Abstract
Objective:
There is an urgent need to identify risk factors for sporadic intracranial aneurysm (IA) development and rupture. A genetic component has long been recognized, but firm conclusions have been elusive given the generally small sample sizes and lack of replication. Genome-wide association studies have overcome some limitations, but the number of robust genetic risk factors for IA remains uncertain.
Methods:
We conducted a comprehensive systematic review and meta-analysis of all genetic association studies (including genome-wide association studies) of sporadic IA, conducted according to Strengthening the Reporting of Genetic Association Studies and Human Genome Epidemiology Network guidelines. We tested the robustness of associations using random-effects and sensitivity analyses.
Results:
Sixty-one studies including 32,887 IA cases and 83,683 controls were included. We identified 19 single nucleotide polymorphisms associated with IA. The strongest associations, robust to sensitivity analyses for statistical heterogeneity and ethnicity, were found for the following single nucleotide polymorphisms: on chromosome 9 within the cyclin-dependent kinase inhibitor 2B antisense inhibitor gene (rs10757278: odds ratio [OR] 1.29; 95% confidence interval [CI] 1.21–1.38; and rs1333040: OR 1.24; 95% CI 1.20–1.29), on chromosome 8 near the SOX17 transcription regulator gene (rs9298506: OR 1.21; 95% CI 1.15–1.27; and rs10958409: OR 1.19; 95% CI 1.13–1.26), and on chromosome 4 near the endothelin receptor A gene (rs6841581: OR 1.22; 95% CI 1.14–1.31).
Conclusions:
Our comprehensive meta-analysis confirms a substantial genetic contribution to sporadic IA, implicating multiple pathophysiologic pathways, mainly relating to vascular endothelial maintenance. However, the limited data for IA compared with other complex diseases necessitates large-scale replication studies in a full spectrum of populations, with investigation of how genetic variants relate to phenotype (e.g., IA size, location, and rupture status).
Intracranial aneurysms (IAs) are present in 2%–5% of the general population; approximately 0.7%–1.9% of cases rupture, causing subarachnoid hemorrhage (SAH).1 The annual incidence of SAH is 8–9 in 100,000, yet because of the high risk of death or severe disability2 and younger age at onset compared with other stroke types (40–65 years),3 SAH has a disproportionately large socioeconomic impact.
SAH has been associated with common modifiable risk factors, including hypertension, smoking, and alcohol intake.4–6 However, a genetic component has long been recognized; for example, first-degree relatives of patients with aneurysmal SAH have a 4- to 7-fold increased risk of being affected compared with the general population.7,8 Candidate gene association studies (CGAS) of sporadic IA have identified numerous potential risk loci, but these studies have been limited by small sample sizes and lack of replication. More recently, genome-wide association studies (GWAS) have identified novel genetic loci strongly associated with IAs,9–14 but these still explain only a fraction of the genetic risk.
Thus, the number of true genetic risk factors and the strength of their associations with IAs remain uncertain. The pooling of data from all genetic association studies allows risk estimates to be determined with more precision than is possible from any single study and allows assessment of robustness to statistical heterogeneity and population ethnicity differences across studies. We therefore undertook a comprehensive systematic review and meta-analysis of all published CGAS and GWAS of sporadic IA, including sensitivity analyses for statistical heterogeneity and ethnicity.
METHODS
Our meta-analysis was conducted in accordance with the Human Genome Epidemiology Network guidelines15 and followed published recommendations to improve the quality of meta-analyses of genetic association studies.16 We assessed the quality of reporting of genotyping on the basis of the Strengthening the Reporting of Genetic Association Studies statement.17
Search strategy.
Electronic databases (PubMed, EMBASE, and Google Scholar) were used to retrieve potentially relevant articles on human genetic studies of IAs and SAH that had been published up to December 2012. Search terms used under the Medical Subject Headings were aneurysm(s), intracranial aneurysm(s), subarachnoid hemorrhage, genetics, SNPs [single nucleotide polymorphisms], polymorphism, GWAS, and linkage and candidate gene(s).
Articles in all languages were searched (see figure 1 for a flow diagram of the search strategy) and translated as necessary. After relevant articles were retrieved, references were also checked for other potentially relevant articles not found in the initial search.
Figure 1. Flow chart illustrates the number of studies evaluated in this meta-analysis.
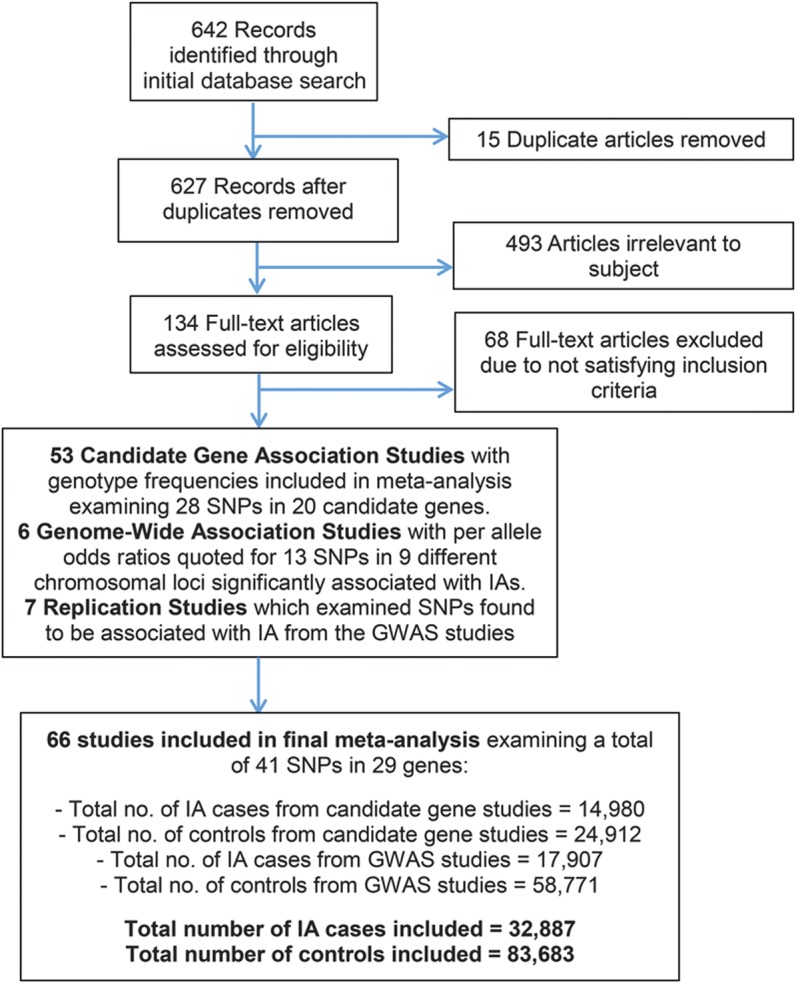
GWAS = genome-wide association studies; IA = intracranial aneurysm; SNP = single nucleotide polymorphism.
Selection criteria.
