Abstract
In vitro culture methods underpin many experimental approaches to biology and drug discovery. The modification of established cell culture methods to make them more biologically relevant or to optimize growth is traditionally a laborious task. Emerging metabolomic technology enables the rapid evaluation of intra- and extracellular metabolites and can be applied to the rational development of cell culture media. In this study, untargeted semiquantitative and targeted quantitative metabolomic analyses of fresh and spent media revealed the major nutritional requirements for the growth of bloodstream form Trypanosoma brucei. The standard culture medium (HMI11) contained unnecessarily high concentrations of 32 nutrients that were subsequently removed to make the concentrations more closely resemble those normally found in blood. Our new medium, Creek's minimal medium (CMM), supports in vitro growth equivalent to that in HMI11 and causes no significant perturbation of metabolite levels for 94% of the detected metabolome (<3-fold change; α = 0.05). Importantly, improved sensitivity was observed for drug activity studies in whole-cell phenotypic screenings and in the metabolomic mode of action assays. Four-hundred-fold 50% inhibitory concentration decreases were observed for pentamidine and methotrexate, suggesting inhibition of activity by nutrients present in HMI11. CMM is suitable for routine cell culture and offers important advantages for metabolomic studies and drug activity screening.
INTRODUCTION
Trypanosoma brucei is the kinetoplastid parasite responsible for human African trypanosomiasis (HAT), a potentially fatal infection of sub-Saharan Africa. Current treatments for HAT are inadequate because of high toxicity and complex administration regimens, and new drugs are urgently required to ensure ongoing control of the disease in the face of emerging reports of drug resistance (1). Newly invigorated efforts to bring novel compounds forward as drugs have led to several promising candidates emerging through screening against parasites in culture (2–5). In vitro continuous culture methods enable the routine screening of compounds for trypanocidal activity against bloodstream form trypanosomes, for example, with resazurin fluorescence-based assays (5–7).
Culture methods for bloodstream form T. brucei were first developed in the 1970s and required feeder layers to support continuous growth (8). This requirement could be overcome by regular supplementation with cysteine (9) and supplementation with mammalian serum (10). Continuous culture was successfully maintained by the addition of reducing and chelating agents (2-mercaptoethanol and bathocuproine sulfonate) to modified Iscove medium with 10% fetal bovine serum (HMI11) (11). HMI11 is now widely used for routine cell culture of laboratory strains of T. brucei and underpins many of the approaches employed in the search for new trypanocidal compounds. Nevertheless, HMI11 is a very rich medium compared to the natural in vivo environment of bloodstream form T. brucei, which may give rise to biochemical or pharmacological activities that are not relevant in vivo.
Metabolomics enables the simultaneous measurement of many metabolites in any system and offers great promise for pharmacological research (12–14). Metabolomic applications are rapidly expanding in the field of postgenomic biology, and it has recently been demonstrated as an untargeted method allowing the determination of mechanisms of drug action (15, 16). Mechanistic knowledge of drug action should allow the rational optimization of trypanocidal compounds to develop new drugs based on either existing drugs or novel chemotypes identified through high-throughput screening. While the availability of in vitro culture methods for T. brucei enables relatively simple evaluation of drug-induced changes in the parasite metabolome, the use of a rich medium (HMI11) raises some practical limitations. Primarily, the high concentration of 35 common metabolites in HMI11 makes it difficult to observe metabolic alterations involving these metabolites. This occurs because relative changes are much less pronounced when there is a large background metabolite pool present and absolute analytical error margins tend to increase along with metabolite intensities. Also, drug-induced metabolic perturbations can be easily masked by compensatory mechanisms if there are high metabolite concentrations available in the culture medium. This issue is not limited to drug-induced perturbations, as studies of genetic manipulation for target validation may also be complicated by excess nutrient availability (17). Indeed, this “metabolite rescue” approach is often used intentionally to generate conditional gene knockouts or to confirm chemical or genetic enzyme inhibition (17, 18). However, the use of a rich culture medium may unintentionally hinder drug discovery efforts that utilize phenotypic screening approaches, as demonstrated for dihydrofolate reductase (DHFR)-thymidylate synthase inhibitors, which demonstrated significant trypanocidal activity only when the medium was depleted of folic acid (17). The same type of masking where highly abundant culture medium components compete with drugs for targets or transporters will also influence screening for other antimetabolites. The ideal culture medium would be one that supports optimal in vitro cell growth while also resembling as closely as possible the in vivo environment of the parasite.
The ability to quantify changes in metabolite levels in growth medium with relative ease offers great potential for the rational optimization of culture media (19, 20). Targeted metabolic profiling approaches may be used to quantify nutrient utilization and to allow comparison of metabolite levels to biologically relevant concentrations. Untargeted metabolomics allows the observation of all of the relative changes in metabolite concentrations in culture media during growth and may highlight a depletion of essential nutrients that would not have been analyzed by traditional targeted approaches.
The aim of this study was to utilize metabolomic technology for the rational development of a simple culture medium optimal for in vitro continuous culture of bloodstream form T. brucei. The improved medium contains biologically relevant concentrations of essential nutrients and avoids supranutritional levels of metabolites that interfere with metabolomic experiments and standard drug activity assays. The new medium was tested for performance by phenotypic assessment, metabolomic analysis, and drug activity assays.
MATERIALS AND METHODS
Parasite culture.
Bloodstream form T. brucei brucei (s427) was cultured in vitro in HMI11 (Gibco) (11), Creek's minimal medium (CMM) (Table 1), or modified media as described later. All media contained 10% fetal calf serum (FCS) Gold (PAA, Piscataway, NJ). Cell culture grade (or high-purity) d-glucose, l-cysteine, and l-glutamine and all other medium components were purchased from Sigma-Aldrich. Five-milliliter cultures were maintained in 25-ml vented flasks (Corning) at 37°C with 5% CO2. Cultures were grown to a maximum density of 4 × 106 cells ml−1 and subcultured by 1-in-100 or 1-in-1,000 dilution every 2 or 3 days, respectively. Cell counts were obtained with a hemocytometer (Neubauer).
Table 1.
Recipe for CMM
| Component | Concna or amt |
|---|---|
| d-Glucose | 10,000 (1,800) |
| l-Glutamine | 1,000 (146) |
| l-Cysteine | 1,000 (121) |
| NaCl | 77,590 (4,500) |
| CaCl2 · 2H2O | 1,490 (219) |
| KCl | 4,400 (330) |
| MgSO4 · 7H2O | 814 (200) |
| NaHCO3 | 35,950 (3,020) |
| HEPESb | 25,032 (5,960) |
| Phenol red | 42 (15) |
| Bathocuproinedisulfonic acid | 52 (28) |
| Mercaptoethanolc | 192, 15 μl |
| Water | 900 ml |
| NaOHd to bring pH to 7.4 | As required |
| FCS Gold (PAA)e | 10%, 100 ml |
Unless stated otherwise, all values are μM (mg liter−1) concentrations. All component concentrations are from the HMI11 recipe (11), except those of d-glucose, l-glutamine, and l-cysteine.
HEPES can be decreased to 10 mM to improve LC-MS compatibility for metabolomic experiments.
Mercaptoethanol should be added under a fume hood.
All components should be dissolved in water, pH adjusted, and sterile filtered.
FCS should be added just prior to use.
Untargeted metabolomic analysis.
Metabolite extraction was performed according to previously described methods (21, 22). A 25-ml volume of cell culture was quenched by rapid cooling to 4°C in a dry ice-ethanol bath. Cell pellets were obtained by centrifugation at 1,250 × g for 10 min, and 5 μl of spent medium was removed and extracted by the addition of 100 μl chloroform-methanol-water (1:3:1). The remaining cell pellet was washed with 1 ml of phosphate-buffered saline (PBS), and the washed pellet (5 × 107 cells) was extracted with 100 μl of chloroform-methanol-water (1:3:1) by mixing for 1 h at 4°C before centrifugation to remove precipitate. The resulting metabolite solution was stored at −80°C until analysis by liquid chromatography-mass spectrometry (LC-MS). Comparative data from HMI11 and CMM cell pellets and spent medium are from five independent biological replicates.
LC-MS analysis utilized hydrophilic interaction chromatography with a ZIC-HILIC column (Sequant) and a formic acid mobile phase and was coupled to high-resolution MS with the Exactive Orbitrap (Thermo) operating in both positive and negative modes with all parameters as previously described (23).
