 | 41 (54.7)r | 56 (74.7)* | 52 (69.3)* |
| 1.9 | 46 (61.3) | 49 (65.3)* | 51 (68.0)* | |
| 1.8 | 40 (53.3) | 42 (56.0) | 39 (52.0) | |
| 1.7 | 34 (45.3)* | 33 (44.0)* | 31 (41.3)* | |
*, value significantly differs from the reference value (r). Agreement between the reference (direct transfer, cutoff of 2.0) and the method evaluated was compared using the two-tailed McNemar test for paired samples. A P value of <0.05 was considered statistically significant.
(ii) Low discrimination at the species level using MALDI-TOF MS.
The analysis of certain streptococcal isolates yielded identification ranking lists with scores of ≥2.0 for more than one streptococcal species, resulting in a consistency category B interpretation and in identification to the genus level only (Streptococcus species). The following identifications were reported by the MALDI-TOF MS: S. anginosus (score of ≥2.0 for S. anginosus/constellatus), S. canis (S. canis/dysgalactiae), S. constellatus (S. constellatus/anginosus), S. dysgalactiae (S. dysgalactiae/pyogenes), S. infantarius (S. equinus/lutetiensis), S. oralis (S. pneumoniae/oralis), S. pyogenes (S. pyogenes/dysgalactiae), and S. vestibularis (S. vestibularis/salivarius). This phenomenon occurred more frequently with the extraction method than with the direct transfer and the direct transfer-formic acid methods. Of note, in 16 of 22 cases, the highest score corresponded to the correct species as identified by phenotypic and molecular methods.
(iii) Misidentifications using MALDI-TOF MS.
All five S. mitis isolates were misidentified as S. pneumoniae using the extraction method. This number was lower for the direct transfer and the direct transfer-formic acid preparations, as these methods generated less species identification calls. One of three S. oralis isolates was identified as S. pneumoniae using the direct transfer-formic acid method. In contrast, all eight S. pneumoniae isolates were correctly identified. The three S. infantarius isolates were misidentified as S. equinus or S. lutetiensis.
(iv) Additional preparation time for the direct transfer-formic acid method.
A comparison of sample preparation time and total time to result for direct transfer and direct transfer-formic acid preparations is shown in Table 2. Sample preparation using the direct transfer method requires two manual steps (transfer of colony material to the MALDI target and overlay with matrix solution), whereas the direct transfer-formic acid method comprises an additional step of formic acid treatment. Using a Bruker Microflex LT and an MSP 96 target (Bruker Daltonik), a maximum of 96 isolates can be analyzed in one run. The relative increase in time for additional formic acid treatment was lower if higher numbers of samples were prepared (preparation time increases of 44.4%, 31.9%, and 22.2% preparing 12, 48, and 96 samples, respectively, i.e., one-eighth, half, and full target capacity per run [Table 2]). With respect to total time to result (including programming the MALDI software and actual measurements), the formic acid method increased working time by 22.6%, 16.4%, and 8.5% for the analysis of 12, 48, and 96 samples per run, respectively.
Table 2.
Preparation and identification times for direct transfer and direct transfer-formic acid methodsc
| Prepn method | No. of samples | Sample prepn time [min (%)]a | Total time to result [min (%)]b |
|---|---|---|---|
| Direct transfer | 12 | 12.4 ± 2.7 | 19.9 ± 2.7 |
| 48 | 26.6 ± 3.3 | 50.1 ± 4.4 | |
| 96 | 47.2 ± 4.0 | 95.5 ± 6.4 | |
| Direct transfer-formic acid | 12 | 17.9 ± 2.4 | 24.4 ± 2.6 |
| 48 | 35.1 ± 3.7 | 58.3 ± 4.5 | |
| 96 | 57.6 ± 5.4 | 103.62 ± 8.20 | |
| Mean increase in time for direct transfer-formic acid prepn | 12 | 5.5 (44.4) | 4.5 (22.6) |
| 48 | 8.5 (32.1) | 8.2 (16.4) | |
| 96 | 10.4 (22.1) | 8.1 (8.5) |
Time for preparing 12, 48, or 96 isolates onto the target and recording their strain numbers.