We included cohort or case-control studies evaluating associations of genetic polymorphisms with proven IA (using CT/MR angiograms or digital subtraction angiography), including unrelated individuals from CGAS and GWAS, in all ethnicities. Where duplicate datasets or cohorts existed, only the largest study was included. All single nucleotide polymorphisms (SNPs) evaluated in at least 2 published studies (CGAS or GWAS) were included in meta-analysis. Exclusion criteria were a history of inherited conditions associated with IAs, e.g., adult polycystic kidney disease or Ehlers-Danlos syndrome.
Data extraction.
Data were extracted by 2 of the authors (V.S.A. and D.J.W.), and differences were resolved by consensus. Where available, genotype frequencies for each SNP were recorded. Where they were unavailable, we estimated the number of cases per genotype category by using published information on risk allele frequencies and odds ratios (ORs) for IA.
Statistical analysis.
For each genetic polymorphism with more than one publication, meta-analysis was performed to determine a pooled OR and 95% confidence intervals (CIs) by using a fixed-effect model (Mantel-Haenszel method). Interstudy heterogeneity p values were measured from Cochran Q test and quantified by using Higgins I2 statistic. SNP associations showing significant interstudy heterogeneity (interstudy heterogeneity p values <0.05 and I2 >50%) were examined under a random-effects (DerSimonian and Laird) model, and, where appropriate, subjected to sensitivity analysis to determine the effect of individual studies on the pooled OR. Ethnic subgroup analysis was also performed where appropriate.
All genetic models (dominant, recessive, and additive) were assessed in studies recording genotype frequencies. Bonferroni correction was applied for multiple testing (p < 0.01). In studies not recording genotype frequencies, per-allele (additive) ORs were pooled with calculation of CIs. Controls were checked to be in Hardy-Weinberg equilibrium for each study before meta-analysis.
Publication bias was assessed by using the Egger regression asymmetry test and visualization of funnel plots. Review Manager (RevMan) version 5.2 from the Cochrane Collaboration (London, UK), 2011, and Comprehensive Meta-Analysis (2.2) from Biostat (Tampa, FL) were used to create the forest plots and perform sensitivity and publication bias analysis.
RESULTS
Our search identified 642 articles, of which 134 articles met initial inclusion criteria. Two studies published in Chinese were translated. After screening for duplication and eligibility, data from 66 case-control studies (60 CGAS and 6 GWAS) were included. Genotype and allele frequencies were reported in 48 publications; the remaining 18 reported additive ORs. In total, 32,887 cases and 83,683 control subjects were included, evaluating 41 polymorphisms in 29 genes (figure 1). The studies encompassed mainly white European, Chinese, and Japanese populations. Risk allele frequencies and control Hardy-Weinberg equilibrium status for each study of all SNPs meta-analyzed are shown in table e-1 on the Neurology® Web site at www.neurology.org.
We identified 19 SNPs (from CGAS and GWAS) that were significantly associated with IAs in at least one genetic model (table 1). The most robust associations were seen in 11 of 12 SNPs discovered from GWAS (table 1), with the strongest associations for loci on chromosome 9 (rs1333040 and rs10757278), chromosome 8 (rs9298506 and rs10958409), and chromosome 4 (rs6841581; figure 2). Other GWAS loci variants associated with IAs included 9p21.3 (rs2891168), 2q33 (rs1429412 and rs700651), 7q13 (rs4628172), 12q22 (rs6538595), and 20p20.1 (rs1132274; table 1 and figures e-1 and e-2).
Table 1.
SNPs associated with IAs
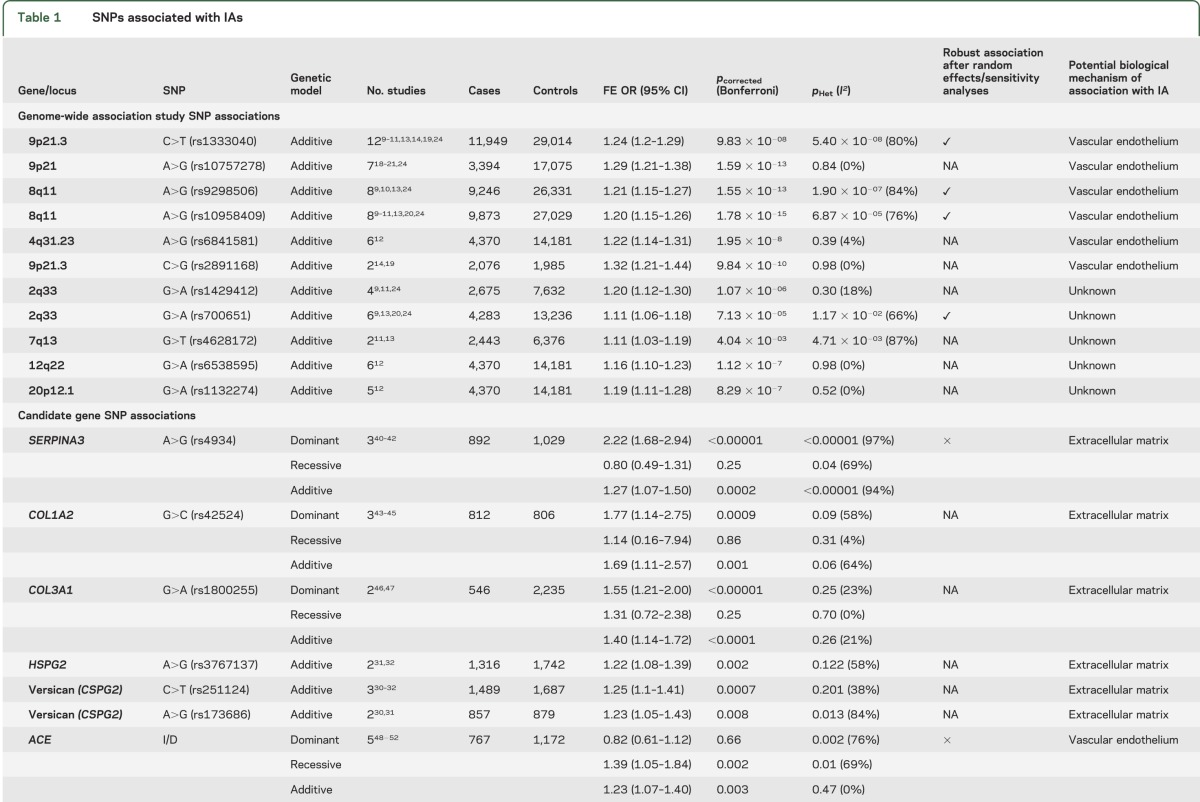
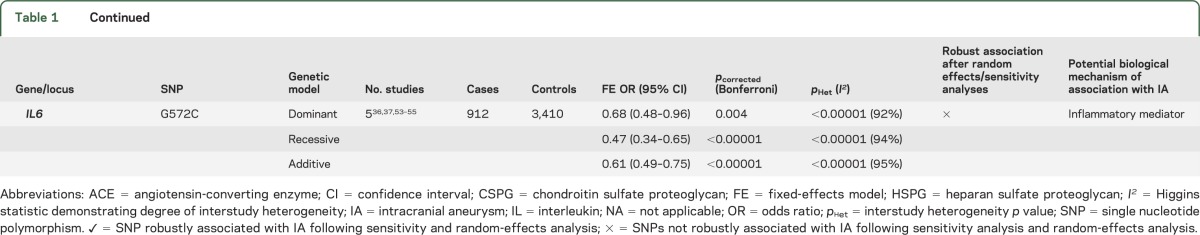
Figure 2. Forest plots (additive model) for genome-wide association studies in all populations.