Metabolomic data analysis was performed with IDEOM (http://mzmatch.sourceforge.net/ideom.php). The IDEOM workflow includes untargeted peak picking by XCMS (Centwave) (24), peak matching and annotation by mzMatch (25), and noise filtering and metabolite identification by IDEOM, with the default parameters (26). Metabolite identification is based on accurate mass and predicted retention time, which is accepted as a level 3 standard of identification by the Metabolomics Standards Initiative (MSI) (27) but should be considered a putative identification. Retention time was verified by authentic standards for the major HMI11 components and other metabolites where possible. These high-confidence (MSI level 1) identifications are highlighted (yellow) in the IDEOM files in the supplemental material; further information to assist with interpretation of the confidence of putative identifications in the supplemental material is available in the IDEOM help file (http://mzmatch.sourceforge.net/ideom/Viewing_results_with_Ideom.pdf). Relative quantitation for untargeted analysis is based on raw peak heights. Comparative analysis is based on relative mean peak height, and statistical analyses used unpaired rank product analysis (28). No normalization was applied to untargeted data, although signal reproducibility was ensured by the routine analysis of six spiked internal standards, median peak heights, total ion current chromatograms, and technical replicates of pooled samples (quality controls).
Targeted and semiquantitative metabolomic analyses.
Targeted quantitative and untargeted semiquantitative metabolomic analyses were done by the same procedure as described for untargeted analysis, with the addition of fully labeled Escherichia coli extract as an internal standard at a 1:4 ratio before the extraction step (29, 30). Fully labeled E. coli was generated by the incubation of E. coli MG1655 with 500 ml of M9 minimal medium and 0.3% (wt/vol) [U-13C]glucose overnight. E. coli metabolites were extracted when the culture reached an optical density at 600 nm of 1.0 by the addition of 8 ml of −20°C acetonitrile-methanol-water (2:2:1) to the cell pellet as previously described (31), and the extract was stored at −80°C until required.
Calibration curves for LC-MS quantification were generated for each metabolite with serial dilutions of freshly prepared authentic standards in 80 μl of methanol-water (1:1) with 20 μl of fully labeled E. coli extract. LC-MS was performed as described for untargeted analysis. Data analysis was performed with LC Quan (Thermo) to manually integrate peak areas, which were normalized on the basis of the coeluting fully labeled analyte from the E. coli extract. Where authentic standards were available, metabolite concentrations were calculated from calibration curves and corrected for dilution. Where authentic standards were not available, semiquantification was performed in IDEOM (26), whereby the raw peak height was normalized to a fully labeled internal standard; for metabolites where no internal standard was detected, normalization was based on two fully labeled internal standards with retention times nearest to that of the metabolite.
Metabolomic analysis of drug action.
Metabolomic analysis of drug action was based on the general method previously described for trypanocidal drugs (15). Pentamidine death curves were obtained for various doses and treatment durations by cell counts of cultures at a parasite density that would provide a suitable biomass for metabolomic analysis (2 × 106 cells ml−1). Drug concentrations were selected that mediated an approximately 20% reduction of parasitemia at 48 h. In order to obtain equivalent parasite cultures for metabolomic sampling, a confluent cell culture was subcultured into five flasks of fresh medium at 4 × 104 cells ml−1 and pentamidine was added to four flasks at set times in order to obtain samples with 48, 24, 12, and 1 h of drug exposure (see Fig. S1 in the supplemental material). All samples, including the untreated sample, were extracted when they reached 2 × 106 cells ml−1. As some growth defect was observed in the 48-h treatment, an additional untreated control was included that started with a lower cell density (2 × 104 cells ml−1) and reached the sampling density (2 × 106 cells ml−1) after 48 h, therefore being exposed to the culture medium for the same time as the samples treated for 48 h. The 0-h control was spiked with pentamidine after metabolism was quenched to 4°C, whereas the 48-h control remained drug free, thus providing an implicit validation of the quenching method, ensuring no ion suppression due to the pentamidine solution and allowing the identification of peaks that arise from the pentamidine solution itself. Four independent biological replicates were analyzed for each time point in HMI11, and three replicates were analyzed for each time point in CMM.
Drug activity screening.
Methotrexate, curcumin, diminazene, methylene blue, 2-deoxyglucose, phloretin, and 5-fluorouracil were purchased from Sigma-Aldrich; acivicin was from Biomol International; eflornithine was a gift from Pere Simarro (World Health Organization); NA42 was a gift from Boris Rodenko (University of Glasgow); nifurtimox was a gift from Vanessa Yardley (London School of Hygiene and Tropical Medicine); suramin was a gift from Mike Turner (University of Glasgow); melarsen oxide was a gift from Alan Fairlamb (University of Dundee); pentamidine was a gift from Pere Simarro (World Health Organization); and DB75 was a gift from Rick Tidwell (University of North Carolina, Chapel Hill).
The activities of trypanocidal compounds were tested in a serial drug dilution assay in order to determine the IC50s (drug concentration causing 50% growth inhibition) with the alamarBlue assay (6). Briefly, 2-fold serial dilutions of the test compounds were prepared in 96-well microtiter plates containing either HMI11 or CMM. Bloodstream form T. brucei parasites in the log phase of growth were diluted in HMI11 or CMM and inoculated into each well to a final concentration of 1 × 104 cells ml−1. After 48 h of incubation at 37°C under a humidified 5% CO2 atmosphere, 20 μl of resazurin (12.5 mg of resazurin [Sigma-Aldrich] in 100 ml of PBS) was added to each well and the cultures were incubated for an additional 24 h. Fluorescence detection was performed with a FLUOstar Optima Microplate Fluorometer (BMG LABTECH GmbH) at excitation and emission wavelengths of 530 and 590 nm, respectively. To assess cell sensitivity to the trypanocides, data were log transformed and the IC50s were determined by regression from the sigmoidal dose-inhibition curve with GraphPad Prism. Three to nine biological replicates were carried out for each of the trypanocidal compounds. Comparative statistics utilized the unpaired Welch t test (α = 0.05).
RESULTS
Trypanosome nutrient utilization from standard growth medium.
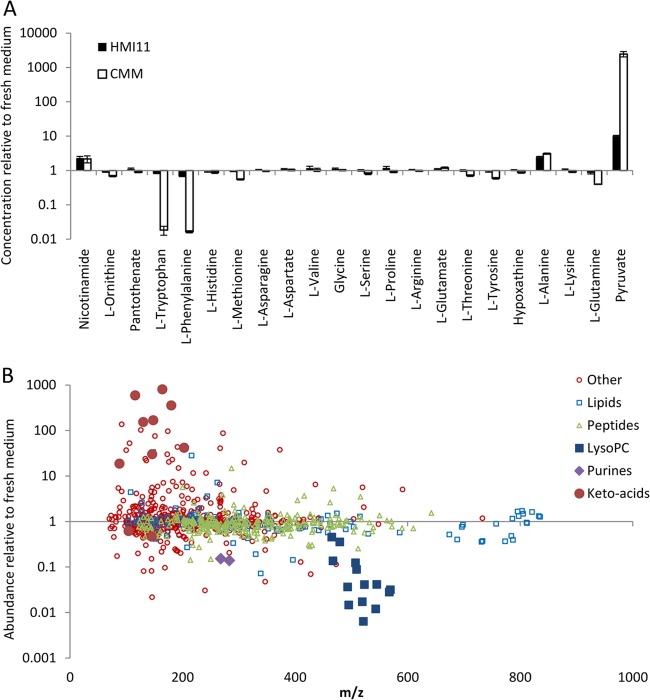
In order to develop an optimal simplified growth medium, it was first necessary to determine which nutrients are significantly consumed by trypanosomes from standard HMI11 medium. Medium samples were collected from fresh and spent HMI11 medium after 56 h of growth during which the trypanosome density increased from 2 × 104 to 4 × 106 cells ml−1, which is the peak density obtained in this medium. These samples were analyzed by a high-resolution LC-MS metabolomic approach that combines the targeted quantitative analysis of selected metabolites with the untargeted semiquantitative analysis of all putative metabolites. This analysis revealed minimal consumption of most nutrients from the medium, although a 32% decrease in phenylalanine was observed (Fig. 1A). Glucose and cysteine also decreased, although absolute quantification of these metabolites by the HILIC-Orbitrap is hindered because of relatively poor chromatography of reducing sugars and oxidation of thiols during sample preparation and autosampler storage (22, 32). Nevertheless, it is well established that bloodstream form trypanosomes in cell culture utilize significant amounts of both glucose (33) and cysteine (34). In addition to the defined components of HMI11, an untargeted metabolomic analysis allowed investigation of the utilization of serum-derived nutrients. While many nutrients were not affected by cell growth, significant depletion of lysophosphatidylcholines (LPCs) and the purine nucleosides inosine and guanosine was observed (Fig. 1B). The nutrients that accumulated the most in spent medium were pyruvate, the major end product of aerobic glycolysis in T. brucei (33), and alanine, the aminated product of pyruvate. Increased levels of several keto acids were also observed, likely because of deamination of the amino acids phenylalanine, tryptophan, tyrosine, valine, (iso)leucine, methionine, and glutamate.
Fig 1.
