Abstract
Ceramide and its metabolites constitute a diverse group of lipids, which play important roles as structural entities of biological membranes as well as regulators of cellular growth, differentiation, and development. The C. elegans genome comprises three ceramide synthase genes; hyl-1, hyl-2, and lagr-1. HYL-1 function is required for synthesis of ceramides and sphingolipids containing very long acyl-chains (≥C24), while HYL-2 is required for synthesis of ceramides and sphingolipids containing shorter acyl-chains (≤C22). Here we show that functional loss of HYL-2 decreases lifespan, while loss of HYL-1 or LAGR-1 does not affect lifespan. We show that loss of HYL-1 and LAGR-1 functions extend lifespan in an autophagy-dependent manner, as knock down of the autophagy-associated gene ATG-12 abolishes hyl-1;lagr-1 longevity. The transcription factors PHA-4/FOXA, DAF-16/FOXO, and SKN-1 are also required for the observed lifespan extension, as well as the increased number of autophagosomes in hyl-1;lagr-1 animals. Both autophagic events and the transcription factors PHA-4/FOXA, DAF-16, and SKN-1 have previously been associated with dietary restriction-induced longevity. Accordingly, we find that hyl-1;lagr-1 animals display reduced feeding, increased resistance to heat, and reduced reproduction. Collectively, our data suggest that specific sphingolipids produced by different ceramide synthases have opposing roles in determination of C. elegans lifespan. We propose that loss of HYL-1 and LAGR-1 result in dietary restriction-induced autophagy and consequently prolonged longevity.
Introduction
Besides being required for the integrity of cellular membranes, sphingolipids, and in particular ceramide and sphingosine-1-phosphate, have emerged as bioactive signalling molecules involved in regulation of cell growth, differentiation, senescence, and apoptosis [1]–[3]. Ceramide is at the central hub of sphingolipid metabolism and is the precursor for complex sphingolipids such as sphingomyelin and glycosphingolipids. Sphingosine-based ceramide species are generated from dihydroceramide in a desaturation step that introduces a 4,5 double bond in the sphingoid base, which constitutes the backbone of all sphingolipids. Ceramide can also be deacylated to sphingosine, which can be phosphorylated by sphingosine kinase to sphingosine-1-phosphate. Synthesis of sphingosine-1-phosphate constitutes the only exit-route from the sphingolipid pathway by the action of sphingosine-1-phosphate lyase, yielding ethanolamine phosphate and hexadecanal, which can be utilized for production of various other lipids. Ceramide is synthesized de novo from palmitate and serine, which through a series of reactions is converted to dihydrosphingosine, which again is acylated to yield dihydroceramide by the action of ceramide synthases. The complexity of sphingolipid metabolism and the biological functions it affects are vast. Each class of sphingolipid has been generally thought of as entities, being regulated and acting in the same way, however, by virtue of their structural diversities each individual molecular sphingolipid species may have distinct regulatory functions in specific cellular pathways. Mammals contain six ceramide synthases, CERS1-6 (formerly named Lass1-6), which are all differentially expressed and show substrate specificity towards subsets of fatty acyl-CoAs, characterized by chain length and degree of saturation and hydroxylation (reviewed in [4]). The fact that targeted knock-down of each CERS results in increased mRNA levels of non-targeted CERS [5], emphasizes the importance of these enzymes in cellular homeostasis.
Several model organisms have been utilized to address sphingolipid metabolism, each offering different advantages. In C. elegans the sphingolipids are of less structural complexity as the sphingoid long chain base exclusively constitutes a C17 iso- or anteiso-branched chain as illustrated in Figure S1. C. elegans comprises three ceramide synthases HYL-1, HYL-2, and LAGR-1, each containing a Lag1p motif required for ceramide synthase activity [6], [7]. It has been shown that radiation-induced apoptosis in the germ line is inhibited in hyl-1(ok976), lagr-1(gk327/gk331), and hyl-1(ok976);lagr-1(gk327) animals, which can be relieved by injection of C16 ceramide [8]. Moreover, mutants of sphingosine kinase-1 (sphk-1(ok1097)) have an elevated germ line death baseline compared to wild type worms and are hypersensitive to radiation-induced apoptosis, effects which are not observed in lagr-1(gk327);sphk-1(ok1097) animals [8]. These observations collectively suggest that specific sphingolipids originating from HYL-1 and/or LAGR-1 ceramide production are involved in germ line radiation-induced apoptosis. It has also been reported that worms lacking ceramide glucosyltransferases (CGTs) arrest at the first larval stage and that this arrest can be rescued by CGT expression in the most anterior and posterior intestinal cells, implying that lack of glycosphingolipids results in starvation-induced growth arrest by impaired feeding [9]. Menuz et al. have recently shown that hyl-2(gnv1/tm2031) animals are sensitive to anoxia while hyl-1(gk203/ok976) animals are more resistant [10]. They also showed that C20-22 ceramides and sphingomyelins are more abundant in hyl-1(ok976) animals while C24-26 ceramides are less abundant. In contrast, hyl-2(gnv1) animals contain less C20-22 ceramides and sphingomyelins but more with C24-26. Tedesco et al. [7] recently showed that RNAi targeted against the Lag1p motif in hyl-1 extended lifespan and reduced mRNA levels of both hyl-1 and hyl-2, while hyl-1 deletion mutants (hyl-1(ok976/gk203)) showed no alterations in lifespan and an increased level of hyl-2 mRNA.
