Abstract
A carbapenem-resistant Alcaligenes faecalis strain was isolated from a surveillance swab of a service member injured in Afghanistan. The isolate was positive for blaNDM by real-time PCR. Species identification was reevaluated on three identification systems but was inconclusive. Genome sequencing indicated that the closest relative was Acinetobacter schindleri and that blaNDM-1 was carried on a plasmid that shared >99% identity with one identified in an Acinetobacter lwoffii isolate. The isolate also carried a novel chromosomally encoded class D oxacillinase.
TEXT
In response to global concerns over the spread of blaNDM (1–3), the Multidrug-resistant Organism Repository and Surveillance Network (MRSN) implemented routine monthly screening for this gene in all carbapenem-resistant Gram-negative organisms in 2010 (4). From this surveillance initiative, we previously described the first incidence of blaNDM-1 in the U.S. military health care system from a strain of Providencia stuartii, isolated from a local Afghan national treated at a military facility in Afghanistan (5). Timely feedback was provided to the submitting facility, which resulted in increased surveillance and enhanced infection control policies.
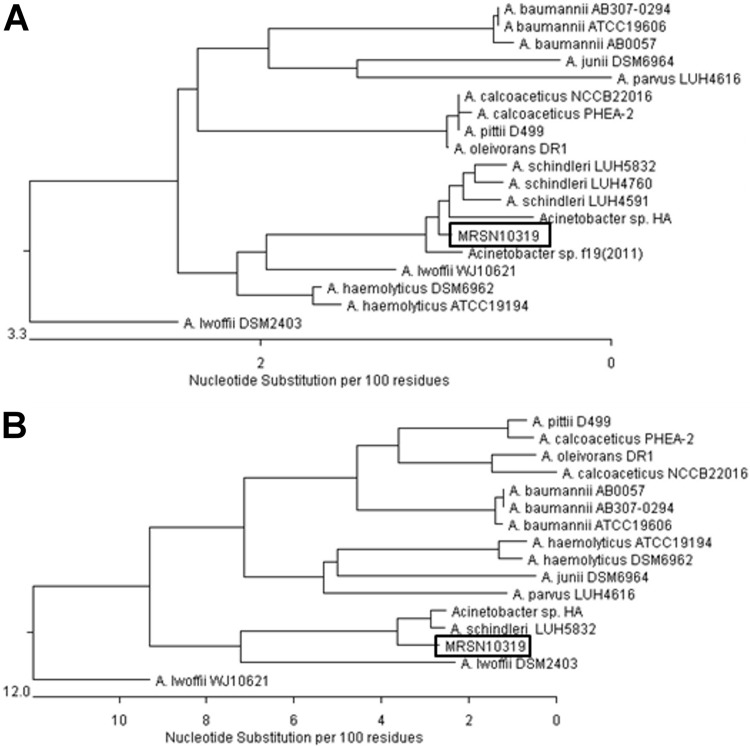
In June 2012, following a blast injury involving shrapnel in Afghanistan, a 22-year-old male received cefazolin for routine prophylaxis and was evacuated to the United States via Germany. During that escalation of care, a routine groin surveillance swab revealed mixed microbial flora. An isolate identified as Alcaligenes faecalis using the BD Phoenix automated microbiology system (BD Diagnostics Systems, Sparks, MD) displayed resistance to all β-lactam antibiotics tested (intermediate to ceftriaxone), including the carbapenems and the monobactam aztreonam (Table 1). A modified Hodge test (MHT) was negative for meropenem. For ertapenem, the MHT was positive but the clover leaf-like growth indentation of Escherichia coli ATCC 25922 was significantly reduced when grown with the test isolate, designated MRSN 10319, compared to its growth alongside the blaKPC-positive control strain Klebsiella pneumoniae ATCC BAA-1705. The isolate was forwarded to the MRSN, a College of American Pathologists (CAP)-certified laboratory, for further evaluation. The identification was reevaluated using three automated identification systems: the Vitek 2 (bioMérieux, Durham, NC), the BD Phoenix, and the Microscan WalkAway (Siemens Healthcare Diagnostics, Inc., Deerfield, IL). The MRSN employs the three most common automated instruments, as these instruments are used throughout the Military Health System (MHS), and discrepancies between their results have been noted (for a comprehensive review, see reference 6). Both Vitek 2 and Microscan identified the organism as Acinetobacter lwoffi, whereas the Phoenix was in agreement with the initial identification of Alcaligenes faecalis. 16S rRNA sequencing was performed (7) and indicated that the isolate shared 99% identity with Acinetobacter schindleri/Acinetobacter johnsonii. Whole-genome sequencing (WGS) using an Ion Torrent PGM (Ion Torrent Systems, Inc., Guilford, CT), which provided 87× coverage of the 16S rRNA gene, demonstrated 99.5% identity to Acinetobacter schindleri 16S rRNA sequences deposited at GenBank (Fig. 1A) (8). Phylogenetic analysis using the rpoB gene sequence confirmed this identification (Fig. 1B).