Relative changes in concentrations of nutrients in spent medium following 56 h of cell growth (to 4 × 106 cells ml−1). (A) Relative changes in the concentrations of targeted metabolites in HMI11 (filled columns) and CMM (open columns). The pyruvate concentration in CMM increased >10,000-fold, from below the limit of detection (1 μM) to 12 mM. (B) Changes in the relative abundance of all putative metabolites in HMI11. Lipids (blue squares) are mostly unchanged, with the exception of LPCs (LysoPC; larger blue filled squares); many peptides (green triangles) are slightly depleted; and most other classes of putative metabolites (red circles) are unchanged, with the notable exception of the amino acid-derived keto acids (larger red filled circles) and the purine nucleosides inosine and guanosine (filled purple diamonds).
Development of minimal growth medium.
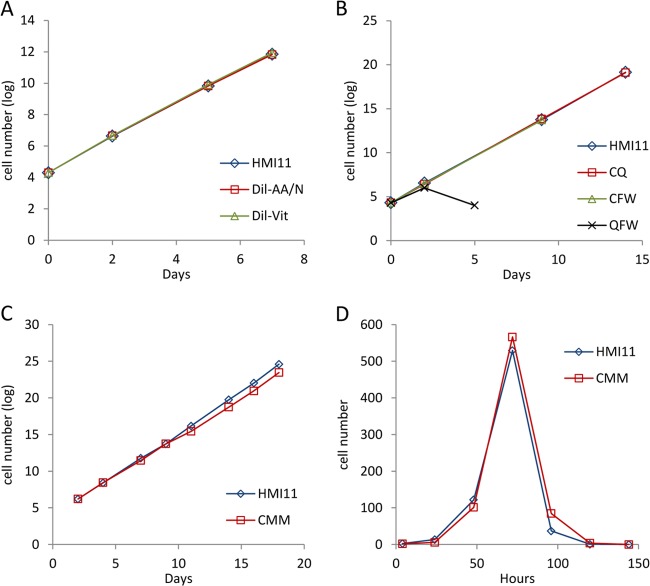
As the initial analysis indicated that no HMI11 components were consumed to exhaustion during a standard cell culture passage, altered HMI medium was prepared containing 10% FCS, basal medium (salts, buffers, and antioxidants), glucose, glutamine, and cysteine according to the HMI11 formula and other components at a 10-fold dilution. Two media were further derived from this by the omission of (i) vitamins or (ii) amino acids and nucleotide precursors. Surprisingly, both media supported cell growth to the same degree as HMI11 (Fig. 2A) and no major nutrient depletion was observed, with the exception of phenylalanine and tryptophan. Final medium optimization was performed with each combination of cysteine, glutamine, phenylalanine, and tryptophan, in addition to glucose and the basal medium. These experiments confirmed that cysteine is essential for growth (34), along with either glutamine or a combination of phenylalanine and tryptophan (Fig. 2B). Therefore, the minimal nutrient combination required for continuous in vitro culture was glucose, cysteine, and glutamine. The concentrations of these three components were then reduced to the final concentrations shown in Table 1 without affecting the growth rate. Our optimal recipe (Table 1), referred to as CMM, supports growth rates identical to those obtained with HMI11 under standard cell culture conditions, and cells enter stationary phase at a density similar to that of cells in HMI11 (Fig. 2). CMM supports normal growth when used for routine cell culture (see Materials and Methods) up to at least 25 passages (after which it was not tested because we routinely cull the cells and begin a new stabilate at that point). All development work was performed with T. brucei brucei strain WT427, and normal growth of strain 2T1, which is commonly used in RNA interference work, was also observed (35). Partial depletion of glutamine, phenylalanine, and tryptophan and accumulation of pyruvate and alanine were observed in spent CMM following a 56-h passage up to a density of 4 × 106 cells ml−1 (Fig. 1A). No significant changes in nutrient concentrations were observed in cell-free CMM controls incubated at 37°C for 56 h. Limited testing of the long-term stability of this medium has been performed. Serum-free CMM is effective for at least 6 months if stored at 4°C, supporting normal growth after the addition of fresh serum. CMM containing 10% FCS is able to support routine cell culture after storage at 4°C for at least 2 months (data not shown).
Fig 2.
Growth curves of T. brucei cultures grown in nutrient-depleted medium. (A) Equivalent growth observed for HMI11 (diamonds) compared to diluted HMI11 without vitamins (Dil-Vit; triangles) or diluted HMI11 without amino acids and nucleotide precursors (Dil-AA/N; squares). (B) Equivalent growth observed in HMI11 (triangles), basal medium with cysteine with glutamine (CQ; squares), or cysteine with phenylalanine and tryptophan (CFW; diamonds) but not in glutamine with phenylalanine and tryptophan (QFW; crosses). (C) Equivalent growth observed in HMI11 and CMM. (D) Short-term growth curves obtained with HMI11 and CMM showing a maximal cell density of around 5 × 106 cells ml−1 and subsequent cell death.
Quantitative measurement of metabolite levels in fresh CMM and HMI11 was done by LC-MS with stable-isotope-labeled internal standards. Interestingly, many of the components that were excluded from the HMI11 recipe were still present in CMM (Table 2), confirming that 10% FCS Gold provides an adequate supply of most of the nutrients essential for trypanosome growth. Importantly, the excessive concentrations of most HMI11 components were avoided in CMM; for example, hypoxanthine and nicotinamide levels in HMI11 are >200-fold higher than necessary. Average metabolite concentrations in healthy adult blood were obtained from the literature with the Human Metabolome Database (www.hmdb.ca) (36), most of which more closely resembled concentrations in CMM than those in HMI11 (Table 2).
Table 2.
Absolute quantification of HMI components in fresh and spent media after 56 h of trypanosome growth to 4 × 106 cells ml−1f
| Component | Exptl concn (μM)a |
Theoretical concn (μM) |
||||||
|---|---|---|---|---|---|---|---|---|
| HMI |
CMM |
HMI11 reciped | CMM recipe | Human bloode | Human CSFe | |||
| Fresh | Spent | Fresh | Spent | |||||
| d-Glucosea | 25,000 | 10,000 | 4,817 | 3,357 | ||||
| l-Cysteinea | 3,349,188 ± 1,426,490 | 661,446 ± 147,585 | 3,987,853 ± 435,789 | 254,831 ± 85,829 | 1,504 | 1,000 | 99 | 0 |
| l-Glutamine | 4,002 ± 238 | 3,382 ± 191 | 727 ± 11 | 289 ± 6 | 3,998 | 1,000 | 550 | 490 |
| l-Phenylalanine | 327 ± 16 | 224 ± 8 | 76 ± 1 | 1.3 ± 0.1 | 400 | 79 | 11 | |
| l-Tryptophan | 87 ± 5 | 73 ± 3 | 21 ± 1 | 0.4 ± 0.1 | 78 | 54 | 6 | |
| l-Tyrosine | 1,013 ± 60 | 951 ± 44 | 194 ± 5 | 115 ± 6 | 574 | 89 | 11 | |
| l-Methionine | 195 ± 8 | 188 ± 7 | 39 ± 1 | 22 ± 1 | 201 | 29 | 4 | |
| l-Arginine | 692 ± 40 | 715 ± 28 | 225 ± 1 | 219 ± 9 | 482 | 110 | 21 | |
| l-Leucinea,b | 48,270,601 ± 13,204,959 | 44,388,168 ± 4,445,117 | 17,478,123 ± 2,079,608 | 4,996,443 ± 584,426 | 801 | 158 | 14 | |
| l-Isoleucinea,b | 801 | 69 | 7 | |||||
| l-Valine | 440 ± 30 | 517 ± 69 | 385 ± 22 | 404 ± 44 | 803 | 234 | 19 | |
| l-Lysine | 1,302 ± 65 | 1,382 ± 56 | 255 ± 4 | 231 ± 9 | 999 | 236 | 27 | |
| l-Threonine | 899 ± 52 | 895 ± 42 | 102 ± 2 | 72 ± 4 | 798 | 155 | 31 | |
| l-Serine | 553 ± 33 | 555 ± 27 | 193 ± 7 | 156 ± 10 | 400 | 145 | 30 | |
| l-Proline | 483 ± 60 | 572 ± 57 | 128 ± 5 | 115 ± 5 | 348 | 190 | 3 | |
| l-Histidine | 241 ± 12 | 225 ± 9 | 52 ± 1 | 46 ± 3 | 271 | 106 | 16 | |
| Glycine | 482 ± 25 | 542 ± 25 | 179 ± 4 | 183 ± 9 | 400 | 266 | 8 | |
| l-Glutamate | 729 ± 39 | 791 ± 32 | 179 ± 3 | 216 ± 10 | 510 | 76 | 13 | |
| l-Asparagine | 276 ± 13 | 284 ± 10 | 91 ± 1 | 88 ± 4 | 189 | 50 | 7 | |
| l-Aspartate | 338 ± 19 | 367 ± 15 | 100 ± 2 | 103 ± 6 | 226 | 21 | 2 | |
| l-Alanine | 514 ± 41 | 1,263 ± 59 | 256 ± 6 | 793 ± 34 | 281 | 448 | 32 | |
| l-Cystinea | 33,127 ± 57,378 | <LODc | 152,042 ± 14,351 | <LODc | 379 | 101 | 0.2 | |
| Pyruvate | 937 ± 17 | 9,408 ± 503 | 4.7 ± 1.2 | 11,525 ± 2,053 | 1,295 | 60 | 98 | |
| Hypoxanthine | 965 ± 47 | 983 ± 37 | 4.4 ± 0.1 | 3.8 ± 0.2 | 1,000 | 8 | 4 | |
| Thymidinea | 154,748 ± 29,103 | 507,275 ± 74,464 | <LODc | <LODc | 161 | 0.2 | ||
| myo-Inositola | 40 | 24 | 138 | |||||
| Cholinea | 7,999,054 ± 1,371,732 | 8,557,781 ± 1,051,370 | 3,691,885 ± 172,624 | 3,698,509 ± 306,766 | 38 | 9 | 6 | |
| Nicotinamide | 4 ± 0.7 | 8.9 ± 1.0 | 0.02 ± 0.001 | 0.04 ± 0.01 | 33 | 0.4 | ||
| Pyridoxala | 420,434 ± 155,128 | 460,607 ± 178,818 | <LODc | 2,726 ± 506 | 24 | 0.3 | ||
| Pantothenate | 26 ± 2 | 29 ± 2 | 2.0 ± 0.1 | 1.8 ± 0.1 | 18 | 4 | 3 | |
| Thiaminea | 3,643,586 ± 61,195 | 3,665,175 ± 218,103 | 3,531,013 ± 41,082 | 3,369,323 ± 26,130 | 15 | 0.1 | 0.07 | |
| Folic acida | 150,002 ± 37,145 | 170,635 ± 65,126 | <LODc | <LODc | 9 | 0.03 | 0.02 | |
| Riboflavina | 55,730 ± 9,975 | 54,715 ± 4,636 | <LODc | <LODc | 1 | 0.3 | 0.4 | |
| Biotina | 10,264 ± 1,217 | 11,764 ± 1,234 | 9,819 ± 1,057 | 10,539 ± 735 | 0.053 | 0.002 | 0.0005 | |
| Cyanocobalamina | 0.010 | 0.0003 | 0.00002 | |||||
Absolute quantification of these metabolites could not be performed by the LC-MS methods and internal standards that were used in this study. Where possible, normalized peak intensities (arbitrary units) are shown in italics to allow comparison of experimental relative abundances.