Preparation time plus time for programming, measuring, and analyzing the data until the final result is shown.
The average time of four people including standard deviation is shown.
(v) Individual score cutoffs for species identification.
Standard cutoffs for species and genus identification are set to 2.0 and 1.7, respectively, by the manufacturer. A reduction of the species cutoff from 2.0 to 1.9 resulted, overall, in a higher rate of species identification for the direct transfer preparation (increase from 63.5% to 73.7%) and for the extraction method (increase from 77.6% to 82.7%). In contrast, identification rates for direct transfer-formic acid preparation were similar for species-level cutoffs of 2.0 and 1.9 (77.6% and 76.3%, respectively). A further reduction of the species-level cutoff to 1.8 and 1.7 resulted in a significant decrease of identification rates (data not shown). This decrease was caused by an increased rate of low discriminations at the species level as described above and not by misidentifications. However, the impact of cutoff adaptations on identification rates varied for individual genera (Table 1), e.g., the species identification rate for aerococci was increased to 100% with all preparation methods applying a MALDI score cutoff of 1.7, whereas for streptococci, a cutoff reduction to 1.7 decreased the species identification rate to 41.3% to 45.3%. Especially the discrimination of the species belonging to the S. anginosus group, the S. bovis/equinus group, the S. mitis group, and the S. salivarius group (16) was significantly reduced when applying species cutoffs below 1.9.
Prospective study: comparison of conventional and MALDI-TOF MS identification in the routine diagnostic laboratory.
In a prospective study from November 2011 to February 2012, 1,619 Gram-positive coccus clinical strains were identified according to the conventional identification algorithm of the Institute of Medical Microbiology, University of Zurich, and by MALDI-TOF MS. The Bruker MALDI Biotyper system was used with the direct transfer-formic acid sample preparation method, applying the Bruker database version 3.1.2.0 (3,995 entries) and a species-level identification cutoff of 2.0. Results are summarized in Table 3. Overall, for 1,548 out of 1,619 isolates (95.6%), conventional and MALDI-TOF MS identification yielded concordant results. A total of 569 isolates were assigned to the same genus/organism group, and 979 isolates were identified to the same species. Discrepancies were observed for 71 (4.6%) isolates.
Table 3.
Prospective study: conventional versus MALDI-TOF MS identification for 1,619 Gram-positive cocci isolated during a 4-month period in the routine diagnostic laboratory
| Organism group/species | No. (%) of congruent identifications by MALDI-TOF MSa (direct transfer-formic acid, species cutoff of 2.0) at the: |
|
|---|---|---|
| Genus/group level | Species levelb | |
| Conventional identification stopped at the genus/group level (n = 609 isolates) | ||
| Coagulase-negative staphylococci | 420 (95) | NA |
| Enterococcus species | 113 (100) | NA |
| Viridans streptococci | 18 (52.9) | NA |
| Streptococcus anginosus group | 18 (90) | NA |
| Total | 569 (93.4) | NA |
| Conventional identification to the species level (n = 1,010) | ||
| Staphylococcus aureus | 567 (100) | 565 (99.6) |
| Streptococcus agalactiae (beta-hemolytic streptococci group B) | 186 (100) | 186 (100) |
| Enterococcus faecium | 106 (100) | 106 (100) |
| Enterococcus faecalis | 57 (100) | 57 (100) |
| Streptococcus dysgalactiae (beta-hemolytic streptococci group C/G) | 24 (100) | 6 (25) |
| Streptococcus pyogenes (beta-hemolytic streptococci group A) | 19 (100) | 18 (94.7) |
| Staphylococcus lugdunensis | 13 (100) | 13 (100) |
| Streptococcus pneumoniae | 10 (100) | 10 (100) |
| Enterococcus avium | 5 (100) | 5 (100) |
| Rare isolatesc | 22 (95.7) | 13 (56.5) |
| Total | 1,009 (99.9) | 979 (96.9) |
MALDI-TOF MS identification using the direct transfer-formic acid protocol and a species-level cutoff of 2.0 with the Bruker database version 3.1.2.0 (containing 3,995 entries).
