Abstract
As the development of molecular serotyping approaches is critical for Salmonella spp., which include >2,600 serovars, we performed an initial evaluation of the ability to identify Salmonella serovars using (i) different molecular subtyping methods and (ii) a newly implemented combined PCR- and sequencing-based approach that directly targets O- and H-antigen-encoding genes. Initial testing was performed using 46 isolates that represent the top 40 Salmonella serovars isolated from human and nonhuman sources, as reported by the U.S. Centers for Disease Control and Prevention and the World Health Organization. Multilocus sequence typing (MLST) was able to accurately predict the serovars for 42/46 isolates and showed the best ability to predict serovars among the subtyping methods tested. Pulsed-field gel electrophoresis (PFGE), ribotyping, and repetitive extragenic palindromic sequence-based PCR (rep-PCR) were able to accurately predict the serovars for 35/46, 34/46, and 30/46 isolates, respectively. Among the methods, S. enterica subsp. enterica serovars 4,5,12:i:−, Typhimurium, and Typhimurium var. 5− were frequently not classified correctly, which is consistent with their close phylogenetic relationship. To develop a PCR- and sequence-based serotyping approach, we integrated available data sources to implement a combination PCR-based O-antigen screening and sequencing of internal fliC and fljB fragments. This approach correctly identified the serovars for 42/46 isolates in the initial set representing the most common Salmonella serovars, as well as for 54/63 isolates representing less common Salmonella serovars. Our study not only indicates that different molecular approaches show the potential to allow for rapid serovar classification of Salmonella isolates, but it also provides data that can help with the selection of molecular serotyping methods to be used by different laboratories.
INTRODUCTION
Salmonellosis is a considerable public health concern, as nontyphoidal Salmonella serovars cause an estimated 93.8 million cases of gastroenteritis globally each year (1). The genus Salmonella is divided into two species, S. enterica and Salmonella bongori. S. enterica is further divided into 6 subspecies, including S. enterica subsp. I (subsp. enterica), II (subsp. salamae), IIIa (subsp. arizonae), IIIb (subsp. diarizonae), IV (subsp. houtenae), and VI (subsp. indica) (2). Serotyping has been the traditional method of subtyping Salmonella below the subspecies level(2, 3). Serotyping can provide valuable information regarding likely pathogen sources (as certain serovars are associated with specific hosts or geographical regions), potential disease severity, and potential antimicrobial resistance of Salmonella isolates. The identification of Salmonella serovars thus remains an important public health diagnostic need. There are >2,600 currently recognized Salmonella serovars, with the majority (>1,500) belonging to S. enterica subsp. enterica, which is also the group of greatest clinical relevance due to its common association with humans and warm-blooded animals (4).
Traditional serotyping is performed according to the White-Kauffmann-Le Minor scheme, which identifies the somatic (O) and flagellar (H) antigens based on the agglutination of bacteria with specific sera (2). Despite its widespread use, traditional serotyping does have a number of drawbacks. Serotyping of Salmonella takes at least 3 days to complete, is labor intensive, requires the maintenance of >250 typing sera and 350 different antigens, and is unable to type rough or mucoid strains. Furthermore, traditional serotyping is often not sensitive enough to provide the level of discrimination needed for food-borne illness outbreak investigations, and it cannot be used to infer phylogenetic relationships. Currently, 46 somatic (O) and 114 flagellar (H) variants of Salmonella have been identified (2). The O antigen is a component of the lipopolysaccharide that is exposed on the bacterial cell surface, and multiple O antigens might be expressed at the same time (5, 6). Genes responsible for O-antigen expression (e.g., sugar transferases, O-antigen flippase [wzx], and polymerase [wzy]) are located within a large regulon called the rfb cluster (5). The comparison of wzx and wzy genes from common serogroups has shown that these genes have little similarity even at the amino acid-sequence level, making wzx and wzy appropriate candidates for serogroup-specific primer design (7, 8). Additional work has shown that sugar synthase genes within the rfb cluster can be targeted to distinguish between common serogroups (9). The genes responsible for the flagellin structure are fliC (phase 1 flagellin) and fljB (phase 2 flagellin). Both fliC and fljB are generally conserved at the terminal ends but are highly variable in the central region that encodes antigens (10, 11). In most Salmonella strains, flagellin expression is coordinately expressed via a phase-variation mechanism (12). A number of studies have utilized the variabilities of the rfb region, fliC, and fljB to identify serovars, typically using probe-based assays or PCR strategies (13–15). While these approaches have been reported to show good concordance with traditional serotyping, the limitations of these methods include problems with the characterization of new or unusual serovars or allelic variants that do not react with existing primers or probes (13–15).
In addition to serovar identification through the use of genetic targets that are directly responsible for O- and H-antigen expression, molecular subtyping methods (e.g., pulsed-field gel electrophoresis [PFGE]) can be used to predict the serovars of Salmonella isolates. In addition to PFGE (16, 17), different ribotyping approaches (18–20), repetitive extragenic palindromic sequence-based PCR (rep-PCR) (21, 22), multilocus sequence typing (MLST) (23, 24), and molecular typing based on genomic markers (25–28) have been investigated for their abilities to replace or complement traditional serotyping. While many of these methods have been able to reliably predict a limited set of serovars, they still lack widespread adoption, likely due to the requirements for specialized equipment, as well as a lack of proven reliability for predicting Salmonella serovars. Furthermore, these methods are based on genomic targets that are not directly responsible for antigen expression, which might lead to serovar misidentification. This is particularly the case for a newly emergent serovar (e.g., S. enterica serovar 4,5,12:i:−), which might be misidentified as the serovar of its evolutionary ancestor (29, 30). To facilitate the further development and implementation of DNA-based approaches for serovar identification of Salmonella isolates, we compared different molecular subtyping methods (i.e., PFGE, rep-PCR, ribotyping, and MLST) and a newly implemented combined PCR-and-sequencing-based approach that directly targets O- and H-antigen-encoding genes for their abilities to predict Salmonella serovars.
MATERIALS AND METHODS
Bacterial isolates.
Salmonella isolates were selected to include representation of (i) the top 20 serovars among U.S. human sources, the top 20 serovars among U.S. nonhuman sources, and the top 20 serovars among nonclinical nonhuman sources (all as reported to the CDC) (31) and (ii) the top 20 serovars among human sources worldwide (as reported to the WHO) (32). This strategy identified a total of 40 serovars (see Table S1 in the supplemental material). Two isolates were chosen to represent the most commonly reported S. enterica subsp. enterica serovars (Typhimurium, Enteritidis, Newport, Heidelberg, Kentucky, and Javiana), and a single isolate of S. Typhimurium var. 5− (formerly S. Typhimurium var. Copenhagen; counted as one of the 40 serovars) was included, for a total of 46 isolates. In addition, we assembled a set of 70 isolates that included all additional 63 serovars present in our laboratory strain collection; these isolates represent less-common (rare) serovars not represented in the top-40 set (see Table S1 in the supplemental material). Finally, seven isolates that included incomplete serovar information (e.g., S. enterica subsp. enterica serovar IIIb 35:Rough) or that were identified as “untypeable” by traditional serotyping were also tested. Detailed isolate information, including all sequence data associated with a given isolate, can be found at www.foodmicrobetracker.com under the isolate ID (e.g., FSL R8-1987).
PFGE.
PFGE with XbaI (Roche Molecular Diagnostics, Pleasanton, CA) was performed according to the CDC PulseNet protocol using a CHEF Mapper (Bio-Rad Laboratories, Hercules, CA) (33). The CDC S. enterica subsp. enterica serovar Braenderup strain H9812 was used as the reference (34). Pictures of PFGE gels were taken with the Gel ChemiDoc system (Bio-Rad Laboratories). BioNumerics version 5.1 (Applied Maths, Austin, TX) was used to analyze the PFGE patterns. Similarity analysis was performed using the Dice coefficient, and clustering was performed using the unweighted-pair group method by arithmetic mean with a 1.5% tolerance limit. PFGE patterns for test isolates were compared against a custom PFGE database available at the Cornell Food Safety Laboratory (FSL). This database included, at the time of analysis, 5,935 isolates representing 170 serovars (this database is available upon request). A serovar was assigned to a given test isolate based on the serovar associated with the isolate that provided the top match in the PFGE pattern comparison, and only PFGE patterns that showed ≤3 band differences with the pattern of the test isolate were considered. If a test isolate did not match any isolate in the database with ≤3 band differences, the serovar for the isolate was considered “unidentified.”
rep-PCR.
Salmonella isolates were cultured on brain heart infusion (BHI) agar for 18 h at 37°C, and the UltraClean microbial DNA isolation kit (MO BIO Laboratories, Solana Beach, CA) was used to extract DNA, according to the manufacturer's instructions. All DNA samples were amplified using the DiversiLab Salmonella kit for DNA fingerprinting (bioMérieux, Inc., Durham, NC), according to the manufacturer's instructions. Analysis of rep-PCR patterns was conducted as described previously (21), using DiversiLab software version 3.4. The “top match” feature of the software was utilized, and a query sample that matched a serovar library entry at >85% was considered to represent a positive identification. At the time of analysis, the rep-PCR database included 313 isolates (309 S. enterica subsp. enterica and 4 S. enterica subsp. arizonae isolates) representing 55 serovars.