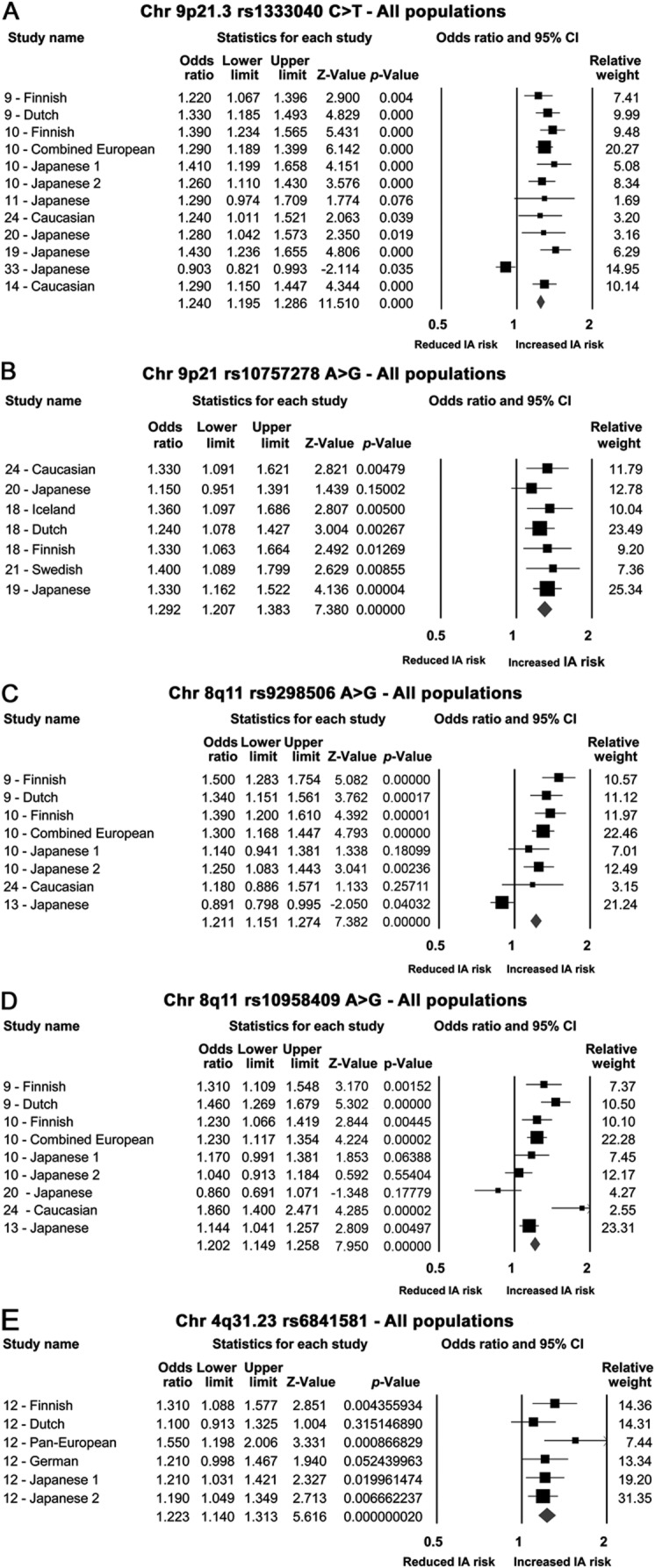
(A) rs1333040 single nucleotide polymorphism (SNP); (B) rs10757278 SNP; (C) rs9298506; (D) rs10958409; (E) rs6841581. CI = confidence interval; IA = intracranial aneurysm.
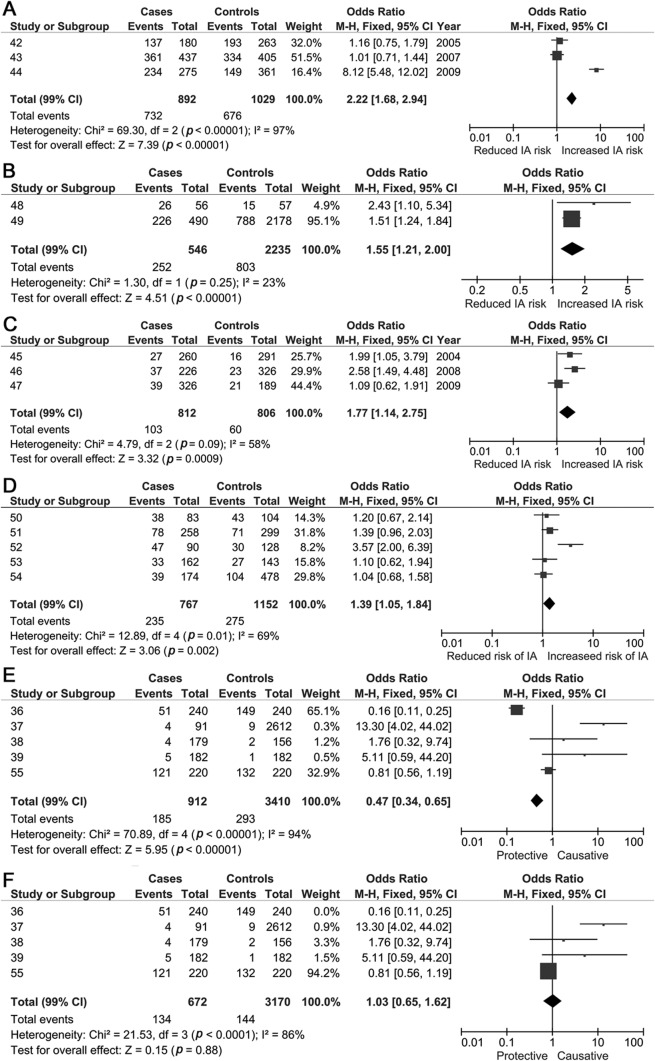
Eight SNPs from CGAS were significantly associated with IAs; SERPINA3 (rs4934) and 2 variants relating to collagen genes (COL1A2 [rs42524 G>C] and COL3A1 [rs1800255 G>A]) showed the most robust associations. The remaining significantly associated CGAS SNPs included heparan sulfate proteoglycan 2 (rs3767137), versican (rs251124 and rs173686), angiotensin-converting enzyme (ACE) I/D, and interleukin 6 (IL6) G572C (table 1, figure 3, and figure e-2).
Figure 3. Forest plots for candidate gene single nucleotide polymorphisms associated with intracranial aneurysms.
(A) Dominant model forest plots for SERPINA3 single nucleotide polymorphism (SNP). (B) COL3A1 (rs1800255) dominant model forest plot. (C) COL1A2 (rs42524) dominant model forest plot. (D) ACE I/D recessive model forest plot. (E) IL6 G572C SNP recessive model forest plot before sensitivity analysis. (F) IL6 G572C SNP recessive model forest plot after sensitivity analysis. CI = confidence interval; IA = intracranial aneurysm; M-H = Mantel-Haenszel.