NA, not applicable. For reasons of comparison with the conventional identification, these isolates were classified according to the MALDI-TOF MS result into the following taxonomic groups: CoNS, staphylococcal species except S. aureus and S. lugdunensis; Enterococcus sp., all enterococci; Streptococcus anginosus group, S. anginosus/constellatus/intermedius; viridans streptococci, alpha-hemolytic streptococci except S. pneumoniae.
Rare isolates (n ≤ 4), including Abiotrophia defective, Aerococcus urinae, Aerococcus viridans, Enterococcus species, Granulicatella adiacens, Micrococcus luteus, Pediococcus acidilactici, Pediococcus pentosaceus, Rothia mucilaginosa, Streptococcus gallolyticus, Streptococcus mitis/oralis, Streptococcus salivarius.
Spectra that did not yield a score of ≥2.0 were reanalyzed using our user-modified Bruker-IMM database, i.e., an extended database combining the commercially available database with in-house reference spectra generated in the retrospective study. Reanalysis increased the total number of congruent results of conventional and MALDI-TOF MS identification from 1,548 (95.6%) to 1,572 (97.1%) (data not shown), leaving 47 (2.9%) discrepancies, which were resolved by 16S rRNA gene analysis (Table 4).
Table 4.
Prospective study: resolution of 47 discrepant results comparing conventional phenotypic and MALDI-TOF MS Gram-positive coccus identifications
| No. of isolates | Conventional identification | MALDI-TOF MS identification (Direct transfer formic acid, species cut-off 2.0) | 16S rRNA gene analysis (gold standard) |
|---|---|---|---|
| No reliable identification with MALDI-TOF MS | |||
| 2 | Coagulase-negative staphylococci | No reliable identification | Staphylococcus cohnii |
| 1 | Coagulase-negative staphylococci | No reliable identification | Staphylococcus warneri/pasteurid |
| Identification at the genus level with MALDI-TOF MS | |||
| 18 | Streptococcus dysgalactiae | Streptococcus sp.a | Streptococcus dysgalactiae |
| 1 | Aerococcus viridans | Aerococcus sp.b | Aerococcus viridans |
| 1 | Coagulase-negative staphylococci | Staphylococcus sp.b | Staphylococcus lugdunensis |
| 1 | Coagulase-negative staphylococci | Staphylococcus sp.b | Staphylococcus epidermidis |
| 1 | Staphylococcus aureus | Staphylococcus sp.b | Staphylococcus aureus |
| 1 | Streptococcus mitis/oralis | Streptococcus sp.b | Streptococcus mitis groupc |
| 1 | Streptococcus pyogenes | Streptococcus sp.a | Streptococcus pyogenes |
| Identification at the genus level with conventional methods | |||
| 3 | Enterococcus sp. | Enterococcus faecium | E. faecium/duransd |
| 1 | Enterococcus sp. | Enterococcus casseliflavus | Enterococcus gallinarum/casseliflavusd |
| Other discrepancies | |||
| 9 | Viridans streptococci | Streptococcus pneumoniae | Streptococcus mitis groupc |
| 3 | Streptococcus mitis/oralis | Streptococcus pneumoniae | Streptococcus mitis groupc |
| 1 | Coagulase-negative staphylococci | Staphylococcus aureus | Staphylococcus aureus |
| 1 | Coagulase-negative staphylococci | Staphylococcus lugdunensis | Staphylococcus lugdunensis |
| 1 | Staphylococcus aureus | Staphylococcus warneri | Mixed culturee |
| 1 | Pediococcus pentosaceus | Enterococcus faecium | Enterococcus faecium |
Identification by MALDI-TOF MS on the genus level (consistency category B), as more than one species yielded a score of ≥2.0.