Leucine and isoleucine peaks overlapped. Peak intensity represents the sum of both metabolites.
A peak was not detected or was below the limit of detection (<LOD), which was set at 1,000 arbitrary units.
Theoretical concentrations of HMI11 components were taken from http://tryps.rockefeller.edu/trypsru2_culture_media_compositions.html.
Theoretical concentrations in human blood and cerebrospinal fluid (CSF) are average values for healthy adults obtained from www.hmdb.ca.
n = 5.
Comparative phenotypic analysis of trypanosomes grown in CMM or HMI11.
Comparison of growth curves and gross morphology revealed no significant phenotypic differences between cells cultured in CMM and those cultured in HMI11 (Fig. 2). To determine cryptic changes in composition, however, a detailed LC-MS metabolomic analysis of intracellular metabolism was performed to investigate the impact of medium composition on cellular metabolism.
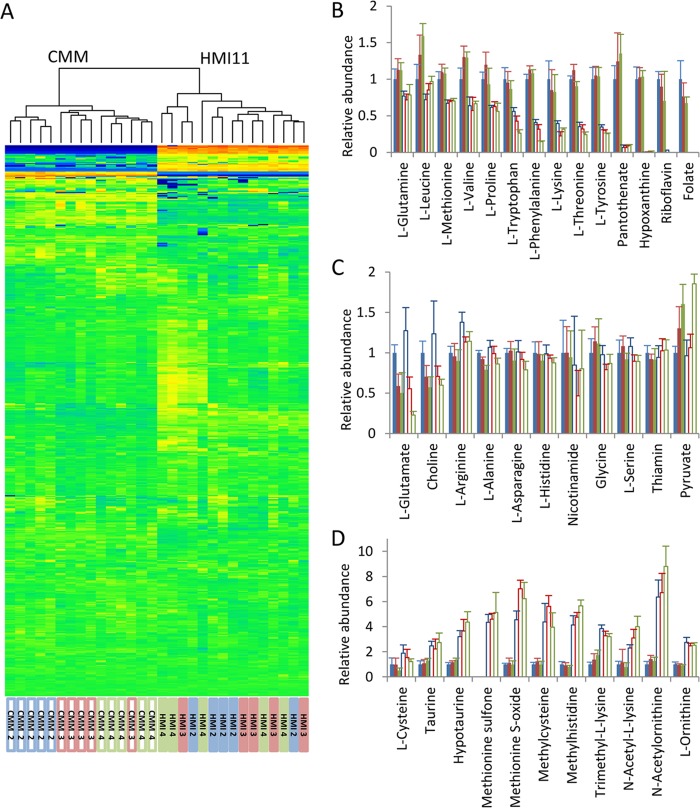
Untargeted metabolomic analysis with the formic acid ZIC-HILIC–Orbitrap platform putatively identified 522 metabolites in cultured bloodstream form T. brucei. Semiquantitative analysis with uniformly 13C-labeled E. coli extract for normalization revealed that 74% of the metabolites were not affected by the culture medium, showing no significant difference in metabolite abundance at any of the cell densities tested (P > 0.05), and only 6% of the metabolites demonstrated ≥3-fold differences across all cell densities (Fig. 3A).
Fig 3.
Comparison of the intracellular T. brucei metabolome in HMI11 versus CMM during the late log growth phase from 2 × 106 to 4 × 106 cells ml−1. (A) Heat map of all putatively identified metabolites (rows) obtained by hierarchical clustering analysis. Samples (columns) cluster according to the growth medium, but most intracellular metabolite levels are unchanged. Heat map data are log transformed and mean scaled. Orange, increased abundance; blue, decreased abundance; green, unchanged abundance. (B) Numerous HMI11 components accumulate to greater levels in HMI11-grown cells (filled columns) than in CMM-grown cells (open columns). (C) Many metabolites were not significantly different between cells grown in the two media, including a number of HMI11 components that were absent from the CMM formula. As the cell density increased, an increase in pyruvate and a decrease in glutamate were observed in both media. (D) Examples of putative metabolites with a higher relative abundance in CMM than in HMI11, showing changes in amino acid metabolism involving methylation, acetylation, and sulfur or redox metabolism. y axes show mean normalized LC-MS peak heights relative to those of cells grown in HMI11 at 2 × 106 cells ml−1. Error bars represent standard deviations. Five biological replicates of each cell density, i.e., 2 × 106 (blue columns), 3 × 106 (red columns), and 4 × 106 (green columns) cells ml−1, were measured.
Many of the metabolites that were less abundant in CMM-grown cells can be explained by direct uptake of the excess nutrients in HMI11 (Fig. 3B), and the lower concentrations of many of these metabolites observed in CMM are closer to physiological concentrations in the parasite's natural habitat, blood. Interestingly, the intracellular concentrations of 11 HMI11 nutrients were not significantly different, even though these nutrients were excluded from the CMM recipe and present at much lower concentrations than in HMI11 medium (Fig. 3C), indicating intrinsic mechanisms of homeostasis of these metabolites in the parasites. Some metabolites were more abundant in cells grown in CMM than in cells grown in HMI11, indicating localized changes in cellular metabolism. These included ornithine, variously acetylated and methylated amino acids, and metabolites related to cysteine and methionine (Fig. 3D).
Analysis of cellular metabolism as the cell density increased from 2 × 106 to 4 × 106 cells ml−1 revealed significant changes in the concentrations of one-fifth of the putatively identified metabolites in both CMM (20%) and HMI11 (18%) (for details, see the IDEOM file in the supplemental material). Intracellular pyruvate increased 2-fold (because of the major role of glycolysis in energy generation). Interestingly, intracellular glutamate levels were depleted by >50% in HMI11-grown cells, despite no decrease in the 730 μM concentration of glutamate in the medium. The same trend was observed for CMM-grown cells, demonstrating substantial metabolic alterations as the cell density increased toward stationary phase, regardless of the growth medium and without any external intervention.
Drug testing with CMM.