Table 1.
Antibiotic susceptibility profile of A. schindleri MRSN 10319
| Antibiotic | MIC (μg/ml), phenotypea | Interpretation (μg/ml)b |
|---|---|---|
| Arbekacin | ≤0.25c | NA |
| Amikacin | ≤8, S | ≤16, 32, ≥64 |
| Ampicillin-sulbactam | >16/8, R | ≤8/4, 16/8, ≥32/16 |
| Aztreonam | >16d | NA |
| Cefepime | >16, R | ≤8, 16, ≥32 |
| Ceftazidime | >16, R | ≤8, 16, ≥32 |
| Ceftriaxone | 32, I | ≤8, 16–32, ≥64 |
| Ciprofloxacin | ≤0.5, S | ≤1, 2, ≥4 |
| Colistin | 0.25,e S | ≤2, —, ≥4 |
| Gentamicin | ≤1, S | ≤4, 8, ≥16 |
| Imipenem | >8, R | ≤4, 8, ≥16 |
| Levofloxacin | ≤1, S | ≤2, 4, ≥8 |
| Meropenem | ≥32, Rf | ≤4, 8, ≥16 |
| Piperacillin-tazobactam | >64/4, R | ≤16/4, 32/4–64/4, ≥128/4 |
| Tetracycline | ≤2, S | ≤4, 8, ≥16 |
| Tobramycin | ≤1, S | ≤4, 8, ≥16 |
| Trimethoprim-sulfamethoxazole | ≤0.5/9.5, S | ≤2/38, —, ≥4/76 |
MICs were determined using three automated systems (see the text), except for arbekacin, colistin, and meropenem. All results were consistent across the three instruments. The MICs and resulting interpretations are presented using the Phoenix output for clarity. R, resistant; I, intermediate; S, susceptible.
As recommended by the Clinical and Laboratory Standards Institute (CLSI) (18). NA, no CLSI interpretative guidelines are available.
MICs for arbekacin represent the average of three independent broth microdilution assays as described previously (19).
No CLSI interpretive guidelines for aztreonam are available for Acinetobacter species.
Average of three independent Etest assays performed as described by the manufacturer (bioMérieux). Etest results were consistent across replicates.
As meropenem is not reported by the BD Phoenix Automated Microbiology System, MICs were performed in triplicate by Etest as described by the manufacturer (bioMérieux). No variation in Etest results was evidenced.
Fig 1.
Dendrograms showing the relationship between A. schindleri MRSN 10319 and other Acinetobacter species based on (A) 16S rRNA and (B) rpoB gene sequences. Acinetobacter species 16S rRNA and rpoB gene sequences were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/pubmed; last accessed 23 August 2012) and aligned using MegAlign (DNASTAR, Madison, MI). Dendrograms based on the numbers of nucleotide changes were generated using the MegAlign program (DNASTAR).
MRSN 10319 was tested by real-time PCR for carbapenemase genes (9) and was positive for blaNDM. The gene was found to contain a single-nucleotide polymorphism (guanine to adenine) at position 468 compared to blaNDM-1, resulting in a synonymous mutation. The gene was located on a 47.3-kb plasmid that shared >99% identity with pNDM-BJ02, a plasmid identified in an isolate of Acinetobacter lwoffi cultured from the urine of a 62-year-old female patient in Beijing in November 2010 (10). Hu and colleagues identified A. lwoffi by Vitek 2 but make no mention of the 16S rRNA sequence to verify this result. In accordance with the discrepancies noted in this report, Dortet and colleagues have noted that the Vitek 2 identifies rare species of Acinetobacter, including A. schindleri, as A. lwoffi (11).
Plasmid pNDM-BJ02 has 46 open reading frames, and there is no plasmid sequence in GenBank that shares more than 15% homology with it. Furthermore, the plasmid cannot be assigned to any of the described incompatibility groups using the PCR replicon typing method developed by Carattoli and colleagues (12). The plasmid harbors a type IV secretion system (T4SS) gene cluster and a single copy of aphA6, which encodes resistance to some aminoglycosides. However, MRSN 10319 was susceptible to all aminoglycosides tested (Table 1). Sequence comparison to pNDM-BJ02 suggests that the original promoter sequence for this gene has been disrupted by an upstream transposition event, as previously noted (10). Hu and colleagues demonstrated that the plasmid had a relatively high transfer frequency (9.1 × 10−3 to 1.3 × 10−2 per donor cell) to E. coli J53 Azir, suggesting that this plasmid has a high propensity for horizontal transmission (10).
Analysis of the chromosomal sequence revealed just a single locus with homology to known antibiotic resistance genes. This locus encoded a class D oxacillinase that shares its closest homology to the recently described blaOXA-237 gene (13) but has 18 amino acid differences and represents a novel blaOXA allele. There is no evidence (i.e., no transposons or insertion sequences) in the surrounding genetic environment to suggest horizontal acquisition of this gene. Based on the antibiotic profile of blaOXA-237 (13) and the lack of any other antibiotic resistance genes, including other β-lactams, aztreonam resistance in this strain is most likely due to this class D oxacillinase. A complete analysis of the MRSN 10319 genome is ongoing and will provide further information.