Identification by MALDI-TOF MS on the genus level (consistency category B), as the best matches yielded scores in the range of ≥1.7 to <2.0 only.
The analysis of the 16S rRNA gene is not suited for the differentiation of the S. mitis group (2). Therefore, S. pneumoniae was excluded by biochemical tests.
No differentiation possible with the analysis of the 16S rRNA gene. The E. gallinarum/casseliflavus isolate showed no pigmentation, indicating that it was E. gallinarum (17).
Repetition of conventional and MALDI-TOF MS identification and analysis of the 16S rRNA gene revealed a mixed culture of S. aureus and S. warneri/pasteuri.
The most important limitations of the MALDI Biotyper system were low discrimination at the species level for isolates of S. dysgalactiae (n = 18) and misidentification of S. mitis group members as S. pneumoniae (n = 12). In contrast, discrimination of CoNS, S. lugdunensis, and S. aureus was highly reliable with MALDI-TOF MS, while single misidentifications of S. aureus and S. lugdunensis as CoNS (n = 3) were observed with conventional methods.
DISCUSSION
Fast and reliable identification of bacteria is mandatory for clinical diagnostics. Efforts to improve laboratory identification algorithms are ongoing. Recently, MALDI-TOF MS has been introduced into diagnostic laboratories and promises to replace or at least complement conventional identification methods (18, 19). The performance of the MALDI-TOF MS systems depends on the sample preparation method, quality of the reference database, and data interpretation algorithms.
Sample preparation method.
Using the manufacturer's standard interpretation criteria (species score cutoff of 2.0), the direct transfer-formic acid and the extraction methods significantly increased identification to the species level (77.6% for both methods) compared to the direct transfer method (64.1%). Our data support findings by McElvania et al. which report that formic acid overlay increases the identification rate of Gram-positive organisms (20). However, the rate of species identification varied among individual genera. For enterococci and staphylococci, the direct transfer and the direct transfer-formic acid methods showed similar performance, whereas species identification rates were increased using the direct transfer-formic acid method for aerococci and streptococci (Table 1). The extraction method performed equally to slightly better than the direct transfer-formic acid method. Streptococci posed an exception, as the species identification rate decreased using the extraction method. This decrease was caused by score values of ≥2.0 for more than one streptococcal species in the MALDI identification ranking list (i.e., species inconsistency), resulting in genus identification only. It occurred most frequently with closely related species of the S. anginosus group, the S. bovis/equinus group, the S. mitis group, and the S. salivarius group (16). Group-level assignment might be an option to circumvent this problem. Both the direct transfer-formic acid and extraction methods require additional preparatory steps compared to direct transfer. In summary, the use of the direct transfer-formic acid method can enhance the identification ability of the Biotyper system for Gram-positive cocci, thereby moderately increasing total time to result (8.5% to 22.6% depending on target load).
Quality of the reference database.
Identification of the clinically most relevant Gram-positive coccus genera, including aerococci, enterococci, staphylococci, and streptococci, was highly reliable in the retrospective and prospective studies, suggesting that the quality of the commercial Bruker database is sufficient for these genera. In contrast, species identification of rare genera, such as Gemella and Granulicatella, was limited. Although more than 90% of these isolates were correctly assigned to the genus level, only half of them could be assigned to the species level in the retrospective part. No misidentification was observed for rare genera. However, the number of isolates from rarely encountered genera was low in this study, and more isolates need to be analyzed to finally assess the performance of the Biotyper system for this group of strains.
Applying our in-house database in the prospective study increased the overall percentage of congruent results of conventional and MALDI-TOF MS identification from 95.6% (commercial database) to 97.1% (in-house database). Thus, the addition of single spectra of rarely isolated species improved the performance of the Biotyper system. Corresponding in-house reference spectra were provided to the manufacturer for the integration of selected data sets in the commercial database.