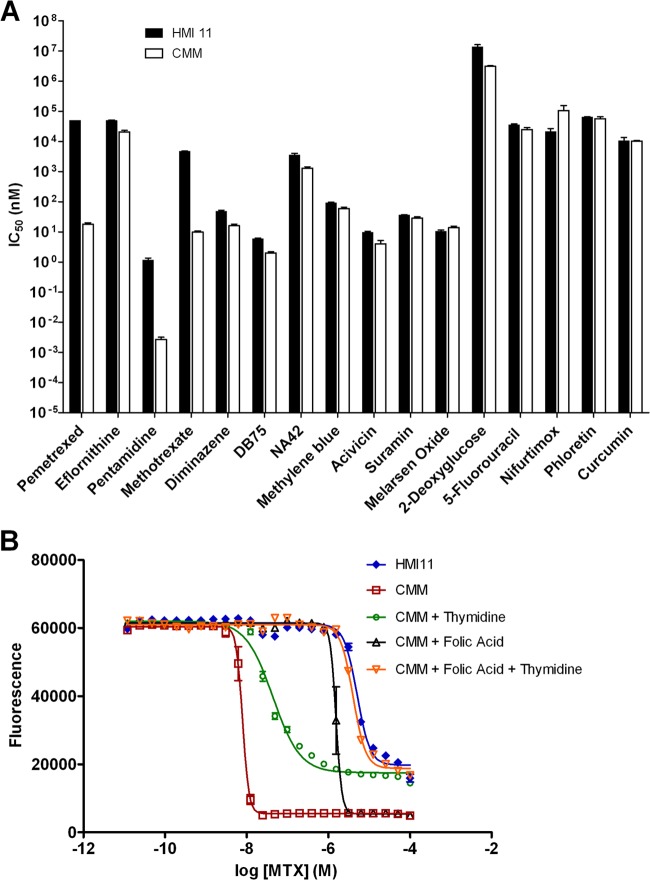
The performance of CMM in drug sensitivity testing was evaluated by standard alamarBlue assay of 16 clinically used and experimental trypanocides. In most cases, the IC50s obtained in CMM were comparable to those obtained in HMI11. Six compounds showed no significant effect of the medium on the IC50, and the differences observed with seven other compounds were <3-fold in magnitude (Fig. 4). Interestingly, three of the compounds tested exhibited >100-fold increases in activity in CMM over that in HMI11. Pentamidine, already the most potent of the trypanocides evaluated in HMI11 (IC50 = 1.2 nM), was remarkably more active in CMM (IC50 = 0.003 nM). Methotrexate, a relatively poorly active compound in HMI11 (IC50 = 4.6 μM), demonstrated good activity in CMM (IC50 = 10 nM). Likewise, pemetrexed was barely active in HMI11 (IC50 > 50 μM) but highly active in CMM (IC50 = 18 nM). Methotrexate and pemetrexed are both known to inhibit DHFR, resulting in inhibition of folate metabolism and subsequent biosynthesis of thymidine nucleotides (37). Hence, the lower activity observed in HMI11 is likely due to rescue by the high concentrations of folate in HMI11 medium compared to CMM. To confirm this, CMM was prepared with additional folate at the same concentration as that found in HMI11. Methotrexate activity in folate-supplemented CMM was reduced >150-fold to 1,500 nM but not completely diminished to the IC50 obtained in HMI11. Additional studies with thymidine-supplemented CMM showed a 4-fold reduction in activity compared to that in CMM, and the combination of CMM with folate and thymidine resulted in methotrexate activity (IC50) equivalent to that in HMI11 (Fig. 4B).
Fig 4.
(A) Trypanocidal activities (IC50s) of standard trypanocidal compounds in HMI11 (filled columns) and CMM (open columns). Large increases in the activities of methotrexate, pentamidine, and pemetrexed in CMM were observed. Increased sensitivities to diminazene, DB75, eflornithine, NA42, and acivicin in CMM were statistically significant (P < 0.05) but relatively minor (2- to 3-fold). (B) The trypanocidal activity of methotrexate (MTX) in CMM was partly inhibited by thymidine (4-fold increase in IC50) and folic acid (150-fold), and activity equivalent to that in HMI11 was observed upon the readdition of both folic acid and thymidine. The data shown are the means ± standard errors of three to nine biological replicates.
The reason for the major increase in pentamidine activity in CMM is unclear, as the mechanism of action of pentamidine remains under debate. The other diamidines tested in this study, diminazene and DB75, showed much smaller increases in activity in CMM than in HMI11 (3.0- and 2.4-fold, respectively). The larger differential observed for pentamidine in these two media suggests a difference in the mechanism of action and/or uptake of pentamidine versus diminazene and DB75. Since it is already known that pentamidine's uptake is more dependent on the HAPT1 and LAPT1 transporters than that of either DB75 (38) or diminazene (39) is, it is not surprising to identify such differences in behavior.
Pentamidine induced changes in the T. brucei metabolome.
In order to further elucidate the mechanism of action of pentamidine, a metabolomic approach was applied to cell cultures with both HMI11 and CMM. Previous work indicated how this approach can be useful in identifying a drug's mode of action when increases in the ornithine concentration and decreases in the putrescine concentration were observed following the treatment of T. brucei with eflornithine (15). In order to determine suitable doses of pentamidine, growth curves were obtained at the higher cell density (2 × 106 cells ml−1) necessary to obtain suitable cell numbers for metabolomic analysis. Under these conditions, a delayed-death phenotype was observed, whereby no effect on parasite growth was observed within the first 24 h, as previously reported for diamidines (40). In HMI11, 5 nM pentamidine was selected as the optimal dose to achieve an approximately 20% reduction in parasite density at 48 h compared to that of untreated cells. In CMM, the same effect was obtained with 0.5 nM pentamidine, in general agreement with a greater potency observed in alamarBlue assays. Higher doses mediated >50% cell death, making it impractical to obtain suitable numbers of intact cells for metabolomic analysis. Drug concentrations were significantly higher than the IC50s obtained by alamarBlue assay, likely because of the higher parasite density required for metabolomic studies and the fact that the alamarBlue readout occurs at 72 h. Further variability, albeit minor, was observed when samples were scaled up to the full volumes required for the metabolomic experiments. Pentamidine at 5 nM in HMI11 mediated approximately 50% cell death after 48 h, whereas minimal trypanocidal activity was observed in 0.5 nM pentamidine in CMM, thus providing data for both lethal and sublethal doses of pentamidine (see the supplemental material).
Since intracellular metabolite concentrations are dependent on cell density (Fig. 3) and cell density is decreased by drug treatment, we were obliged to develop a novel sampling methodology to ensure that all cultures were sampled at the same cell density (see the supplemental material). This approach requires sampling at different times; therefore, two untreated controls were included to allow for potential time-dependent metabolic fluctuations, one starting at the same cell density as treated cultures but sampled earlier and a second starting at a lower cell density and sampled at 48 h. The two control samples allowed for the inclusion of a completely drug-free control and a control with drug added after quenching, which is essential for the interpretation of drug (or contaminant)-derived features in untargeted LC-MS data.
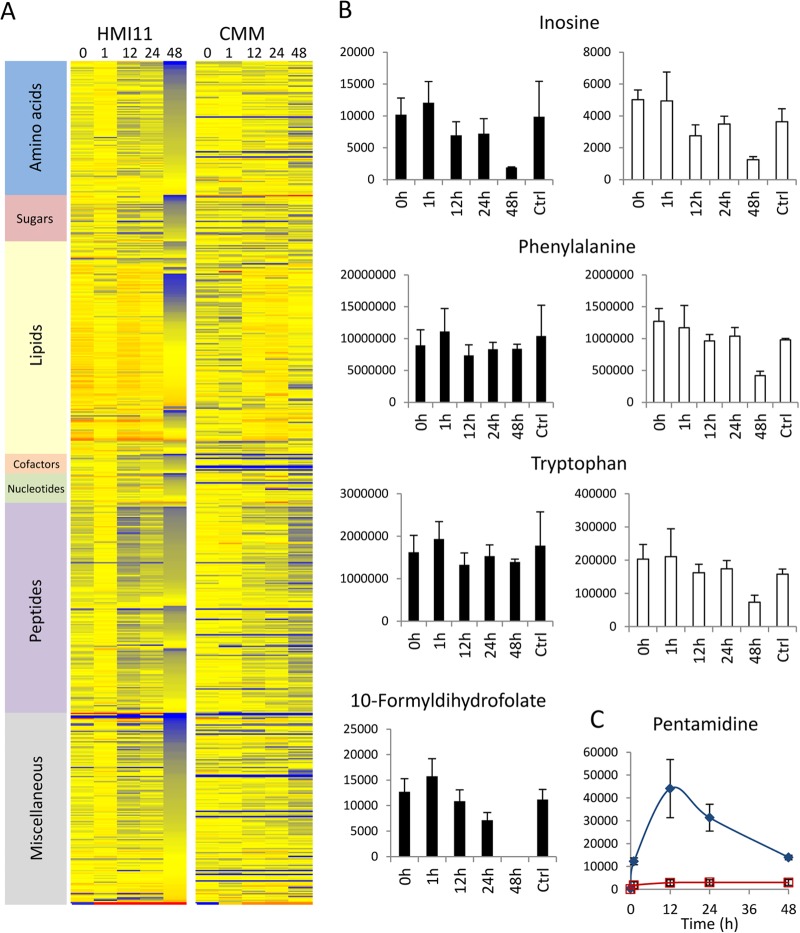
Analysis of intracellular metabolite levels after 0-, 1-, 12-, and 24-h incubations with pentamidine revealed no major (>2-fold) reproducible drug-induced changes in the metabolome (Fig. 5A). The most significant change was a 44% decrease in a metabolite putatively identified as 10-formyldihydrofolate at 24 h (Fig. 5B). This metabolite was completely depleted from HMI11 (below the limit of detection) at 48 h but not observed at any time point in cells grown in CMM (below the limit of detection). This observation suggests that pentamidine may interfere with folate metabolism. However, supplementing CMM with folate had no impact on measured activity (IC50 = 0.005 nM; P > 0.05), indicating that folate metabolism is not the primary target of drug action.