This report highlights the limitations of automated identification systems when working with unusual species. Commonly used clinical laboratory identification systems do not include A. schindleri or A. johnsonii on identification panels (manufacturer literature), which can lead to erroneous identification. Due to the high correlation between blaNDM carriage and the Enterobacteriaceae, surveillance strategies for this gene have primarily focused on this group of bacteria. However, given the association of this gene with highly promiscuous plasmids, as well as documented horizontal dissemination of this gene, it is critical that surveillance efforts continue to test all carbapenem-resistant Gram-negative organisms for blaNDM. A number of good techniques exist for detecting NDM-producing Enterobacteriaceae (14), but false-negative and weakly positive results have been observed in this family with the popular MHT (15, 16). Detection in Acinetobacter species remains a challenge due to the potential failure of many techniques, including the MHT and Etest MBL strip (17). Bonnin and colleagues have suggested that for carbapenem-resistant Acinetobacter baumannii, isolates should first be screened using EDTA inhibition-based techniques, followed by further PCR-based techniques in a reference laboratory (17). We suggest that the same method be applied to all other carbapenem-resistant Acinetobacter species isolated from clinical specimens.
Nucleotide sequence accession number.
The novel blaOXA locus described in this study has been assigned the designation blaOXA278 (http://www.lahey.org/studies/; last accessed March 2013), and the complete gene sequence has been deposited at GenBank (accession number KC771279).
ACKNOWLEDGMENTS
Major funding for this project was provided by the Global Emerging Infections Surveillance and Response System (GEIS; grant number C070912WR) and Defense Medical Research and Development Program (DMRDP; grant number D61I10J258).
The material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of Defense.
Footnotes
Published ahead of print 3 April 2013
REFERENCES
- 1. Bonomo RA. 2011. New Delhi metallo-beta-lactamase and multidrug resistance: a global SOS? Clin. Infect. Dis. 52:485–487 [DOI] [PubMed] [Google Scholar]
- 2. Hammerum AM, Toleman MA, Hansen F, Kristensen B, Lester CH, Walsh TR, Fuursted K. 2010. Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect. Dis. 10:829–830 [DOI] [PubMed] [Google Scholar]
- 3. Walsh TR. 2010. Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36(Suppl 3):S8–S14 [DOI] [PubMed] [Google Scholar]
- 4. Waterman P, Kwak Y, Clifford R, Julius M, Onmus-Leone F, Tsurgeon C, Riley M, Black C, McGann P, Lesho E. 2012. A multidrug-resistance surveillance network: 1 year on. Lancet Infect. Dis. 12:587–588 [DOI] [PubMed] [Google Scholar]
- 5. McGann P, Hang J, Clifford RJ, Yang Y, Kwak YI, Kuschner RA, Lesho EP, Waterman PE. 2012. Complete sequence of a novel 178-kilobase plasmid carrying bla(NDM-1) in a Providencia stuartii strain isolated in Afghanistan. Antimicrob. Agents Chemother. 56:1673–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Winstanley T, Courvalin P. 2011. Expert systems in clinical microbiology. Clin. Microbiol. Rev. 24:515–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nemec A, De Baere T, Tjernberg I, Vaneechoutte M, van der Reijden TJ, Dijkshoorn L. 2001. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 51:1891–1899 [DOI] [PubMed] [Google Scholar]
- 9. Milillo M, Kwak YI, Snesrud E, Waterman PE, Lesho E, McGann P. 2013. Rapid and simultaneous detection of blaKPC and blaNDM using multiplex real-time PCR. J. Clin. Microbiol. 51:1247–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 56:1698–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dortet L, Legrand P, Soussy CJ, Cattoir V. 2006. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J. Clin. Microbiol. 44:4471–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 13. Higgins PG, Perez-Llarena FJ, Zander E, Fernandez A, Bou G, Seifert H. 25 February 2013. OXA-235, a novel class D beta-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. doi:10.1128/AAC.02413-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nordmann P, Poirel L, Carrer A, Toleman MA, Walsh TR. 2011. How to detect NDM-1 producers. J. Clin. Microbiol. 49:718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob. Agents Chemother. 55:1274–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Girlich D, Poirel L, Nordmann P. 2012. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 50:477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bonnin RA, Naas T, Poirel L, Nordmann P. 2012. Phenotypic, biochemical, and molecular techniques for detection of metallo-beta-lactamase NDM in Acinetobacter baumannii. J. Clin. Microbiol. 50:1419–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing. M100-S22 CLSI, Wayne, PA [Google Scholar]
- 19. Zapor MJ, Barber M, Summers A, Miller GH, Feeney LA, Eberly LE, Wortmann G. 2010. In vitro activity of the aminoglycoside antibiotic arbekacin against Acinetobacter baumannii-calcoaceticus isolated from war-wounded patients at Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 54:3015–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]