Two important limitations for the identification with the Bruker MALDI Biotyper were observed for the S. mitis group and S. dysgalactiae. As reported previously (3, 11, 21), the Biotyper system's ability to discriminate members of the S. mitis group was limited, and several isolates of S. mitis and S. oralis were misidentified as S. pneumoniae. Members of the S. mitis group, including S. pneumoniae, are closely related, and even the discriminatory power of the 16S rRNA gene is too low for reliable differentiation (2). However, due to clinical reasons, the differentiation of S. pneumoniae is of critical importance, and additional tests are required in case of a MALDI-TOF MS S. pneumoniae identification, either by biochemical methods such as optochin susceptibility and/or bile solubility or by molecular methods such as recA gene sequence analysis (22). Cherkaoui et al. (23) reported high confidence identification to the species level for beta-hemolytic streptococci. In contrast, this study showed a low identification rate for S. dysgalactiae. Identification rankings frequently listed not only S. dysgalactiae but also S. pyogenes, both with score values of ≥2.0 in the same analysis, resulting in a consistency category B interpretation (genus identification only). The reason for this discrepancy is, most likely, the use of an earlier version (2.0) of the Bruker Biotyper platform by Cherkaoui et al. (4), which did not yet include consistency category interpretation rules. S. dysgalactiae and S. pyogenes are genetically closely related and have overlapping infection spectra (24). Nevertheless, differentiation of these two species is required in the clinical environment. In-depth analysis showed that the first matching hit was always concordant with species identification as determined by 16S rRNA gene analysis (data not shown). Species consistency criteria may, thus, be modified for S. dysgalactiae and S. pyogenes, accepting the result with the highest score as species identification.
As reported by others (25), we encountered difficulties in discriminating members of the Streptococcus bovis/equinus complex, especially of S. equinus, S. infantarius, and S. lutetiensis, when using MALDI-TOF MS. The differentiation of these species is challenging, and their taxonomy is under constant change (22). Future analysis has to show the clinical importance of discriminating these species.
Data interpretation algorithms.
A reduction of the species cutoff from 2.0 to 1.9 increased overall species identification rates (63.5% to 73.7% and 77.6% to 82.7% for the direct transfer and extraction methods, respectively), whereas the direct transfer-formic acid identification rate remained almost constant (77.6% compared to 76.3%, applying species-level score cutoffs of 2.0 and 1.9, respectively). Of note, the impact of lower cutoffs on species identification rates differed for individual genera (Table 1). An adaptation of the score value depending on individual genera may thus avoid unnecessary further biochemical and molecular testing, thereby increasing the efficiency of the diagnostic workflow. However, in contrast to sample preparation and reference database, the interpretation algorithms of the Biotyper system are not open to the user, and the use of a modified score value would thus mean a considerable manual effort.
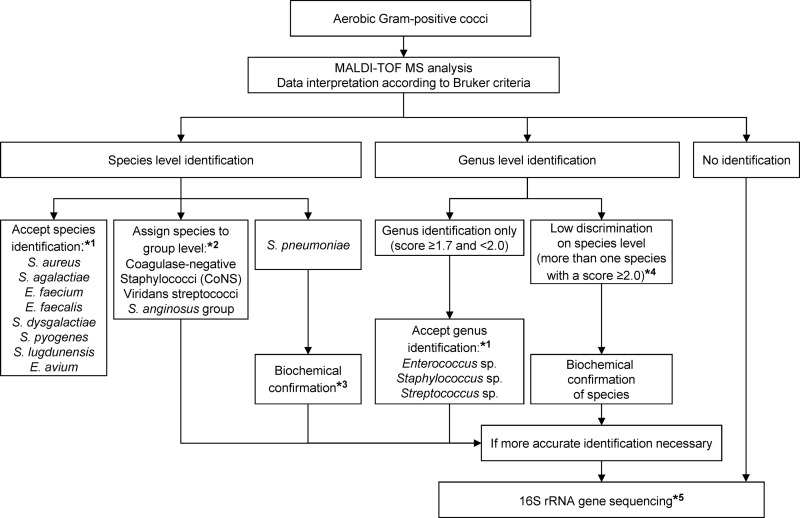
Five conclusions can be drawn from this study: (i) despite some limitations regarding differentiation of the S. mitis group from S. pneumoniae and the identification of S. dysgalactiae (beta-hemolytic streptococci group C/G), this study confirms that MALDI-TOF MS is ready to be implemented in the clinical laboratory for the identification of Gram-positive cocci using standard protocols of the manufacturer; (ii) species consistency criteria could be modified for S. dysgalactiae and S. pyogenes, accepting the result with the highest score as species identification; (iii) the use of the direct transfer-formic acid method improves identification rates of the Biotyper system for Gram-positive cocci and is feasible for the routine clinical laboratory; (iv) expanding the commercial database by generating one's own reference spectra further improves rates of species identification, especially for more rarely isolated genera/species; (v) based on the findings of this study, we suggest a practical algorithm for the identification of Gram-positive cocci in routine diagnostics complementing MALDI-TOF MS with phenotypic and molecular methods (Fig. 1). The algorithm is based on the current manufacturer's system setup (i.e., score value cutoffs, consistency rules) to ensure feasibility for routine purposes, and it covers the most frequently found genera and species and can easily be updated.