Ribotyping.
Automated ribotyping with the restriction enzyme PvuII was performed using the RiboPrinter microbial characterization system and reagents from the DuPont Qualicon ribotyping kit, according to the manufacturer's instructions (DuPont Qualicon, Wilmington, DE). Using the RiboPrinter software, PvuII patterns were compared against the DuPont Salmonella PvuII database, which at the time of analysis included 592 isolates representing 227 serovars. The top match was used to predict the serovar of a tested isolate; if no pattern in the DuPont database matched with >70% similarity, the isolate serovar was reported as “unidentified.”
MLST.
Partial sequencing of seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) was performed as described previously (35) at the Cornell University Life Sciences Core Laboratories Center (Ithaca, NY). Sequences were assembled and analyzed using Lasergene 7.2.1 software (DNAStar). Allelic type (AT) and sequence type (ST) numbers were assigned by submitting the sequences and strain information to the Salmonella MLST website (http://mlst.ucc.ie/mlst/dbs/Senterica). When a sequence from a Salmonella isolate matched an existing ST in the database, the serovar information for the existing ST was assigned to our query. For new STs, the nearest ST (matching 6/7 ATs) was used to assign a serovar, and all new ATs (including corresponding electropherograms) and STs were submitted to the MLST database. All sequences for the 7-gene MLST are available at www.foodmicrobetracker.com.
DNA preparation for PCR.
For PCR amplification of the O serogroups, fliC, and fljB, total genomic DNA was extracted from 1 ml of overnight culture in BHI agar, according to the instructions in the QIAamp DNeasy kit (Qiagen Inc., Valencia, CA). DNA concentrations were determined using NanoDrop 1000 (Thermo Scientific, Wilmington, DE) and were standardized to 25 ng/μl. The purity of all genomic DNA preparations was verified using A260/A280 ratios.
PCR detection of O serogroups.
PCR detection of serogroups was performed using (i) a multiplex PCR that identifies the serogroups O:4, O:7, O:8, O:9, and O:3,10 (19) and (ii) two separate single PCRs that identify serogroups O:13 (serogroup set 1) (12) and O:18 (serogroup set 2) (13). PCRs were performed using previously published primers (Table 1) and under optimized PCR conditions (see Table S2 in the supplemental material). PCR products were separated by agarose gel electrophoresis using Tris-acetate-EDTA buffer and were visualized by staining with 0.005% ethidium bromide. PCR products obtained from selected O-antigen PCRs were also sequenced using standard methods, as detailed below.
Table 1.
Summary of primers used to determine Salmonella serovars
| Gene target (serogroup) | Designation | Amplicon | Forward primer | Forward sequence (5′–3′) | Reverse primer | Reverse sequence (5′–3′) | Reference |
|---|---|---|---|---|---|---|---|
| wzx (O:4 [B]) | Multiplex PCR 1 | 230 | F-wzxB | GGC ATA TAT TTC TGT ATT CGC G | R-wzxB | GCC TTA ATT AAG TAA GTT AGT GGA AGC | Herrera-León et al. (8) |
| wzx (O:7 [C1]) | Multiplex PCR 1 | 483 | F-wzxC1 | CAG TAG TCC GTA AAA TAC AGG GTG G | R-wzxC1 | CAA TGC TAT AAA TAC TGT GTT AAA TTG C | Herrera-León et al. (8) |
| wzx (O:8 [C2-C3]) | Multiplex PCR 1 | 154 | F-wzxC2 | ACT GAA GGT GGT ATT TCA TGG G | R-wzxC2 | AAG ACA TCC CTA ACT GCC CTG C | Herrera-León et al. (8) |
| tyv (O:9 [D]) | Multiplex PCR 1 | 615 | F-tyvD | GAG GAA GGG AAA TGA AGC TTT T | R-tyvD | TAG CAA ACT GTC TCC CAC CAT AC | Herrera-León et al. (8) |
| wzx (O:3,10 [E1]) | Multiplex PCR 1 | 345 | F-wzxE1 | TAA AGT ATA TGG TGC TGA TTT AAC C | R-wzxE1 | GTT AAA ATG ACA GAT TGA GCA GAG | Herrera-León et al. (8) |
| wzy (O:13 [G]) | Serogroup set 1 | 90 | O13-wzyF | CTC TTG ATG AAT GTT ATT A | O13-wzyR | GTT AAC CCC TCC TAA TA | Fitzgerald et al. (52) |
| wzx (O:18 [K]) | Serogroup set 2 | 360 | O18F | CTC TAG GAT CAA CTG AAG GTG GTC | O18R | CAA CCC AGC AAT AAA GCA GAA | Fitzgerald et al. (53) |
| fliC | fliC set 1a | ∼1,520 | FL_START | ATG GCA CAA GTC ATT AAT AC | rFSa1 | TTA ACG CAG TAA AGA GAG GAC | Mortimer et al. (36) |
| fliC | fliC set 2a | ∼1,520 | sefliCforatg | CCG AAT TCA TGG CAC AAG TCA TTA ATA CAA AC | sefliCrevstop | CCG GAT CCT TAA CGC AGT AAA GAG AGG ACG T | Imre et al. (37) |
| fljB | fljB set 1a | ∼1,520 | fljBatgfor | CCG AAT TCA TGG CAC AAG TAA TCA ACA CTA A | fljBstoprev | CGG GAT CCT TAA CGT AAC AGA GAC AGC ACG | Imre et al. (37) |
| fljB | fljB set 2a | ∼1,600 | MR-22 fljBF | GGC ACA AGT AAT CAA CAC TAA CA | MR-23 fljBR | CAT TTA CAG CCA TAC ATT CCA TA | Current study |
| fliC or fljBb | Sequencing set 1 | ∼887 | MR-1 forward | AAC AAC AAC CTG CAG CGT GTG | MR-2 reverse | GTC GGA ATC TTC GAT ACG GCT AC | Current study |
When used for sequencing, these primers did not provide full double coverage of the internal variable region of fliC or fljB.
Primers MR-1 forward and MR-2 reverse were used exclusively for sequencing of fliC and fljB PCR products. These primers provided double coverage of the internal variable region of fliC and fljB.
PCR amplification and sequencing of genes encoding H1 and H2 antigens.
Amplifications of fliC and fljB were performed using previously described (36, 37) primers (Table 1, fliC set 1 or 2, fljB set 1) and under optimized PCR conditions (see Table S2 in the supplemental material). We also designed an alternative set of fljB PCR primers (Table 1, fljB set 2) that was used for the amplification of an approximately 1,600-nucleotide (nt) fragment (see Table S2 in the supplemental material for PCR conditions). This set was designed because the previously described set of fljB primers (Table 1, fljB set 1) did not allow for a reliable amplification of fljB, predominately among the isolates representing rare serovars (see Table S1 in the supplemental material, which details the primers that were used for each isolate). Prior to sequencing, all PCR products were treated with exonuclease I and shrimp alkaline phosphatase, according to the manufacturer's instructions (Affymetrix, Cleveland, Ohio). As sequencing with the previously published fliC or fljB primers only provided single coverage of the PCR product, the newly designed primers MR-1_forward and MR-2_reverse were used to obtain double coverage of the variable internal regions in fliC or fljB (Table 1). Sequencing was carried out on the Applied Biosystems automated 3730 DNA analyzer using BigDye Terminator chemistry at the Cornell University Life Sciences Core Laboratories Center. Sequences were assembled and analyzed using Lasergene 7.2.1 software (DNAStar, Madison, WI). BLASTn search analysis was used to compare fliC and fljB sequences with those in GenBank (38) and to infer the type of fliC or fljB antigens. Alignment of fliC and fljB amino acid sequences was performed using the FFT-NS-i method in Multiple Alignment using Fast Fourier Transform (MAFFT) (39), and cluster analysis was performed using the maximum-likelihood (ML) algorithm in RAxML (40) with rapid bootstrapping (100 bootstrap replicates). Amino acid sequence distances (p distances) were calculated using MEGA version 5.0.5 (41). The use of amino acid sequences was chosen to allow for a more reliable alignment of the highly divergent fliC and fljB genes.
Traditional serotyping.
Immunological serotyping was completed by either the New York State Department of Health or the National Veterinary Services Laboratory (Ames, IA) using the Salmonella latex test, according to the manufacturer's instructions (Oxoid, Ogdensburg, NY) (2).
RESULTS
PFGE.
PFGE patterns were generated for all 46 isolates tested and then were compared to a custom database that included PFGE patterns for the isolates representing 170 serovars, including all 40 serovars evaluated here. Using the methods detailed above, serovars were predicted correctly for 35/46 (75%) isolates (Table 2). Among the 11 isolates that were not accurately predicted, 3 isolates were predicted to represent serovars that were not congruent with traditional serotyping: one S. Typhimurium isolate matched S. 4,5,12:i:− (0-band difference), one S. enterica subsp. enterica serovar Saintpaul isolate matched S. Typhimurium (2-band difference), and one S. Typhimurium var. 5− isolate matched S. Typhimurium (0-band difference) and S. Typhimurium var. 5− (0-band difference) (see Table S3 in the supplemental material). No serovar could be assigned for 8/46 isolates, as their PFGE patterns differed by >3 bands from all isolates in the database; these isolates represented S. enterica subsp. enterica serovar Choleraesuis, S. enterica subsp. enterica serovar Give, S. enterica subsp. enterica serovar Mississippi, S. enterica subsp. enterica serovar Orion var. 15+,34+, S. enterica subsp. enterica serovar Reading, S. enterica subsp. enterica serovar Virchow, S. enterica subsp. enterica serovar Weltevreden, and S. enterica subsp. enterica serovar Worthington (Table 2).