Fig 5.
Pentamidine-induced changes in the metabolome over a 48-h time course in CMM versus HMI11. (A) Heat map of changes in the abundance of all putative metabolites relative to that in untreated controls (Ctrl) during the 48-h time course in HMI11 and CMM. Red, increased; blue, decreased; yellow, unchanged. (B) Inosine decreased significantly after 48 h of treatment in both media. Decreases in phenylalanine and tryptophan could be observed only in CMM. 10-Formyldihydrofolate was detected only in HMI-grown cells. (C) The relative levels of pentamidine were approximately 10-fold higher in HMI11 (blue filled diamonds) than in CMM (red open squares), in agreement with the 10-fold higher dosage. y axes show raw LC-MS peak heights in arbitrary units. The data are the means ± standard deviations of three (CMM) or four (HMI11) biological replicates.
Numerous metabolites were significantly depleted after 48 h of incubation with pentamidine in HMI11, including nucleotides, lipids, carbohydrates, and amino acid metabolites (Fig. 5A). Approximately 50% cell death was apparent under these conditions, and it is therefore likely that these were nonspecific changes in metabolism in response to cell death and metabolite leakage. None of these changes were observed after 48 h with the sublethal dose of pentamidine in CMM, with the exception of inosine. In addition to inosine, the only significant changes in known metabolites in CMM-grown cells were decreased levels of phenylalanine and tryptophan (Fig. 5B). Changes in these aromatic amino acids were not observed in HMI11, most likely because of the buffer provided by the elevated concentrations present in the growth medium. Unlike eflornithine (15), the metabolic profiles observed for pentamidine-treated cells in HMI11 and CMM indicate that this drug is unlikely to act through the inhibition of specific metabolic pathways. The lower potency of pentamidine in HMI11 than in CMM did not appear to be due to altered uptake or accumulation of pentamidine within the cell. The pentamidine levels in cells in HMI11 were approximately 10-fold higher than those in cells in CMM, in agreement with the 10-fold difference in the initial pentamidine concentrations (Fig. 5C).
DISCUSSION
Here, we present a rational methodology, based on the analysis of metabolite utilization, for the development of cell culture media for specific applications. In the field of drug development, an optimal culture medium is one that mimics the in vivo biological environment as closely as possible while supporting a phenotype that enables meaningful biological studies. In the case of bloodstream form T. brucei, a fully defined medium is not practical and calf serum is generally used to supply many of the nutrients that this parasite salvages from its mammalian host. Attempts to standardize media with either dialyzed serum (41) or bovine serum proteins (42) have required many additional nutrients, and the formulation of new media has been a largely empirical and time-consuming endeavor. The commonly used HMI11 formula successfully supports continuous T. brucei culture and is based on Iscove's medium, which contains all of the major nutrients required for energy generation and macromolecule synthesis (11). However, the concentration of many components is significantly higher than the normal concentrations found in human blood or cerebrospinal fluid, the natural in vivo environment of the stage of T. brucei responsible for clinical pathogenesis (Table 2).
The metabolomics-based approach described here allowed rapid evaluation of the actual nutrient utilization under standard cell culture conditions. The relative consumption and production of 35 HMI11 components and hundreds of serum-derived nutrients were measured in a single experiment. Most of the HMI11 components were not depleted during cell growth, allowing the development of a simplified medium. Targeted metabolomic analysis revealed that many essential nutrients are present in fresh 10% FCS at concentrations close to the average levels reported in human blood, and these concentrations are sufficient to support growth rates in cell culture equivalent to those in HMI11. It is well established that bloodstream form T. brucei acquires energy exclusively from glycolysis (33) and that cysteine and a reducing agent are required to maintain a cell culture in the absence of feeder cells (9, 11). In addition to glucose and cysteine, the only other nutrient that was not supplied in sufficient quantity by 10% FCS was glutamine. Interestingly, the glutamine in the CMM recipe could be replaced with the aromatic amino acids phenylalanine and tryptophan. The significant accumulation of amino acid-derived keto acids in spent medium is in agreement with previous observations of accumulation of hydrophobic keto acids in the plasma and urine of infected rodents (43, 44). This suggests a crucial role for hydrophobic amino acids in the provision of amino groups, possibly for methionine recycling (45). However, methionine depletion from the medium suggests that methionine acquisition occurs by uptake from the medium to a significant degree (46), while accumulation of the corresponding keto acid in spent medium suggests that methionine itself is an amino donor for as-yet-uncharacterized substrates. The significant accumulation of alanine in the spent medium suggests that transamination of pyruvate to alanine may be a primary role of amino acid-derived nitrogen.
When FCS was replaced with dialyzed serum, CMM did not support growth, confirming that some of the nutrients supplied by FCS are essential for growth. One potential disadvantage of this reliance on FCS is the risk of batch variability; hence, the formula was developed with FCS Gold (PAA, Piscataway, NJ), which is a standardized reconstituted serum, to avoid the risk of interbatch variation.
Indeed, one tested batch of tetracycline-free serum (Biosera) failed to support optimal growth with CMM and LC-MS analysis revealed amino acid levels lower than those in FCS Gold (data not shown). The addition of six amino acids (Tyr, Phe, Trp, Leu, Met, and Arg) at 100 μM returned the growth rates to normal.
The untargeted metabolomic approach allowed rapid analysis of the serum components that are extensively consumed during in vitro growth. Many small peptides were depleted from the medium, suggesting an active mechanism for the salvage of amino acids from peptides or proteins in the extracellular environment. While these mechanisms may not be essential for growth in rich culture medium or even in human blood, they may play an important role in nutrient-starved environments such as cerebrospinal fluid in central nervous system stage infection or during the insect stage of the parasite life cycle. Also of significance was the depletion of LPCs. Extensive utilization of LPCs is in agreement with the lipid salvage mechanisms previously described for bloodstream form trypanosomes (47). So dramatic was their loss that we investigated whether this might represent the growth-limiting determinant of bloodstream form trypanosome growth in vitro. However, addition of fresh LPCs did not extend growth. No known metabolite was completely removed from the medium by the time cells had ceased to proliferate, nor were novel metabolites that might be toxic detected; hence, a simple solution to the relatively poor growth capacity of T. brucei bloodstream forms in vitro has not been found. Purine salvage is essential in trypanosomatids that lack purine biosynthesis pathways, which explains the observed depletion of inosine and guanosine from the medium. These results suggest a preference for the uptake of purine nucleosides rather than free purine bases, as hypoxanthine levels were not significantly depleted in either hypoxanthine-rich (HMI11) or hypoxanthine-depleted (CMM) medium. Depletion of inosine and guanosine as the cells reached stationary phase suggested that a lack of purine nucleosides may be growth limiting, but addition of inosine (100 μM) to HMI11 was unable to support growth to higher cell densities, and it is likely that other factors such as pH, redox, or quorum-sensing mechanisms prevent growth beyond 5 × 106 cells ml−1.
The benefits of CMM for metabolomic studies were demonstrated by the study of pentamidine action. The decreased number of LC-MS artifacts was exemplified by hypoxanthine, which had 56 peaks annotated as “related peaks” in HMI11 according to the algorithm in mzMatch (48), compared to just 7 in CMM. Overall, the number of detected peaks from cells grown in CMM was only 5% lower than that from cells grown in HMI11, confirming that the overall compositions of cells from the two media are very similar. The major benefit of CMM was the ability to detect changes in metabolites that are usually abundant in the medium. No changes in hydrophobic amino acids were observed in pentamidine-treated cells in HMI11, whereas the lower levels of phenylalanine and tryptophan in CMM allowed the detection of significant changes in the intracellular concentrations of these metabolites.