Fig 1.
Algorithm for the identification of Gram-positive cocci in routine diagnostics using MALDI-TOF MS. Recommendations are based on the results of the prospective study, including 1,619 clinical isolates. *1, species and genera with less than five isolates were not integrated in this algorithm, as the numbers are too low to give a proper recommendation. However, all rare isolates were correctly identified by MALDI-TOF MS. It is suggested that these isolates are identified by 16S rRNA gene sequencing until sufficient data are available to update the approved lists. *2, group assignment as follows: CoNS, all staphylococcal species except S. aureus and S. lugdunensis; viridans streptococci, alpha-hemolytic streptococcal species except S. pneumoniae; S. anginosus group, S. anginosus/constellatus/intermedius. *3, biochemical confirmation of S. pneumoniae by, e.g., testing of bile solubility and optochin susceptibility. *4, observed for the following streptococci: S. anginosus/constellatus, S. dysgalactiae/pyogenes, S. dysgalactiae/canis, S. equinus/lutetiensis, S. pneumoniae/oralis, and S. vestibularis/salivarius. *5, 16S rRNA gene sequencing and analysis as outlined by Bosshard et al. (1).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the laboratory technicians of the Institute of Medical Microbiology, University of Zurich, Zurich, Switzerland, for their dedicated help. The Institute of Medical Microbiology, University of Zurich, Switzerland, collaborates with Bruker Daltonik GmbH (Bremen, Germany) for the purpose of improving the commercially available Bruker database. We state that Bruker Daltonik did not have any influence on data collection and interpretation of this study.
This study was supported by the University of Zurich.
Footnotes
Published ahead of print 3 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02654-12.
REFERENCES
- 1. Bosshard PP, Abels S, Altwegg M, Böttger EC, Zbinden R. 2004. Comparison of conventional and molecular methods for identification of aerobic catalase-negative Gram-positive cocci in the clinical laboratory. J. Clin. Microbiol. 42:2065–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zbinden A, Kohler N, Bloemberg GV. 2011. recA-based PCR assay for accurate differentiation of Streptococcus pneumoniae from other viridans streptococci. J. Clin. Microbiol. 49:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Veen SQ, Claas EC, Kuijper EJ. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, François P, Schrenzel J. 2010. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neville SA, Lecordier A, Ziochos H, Chater MJ, Gosbell IB, Maley MW, van Hal SJ. 2011. Utility of matrix-assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 49:2980–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dubois D, Grare M, Prere MF, Segonds C, Marty N, Oswald E. 2012. Performances of the MALDI-TOF mass spectrometry system VITEK MS for the rapid identification of bacteria in routine clinical microbiology. J. Clin. Microbiol. 50:2568–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan EK, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. 2012. Prospective evaluation of a MALDI-TOF MS system in a hospital clinical microbiology laboratory for the identification of bacteria and yeasts: a bench-by-bench study to assess the impact on time-to-identification (TTI) and cost-effectiveness. J. Clin. Microbiol. 50:3301–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sogawa K, Watanabe M, Sato K, Segawa S, Miyabe A, Murata S, Saito T, Nomura F. 2011. Rapid identification of microorganisms by mass spectrometry: improved performance by incorporation of in-house spectral data into a commercial database. Anal. Bioanal. Chem. 403:1811–1822 [DOI] [PubMed] [Google Scholar]