Table 2.
Comparison of DNA based subtyping methods used to predict the top 40 Salmonella serovars evaluated in this study
| DNA-based subtyping method | No. of isolates for which the serovar was identified correctly (n = 46) (%) | Incorrectly identified S. enterica serovars (no.) | S. enterica serovars not identified (no.) |
|---|---|---|---|
| MLST | 42 (91) | 4,5,12:i:− (1), Typhimurium var. 5− (1) | Orion var. 15+,34+ (1)a, Reading (1)a |
| Molecular serotyping | 42 (91) | Choleraesuis (1), Senftenberg (1), Typhimurium (1), Typhimurium var. 5− (1) | (0) |
| PFGE | 35 (76) | Saintpaul (1), Typhimurium (1), Typhimurium var. 5− | Choleraesuis (1)b, Give (1)b, Mississippi (1)b, Orion var. 15+,34+ (1)b, Reading (1)b, Virchow (1)b, Weltevreden (1)b, Worthington (1)b |
| Rep-PCR | 30 (65) | Derby (1), Infantis (1), Kentucky (2), Muenster (1), Paratyphi B. var. Java (1), Reading (1), Senftenberg (1), Stanley (1), Typhimurium (1), Virchow (1) | Give (1)d, Javiana (1)c, Orion var. 15+,34+ (1)d, Typhimurium var. 5− (1)d, Weltevreden (1)d |
| Ribotyping | 34 (74) | 4,5,12:i:− (1), Braenderup (1), Give (1), Javiana (1), Muenster (1), Orion var. 15+,34+ (1), Uganda (1) | Blockley (1)e, Dublin (1)e, Montevideo (1)f, Typhi (1)f, Typhimurium var. 5− (1)e |
For unidentified serovars, traditional serotyping information was not available for the most similar isolate(s) in the MLST database.
PFGE patterns for the most similar patterns differed by >3 bands; thus, a serovar could not determined.
DiversiLab percent identity to library strains was <85%.
Serovar not in DiversiLab library at time of analysis.
Serovar was not in ribotype database at time of analysis.
Serovar could not be assigned, as ribotype pattern did not match existing pattern in database at >70%.
rep-PCR.
rep-PCR patterns were generated on the DiversiLab system for all 46 isolates tested. Overall, the DiversiLab rep-PCR system accurately predicted 30/46 (65%) serovars tested when applying an 85% similarity cutoff (Table 2). Of the remaining 16 isolates, 11/16 had rep-PCR patterns that matched an existing pattern in the rep-PCR library at >85% identity, but the assigned serovar was not congruent with traditional serotyping (Table 2). Among the 5 isolates that had rep-PCR patterns with <85% identity to patterns in the DiversiLab library, four represented serovars were not included in the library (S. Give, S. Orion var. 15+,34+, S. Typhimurium var. 5−, and S. Weltevreden; Table 2). While rep-PCR patterns for 5 S. Javiana isolates were in the DiversiLab library, one S. Javiana isolate tested (FSL S5-406) did not match an existing pattern at >85% identity (top match was S. Mississippi at 72.3% identity) (see Table S3 in the supplemental material).
Ribotyping.
Automated ribotyping produced ribotype patterns for all 46 isolates. A total of 34/46 (74%) serovars predicted by ribotyping were congruent with traditional Salmonella serotyping results. Of the 12 serovars that were not accurately predicted, 7 isolates had ribotype patterns that matched database patterns with >70% identity, but the assigned serovars were not congruent with traditional serotyping results (Table 2). Ribotype patterns for S. enterica subsp. enterica serovar Montevideo (isolate FSL S5-630) and S. enterica subsp. enterica serovar Typhi (isolate FSL R6-540) did not match any existing patterns in the database at >70% similarity and thus could not be assigned a serovar, and both S. Montevideo and S. Typhi ribotype patterns were available in the database (see Table S2 in the supplemental material). An additional 3 isolates did not match any existing patterns at >70% and the database did not contain those serovars (i.e., S. enterica subsp. enterica serovar Blockley [isolate FSL S5-648], S. enterica subsp. enterica serovar Dublin [isolate FSL S5-439], and S. Typhimurium var. 5− [isolate FSL S5-786]) (Table 2).
MLST.
The Max Planck 7-gene MLST scheme was able to accurately predict serovars for 42/46 (91%) isolates (Table 2). Two isolates, representing S. 4,5,12:i:− and S. Typhimurium var. 5− (isolates FSL S5-580 and FSL S5-786, respectively), were identified as S. Typhimurium. An additional 2 isolates representing S. Orion var. 15+,34+ (isolate FSL R8-3408) and S. Reading (isolate FSL R8-1987) could not be identified; isolates representing the corresponding STs in the MLST database lacked serovar information. Among the 322 partial housekeeping gene sequences submitted, new ATs were identified for S. Javiana (for isolate FSL S5-406, hisD AT520), S. enterica subsp. enterica serovar Oranienburg (for isolate FSL S5-642, hemD AT315), and S. Give (for isolate FSL S5-487, sucA AT397). A total of 6 new STs were identified for isolates representing S. Javiana (ST1674), S. Montevideo (ST1677), S. Oranienburg (ST1675), S. Dublin (ST1673), S. enterica subsp. enterica serovar Uganda (ST1676), and S. Give (ST1678) (see Table S4 in the supplemental material).
PCRs targeting O-antigen genes allowed for the reliable identification of clinically important Salmonella serogroups, but specific primers for less-common O antigens need to be developed.
PCRs targeting O-antigen genes were used to determine the serogroups of 46 isolates representing clinically important S. enterica subsp. enterica serovars and 70 less-common S. enterica serovars (see Table S1 in the supplemental material). Based on traditional serotyping data, these PCRs were expected to allow for the identification of the O groups for 44/46 isolates representing common serovars and 40/64 isolates representing less-common serovars, for a total of 84/110 isolates (Table 3). PCR-based serogroup results were congruent with immunological serotyping data for all 84 of these isolates, including 44 isolates representing common serovars. Correctly identified serogroups included O:4 (n = 21), O:7 (n = 15), O:8 (n = 16), O:9 (n = 11), O:3,10 (n = 9), O:13 (n = 11), and O:18 (n = 1) (Table 3). Sequencing of selected O-group PCR products revealed limited diversity within a given O group. For example, a 532-nt partial tyvD sequence obtained from six O:9 isolates showed only 4 polymorphic nucleotides, all present in the same isolate (see Fig. S1 in the supplemental material). Also, the sequencing of a 402-nt wzx fragment in one E4 (O:1,3,19) and seven E1 (O:3,10) isolates revealed limited diversity and no polymorphisms that could differentiate E4 from E1 isolates (see Fig. S2 in the supplemental material).
Table 3.
Results of O-group determination using serogroup specific PCRs
| Serogroup | No. of isolates within serogroupa | No. of isolates with a positive PCR result for O group |
No. of isolates with neg. PCR result | ||||||
|---|---|---|---|---|---|---|---|---|---|
| O:4 (B) | O:7 (C1) | O:8 (C2-C3) | O:9 (D1) | O:3,10 (E1) | O:13 (G) | O:18 (K) | |||
| O groups with primers for detection | |||||||||
| O:4 (B) | 21 | 21 | 0 | ||||||
| O:7 (C1) | 15 | 15 | 0 | ||||||
| O:8 (C2-C3) | 16 | 16 | 0 | ||||||
| O:9 (D1) | 11 | 11 | 0 | ||||||
| O:3,10 (E1) | 9 | 9 | 0 | ||||||
| O:13 (G) | 11 | 11 | 0 | ||||||
| O:18 (K) | 1 | 1 | 0 | ||||||
| O groups lacking primers for detection | |||||||||
| O:2 (A) | 1 | 1 | |||||||
| O:9,46 (D2) | 1 | 1b | 0 | ||||||
| O:1,3,19 (E4) | 1 | 1c | 0 | ||||||
| O:11 (F) | 5 | 5d | 0 | ||||||
| O:6,14 (H) | 1 | 1 | |||||||
| O:16 (I) | 4 | 4 | |||||||
| O:28 (M) | 2 | 2 | |||||||
| O:30 (N) | 1 | 1 | |||||||
| O:35 (O) | 4 | 4 | |||||||
| O:38 (P) | 1 | 1 | |||||||
| O:39 (Q) | 1 | 1 | |||||||
| O:40 (R) | 2 | 2 | |||||||
| O:51 | 1 | 1 | |||||||
| O:54 | 1 | 1e | 0 | ||||||
| Untypeable | 6 | 1 | 2 | 3 | |||||
Serogroups for 116 isolates determined by immunological serotyping. Among the 116 isolates, 95 yielded positive PCR results with our primers (Table 1). The remaining 21 isolates did not show amplification with any of the 7 O-group primer sets tested.