In a recent study of eflornithine action, we demonstrated significant changes in the concentrations of the substrate and product of the target enzyme, ornithine decarboxylase, within hours of treatment with either lethal or sublethal doses (15). In contrast, no such changes were observed during the early stages of pentamidine treatment, suggesting that the trypanocidal activity of pentamidine is not mediated by the direct inhibition of a single enzyme. Major changes in metabolite levels were apparent only after 48 h and 50% cell death, where metabolites from multiple metabolic pathways were perturbed. In contrast, few major changes were observed after 48 h of sublethal pentamidine treatment in CMM, except for decreased levels of inosine, phenylalanine, and tryptophan. This work has identified those three metabolites as important nutrients for cell growth, and depletion of those metabolites suggests increased metabolic turnover in treated cells.
Previous studies have linked pentamidine's action to the inhibition of various enzymes, including S-adenosylmethionine decarboxylase (49). However, in agreement with other previous work (50), no effect on polyamine abundance was observed following pentamidine treatment. An in vivo study linked pentamidine activity to the accumulation of arginine and lysine (50); however, our results showed no effect on basic amino acids in either HMI11 or CMM. Overall, the metabolomic data do not support a metabolic mode of action of pentamidine, and this finding is supported by recently published RITseq data that did not find any relationships between metabolic enzymes and resistance to pentamidine (51). Pentamidine is known to bind DNA, and it is likely that the mode of action is related to interactions with nucleic acids (52) or mitochondrial membrane activity (53). While a metabolomic approach is generally not suitable for the investigation of modes of action involving macromolecules, the drug-induced depletion of inosine and 10-formyldihydrofolate indirectly supports a link between nucleotide turnover and pentamidine activity.
CMM was successfully applied to the determination of in vitro drug efficacy in trypanosomes by the standard alamarBlue assay. While many of the drugs tested displayed comparable potencies in both CMM and HMI11, there were large differences observed for pentamidine and the antifolates. These differences imply that components of standard HMI11 medium inhibit the action of these drugs by inhibition of drug uptake and/or inhibition of action by direct interaction with the target or by metabolite rescue. The increased activity of antifolates in folate-depleted medium has been reported previously in T. brucei (17), and the increased IC50 observed when folate and thymidine were increased from CMM to HMI11 concentrations confirms the inhibitory effect of these metabolites on methotrexate activity. Antifolates demonstrate poor activity in standard drug screening approaches with rich medium, which might indicate that folate metabolism would be an unsuitable drug target in trypanosomatids. Nevertheless, DHFR has recently been confirmed as a validated drug target in T. brucei, and it is suggested that HMI medium is unsuitable for phenotypic drug screening of antifolates (17). This phenomenon is likely to span many areas of metabolism, and it is likely that high-throughput screening approaches using HMI11 have underestimated the potency of many trypanocidal compounds, particularly those that act by metabolic inhibition or rely on uptake by metabolite transporters. CMM allows the screening of trypanocidal activity by established methods but in a medium that more closely represents metabolite concentrations in human blood (Table 2). This should allow the identification of novel compounds with trypanocidal activity that might have otherwise gone unnoticed if they were subject to inhibition by the high concentrations of nutrients in standard HMI11.
In addition to the improved sensitivity of drug activity testing in CMM, the exclusion of 35 additives from HMI11 substantially decreases the cost associated with regular trypanosome culture. The physicochemical properties of all of the components allow the rapid preparation of large volumes, and unlike those of HMI11, all of the components dissolve rapidly (within minutes). The only practical precautions, as with HMI11, are that 2-mercaptoethanol should be added under a fume hood and the usual care should be taken during pH adjustment.
In summary, this study has demonstrated that metabolomics is a useful tool for the rational optimization of cell culture medium. Furthermore, it offers a rapid and unbiased method for the analysis of cellular metabolism in different culture media. The minimal medium (CMM) developed by this approach supports optimal in vitro growth while providing a more biologically relevant culture medium for experimental studies of T. brucei biology. In particular, improved sensitivity has been demonstrated for compound activity screening, which should substantially improve drug discovery efforts for HAT treatment.
Supplementary Material
ACKNOWLEDGMENTS
D.J.C. is supported by an NHMRC postdoctoral training fellowship. This work was partly supported by the Wellcome Trust through The Wellcome Trust Centre for Molecular Parasitology, which is supported by core funding from the Wellcome Trust (grant 085349).
Metabolomic analysis was provided by Glasgow Polyomics, and we are grateful to Karl Burgess for assistance. D.K.H., F.R., M.P.B., and K.R.M. are supported by a SysMO/BBSRC grant (the Silicotryp).
Footnotes
Published ahead of print 9 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00044-13.
REFERENCES
- 1. Barrett MP, Vincent IM, Burchmore RJ, Kazibwe AJ, Matovu E. 2011. Drug resistance in human African trypanosomiasis. Future Microbiol. 6:1037–1047 [DOI] [PubMed] [Google Scholar]
- 2. Brun R, Don R, Jacobs RT, Wang MZ, Barrett MP. 2011. Development of novel drugs for human African trypanosomiasis. Future Microbiol. 6:677–691 [DOI] [PubMed] [Google Scholar]
- 3. Jacobs RT, Nare B, Wring SA, Orr MD, Chen D, Sligar JM, Jenks MX, Noe RA, Bowling TS, Mercer LT, Rewerts C, Gaukel E, Owens J, Parham R, Randolph R, Beaudet B, Bacchi CJ, Yarlett N, Plattner JJ, Freund Y, Ding C, Akama T, Zhang YK, Brun R, Kaiser M, Scandale I, Don R. 2011. SCYX-7158, an orally-active benzoxaborole for the treatment of stage 2 human African trypanosomiasis. PLoS Negl. Trop. Dis. 5:e1151 doi:10.1371/journal.pntd.0001151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mäser P, Wittlin S, Rottmann M, Wenzler T, Kaiser M, Brun R. 2012. Antiparasitic agents: new drugs on the horizon. Curr. Opin. Pharmacol. 12:562–566 [DOI] [PubMed] [Google Scholar]
- 5. Bowling T, Mercer L, Don R, Jacobs R, Nare B. 2012. Application of a resazurin-based high-throughput screening assay for the identification and progression of new treatments for human African trypanosomiasis. Int. J. Parasitol. Drugs Drug Resist. 2:262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Räz B, Iten M, Grether-Buhler Y, Kaminsky R, Brun R. 1997. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T. b. rhodesiense and T. b. gambiense) in vitro. Acta Trop. 68:139–147 [DOI] [PubMed] [Google Scholar]
- 7. Sykes ML, Avery VM. 2009. Development of an Alamar Blue viability assay in 384-well format for high throughput whole cell screening of Trypanosoma brucei brucei bloodstream form strain 427. Am. J. Trop. Med. Hyg. 81:665–674 [DOI] [PubMed] [Google Scholar]
- 8. Hirumi H, Hirumi K, Nelson RT, Bwayo JJ. 1980. Present status of the cultivation of African trypanosomes, p 165–200 In Rowe S, Hirumi H. (ed), Tropical diseases research series, vol 3 Schwabe & Co., Basel, Switzerland [Google Scholar]
- 9. Duszenko M, Ferguson MA, Lamont GS, Rifkin MR, Cross GA. 1985. Cysteine eliminates the feeder cell requirement for cultivation of Trypanosoma brucei bloodstream forms in vitro. J. Exp. Med. 162:1256–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baltz T, Baltz D, Giroud C, Crockett J. 1985. Cultivation in a semi-defined medium of animal infective forms of Trypanosoma brucei, T. equiperdum, T. evansi, T. rhodesiense and T. gambiense. EMBO J. 4:1273–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hirumi H, Hirumi K. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985–989 [PubMed] [Google Scholar]
- 12. Creek DJ, Anderson J, McConville MJ, Barrett MP. 2012. Metabolomic analysis of trypanosomatid protozoa. Mol. Biochem. Parasitol. 181:73–84 [DOI] [PubMed] [Google Scholar]
- 13. Drexler DM, Reily MD, Shipkova PA. 2011. Advances in mass spectrometry applied to pharmaceutical metabolomics. Anal. Bioanal. Chem. 399:2645–2653 [DOI] [PubMed] [Google Scholar]