SNPs demonstrating statistical heterogeneity.
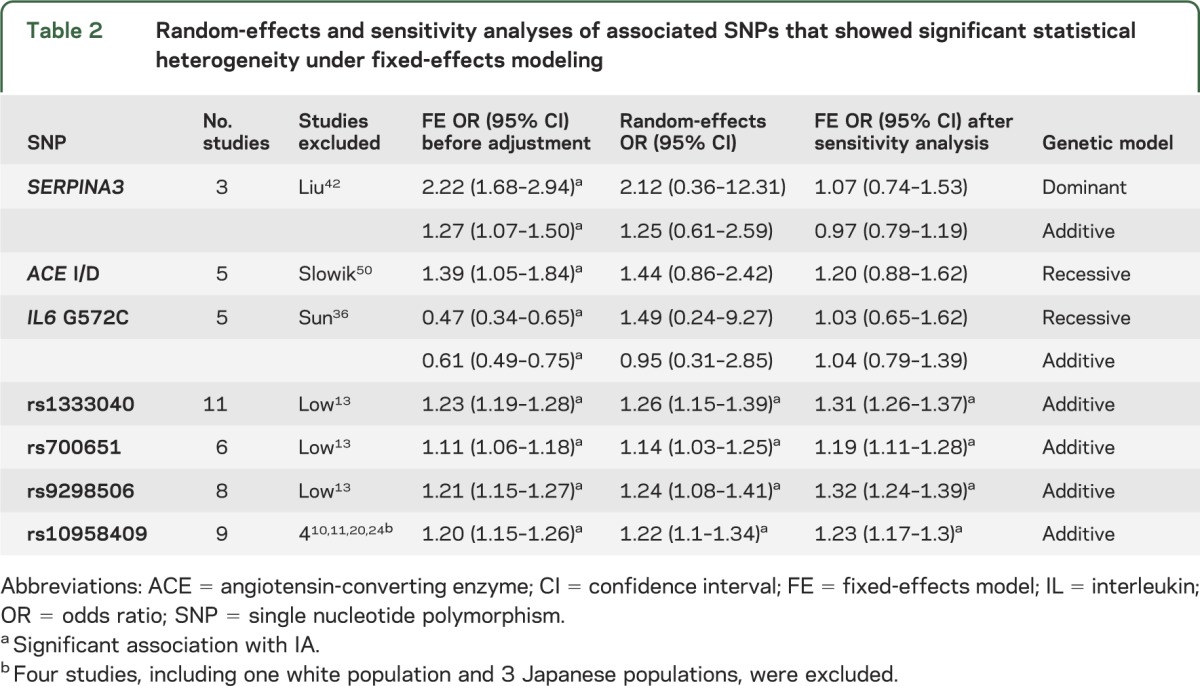
Nine of the 19 significantly associated SNPs (SERPINA3, ACE I/D, IL6 G572C, chondroitin sulfate proteoglycan 2 [rs173686, rs1333040, rs700651, rs9298506, rs10958409, and rs4628172]) showed significant statistical heterogeneity; random-effects and sensitivity analyses were possible only for the 7 variants reported in more than 2 publications (table 2). After sensitivity analysis, the SERPINA3, ACE I/D, and IL6 G572C SNPs (figure 3F) derived from CGAS no longer showed an association with IA.
Table 2.
Random-effects and sensitivity analyses of associated SNPs that showed significant statistical heterogeneity under fixed-effects modeling
Ethnicity analysis.
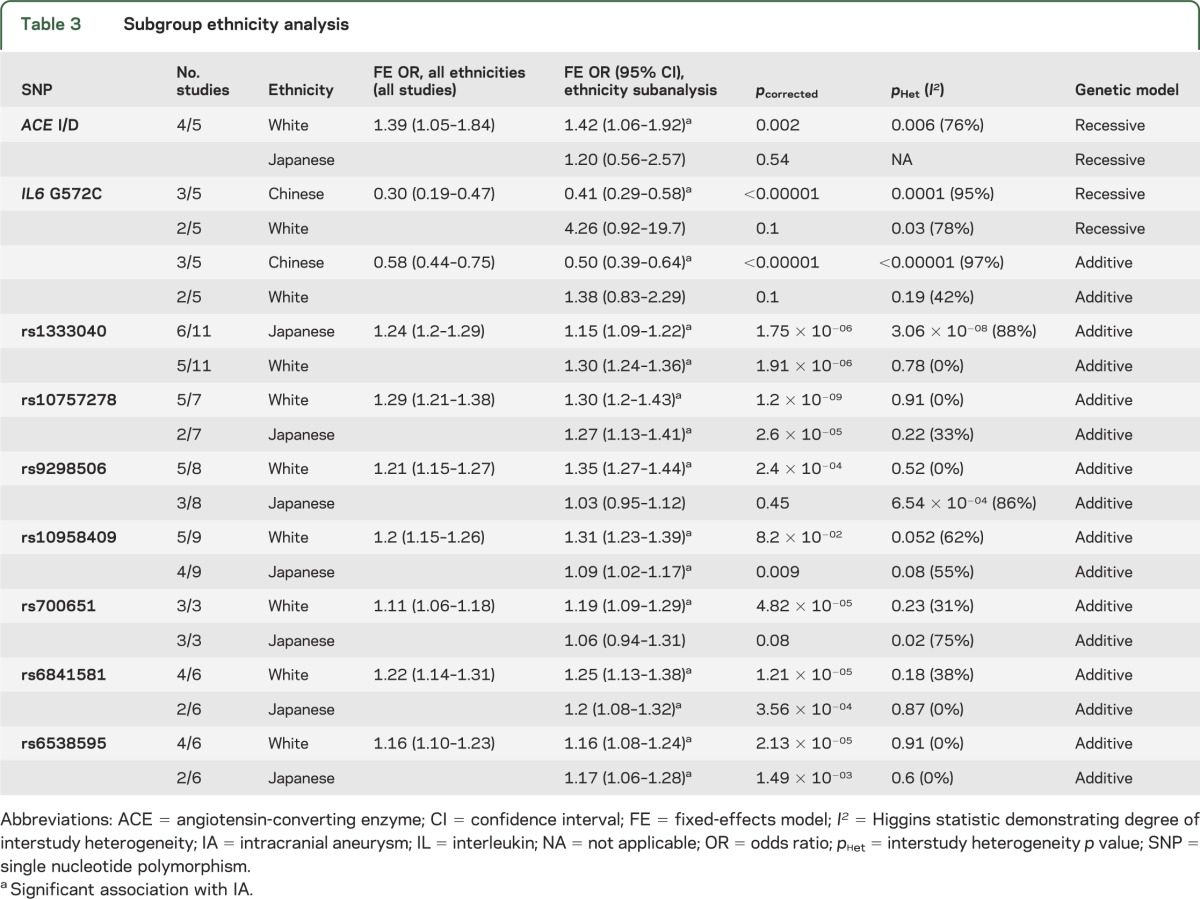
Nine SNPs (2 from CGAS and 7 from GWAS) were subjected to subgroup analysis to determine the effect of ethnicity (table 3 and figures e-3 to e-6). Associations with IA in Japanese, Chinese, and white populations were compared for each SNP, after which 5 of these 9 SNPs (all from GWAS) remained robustly associated with IA in all groups (table 3).