Analysis of serogroup O:9 primers revealed primer match to S. Baildon (O:9,46).
Primer design was based on Salmonella sequences representing serogroups O:3,10 and O:1,3,19.
Analysis of serogroup O:7 primers revealed a nonspecific primer match to S. Rubislaw (O:11).
Factor O:54 is plasmid-controlled and might mask factors O:6,7,14 (C1) for S. Montevideo.
By traditional serotyping, 26 isolates represented O groups that were not targeted by the O-group PCR assays used. Among these 26 isolates, 18 did not yield PCR products with any of the O-group PCRs evaluated (Table 3). PCR inhibition could be excluded because DNA purity was confirmed by A260/A280 ratios and these genomic DNA preparations showed amplification with other PCR primers. However, 8 isolates each yielded a positive PCR result with one primer set; for these isolates, including O groups O:11 (n = 5), O:9,46 (n = 1), O:1,3,19 (n = 1), and O:54 (n = 1), PCR-based serogroups were not congruent with traditional typing. All five O:11 isolates were positive with O:7 primers (Table 3). We subsequently found that the serogroup O:7 forward (22/22 nt) and reverse (23/23 nt) primers matched tyv (an O-antigen gene present in the rfb region) in S. enterica subsp. enterica serovar Rubislaw (O:11), with a predicted amplicon size (615 nt) that matched the size expected for O:7. The only isolate representing serogroup O:9,46 was positive with the O:9 primers. The serogroup O:9 forward (24/25 nt) and reverse (28/29 nt) primers matched tyv in S. enterica subsp. enterica serovar Baildon (O:9,46). Sequencing and alignment of tyvD in serogroup O:9 revealed that this gene is highly conserved (see Fig. S1 in the supplemental material). The one isolate of serogroup O:1,3,19 was positive with the O:3,10 primers, and sequencing and alignment revealed that wzx was highly conserved between the two serogroups (see Fig. S2 in the supplemental material) and the primers had been designed to detect both O:3,10 and O:1,3,19 (8). S. Montevideo (serogroup O:54) was detected by O:7 primers; this exception was not completely unexpected, as Montevideo serogroup expression is plasmid controlled and might mask factor O:7 (42).
Among the 7 isolates that could not be classified by immunological serotyping, three isolates yielded positive results with one of the O-group primer sets used here. These isolates were classified as serogroups O:3,10 (isolate FSL R8-2289) and O:18 (isolates FSL R6-592 and FSL R8-904) (Table 4). The remaining 4 untypeable isolates (FSL R8-3567, FSL A4-524, FSL R8-143, and FSL R8-756) did not yield PCR products with any of the O-group primer sets used.
Table 4.
Molecular serotyping results for serologically untypeable isolatesa
| Isolate | Immunological serotyping resultse |
Molecular serotyping results |
||||||
|---|---|---|---|---|---|---|---|---|
| Serogroup | H1 antigen | H2 antigen | Salmonella serovar | Serogroupc | H1 antigen(s) | H2 antigen(s) | Salmonella serovarb | |
| FSL R8-3567 | O:35 (O) | NA | NA | IIIb 35:Rough | ND | l,v | 1,5 | Requires O-antigen identification |
| FSL R6-592 | NA | NA | NA | Untypeable | O:18 (K) | z4,z23 | S. I, II, or IIIa 18:z4,z23:− | |
| FSL R8-904 | NA | NA | NA | Untypeable | O:18 (K) | z4,z23 | S. I, II, or IIIa 18:z4,z23:− | |
| FSL R8-2289 | NA | NA | NA | Untypeable | O:3,10 (E1)d | g,[s],t | S. II 3,10:g,[s],t:− | |
| FSL A4-524 | NA | NA | NA | Untypeable | ND | y | 1,7 | Requires O-antigen identification |
| FSL R8-143 | NA | NA | NA | Untypeable | ND | z52 | 1,7 | Requires O-antigen identification |
| FSL R8-756 | NA | NA | NA | Untypeable | ND | k | 1,5,7 | Requires O-antigen identification |
Represents all isolates where immunological determination of antigens was inhibited by strain phenotype (e.g., rough, mucoid, or nonmotile).
Species other than S. enterica subsp. enterica are designated by the following symbols: II for serovars of S. enterica subsp. salamae; IIIa for serovars of S. enterica subsp. arizonae.
ND indicates serogroup was not detected with primer sets tested in this study (i.e., primer sets for detection of O:4, O:7, O:8, O:9, O:3,10, O:13, and O:18).
Serogroup primers for O:3,10 (E1) were also found to detect serogroup O:1,3,19 (E4).
NA, not available.
fliC and fljB sequencing allows for H1 and H2 antigen prediction that is congruent with serological typing.
Among the 109 isolates with serovar information that were tested, 28 H1 antigens and 15 unique H2 antigens were represented. Flagellar antigens for these isolates were identified through a molecular approach that included amplification of fliC and fljB, encoding H1 and H2, respectively, and sequencing to obtain coverage of the internal variable region. The results for PCR- and sequence-based determination of H1 antigens were congruent with traditional serotyping for all 109 isolates (see Table S1 in the supplemental material), while H2-antigen determination was congruent with traditional serotyping for 104/109 isolates. Isolates for which molecular and traditional H2-antigen determinations did not match included 2 from the isolate set representing the 40 most common serovars, as well as three isolates from the set representing less-common serovars (Table 5). Specifically, for one S. Typhimurium isolate (FSL S5-433), we obtained a PCR product but were unable to sequence the product, and for one S. Choleraesuis isolate, no PCR product was obtained with the fljB primers. In addition, for an S. enterica subsp. enterica serovar Corvallis isolate, sequencing determined the H2 antigens to be 1,5, while immunological serotyping indicated [z6], and for an S. enterica subsp. enterica serovar Wandsworth isolate, sequencing determined the H2 antigens to be 1,7, while traditional serotyping indicated a 1,2 H2 antigen (Table 5). Finally, for one S. enterica subsp. enterica serovar Wangata isolate, no fljB PCR product could be obtained (Table 5). While H1 antigens could be determined by molecular serotyping for all seven untypeable isolates tested here, the H2-antigen-encoding gene was only amplified for four isolates, which were identified as 1,5 (n = 1), 1,7 (n = 2), and 1,5,7 (n = 1) (Table 4).
Table 5.
Discrepancies between traditional and molecular serotyping of O, H1, and H2 antigens
| Isolate set | Isolate | Discrepancy | Immunological serotyping results |
Molecular serotyping results |
Conclusion | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Serogroup | H1 antigen(s) | H2 antigen(s) | S. enterica serovar | Serogroup | H1 antigen(s) | H2 antigen(s) | S. enterica serovar | ||||
| Top 40 | FSL S5-433a | H2 antigen | O:4 (B) | i | 1,2 | Typhimurium | O:4 (B) | i | *b | 4,5,12:i:− | fljB sequencing failureb |
| FSL R8-3632c | H2 antigen | O:7 (C1) | c | 1,5 | Choleraesuis | O:7 (C1) | c | * | 6,7:c:− | fljB primer exception | |
| FSL S5-658 | Serogroup | O:1,3,19 (E4) | g,[s],t | Senftenberg | O:3,10 (E1)d | g,[s],t | * | Westhampton | Nonspecific serogroup primer | ||
| Rare 70 | FSL R8-092 | H2 antigen | O:8 (C2-C3) | z4, z23 | [z6] | Corvallis | O:8 (C2-C3) | z4, z23 | 1,5 | 6,8:z4,z23:1,5 | H2 identification error |
| FSL R8-1542 | H2 antigen | O:9 (D1) | z4, z23 | 1,7 | Wangata | O:9 (D1) | z4, z23 | * | 9,12:z4,z23:− | fljB primer exception | |
| FSL R6-199 | Serogroup | O:9,46 (D2) | a | e,n,x | Baildon | O:9 (D1)e | a | e,n,x | Lomalinda | Nonspecific serogroup primere | |
| FSL R6-526 | H2 antigen | O:39 (Q) | b | 1,2 | Wandsworth | NAf | b | 1,7 | Incompletef | H2 identification error | |
| FSL A4-595 | Serogroup | O:11 (F) | k | e,n,x,[z15] | Kisarawe | O:7(C1) | k | e,n,x | Singapore | Nonspecific serogroup primerg | |
| FSL R8-3524 | Serogroup | O:11 (F) | i | 1,2 | Aberdeen | O:7 (C1) | i | 1,2 | Augustenborg | Nonspecific serogroup primerg | |
| FSL R8-3555h | Serogroup | O:11 (F) | a | e,n,z15 | Luciana | O:7 (C1) | a | e,n,z15 | 6,7:a:e,n,z15 | Nonspecific serogroup primerg | |
| FSL S5-477 | Serogroup | O:11 (F) | r | e,n,x | Rubislaw | O:7 (C1) | r | e,n,x | 6,7:r:e,n,x | Nonspecific serogroup primerg | |
| FSL S5-654 | Serogroup | O:11 (F) | z | z6 | Nyanza | O:7 (C1) | z | z,6 | S. enterica subsp. II | Nonspecific serogroup primerg | |
Repeated immunological serotyping confirmed the isolate was S. Typhimurium.