- 10. Christensen JJ, Dargis R, Hammer M, Justesen US, Nielsen XC, Kemp M, Danish MALDI-TOF MS Study Group 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis of Gram-positive, catalase-negative cocci not belonging to the Streptococcus or Enterococcus genus and benefits of database extension. J. Clin. Microbiol. 50:1787–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of Gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haigh J, Degun A, Eydmann M, Millar M, Wilks M. 2011. Improved performance of bacterium and yeast identification by a commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry system in the clinical microbiology laboratory. J. Clin. Microbiol. 49:3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sedo O, Sedlacek I, Zdrahal Z. 2011. Sample preparation methods for MALDI-MS profiling of bacteria. Mass Spectrom. Rev. 30:417–434 [DOI] [PubMed] [Google Scholar]
- 14. Freiwald A, Sauer S. 2009. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 4:732–742 [DOI] [PubMed] [Google Scholar]
- 15. Fournier R, Wallet F, Grandbastien B, Dubreuil L, Courcol R, Neut C, Dessein R. 2012. Chemical extraction versus direct smear for MALDI-TOF mass spectrometry identification of anaerobic bacteria. Anaerobe 18:294–297 [DOI] [PubMed] [Google Scholar]
- 16. Facklam R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. J. Clin. Microbiol. 15:613–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Facklam RR, Collins MD. 1989. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 27:731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Croxatto A, Prod'hom G, Greub G. 2012. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 36:380–407 [DOI] [PubMed] [Google Scholar]
- 19. Demirev PA, Fenselau C. 2008. Mass spectrometry for rapid characterization of microorganisms. Annu. Rev. Anal. Chem. 1:71–93 [DOI] [PubMed] [Google Scholar]
- 20. McElvania TeKippe E, Shuey S, Winkler DW, Butler MA, Burnham CD. 20 February 2013. Optimizing identification of clinically relevant Gram-positive organisms using the Bruker Biotyper MALDI-TOF MS system. J. Clin. Microbiol. [Epub ahead of print.] doi:10.1128/JCM.02680-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. López Roa P, Sánchez Carrillo C, Marín M, Romero F, Cercenado E, Bouza E. 10 March 2012. Value of matrix-assisted laser desorption ionization–time of flight for routine identification of viridans group streptococci causing bloodstream infections. Clin. Microbiol. Infect. [Epub ahead of print.] doi:10.1111/j.1469-0691.2012.03837.x [DOI] [PubMed] [Google Scholar]
- 22. Hinse D, Vollmer T, Erhard M, Welker M, Moore ER, Kleesiek K, Dreier J. 2011. Differentiation of species of the Streptococcus bovis/equinus-complex by MALDI-TOF mass spectrometry in comparison to sodA sequence analyses. Syst. Appl. Microbiol. 34:52–57 [DOI] [PubMed] [Google Scholar]
- 23. Cherkaoui A, Emonet S, Fernandez J, Schorderet D, Schrenzel J. 2011. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid identification of beta-hemolytic streptococci. J. Clin. Microbiol. 49:3004–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jensen A, Kilian M. 2012. Delineation of Streptococcus dysgalactiae, its subspecies, and its clinical and phylogenetic relationship to Streptococcus pyogenes. J. Clin. Microbiol. 50:113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Romero B, Morosini MI, Loza E, Rodríguez-Baños M, Navas E, Cantón R, Campo RD. 2011. Reidentification of Streptococcus bovis isolates causing bacteremia according to the new taxonomy criteria: still an issue? J. Clin. Microbiol. 49:3228–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.