Table 3.
Subgroup ethnicity analysis
Publication bias.
Inspection of funnel plots and calculation of Egger p values confirmed that none of the SNPs (CGAS or GWAS) associated with IAs (table 1) showed publication bias. See figures e-7 to e-11 for funnel plots of all SNPs (CGAS and GWAS) associated with IA.
CGAS not confirming associations with IAs.
Twenty-two candidate SNPs reported in more than one study were not associated with IAs after meta-analysis (table e-2), including the following: endothelial nitric oxide synthase variable number tandem repeat 4a/4b, endothelial nitric oxide synthase T786C, endothelial nitric oxide synthase G894T, endoglin intron 7 Wt/Ins, matrix metalloproteinase (MMP)-3 5A/6A, elastin intron 20T>C, elastin intron 23C>T, IL-1β C5111T, IL-6 G174C, plasminogin activator inhibitor-1 4G/5G, glycoprotein IIIa A1/A2, factor XIII valine/leucine, APOE ε2 and ε4, COL4A1 C>T (rs3783107), fibrillin-2 C>G (rs331079), MMP-2 C>T (rs243865), MMP-9 C1562T (rs3918242), COL3A1 (rs2138533C>T and rs11887092A>G), rs10757272C>T, and rs11661542A>C.
DISCUSSION
This comprehensive systematic review and meta-analysis has identified 19 SNPs associated with IAs among 32,887 cases and 83,683 controls. The variety of genes implicated by these SNPs suggests that multiple pathophysiologic pathways, mainly involved in vascular endothelial maintenance and extracellular matrix integrity, are likely to contribute to IA development and rupture. The most robust SNPs identified arose from variants on chromosomes 4, 8, and 9, derived from GWAS and confirmed by subsequent replication case-control studies.
Genetic variants associated with IAs identified by GWAS.
SNPs with potential roles in vascular endothelial maintenance.
The SNP rs1333040 located on chromosome 9p21.3 in the cyclin-dependent kinase inhibitor 2B antisense gene was strongly associated with IAs. Two major linkage disequilibrium (LD) blocks exist within the 9p21.3 locus: one associated with vascular diseases and the other associated with diabetes. This locus is associated with a wide spectrum of arterial disorders, including coronary artery disease and abdominal aortic aneurysms,18 raising the possibility that there are as yet unidentified common processes involved in all these common cardiovascular pathologies. The cyclin-dependent kinase inhibitor 2B antisense gene, on intron 12, is associated with cell cycle signaling, although its exact biological roles are not fully understood.9,10 Association of the rs1333040 SNP with IAs was found to be independent of smoking and hypertension in one study.19 In multivariable analysis, this SNP was preferentially associated with posterior communicating artery aneurysms, providing preliminary evidence that aneurysm site could be related to genetic factors.19 However, larger cohorts will need to be studied in addition to scanning the entire region to check which SNPs are in LD with the rs1333040-T allele and whether they have any cumulative (epistatic) effect on IA phenotype, including location.
The SNP rs10757278 on chromosome 9p21.3, located in a noncoding RNA region called ANRIL (antisense noncoding RNA in the INK4 locus), showed robust association with IAs, but its function remains unclear. Moreover, the observed association of rs10757278 with IA may arise from moderate LD with the known IA risk variant rs1333040 SNP (r = 0.59), from which it is approximately 41 kbp upstream.20 However, these 2 SNPs were not in strong LD in a Japanese population, raising the possibility of a smaller LD block associated with IA. In a Japanese study,19 haplotype analysis of both risk alleles of these SNPs demonstrated significant association with IAs (OR 1.56; 95% CI 1.32–1.85; p = 2.9 × 10−07). Functional studies show altered ANRIL expression as a result of variations within the 9p21.3 region,14 with reduced expression of RNA encoded by CDKN2A and CDKN2B in mice14; these 2 genes, adjacent to the 9p21 region, encode tumor-suppressor proteins (p14–16), which are involved in cell proliferation.21 Moreover, p14 levels correlate with ANRIL activity and modulate MMP-3 levels, which may influence extracellular matrix repair.21
In the most recent GWAS14 in a Caucasian population of familial and sporadic IA, 6 SNPs, also located in the 9p21.3 region, were associated with IA; one of these (rs6475606) achieved GWAS-level statistical significance (OR 1.34; p = 4.29 × 10−07) for association with sporadic and familial IAs. Association with the rs1333040 SNP was also confirmed in this study,14 further strengthening association of this locus with IA.
Two SNPs on chromosome 8q (rs10958409 and rs9298506), surrounding the SOX17 gene, were studied in European and Japanese populations; rs10958409 was associated in both populations, but the rs9298506 SNP was associated with IA in the European but not Japanese population. SOX17 plays an important role in generation and maintenance of stem cells of endothelial and hematopoietic lineages, crucial in the formation and maintenance of vascular endothelium.22,23 Furthermore, SOX17 knockout mice show multiple vascular abnormalities reflecting defective endothelial remodeling.22 Defective stem cells involved in vascular maintenance are thus plausible targets in IA pathogenesis. The heterogeneity observed in the meta-analysis of these 2 SNPs may partly be explained by the difference in risk allele frequencies in the white population (table e-1)24 compared with the other populations studied (rs10958409, risk allele frequency 0.88 vs approximately 0.2; rs9298506, risk allele frequency 0.17 vs approximately 0.80). Further support implicating chromosome 8 loci was shown in a recent GWAS,14 in which a novel SNP, also in the region of the SOX17 gene, was significantly associated with IAs (rs1072737: OR 1.22; 95% CI 1.07–1.39), independent of smoking. For both SOX17 loci, in the most recent GWAS,14 smoking was an independent risk factor, acting in a multiplicative manner (chromosome 8: rs1072737; OR 2.12; 95% CI 1.76–2.56 and chromosome 9: rs6475606; OR 2.11; 95% CI 1.77–2.52).
A further recent GWAS12 found the SNP rs6841581A>G on chromosome 4q31.23, coding for the endothelin receptor type A (EDNRA) gene, was significantly associated with IA in Dutch, Finnish, and Japanese populations (OR 1.22; 95% CI 1.14–1.31; p = 1.95 × 10−8). Another GWAS13 in a Japanese population showed that the rs6842241 SNP, located at 4q31.22 near the EDNRA gene, was significantly associated with IA (OR 1.25; 95% CI 1.16–1.34; p = 9.58 × 10−09). EDNRA is a G-protein–coupled receptor for endothelins, which modulate vasoconstriction and vasodilatation after hemodynamic insult. Endothelin-1—the predominant isoform in vascular smooth-muscle cells—activates EDNRA. Endothelin signaling is activated at the site of vascular injury,12,25 causing cell proliferation; indeed, after SAH, EDNRA and endothelin-1 protein levels are both increased in CSF.26–28 Alternatively, downregulation of EDNRA signaling could lead to defective vascular repair, allowing aneurysms to develop after injury, a hypothesis supported by functional analysis of EDNRA variants showing lower transcriptional activity for the rs6841581 risk allele (G).13
SNPs with unknown function or not examined in more than 2 studies.