Primers amplified PCR product of expected size. Asterisks indicate that sequencing quality was noisy and deteriorated and could not be interpreted.
Repeated immunological serotyping confirmed the isolate was S. Choleraesuis.
Primers targeting serogroup O:3,10 also detected serogroup O:1,3,19.
Primers targeting serogroup O:9 also detected serogroup O:9,46.
NA indicates primers were not available for the detection of serogroup O:39, which led to an incomplete molecular method-based serovar result.
Primers targeting serogroup O:7 also detected serogroup O:11.
Isolate chosen to represent five serogroup O:11 (F) isolates with O-group discrepancy. Repeated immunological serotyping confirmed the isolate was S. Luciana (serogroup O:11).
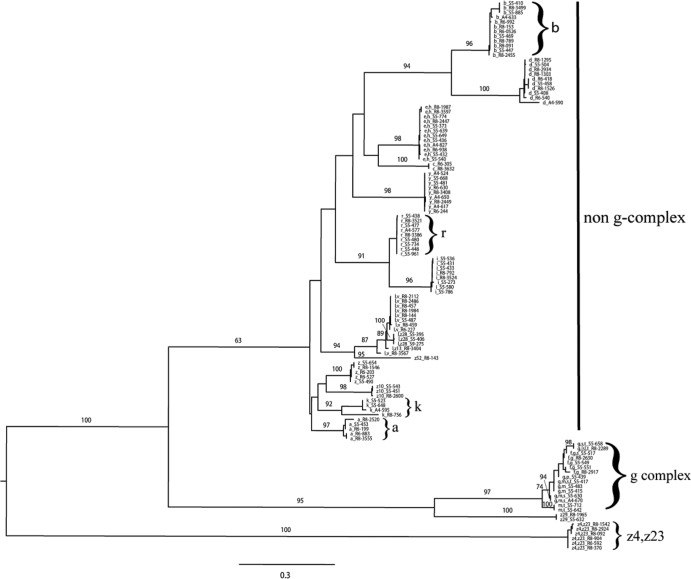
Cluster analysis performed on the 116 partial fliC amino acid sequences obtained here (Fig. 1) showed three distinct clades that represented (i) the g complex with g or m,t antigenic factors, (ii) the z4,z23 antigenic group, and (iii) a large cluster with predominately single antigens (e.g., a or b, those described previously as the “non-g-complex”) (36). The tree also included a large number of well-supported nodes (bootstrap values, >90) within these clades, typically supporting branches that included sequences for a given H1 antigen (a total of 26 unique antigenic factors were represented in this tree). Most fliC antigenic groups represented highly homologous sequences; for example, the sequence similarities within antigenic group r were >99%. However, not all fliC antigen groups were as homologous; for example, the sequence similarities for antigenic group k ranged from 74.1% to 100%. Despite this, the k antigenic group represented a clearly defined clade.
Fig 1.
Midpoint-rooted maximum-likelihood phylogenetic tree of partial fliC amino acid sequences from 116 Salmonella isolates representing 46 common, 63 uncommon, and 7 untypeable serovars. The scale represents the estimated number of amino acid substitutions per site. Numerical values represent the percentage of bootstrap replications that support the respective node. Bootstrap values of >60 are shown for major clades. Each label shows the H1 antigen followed by Food Safety Laboratory (FSL) number; e.g., b_S5-410 indicates H1 antigen b, isolate FSL S5-410.
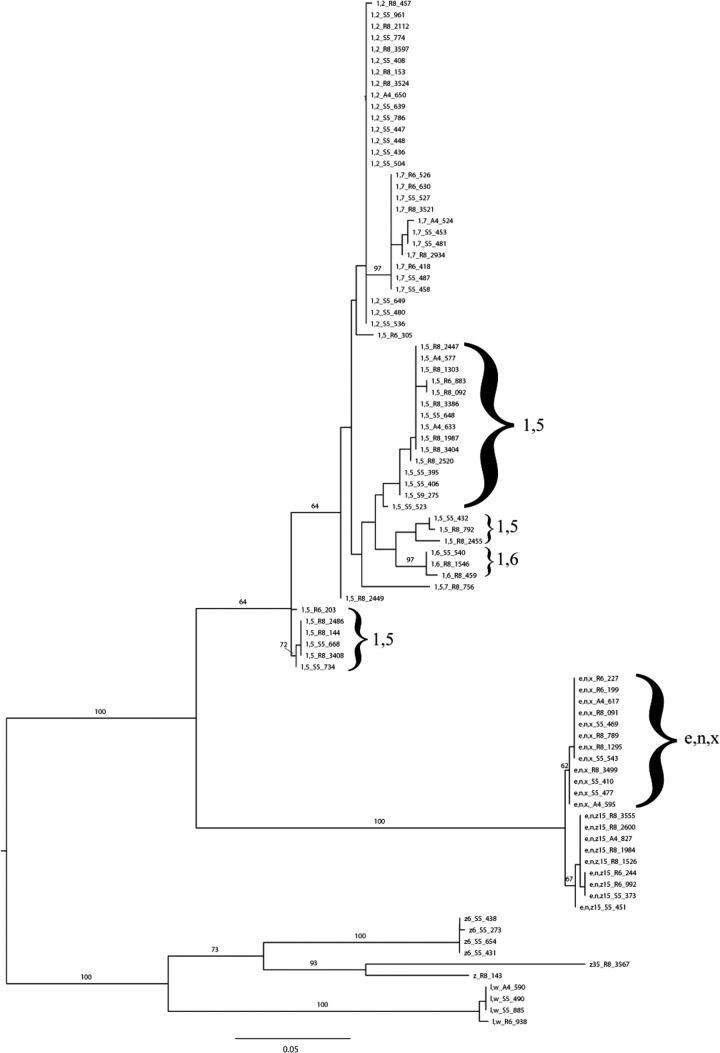
Cluster analysis of 90 fljB partial amino acid sequences (Fig. 2) also showed that the majority of the 11 unique antigenic factors (represented by 32 isolates representing common serovars, 54 isolates representing rare serovars, and 4 untypeable isolates) grouped into well-defined clades, with many antigenic groups displaying a high level of amino acid homology. For example, the partial amino acid sequence similarities for antigenic group e,n,x ranged from 99.5 to 100%. Antigenic group 1,5 showed the lowest level of homology, as its sequence similarities ranged from 88.4 to 100%, and even though this group is paraphyletic with the amino acid sequences for antigenic factors 1,6, fljB sequencing still allowed for antigen determination that was congruent with traditional serotyping. Overall, phylogenetic trees based on partial amino acid sequences for fliC and fljB display clearly defined clusters that allow for the identification of antigenic groups, indicating their potential for sequence-based identification of H1 and H2 antigens, respectively.
Fig 2.
Midpoint-rooted maximum-likelihood phylogenetic tree of 90 partial fljB amino acid sequences from Salmonella isolates representing 32 common, 54 uncommon, and 4 untypeable serovars. The scale represents the estimated number of amino acid substitutions per site. Numerical values represent the percentage of bootstrap replications that support the respective node. Bootstrap values of >60 are shown for major clades. Each label shows the H2 antigen followed by the Food Safety Laboratory (FSL) number; e.g., 1,2_R8-457 indicates H2 antigen 1,2, isolate FSL R8-457.
Comparison of DNA-based subtyping methods and their ability to predict serovars.
Based on the 46 isolates representing the 40 most common Salmonella serovars, the predictive ability of DNA-based subtyping methods evaluated in this study ranged from 30/46 (65%, rep-PCR) to 42/46 (91%, MLST and molecular serotyping) (Table 2). Salmonella serovars 4,5,12:i:−, Typhimurium, and Typhimurium var. 5− represented the 3 serovars for which molecular methods were most frequently unable to predict a serovar that was congruent with traditional serotyping.
DISCUSSION
Salmonella serotyping remains a critical component of Salmonella surveillance efforts, as it facilitates the rapid identification and source tracking of salmonellosis outbreaks, particularly if rapid access to molecular subtyping, such as PFGE, is not available. Traditional serotyping not only provides subtyping data that allow for worldwide comparison, which has facilitated the detection of a number of salmonellosis outbreaks with international scope (43–45), but it also facilitates comparison with historical data sets, since serotyping has been in use for about 70 years. As new methods for serotyping and subtyping of Salmonella are developed, it is thus important that these methods can be referenced and correlated to serovars according to the existing White-Kauffmann-Le Minor scheme, both to maintain the continuity of serovar data and to facilitate communication with laboratories that use traditional serotyping approaches. Conceptually, molecular approaches to the serotyping of Salmonella might use either (i) characterization of genetic targets that are directly responsible for O- and H-antigen expression or (ii) genetic characterization of Salmonella through banding- or sequence-based subtyping methods (targeting genes unrelated to O- and H-antigen expression), followed by serovar prediction through comparison with databases that contain references patterns for isolates with traditional serovar information.
Our study indicates that (i) serovar prediction based on banding-pattern-based methods (i.e., PFGE, rep-PCR, and ribotyping) and DNA-sequence typing schemes (i.e., MLST) is feasible for most serovars but requires large and comprehensive databases and that (ii) sequence-based serotyping provides an alternative method to SNP- or microarray-based O- and H-antigen determination or subtyping-based serovar prediction.