- 14. Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal B, van Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S. 2009. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics 5:435–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vincent IM, Creek DJ, Burgess K, Woods DJ, Burchmore RJS, Barrett MP. 2012. Untargeted metabolomics reveals a lack of synergy between nifurtimox and eflornithine against Trypanosoma brucei. PLoS Negl. Trop. Dis. 6:e1618 doi:10.1371/journal.pntd.0001618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kwon YK, Lu W, Melamud E, Khanam N, Bognar A, Rabinowitz JD. 2008. A domino effect in antifolate drug action in Escherichia coli. Nat. Chem. Biol. 4:602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sienkiewicz N, Jarosławski S, Wyllie S, Fairlamb AH. 2008. Chemical and genetic validation of dihydrofolate reductase-thymidylate synthase as a drug target in African trypanosomes. Mol. Microbiol. 69:520–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geary TG, Divo AA, Bonanni LC, Jensen JB. 1985. Nutritional requirements of Plasmodium falciparum in culture. III. Further observations on essential nutrients and antimetabolites. J. Eukaryot. Microbiol. 32:608–613 [DOI] [PubMed] [Google Scholar]
- 19. Bradley SA, Ouyang A, Purdie J, Smitka TA, Wang T, Kaerner A. 2010. Fermentanomics: monitoring mammalian cell cultures with NMR spectroscopy. J. Am. Chem. Soc. 132:9531–9533 [DOI] [PubMed] [Google Scholar]
- 20. Zang L, Frenkel R, Simeone J, Lanan M, Byers M, Lyubarskaya Y. 2011. Metabolomics profiling of cell culture media leading to the identification of riboflavin photosensitized degradation of tryptophan causing slow growth in cell culture. Anal. Chem. 83:5422–5430 [DOI] [PubMed] [Google Scholar]
- 21. Saunders EC, Ng WW, Chambers JM, Ng M, Naderer T, Kramer JO, Likic VA, McConville MJ. 2011. Isotopomer profiling of Leishmania mexicana promastigotes reveals important roles for succinate fermentation and aspartate uptake in tricarboxylic acid cycle (TCA) anaplerosis, glutamate synthesis, and growth. J. Biol. Chem. 286:27706–27717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. t'Kindt R, Jankevics A, Scheltema RA, Zheng L, Watson DG, Dujardin JC, Breitling R, Coombs GH, Decuypere S. 2010. Towards an unbiased metabolic profiling of protozoan parasites: optimisation of a Leishmania sampling protocol for HILIC-Orbitrap analysis. Anal. Bioanal. Chem. 398:2059–2069 [DOI] [PubMed] [Google Scholar]
- 23. Creek DJ, Jankevics A, Breitling R, Watson DG, Barrett MP, Burgess KE. 2011. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography-mass spectrometry: improved metabolite identification by retention time prediction. Anal. Chem. 83:8703–8710 [DOI] [PubMed] [Google Scholar]
- 24. Tautenhahn R, Bottcher C, Neumann S. 2008. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinf. 9:504 doi:10.1186/1471-2105-9-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scheltema RA, Jankevics A, Jansen RC, Swertz MA, Breitling R. 2011. PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 83:2786–2793 [DOI] [PubMed] [Google Scholar]
- 26. Creek DJ, Jankevics A, Burgess KEV, Breitling R, Barrett MP. 2012. IDEOM: an Excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics 28:1048–1049 [DOI] [PubMed] [Google Scholar]
- 27. Sumner L, Amberg A, Barrett D, Beale M, Beger R, Daykin C, Fan T, Fiehn O, Goodacre R, Griffin J, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane A, Lindon J, Marriott P, Nicholls A, Reily M, Thaden J, Viant M. 2007. Proposed minimum reporting standards for chemical analysis. Metabolomics 3:211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573:83–92 [DOI] [PubMed] [Google Scholar]
- 29. Mashego MR, Wu L, Van Dam JC, Ras C, Vinke JL, Van Winden WA, Van Gulik WM, Heijnen JJ. 2004. MIRACLE: mass isotopomer ratio analysis of U-13C-labeled extracts. A new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnol. Bioeng. 85:620–628 [DOI] [PubMed] [Google Scholar]
- 30. Wu L, Mashego MR, van Dam JC, Proell AM, Vinke JL, Ras C, van Winden WA, van Gulik WM, Heijnen JJ. 2005. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal. Biochem. 336:164–171 [DOI] [PubMed] [Google Scholar]
- 31. Winder CL, Dunn WB, Schuler S, Broadhurst D, Jarvis R, Stephens GM, Goodacre R. 2008. Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites. Anal. Chem. 80:2939–2948 [DOI] [PubMed] [Google Scholar]
- 32. Zhang T, Creek DJ, Barrett MP, Blackburn G, Watson DG. 2012. Evaluation of coupling reversed phase, aqueous normal phase, and hydrophilic interaction liquid chromatography with Orbitrap mass spectrometry for metabolomic studies of human urine. Anal. Chem. 84:1994–2001 [DOI] [PubMed] [Google Scholar]
- 33. Haanstra JR, van Tuijl A, van Dam J, van Winden W, Tielens AG, van Hellemond JJ, Bakker BM. 2012. Proliferating bloodstream-form Trypanosoma brucei use a negligible part of consumed glucose for anabolic processes. Int. J. Parasitol. 42:667–673 [DOI] [PubMed] [Google Scholar]
- 34. Duszenko M, Mühlstädt K, Broder A. 1992. Cysteine is an essential growth factor for Trypanosoma brucei bloodstream forms. Mol. Biochem. Parasitol. 50:269–273 [DOI] [PubMed] [Google Scholar]
- 35. Alsford S, Kawahara T, Glover L, Horn D. 2005. Tagging a T. brucei rRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol. Biochem. Parasitol. 144:142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 37:D603–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zaboikin M, Srinivasakumar N, Schuening F. 2006. Gene therapy with drug resistance genes. Cancer Gene Ther. 13:335–345 [DOI] [PubMed] [Google Scholar]
- 38. Lanteri CA, Stewart ML, Brock JM, Alibu VP, Meshnick SR, Tidwell RR, Barrett MP. 2006. Roles for the Trypanosoma brucei P2 transporter in DB75 uptake and resistance. Mol. Pharmacol. 70:1585–1592 [DOI] [PubMed] [Google Scholar]
- 39. de Koning HP, Anderson LF, Stewart M, Burchmore RJ, Wallace LJ, Barrett MP. 2004. The trypanocide diminazene aceturate is accumulated predominantly through the TbAT1 purine transporter: additional insights on diamidine resistance in African trypanosomes. Antimicrob. Agents Chemother. 48:1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ward CP, Wong PE, Burchmore RJ, de Koning HP, Barrett MP. 2011. Trypanocidal furamidine analogues: influence of pyridine nitrogens on trypanocidal activity, transport kinetics, and resistance patterns. Antimicrob. Agents Chemother. 55:2352–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuettel S, Wadum MC, Güther ML, Mariño K, Riemer C, Ferguson MA. 2012. The de novo and salvage pathways of GDP-mannose biosynthesis are both sufficient for the growth of bloodstream-form Trypanosoma brucei. Mol. Microbiol. 84:340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hirumi H, Martin S, Hirumi K, Inoue N, Kanbara H, Saito A, Suzuki N. 1997. Cultivation of bloodstream forms of Trypanosoma brucei and T. evansi in a serum-free medium. Trop. Med. Int. Health 2:240–244 [DOI] [PubMed] [Google Scholar]
- 43. Wang Y, Utzinger J, Saric J, Li JV, Burckhardt J, Dirnhofer S, Nicholson JK, Singer BH, Brun R, Holmes E. 2008. Global metabolic responses of mice to Trypanosoma brucei brucei infection. Proc. Natl. Acad. Sci. U. S. A. 105:6127–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seed JR, Sechelski J, Hall JE. 1987. Further biochemical characterization of chronic Trypanosoma brucei gambiense-Microtus montanus infection. Am. J. Trop. Med. Hyg. 37:314–319 [DOI] [PubMed] [Google Scholar]
- 45. Berger BJ, Dai WW, Wang H, Stark RE, Cerami A. 1996. Aromatic amino acid transamination and methionine recycling in trypanosomatids. Proc. Natl. Acad. Sci. U. S. A. 93:4126–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hasne MP, Barrett MP. 2000. Transport of methionine in Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 111:299–307 [DOI] [PubMed] [Google Scholar]
- 47. Smith TK, Bütikofer P. 2010. Lipid metabolism in Trypanosoma brucei. Mol. Biochem. Parasitol. 172:66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scheltema R, Decuypere S, Dujardin J, Watson D, Jansen R, Breitling R. 2009. Simple data-reduction method for high-resolution LCMS data in metabolomics. Bioanalysis 1:1551–1557 [DOI] [PubMed] [Google Scholar]
- 49. Bitonti AJ, Dumont JA, McCann PP. 1986. Characterization of Trypanosoma brucei brucei S-adenosyl-l-methionine decarboxylase and its inhibition by Berenil, pentamidine and methylglyoxal bis(guanylhydrazone). Biochem. J. 237:685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berger BJ, Carter NS, Fairlamb AH. 1993. Polyamine and pentamidine metabolism in African trypanosomes. Acta Trop. 54:215–224 [DOI] [PubMed] [Google Scholar]
- 51. Alsford S, Eckert S, Baker N, Glover L, Sanchez-Flores A, Leung KF, Turner DJ, Field MC, Berriman M, Horn D. 2012. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature 482:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW. 2008. Antiparasitic compounds that target DNA. Biochimie 90:999–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lanteri CA, Tidwell RR, Meshnick SR. 2008. The mitochondrion is a site of trypanocidal action of the aromatic diamidine DB75 in bloodstream forms of Trypanosoma brucei. Antimicrob. Agents Chemother. 52:875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.