Five SNPs from GWAS do not correspond to genes with any known functions relevant to IA development. Two of these SNPs (rs1429412 and rs700651), at locus 2q33, flank the BOLL gene (involved in germ cell development) and the phospholipase C–like 1 gene (involved in intracellular cascade reactions with abolished phospholipase C activity). Phospholipase C–like 1 lies close to the vascular endothelial growth factor receptor-2, which is involved in CNS angiogenesis and is a marker for endothelial progenitor cells. Three further SNPs on chromosome 2 were associated with familial IAs: rs11693075 (close to the histone deacetylase complex SAP130 gene), rs1897472, and rs3769801 (both located within the phosphodiesterase-1 gene).14
One associated SNP located on chromosome 7 (rs4628172 G>T) is within the TMEM195 gene, which encodes transmembrane proteins, possibly involved in fatty acid biosynthesis and iron binding.11,13 Further associated SNPs from the latest GWAS include variants on chromosome 12q22 (rs6538595, rs11112585, and rs2374513) within the C12orf75 gene.14 The rs1132274 G>T SNP on chromosome 20p12.1 was also associated with IA.10 However, none of these SNPs yet has known clear functional relevance to IA.
Three further loci not included in this meta-analysis (because they were reported in only one study) showed significant association with IAs.10 These loci included 18q11.2 (rs11661542), 10q24.32 (rs12413409), and 13q13.2 (rs9315204), but their mechanisms of association remain unclear.
Genetic variants associated with IAs identified by CGAS.
Eight SNPs from CGAS were associated with IAs in our meta-analysis, albeit with significant statistical heterogeneity. These SNPs relate to genes with plausible relevance to IA biology, including those important for extracellular matrix integrity, inflammatory mediators, and vascular endothelium maintenance. Extracellular matrix gene variants may predispose to IA development by weakening arterial walls. In our meta-analysis, 6 such SNPs showed significant associations with IA: COL1A2 (rs42524), COL3A1 (rs1800255), SERPINA3, versican (rs251124 and rs173686), and heparan sulfate proteoglycan 2 (rs3767137). The COL1A2 rs42524 SNP is located on chromosome 7q22.1 close to the elastin gene—a protein component of arterial walls, previously associated with IAs in a linkage-association study.29 Collagen type 1 is usually found in the adventitial layer of cerebral arteries and provides tensile strength, making it a plausible factor in IA development. Nevertheless, caution is needed in generalizing this result as all 3 studies identified were in Far East populations; for robust association to be confirmed, this SNP has to be examined in other populations, including Europe, because risk allele frequencies may differ. Collagen type 3 (COL 3A1) was investigated in 3 Chinese populations; rs1800255 (G>A) was the only SNP significantly associated with IA, but replication in other ethnicities and larger cohorts is required to confirm these findings.
We also found associations between 2 versican genetic variants (rs251124 and rs173686) and IAs in Dutch, Japanese, and Chinese populations.30–32 In a previous linkage study on a Japanese cohort, chromosome 5q14.3, very close to the versican gene, was implicated in IA.30 In vivo studies on abdominal aortic aneurysm samples found reduced versican messenger RNA expression,33,34 which could reflect a decline in smooth-muscle cells associated with weakness in the vascular wall. Moreover, a GWAS35 on thoracic and aortic dissections showed significant association with the 5q13–14 locus, corresponding to the versican gene.
The proinflammatory cytokine IL6 G572C SNP had an overall protective association with IAs in a recessive model. However, there was significant statistical heterogeneity, and sensitivity analysis revealed no association once one Chinese study36 was excluded, which may partly reflect the greater risk allele frequency (0.79 vs 0.05–0.09) and inclusion of only unruptured IAs in this study.36 Another source of heterogeneity may be sex, because one other study37 included only male controls, and this may not be appropriate because SAH has a female predominance. Therefore, the overall association of this SNP with IA is not robust. Sustained abnormal vascular remodeling in association with inflammation is hypothesized to be a key mechanism in IA development.22 IL-6 can injure vessel walls by inhibiting collagen synthesis, thus increasing wall fragility and increasing the risk of aneurysmal dilatation. IL-6 levels are increased in the CSF and plasma after SAH,38,39 but whether this is directly related to the G572C SNP remains uncertain because in these studies patients were not genotyped for the mutant allele; moreover, IL-6 may be released as part of a systemic inflammatory response in the acute phase after SAH.
Strengths of our study.
Our systematic review and meta-analysis is the largest and most comprehensive to date on genetic variants associated with IAs. We assessed strength of association by using multiple genetic models and carefully assessed the robustness of all associations by performing random-effects, sensitivity, and ethnic subgroup analyses. We conducted our analysis and reporting according to quality guidelines for genetic association studies, including Human Genome Epidemiology Network15 and Strengthening the Reporting of Genetic Association Studies.17
Limitations of our study.
Our assessment of interstudy heterogeneity by visual inspection of the forest plots and sensitivity analyses only partly addresses potential sources of heterogeneity between studies. Further individual patient data are required on other factors that could also contribute to heterogeneity, such as sex proportions, age, genotyping methods or errors, disease phenotype (aneurysm size, location, and rupture status), and environmental risk factor status. Our meta-analysis was only able to pool reported positively associated SNPs from the 6 GWAS studies, but data on many SNPs sequenced not reaching the threshold for GWAS statistical significance were not available, thus limiting our meta-analysis to 41 SNPs in total. Indeed, for many of the candidate-gene studies, pooling all available data from GWAS might allow more robust risk estimates to be calculated; despite 8 candidate gene SNPs being associated with IAs, on average only 2.7 publications were available for each variant.
We have identified 19 candidate SNPs associated with IAs, of which 16 were robust to random-effects or sensitivity analyses. The ORs suggest that the genetic contribution to aneurysm development is substantial, but the variants identified implicate multiple pathophysiologic mechanisms, particularly involving vascular endothelium maintenance. Nevertheless, the amount of data on genetic associations with IAs remains much smaller than for other complex diseases, so further large replication studies in a full spectrum of populations are required, ideally using GWAS or exome sequencing to identify rarer functional variants. A limitation of most studies included is that both ruptured and unruptured IAs were analyzed together; because only a minority of IAs are destined to rupture, future studies should address genetic risk factors specifically related to rupture status. Another important area for future research is to explore how genetic factors are related to aneurysm location, size, or morphology, because there may be specific genetic associations for particular aneurysm phenotypes (which may also relate to rupture risk). Finally, validation studies of risk models in prospective cohorts (e.g., of unruptured IAs followed up for rupture risk) would be helpful in determining the relevance of genetic risk loci in addition to well-established risk factors for overall disease phenotype, to ultimately reduce the devastating burden of aneurysmal SAH.