Serovar prediction based on banding-pattern-based methods and DNA-sequence-typing schemes is feasible for most serovars but requires large and comprehensive databases.
For banding-pattern-based subtyping methods, the ability to correctly predict serovars ranged from 65% to 76% for isolates representing the 40 most common Salmonella serovars, and by comparison, MLST correctly predicted the serovars of 91% of these isolates. Previous studies typically only tested the ability of one or a few subtyping methods to predict serovars in isolates representing limited diversity and a small number of serovars (19, 21, 22, 46–48). For example, Gaul et al. (47) compared one banding-pattern method, PFGE, to traditional serotyping on a collection of 674 swine Salmonella isolates. Future large-scale studies will be necessary, though, to determine whether other subtyping methods that do not target genetic targets that are directly responsible for O- and H-antigen expression (e.g., 20, 27, 28) can provide reliable serovar prediction.
In general, if subtyping data are to be used for serovar prediction, large and comprehensive libraries of subtype patterns are needed. While it would be ideal for these databases to represent at least a majority of the >2,600 Salmonella serovars, a database that includes the majority of typically encountered serovars might be sufficient for many applications. We specifically observed that in some cases, common serovars could not be identified due to database limitations; i.e., the serovar was not available in the database. In contrast to most databases for banding-pattern methods, which are typically proprietary (e.g., for automated ribotyping, rep-PCR) or have restricted access (e.g., PulseNet), MLST is characterized by the availability of open-source databases (http://pubmlst.org/databases.shtml), with continuous community additions of subtype data. Among the subtype methods evaluated, PFGE and MLST have the largest databases, even though the PulseNet PFGE database could not be used for the study reported here, as it is not publicly available. However, the PFGE database is available for PulseNet laboratories, an important group of end users for molecular serotyping methods. One recent study indeed indicates that the PulseNet database might be a valuable tool for predicting Salmonella serovars based on PFGE data (16). While the Salmonella MLST database is large (it included >5,700 Salmonella isolates and >600 serovars as of 15 October 2012), a recent study suggested that the reliable MLST-based prediction of Salmonella serovars might remain challenging (24). In particular, this study showed that a number of phylogenetic groups (e-BURST groups) contained multiple serovars and that many serovars are distributed among distinct e-BURST groups, suggesting polyphyletic origins. In our study, rather than using phylogenetic groupings to predict serovars, we used perfect ST matches to isolates in the MLST database to predict serovars; data for closely related isolates (matches in 6 of 7 ATs) were used to predict the serovar for a query isolate only in cases where no perfect ST match was available. While this approach is more pragmatic and might be more likely to not yield a “match” that allows for serovar prediction, based on our data, it shows a good ability to predict serovars. Importantly, traditional serotyping of Salmonella has been estimated to allow for correct serovar identification with about 92% to 95% of isolates (25), suggesting that at least for the isolate set used here, the accuracy of MLST for prediction of serovars is in the same range as that expected for traditional serotyping. For example, Wattiau et al. (49) reported that 90.8% of 754 S. enterica subsp. enterica isolates were correctly serotyped by classical methods, with 9.1% of isolates showing no results with classical serotyping due to strain autoagglutination or lack of antigen expression.
While the development of larger databases for subtyping methods might allow for some improvements with regard to their ability to correctly predict Salmonella serovars, there are inherent limitations to serovar prediction by subtyping methods, for example, those detailed by Achtman et al. (24) for MLST-based prediction of serovars. Our data specifically support that many subtyping methods are likely not able to correctly identify and differentiate the closely related Salmonella serovars Typhimurium (4,5,12:i:1,2), 4,5,12:i:−, and Typhimurium var. 5−. This is consistent with recent studies (16, 47) that also showed that the majority of isolates for which serovars were not correctly predicted by PFGE belonged to S. 4,5,12:i:−; in one study, 135 misclassified S. 4,5,12:i:− isolates were predicted to either be S. Typhimurium (95 isolates) or S. Typhimurium var. 5− (40 isolates) (16). Similar limitations with closely related Salmonella serovars have been reported when evaluating ribotyping; in one study, 20 S. 4,5,12:i:− isolates were predicted to be S. Typhimurium (19). While rep-PCR was reported to correctly predict S. 4,5,12:i:− in one study with three S. 4,5,12:i:− isolates (21), our study identified problems with the correct prediction of S. 4,5,12:i:− across banding-pattern-based subtyping methods. This is consistent with the observation that strains of this serovar appear to represent multiple independent emergence events from S. Typhimurium ancestors. In addition, previous studies have also shown that subtyping methods can, in some instances, not correctly predict serovars that differ by one or two antigens, such as (i) S. Newport (subsp. I 6,8,20:e,h:1,2) and S. enterica subsp. enterica serovar Bardo (subsp. I 8:e,h:1,2) (50) or (ii) S. enterica subsp. enterica serovar Hadar (subsp. I 6,8:z10:e,n,x) and S. enterica subsp. enterica serovar Istanbul (subsp. I 8:z10:e,n,x) (21). On the other hand, Salmonella characterization methods that allow for phylogeny reconstruction (e.g., MLST) might provide some advantages over traditional or molecular serotyping approaches, which do not provide phylogenetic information. For example, MLST can differentiate isolates into distinct phylogenetic groups even if they represent the same serovar, and this has been demonstrated for a number of polyphyletic serovars (51).
Sequence-based serotyping provides an alternative method to SNP- or microarray-based O- and H-antigen determination.
Methods that directly characterize genetic targets that are responsible for O and H antigens conceptually represent an attractive opportunity for molecular serotyping, which should address a number of the drawbacks of serovar prediction based on molecular subtyping methods. To date, some methods have been developed that use primers and probes in various assay formats to detect specific O-, H1-, and H2-antigen markers (within the rfb cluster, fliC, and fljB, respectively), including a Luminex-based system (15, 52) and ArrayTube genoserotyping tool (13). In initial evaluations, these methods have demonstrated good congruency with traditional serotyping. For example, the Luminex-based system developed by the CDC allowed accurate O-group prediction for 362/384 isolates (94.3%) representing 6 common O groups (52) and accurate H-antigen prediction for 461/500 isolates (92.2%) (15). In a smaller study, the ArrayTube genoserotyping tool allowed for correct serovar prediction for 76/100 (76%) isolates (13). While these methods offer the potential for rapid results, ease of use, and high-throughput molecular serovar prediction, including for both rough and mucoid strains, these methods can currently only identify a portion of the >1,500 S. enterica subsp. I serovars. For example, the most recently described Luminex assay was not able to determine H antigens for 46/500 isolates due to a limited number of probes (15), and the ArrayTube genoserotyping tool is currently only able to detect 41/114 flagellar antigens (13). While both of these approaches appear to work reasonably well for the identification of common serovars where sufficient genetic information (e.g., full-genome sequence data) is available for design of appropriate reagents (i.e., primers and probes), difficulties are likely encountered when these systems are challenged with isolates representing rare serovars that were not used for the design of the primers or probes. Examples of specific concerns include (i) no reaction with primer and probes because the genes encoding O or H antigens are not targeted by primer and probes and (ii) false-positive results for a given O or H antigen if primers and probes target a region that is conserved between common and rare antigens that were not considered in the assay design. An additional concern for all methods that target the genes responsible for antigen expression includes the potential for detection of alleles (primarily fljB) that might not be expressed due to a mutation in the phase-variation mechanism, even though the genes are still present in the genome (11); this would lead to misclassification of monophasic serovars.
In contrast to molecular serotyping systems that rely on primers and probes to identify genes that determine or indirectly correlate to the antigenic formula for Salmonella isolates, we implemented an approach that combines (i) PCR-based detection of genes that are specific for a given O antigen based on previous studies that used PCR to identify major O-antigen groups (8, 53) and (ii) PCR amplification of fliC and fljB, followed by sequencing of the internal variable region of these genes to allow for H1- and H2-antigen determination. Overall, this approach allowed for the correct identification of 91% of the isolates representing the 40 most common serovars and of 86% of the isolates representing less-common serovars. While the sequencing of fliC and fljB has previously been used to discover target sequences for the development of probe-based molecular-serotyping approaches, we are not aware of any comprehensive studies that used the sequencing of these two genes as the primary approach for molecular serotyping. While our data suggest that PCR-based O-antigen typing along with fliC and fljB sequencing presents a viable approach for molecular serotyping, some challenges remain in developing this method so that it can be used broadly and allow for serotyping of a wide range of Salmonella serovars. For one, our current method only detects 7 common O antigens, with some primers showing a positive reaction with two antigens. This causes some false-positive results, including one primer set that yields positive results with both serogroup O:3,10 (E1) and O:1,3,19 (E4) isolates (8) and a set that yields positive results with both the serogroups O:7 (C1) and O:11 (F). The design of better PCR primers and approaches that use PCR and subsequent sequencing of target genes that contribute to O-antigen expression should, in the future, be able to address this issue. Specifically, as full-genome sequences for isolates representing additional O groups become available (54, 55), the design of primers capable of detecting all 46 Salmonella serogroups should be feasible. Without full-genome sequences and the ability to compare rfb clusters, the ability to design new robust O-antigen primers for serogroup determination is limited. With regard to the identification of H1 and H2 antigens, the design of primer sets that allow for the reliable amplifications of fliC and fljB remains a challenge. These genes include internal variable and external conserved regions, which represent a challenge in the design of primers that only amplify the target gene (i.e., either fliC or fljB) and allow for reliable amplification among diverse serovars. For example, we found that previously reported fljB primers failed to amplify fljB in a number of isolates representing less-common serovars. Although the majority of isolates evaluated here allowed for successful fliC and fljB amplification, even with the new set of fljB primers designed here, we found a few exceptions, including the inability to amplify fljB in one S. Choleraesuis isolate. This supports the need to develop additional or improved primers. Again, the availability of full-genome sequences for additional serovars should help in the design of improved primers for fliC and fljB amplification, even though the use of more than one primer set might be necessary to allow for amplification in isolates representing diverse serovars. Additional testing of clinical and nonclinical isolates, including a blinded analysis, will be necessary to fully validate the current set of primers, plus any newly developed primers, for molecular serotyping. Genome sequences should also facilitate the development of PCR-based approaches for the detection of rare flagellar antigens that are encoded by other genes (56). Finally, the development of robust and large fliC and fljB sequence databases will be necessary to allow for broad use of the sequencing-based molecular serotyping approaches described here. To this end, we have deposited the fliC and fljB sequence data reported here in the public Food Microbe Tracker database (www.foodmicrobetracker.com) (57).
Implementation of the PCR- and sequencing-based molecular serotyping scheme detailed here should be financially feasible for most laboratories. In a clinical setting, DNA would be isolated from Salmonella isolates and then used for up to 3 PCRs for serogroup determination (in a multiplex format) and 2 PCRs for fliC and fljB determination. PCR products for fliC and fljB would then be purified and sequenced. The resulting sequences could be queried against a publicly available or an in-house database, and antigens for the Salmonella isolate could be assigned. As the cost of 7-gene MLST has been estimated at <$35 per isolate (24) when typing ∼200 isolates per week, the molecular serotyping procedure described here, which involves sequencing of 2 genes plus up to three multiplex PCRs, should only cost about $15 to $20 per isolate. This is less expensive than traditional serotyping (estimated at $35 to $185 per isolate) (20).
Conclusions.
As a variety of efforts are under way to replace or supplement the traditional serotyping of Salmonella with molecular methods, many laboratories are faced with decisions as to which technologies or approaches to implement. Current approaches use either serovar prediction based on molecular subtyping data or the direct characterization of genes affecting O- or H-antigen expression. Among the methods evaluated here, sequencing-based approaches, including (i) MLST and (ii) a combination of a PCR-based O-antigen screening and sequencing of internal fliC (H1 antigen) and fljB (H2 antigen) fragments, provided the best serovar prediction. Both of these methods also use equipment that can be used for a variety of applications, compared to the more specialized equipment that is used for many banding-pattern-based subtyping (e.g., ribotyping, rep-PCR) or other molecular serotyping methods that were not evaluated here (e.g., PremiTest [49], Luminex [15, 52], or ArrayTube genoserotyping [13]). This might favor the implementation of PCR- and sequencing-based methods in some laboratories, particularly as advances in sequencing technology might make these methods more attractive. Sequencing-based approaches for serovar determination can also be easily integrated into whole-genome sequencing-based approaches for Salmonella characterization, which are emerging as a viable alternative to other subtyping methods (58, 59). Sequence data for fliC, fljB, genes in the rfb cluster, and genes targeted by MLST can be rapidly extracted from full-genome sequence data and used for serovar identification. In the future, given sufficient databases, the clustering of full-genome data should also allow for accurate serovar classification in all but a few cases (e.g., where a recent genetic change occurred in a gene that is responsible for serovar expression). Our data also indicate that banding-pattern-based subtyping methods might have the potential to allow for serovar prediction which might be adequate under some conditions, particularly for users that have or can develop larger databases that contain subtype patterns for isolates representing diverse serovars or at least the serovars typically encountered by a given laboratory. In addition, the combination of multiple molecular and possibly traditional serotyping approaches will facilitate improved serovar classification of Salmonella.
Importantly, the combination of a PCR-based O-antigen screening and sequencing of internal fliC and fljB fragments reported here allows for continuity with traditional serotyping data. While some authors have proposed that MLST-based approaches should fully replace serotyping (24), we believe that compatibility with traditional serovar data is critical for Salmonella characterization, at least in the medium-term future. On the other hand, a combination of MLST or other phylogenetic characterization methods with the molecular serotyping approach described will provide considerable advantages over using only one of these approaches.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the USDA special research grants 2006-34459-16952 and 2010-34459-20756.
We thank Sherry Roof and Emily Wright for their assistance with the subtyping of Salmonella isolates.
Footnotes
Published ahead of print 3 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03201-12.
REFERENCES
- 1. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness” Studies 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50:882–889 [DOI] [PubMed] [Google Scholar]
- 2. Grimont P, Weill FX. 2007. Antigenic formulae of the Salmonella serovars, 9th ed WHO Collaborating Centre for Reference and Research on Salmonella, Geneva, Switzerland [Google Scholar]
- 3. Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PA, Weill FX. 2010. Supplement 2003–2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 161:26–29 [DOI] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention 2011. Salmonella surveillance: annual summary, 2011. U.S.. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 5. Samuel GG, Reeves PP. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503–2519 [DOI] [PubMed] [Google Scholar]
- 6. Reeves PR, Hobbs M, Valvano MA, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz CR, Rick PD. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495–503 [DOI] [PubMed] [Google Scholar]
- 7. Fitzgerald C, Sherwood R, Gheesling LL, Brenner FW, Fields PI. 2003. Molecular analysis of the rfb O antigen gene cluster of Salmonella enterica serogroup O:6,14 and development of a serogroup-specific PCR assay. Appl. Environ. Microbiol. 69:6099–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herrera-León S, Ramiro R, Arroyo M, Díez R, Usera MA, Echeita MA. 2007. Blind comparison of traditional serotyping with three multiplex PCRs for the identification of Salmonella serotypes. Res. Microbiol. 158:122–127 [DOI] [PubMed] [Google Scholar]
- 9. Luk JM, Kongmuang U, Reeves PR, Lindberg AA. 1993. Selective amplification of abequose and paratose synthase genes (rfb) by polymerase chain reaction for identification of Salmonella major serogroups (A, B, C2, and D). J. Clin. Microbiol. 31:2118–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joys TM. 1985. The covalent structure of the phase-1 flagellar filament protein of Salmonella Typhimurium and its comparison with other flagellins. J. Biol. Chem. 260:15758–15761 [PubMed] [Google Scholar]
- 11. McQuiston JR, Parrenas R, Ortiz-Rivera M, Gheesling L, Brenner F, Fields PI. 2004. Sequencing and comparative analysis of flagellin genes fliC, fljB, and flpA from Salmonella. J. Clin. Microbiol. 42:1923–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Silverman M, Zieg J, Hilmen M, Simon M. 1979. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc. Natl. Acad. Sci. U. S. A. 76:391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franklin K, Lingohr EJ, Yoshida C, Anjum M, Bodrossy L, Clark CG, Kropinski AM, Karmali MA. 2011. Rapid genoserotyping tool for classification of Salmonella serovars. J. Clin. Microbiol. 49:2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshida C, Franklin K, Konczy P, McQuiston JR, Fields PI, Nash JH, Taboada EN, Rahn K. 2007. Methodologies towards the development of an oligonucleotide microarray for determination of Salmonella serotypes. J. Microbiol. Methods 70:261–271 [DOI] [PubMed] [Google Scholar]
- 15. McQuiston JR, Waters RJ, Dinsmore BA, Mikoleit ML, Fields PI. 2011. Molecular determination of H antigens of Salmonella by use of a microsphere-based liquid array. J. Clin. Microbiol. 49:565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zou W, Lin WJ, Hise KB, Chen HC, Keys C, Chen JJ. 2012. Prediction system for rapid identification of Salmonella serotypes based on pulsed-field gel electrophoresis fingerprints. J. Clin. Microbiol. 50:1524–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kérouanton A, Marault M, Lailler R, Weill FX, Feurer C, Espié E, Brisabois A. 2007. Pulsed-field gel electrophoresis subtyping database for foodborne Salmonella enterica serotype discrimination. Foodborne Pathog. Dis. 4:293–303 [DOI] [PubMed] [Google Scholar]
- 18. Esteban E, Snipes K, Hird D, Kasten R, Kinde H. 1993. Use of ribotyping for characterization of Salmonella serotypes. J. Clin. Microbiol. 31:233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bailey JS, Fedorka-Cray PJ, Stern NJ, Craven SE, Cox NA, Cosby DE. 2002. Serotyping and ribotyping of Salmonella using restriction enzyme PvuII. J. Food Prot. 65:1005–1007 [DOI] [PubMed] [Google Scholar]
- 20. Guard J, Sanchez-Ingunza R, Morales C, Stewart T, Liljebjelke K, Kessel J, Ingram K, Jones D, Jackson C, Fedorka-Cray P, Frye J, Gast R, Hinton A., Jr 2012. Comparison of dkgB-linked intergenic sequence ribotyping to DNA microarray hybridization for assigning serotype to Salmonella enterica. FEMS Microbiol Lett. 337:61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wise MG, Siragusa GR, Plumblee J, Healy M, Cray PJ, Seal BS. 2009. Predicting Salmonella enterica serotypes by repetitive sequence-based PCR. J. Microbiol. Methods 76:18–24 [DOI] [PubMed] [Google Scholar]