Supplementary Material
GLOSSARY
- ACE
angiotensin-converting enzyme
- ANRIL
antisense noncoding RNA in the INK4 locus
- CI
confidence interval
- CGAS
candidate gene association study
- EDNRA
endothelin receptor type A
- GWAS
genome-wide association studies
- IA
intracranial aneurysm
- IL
interleukin
- LD
linkage disequilibrium
- MMP
matrix metalloproteinase
- OR
odds ratio
- SAH
subarachnoid hemorrhage
- SNP
single nucleotide polymorphism
Footnotes
Supplemental data at www.neurology.org
AUTHOR CONTRIBUTIONS
Dr. Alg designed the study, performed the literature search, acquired and carried out the statistical analysis of the data, and drafted the manuscript. Dr. Sofat analyzed and interpreted the data and revised the manuscript. Dr. Houlden supervised the study, analyzed and interpreted the data, and provided revisions to the manuscript. Dr. Werring conceived and supervised the study, checked the literature search, interpreted the results, and provided critical revisions to the manuscript.
STUDY FUNDING
Supported and funded by the Stroke Association (TSA 2007/08). Dr. Alg receives research support from the Stroke Association; Dr. Werring receives research support from the Department of Health/Higher Education Funding Council for England (Clinical Senior Lectureship Award). Part of this work was undertaken at UCLH/UCL, which received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme.
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Rinkel GJ, Djibuti M, Algra A, van Gijn J. Prevalence and risk of rupture of intracranial aneurysms: a systematic review. Stroke 1998;29:251–256 [DOI] [PubMed] [Google Scholar]
- 2.Rinkel GJ, Algra A. Long-term outcomes of patients with aneurysmal subarachnoid haemorrhage. Lancet Neurol 2011;10:349–356 [DOI] [PubMed] [Google Scholar]
- 3.Vlak MH, Rinkel GJ, Greebe P, van der Bom JG, Algra A. Trigger factors for rupture of intracranial aneurysms in relation to patient and aneurysm characteristics. J Neurol 2012;259:1298–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke M. Systematic review of reviews of risk factors for intracranial aneurysms. Neuroradiology 2008;50:653–664 [DOI] [PubMed] [Google Scholar]
- 5.Feigin VL, Rinkel GJ, Lawes CM, et al. Risk factors for subarachnoid hemorrhage: an updated systematic review of epidemiological studies. Stroke 2005;36:2773–2780 [DOI] [PubMed] [Google Scholar]
- 6.Juvela S. Risk factors for aneurysmal subarachnoid hemorrhage. Stroke 2002;33:2152–2153; author reply 2152–2153 [DOI] [PubMed] [Google Scholar]
- 7.Krischek B, Inoue I. The genetics of intracranial aneurysms. J Hum Genet 2006;51:587–594 [DOI] [PubMed] [Google Scholar]
- 8.Bromberg JE, Rinkel GJ, Algra A, et al. Subarachnoid haemorrhage in first and second degree relatives of patients with subarachnoid haemorrhage. BMJ 1995;311:288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilguvar K, Yasuno K, Niemela M, et al. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat Genet 2008;40:1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasuno K, Bilguvar K, Bijlenga P, et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat Genet 2010;42:420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akiyama K, Narita A, Nakaoka H, et al. Genome-wide association study to identify genetic variants present in Japanese patients harboring intracranial aneurysms. J Hum Genet 2010;55:656–661 [DOI] [PubMed] [Google Scholar]
- 12.Yasuno K, Bakircioglu M, Low SK, et al. Common variant near the endothelin receptor type A (EDNRA) gene is associated with intracranial aneurysm risk. Proc Natl Acad Sci USA 2011;108:19707–19712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Low SK, Takahashi A, Cha PC, et al. Genome-wide association study for intracranial aneurysm in the Japanese population identifies three candidate susceptible loci and a functional genetic variant at EDNRA. Hum Mol Genet 2012;21:2102–2110 [DOI] [PubMed] [Google Scholar]
- 14.Foroud T, Koller DL, Lai D, et al. Genome-wide association study of intracranial aneurysms confirms role of Anril and SOX17 in disease risk. Stroke 2012;43:2846–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Little JH. The HuGENet™ HuGE Review Handbook, version 1.0. HuGENet 2006:59. Available at http://www.hugenet.ca Accessed February 1, 2012. [Google Scholar]
- 16.Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The quality of meta-analyses of genetic association studies: a review with recommendations. Am J Epidemiol 2009;170:1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little J, Higgins JP, Ioannidis JP, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE Statement. Hum Genet 2009;125:131–151 [DOI] [PubMed] [Google Scholar]
- 18.Helgadottir A, Thorleifsson G, Magnusson KP, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet 2008;40:217–224 [DOI] [PubMed] [Google Scholar]
- 19.Nakaoka H, Takahashi T, Akiyama K, et al. Differential effects of chromosome 9p21 variation on subphenotypes of intracranial aneurysm: site distribution. Stroke 2010;41:1593–1598 [DOI] [PubMed] [Google Scholar]
- 20.Hashikata H, Liu W, Inoue K, et al. Confirmation of an association of single-nucleotide polymorphism rs1333040 on 9p21 with familial and sporadic intracranial aneurysms in Japanese patients. Stroke 2010;41:1138–1144 [DOI] [PubMed] [Google Scholar]
- 21.Olsson S, Csajbok LZ, Jood K, Nylen K, Nellgard B, Jern C. Association between genetic variation on chromosome 9p21 and aneurysmal subarachnoid haemorrhage. J Neurol Neurosurg Psychiatry 2011;82:384–388 [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto T, Meng H, Young WL. Intracranial aneurysms: links among inflammation, hemodynamics and vascular remodeling. Neurol Res 2006;28:372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto T, Pittet JF. Angiopoietin-2: modulator of vascular permeability in acute lung injury? PLoS Med 2006;3:e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deka R, Koller DL, Lai D, et al. The relationship between smoking and replicated sequence variants on chromosomes 8 and 9 with familial intracranial aneurysm. Stroke 2010;41:1132–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Douglas SA, Louden C, Vickery-Clark LM, Feuerstein GZ, Ohlstein EH. Expression of endothelin-1, endothelin-3, endothelin-converting enzyme-1, and endothelin-A and endothelin-B receptor mRNA after angioplasty-induced neointimal formation in the rat. Circ Res 1996;78:322–328 [DOI] [PubMed] [Google Scholar]