- 22. Chenu JW, Cox JM, Pavic A. 2011. Classification of Salmonella enterica serotypes from Australian poultry using repetitive sequence-based PCR. J. Appl. Microbiol. 112:185–196 [DOI] [PubMed] [Google Scholar]
- 23. Kotetishvili M, Stine OC, Kreger A, Morris JG, Sulakvelidze A. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Achtman MM, Wain JJ, Weill FX, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S, S. Enterica MLST Study Group 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8:e1002776 doi:10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wattiau P, Weijers T, Andreoli P, Schliker C, Veken HV, Maas HM, Verbruggen AJ, Heck ME, Wannet WJ, Imberechts H, Vos P. 2008. Evaluation of the Premi Test Salmonella, a commercial low-density DNA microarray system intended for routine identification and typing of Salmonella enterica. Int. J. Food Microbiol. 123:293–298 [DOI] [PubMed] [Google Scholar]
- 26. Kim S, Frye JG, Hu J, Fedorka-Cray PJ, Gautom R, Boyle DS. 2006. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J. Clin. Microbiol. 44:3608–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arrach N, Porwollik S, Cheng P, Cho A, Long F, Choi SH, McClelland M. 2008. Salmonella serovar identification using PCR-based detection of gene presence and absence. J. Clin. Microbiol. 46:2581–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leader BT, Frye JG, Hu J, Fedorka-Cray PJ, Boyle DS. 2009. High-throughput molecular determination of Salmonella enterica serovars by use of multiplex PCR and capillary electrophoresis analysis. J. Clin. Microbiol. 47:1290–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soyer Y, Switt AM, Davis MA, Maurer J, McDonough PL, Schoonmaker-Bopp DJ, Dumas NB, Root T, Warnick LD, Gröhn YT, Wiedmann M. 2009. Salmonella enterica serotype 4,5,12:i:-, an emerging Salmonella serotype that represents multiple distinct clones. J. Clin. Microbiol. 47:3546–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moreno Switt AI, Soyer Y, Warnick LD, Wiedmann M. 2009. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i:-. Foodborne Pathog. Dis. 6:407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Centers for Disease Control and Prevention 2006. Salmonella surveillance: annual summary, 2006. U.S.. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 32. Galanis E, Lo Fo Wong Lo DM, Patrick ME, Binsztein N, Cieslik A, Chalermchikit T, Aidara-Kane A, Ellis A, Angulo FJ, Wegener HC, World Health Organization Global Salm-Surv 2006. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg. Infect. Dis. 12:381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 34. Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kidgell C, Reichard U, Wain J, Linz B, Torpdahl M, Dougan G, Achtman M. 2002. Salmonella Typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39–45 [DOI] [PubMed] [Google Scholar]
- 36. Mortimer CKB, Peters TM, Gharbia SE, Logan JMJ, Arnold C. 2004. Towards the development of a DNA-sequence based approach to serotyping of Salmonella enterica. BMC Microbiol. 4:31 doi:10.1186/1471-2180-4-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imre A, Olasz F, Nagy B. 2005. Development of a PCR system for the characterisation of Salmonella flagellin genes. Acta Vet. Hung. 53:163–172 [DOI] [PubMed] [Google Scholar]
- 38. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 39. Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286–298 [DOI] [PubMed] [Google Scholar]
- 40. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 41. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Popoff MY, Le Minor L. 1985. Expression of antigenic factor O:54 is associated with the presence of a plasmid in Salmonella. Ann. Inst. Pasteur Microbiol. 136B:169–179 [DOI] [PubMed] [Google Scholar]
- 43. Werber D, Dreesman J, Feil F, van Treeck U, Fell G, Ethelberg S, Hauri AM, Roggentin P, Prager R, Fisher IST, Behnke SC, Bartelt E, Weise E, Ellis A, Siitonen A, Andersson Y, Tschäpe H, Kramer MH, Ammon A. 2005. International outbreak of Salmonella Oranienburg due to German chocolate. BMC Infect. Dis. 5:7 doi:10.1186/1471-2334-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elviss NC, Little CL, Hucklesby L, Sagoo S, Surman-Lee S, de Pinna E, Threlfall EJ, Food, Water and Environmental Surveillance 2009. Microbiological study of fresh herbs from retail premises uncovers an international outbreak of salmonellosis. Int. J. Food Microbiol. 134:83–88 [DOI] [PubMed] [Google Scholar]
- 45. Nicolay N, Thornton L, Cotter S, Garvey P, Bannon O, McKeown P, Cormican M, Fisher I, Little C, Boxall N, De Pinna E, Peters TM, Cowden J, Salmon R, Mason B, Irvine N, Rooney P, O'Flanagan D. 2011. Salmonella enterica serovar Agona European outbreak associated with a food company. Epidemiol. Infect. 139:1272–1280 [DOI] [PubMed] [Google Scholar]
- 46. Weigel RM, Qiao B, Teferedegne B, Suh DK, Barber DA, Isaacson RE, White BA. 2004. Comparison of pulsed field gel electrophoresis and repetitive sequence polymerase chain reaction as genotyping methods for detection of genetic diversity and inferring transmission of Salmonella. Vet. Microbiol. 100:205–217 [DOI] [PubMed] [Google Scholar]
- 47. Gaul SB, Wedel S, Erdman MM, Harris DL, Harris IT, Ferris KE, Hoffman L. 2007. Use of pulsed-field gel electrophoresis of conserved XbaI fragments for identification of swine Salmonella serotypes. J. Clin. Microbiol. 45:472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zou W, Lin WJ, Foley SL, Chen CH, Nayak R, Chen JJ. 2010. Evaluation of pulsed-field gel electrophoresis profiles for identification of Salmonella serotypes. J. Clin. Microbiol. 48:3122–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wattiau P, Van Hessche M, Schlicker C, Vander Veken H, Imberechts H. 2008. Comparison of classical serotyping and PremiTest assay for routine identification of common Salmonella enterica serovars. J. Clin. Microbiol. 46:4037–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Soyer Y, Alcaine SD, Schoonmaker-Bopp DJ, Root TP, Warnick LD, McDonough PL, Dumas NB, Gröhn YT, Wiedmann M. 2010. Pulsed-field gel electrophoresis diversity of human and bovine clinical Salmonella isolates. Foodborne Pathog. Dis. 7:707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alcaine SD, Soyer Y, Warnick LD, Su WL, Sukhnanand S, Richards J, Fortes ED, McDonough P, Root TP, Dumas NB, Gröhn Y, Wiedmann M. 2006. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl. Environ. Microbiol. 72:7575–7585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fitzgerald C, Collins M, van Duyne S, Mikoleit M, Brown T, Fields P. 2007. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 45:3323–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fitzgerald C, Gheesling L, Collins M, Fields PI. 2006. Sequence analysis of the rfb loci, encoding proteins involved in the biosynthesis of the Salmonella enterica O17 and O18 antigens: serogroup-specific identification by PCR. Appl. Environ. Microbiol. 72:7949–7953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. den Bakker HC, Moreno Switt AI, Govoni G, Cummings CA, Ranieri ML, Degoricija L, Hoelzer K, Rodriguez-Rivera LD, Brown S, Bolchacova E, Furtado MR, Wiedmann M. 2011. Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 12:425 doi:10.1186/1471-2164-12-425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Allard M, Luo Y, Strain E, Li C, Keys CE, Son I, Stones R, Musser SM, Brown EW. 2012. High resolution clustering of Salmonella enterica serovar Montevideo strains using a next-generation sequencing approach. BMC Genomics 13:32 doi:10.1186/1471-2164-13-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mehta G, Arya SC. 2002. Capsular Vi polysaccharide antigen in Salmonella enterica serovar Typhi isolates. J. Clin. Microbiol. 40:1127–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vangay P, Fugett EB, Sun Q, Wiedmann M. 2013. Food microbe tracker: a web-based tool for storage and comparison of food-associated microbes. J. Food Prot. 76:283–294 [DOI] [PubMed] [Google Scholar]
- 58. Bakker HC, Switt AI, Cummings CA, Hoelzer K, Degoricija L, Rodriguez-Rivera LD, Wright EM, Fang R, Davis M, Root T, Schoonmaker-Bopp D, Musser KA, Villamil E, Waechter H, Kornstein L, Furtado MR, Wiedmann M. 2011. A whole-genome single nucleotide polymorphism-based approach to trace and identify outbreaks linked to a common Salmonella enterica subsp. enterica serovar Montevideo pulsed-field gel electrophoresis type. Appl. Environ. Microbiol. 77:8648–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Diaz-Sanchez S, Hanning I, Pendleton S, D'Souza D. 2013. Next-generation sequencing: the future of molecular genetics in poultry production and food safety. Poult. Sci. 92:562–572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.