- 26.Suzuki H, Sato S, Suzuki Y, Takekoshi K, Ishihara N, Shimoda S. Increased endothelin concentration in CSF from patients with subarachnoid hemorrhage. Acta Neurol Scand 1990;81:553–554 [DOI] [PubMed] [Google Scholar]
- 27.Kraus GE, Bucholz RD, Yoon KW, Knuepfer MM, Smith KR., Jr Cerebrospinal fluid endothelin-1 and endothelin-3 levels in normal and neurosurgical patients: a clinical study and literature review. Surg Neurol 1991;35:20–29 [DOI] [PubMed] [Google Scholar]
- 28.Itoh S, Sasaki T, Asai A, Kuchino Y. Prevention of delayed vasospasm by an endothelin ETA receptor antagonist, BQ-123: change of ETA receptor mRNA expression in a canine subarachnoid hemorrhage model. J Neurosurg 1994;81:759–764 [DOI] [PubMed] [Google Scholar]
- 29.Onda H, Kasuya H, Yoneyama T, et al. Genomewide-linkage and haplotype-association studies map intracranial aneurysm to chromosome 7q11. Am J Hum Genet 2001;69:804–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H, Zhang D, Zhao J. Chondroitin sulfate proteoglycan 2 (CSPG2) gene polymorphisms rs173686 and rs251124 are not associated with intracranial aneurysms in Chinese Han nationality. Ups J Med Sci 2007;112:289–295 [DOI] [PubMed] [Google Scholar]
- 31.Ruigrok YM, Rinkel GJ, Wijmenga C. The versican gene and the risk of intracranial aneurysms. Stroke 2006;37:2372–2374 [DOI] [PubMed] [Google Scholar]
- 32.Ruigrok YM, Rinkel GJ, Wijmenga C, et al. Association analysis of genes involved in the maintenance of the integrity of the extracellular matrix with intracranial aneurysms in a Japanese cohort. Cerebrovasc Dis 2009;28:131–134 [DOI] [PubMed] [Google Scholar]
- 33.Yang F, Zhao P, Zhang Y, et al. Relationship between chondroitin sulfate proteoglycan and coronary atherosclerosis in the youth. Chin Med J (Engl) 1996;109:162–167 [PubMed] [Google Scholar]
- 34.Handley CJ, Samiric T, Ilic MZ. Structure, metabolism, and tissue roles of chondroitin sulfate proteoglycans. Adv Pharmacol 2006;53:219–232 [DOI] [PubMed] [Google Scholar]
- 35.Breimer LH. Genetics of aneurysmal disease: the dogs that did not bark. Angiology 2010;61:233–237 [DOI] [PubMed] [Google Scholar]
- 36.Sun H, Zhang D, Zhao J. The interleukin-6 gene -572G>C promoter polymorphism is related to intracranial aneurysms in Chinese Han nationality. Neurosci Lett 2008;440:1–3 [DOI] [PubMed] [Google Scholar]
- 37.Morgan L, Cooper J, Montgomery H, Kitchen N, Humphries SE. The interleukin-6 gene-174G>C and -572G>C promoter polymorphisms are related to cerebral aneurysms. J Neurol Neurosurg Psychiatry 2006;77:915–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaetani P, Tartara F, Pignatti P, et al. Cisternal CSF levels of cytokines after subarachnoid hemorrhage. Neurol Res 1998;20:337–342 [DOI] [PubMed] [Google Scholar]
- 39.Osuka K, Suzuki Y, Tanazawa T, et al. Interleukin-6 and development of vasospasm after subarachnoid haemorrhage. Acta Neurochir (Wien) 1998;140:943–951 [DOI] [PubMed] [Google Scholar]
- 40.Slowik A, Borratynska A, Turaj W, et al. Alpha1-antichymotrypsin gene (SERPINA3) A/T polymorphism as a risk factor for aneurysmal subarachnoid hemorrhage. Stroke 2005;36:737–740 [DOI] [PubMed] [Google Scholar]
- 41.Krischek B, Akagawa H, Tajima A, et al. The alanine/threonine polymorphism of the alpha-1-antichymotrypsin (SERPINA3) gene and ruptured intracranial aneurysms in the Japanese population. Cerebrovasc Dis 2007;23:46–49 [DOI] [PubMed] [Google Scholar]
- 42.Liu W, Zhu Y, Ge M, Pang Q, Yu Y. Polymorphism rs4934 of SERPINA3 and sporadic intracranial aneurysms in the Chinese population. Cerebrovasc Dis 2010;29:68–72 [DOI] [PubMed] [Google Scholar]
- 43.Yoneyama T, Kasuya H, Onda H, et al. Collagen type I alpha2 (COL1A2) is the susceptible gene for intracranial aneurysms. Stroke 2004;35:443–448 [DOI] [PubMed] [Google Scholar]
- 44.Zhu Y, Li W, Ge M, et al. Polymorphism rs42524 of COL1A2 and sporadic intracranial aneurysms in the Chinese population. J Neurosurg 2008;109:1060–1064 [DOI] [PubMed] [Google Scholar]
- 45.Joo SP, Kim TS, Lee IK, et al. The role of collagen type I alpha2 polymorphisms: intracranial aneurysms in Koreans. Surg Neurol 2009;72:48–53 [DOI] [PubMed] [Google Scholar]
- 46.Hua T, Zhang D, Zhao YL, Wang S, Zhao JZ. Correlation of COL3A1 gene with type III collagen stability in intracranial aneurysm [in Chinese]. Zhonghua Yi Xue Za Zhi 2008;88:445–448 [PubMed] [Google Scholar]
- 47.Chen J, Zhu Y, Jiang Y, et al. A functional variant of the collagen type III alpha1 gene modify risk of sporadic intracranial aneurysms. Hum Genet 2012;131:1137–1143 [DOI] [PubMed] [Google Scholar]
- 48.Takenaka K, Yamakawa H, Sakai H, et al. Angiotensin I-converting enzyme gene polymorphism in intracranial saccular aneurysm individuals. Neurol Res 1998;20:607–611 [DOI] [PubMed] [Google Scholar]
- 49.Keramatipour M, McConnell RS, Kirkpatrick P, et al. The ACE I allele is associated with increased risk for ruptured intracranial aneurysms. J Med Genet 2000;37:498–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slowik A, Borratynska A, Pera J, et al. II genotype of the angiotensin-converting enzyme gene increases the risk for subarachnoid hemorrhage from ruptured aneurysm. Stroke 2004;35:1594–1597 [DOI] [PubMed] [Google Scholar]
- 51.Pannu H, Kim DH, Seaman CR, Van Ginhoven G, Shete S, Milewicz DM. Lack of an association between the angiotensin-converting enzyme insertion/deletion polymorphism and intracranial aneurysms in a Caucasian population in the United States. J Neurosurg 2005;103:92–96 [DOI] [PubMed] [Google Scholar]
- 52.Staalso JM, Nielsen M, Edsen T, et al. Common variants of the ACE gene and aneurysmal subarachnoid hemorrhage in a Danish population: a case-control study. J Neurosurg Anesthesiol 2011;23:304–309 [DOI] [PubMed] [Google Scholar]
- 53.Fontanella M, Rainero I, Gallone S, et al. Interleukin 6 gene polymorphisms are not associated with aneurysmal subarachnoid haemorrhage in an Italian population. J Neurol Neurosurg Psychiatry 2008;79:471–473 [DOI] [PubMed] [Google Scholar]
- 54.Zhang G, Tu Y, Feng W, Huang L, Li M, Qi S. Association of interleukin-6-572G/C gene polymorphisms in the Cantonese population with intracranial aneurysms. J Neurol Sci 2011;306:94–97 [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, Sun J, Wu C, Cao X, He M, You C. The interleukin-6-572G/C gene polymorphism and the risk of intracranial aneurysms in a Chinese population. Genet Test Mol Biomarkers 2012;16:822–826 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.