In the present study we have further examined the role of ceramide synthases in longevity and find that simultaneous deletion of hyl-1 and lagr-1 results in a lifespan extension, which depends on autophagy and the transcriptions factors PHA-4, DAF-16, and SKN-1. We also find that hyl-1;lagr-1 animals have an altered sphingolipid profile and that the extended lifespan can be normalized by sphk-1 RNAi, indicating the composition and/or levels of certain sphingolipids are key effectors in the lifespan extension.
Results
The Autophagy-associated Gene ATG-12, and the Transcription Factors PHA-4, DAF-16, and SKN-1 are Required for hyl-1;lagr-1 Longevity
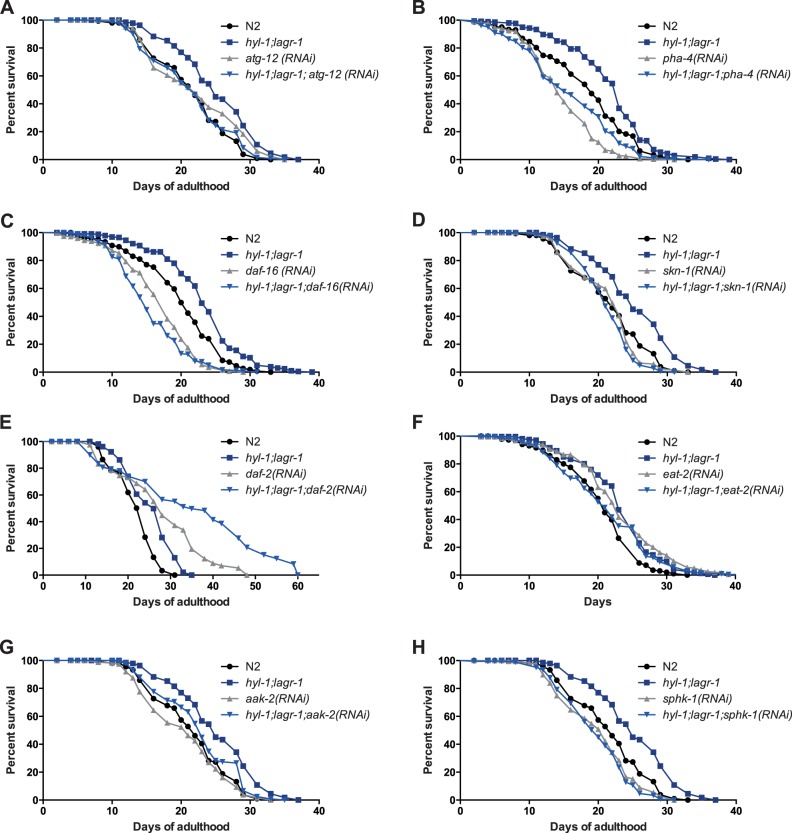
To address the role of ceramide synthases in C. elegans lifespan we obtained strains carrying a loss of function allele of each of the genes encoding a ceramide synthase. After backcrossing we made all possible combinations of ceramide synthase mutant strains, resulting in a collection of ceramide synthase mutants; hyl-1(ok976), hyl-2(ok1766), lagr-1(gk331), hyl-1;lagr-1(ok976;gk331), and hyl-2;lagr-1 (ok1766;gk331). In accordance with previous findings, we found that hyl-1;hyl-2 mutants are unviable and cannot be obtained by genetic crossing [10]. We first examined the lifespan of each of these strains under standard culture conditions. In contrast to previous observations [10], we found that hyl-2 and hyl-2;lagr-1 animals live significantly shorter than wild type animals (Figure S2), while hyl-1;lagr-1 animals live significantly longer compared to wild type animals (Figure 1 and Figure S2). In an effort to identify the mechanism(s) underlying the hyl-1;lagr-1 lifespan extension, we asked whether functional loss of HYL-1 and LAGR-1 affected the expression of key longevity genes including the insulin receptor daf-2, the transcription factors daf-16, pha-4, and skn-1, the autophagy genes atg-3, atg-12, atg-18, and lgg-1 as well as the catalytic subunit of the AMP-dependent kinase aak-2. Among the genes examined, we only found a small, yet significant increase in pha-4 expression, while atg-12 expression was significantly reduced in hyl-1;lagr-1 animals (Figure S3). Despite the small differences in their expression level, we hypothesized that these factors could be involved in the life span extension of hyl-1;lagr-1 animals, and thus knocked down these longevity genes by RNAi. Consistently, hyl-1;lagr-1 animals lived significantly longer than wild type animals when fed empty vector control bacteria (Figure 1, Table 1, P<0.0001). Autophagy is a recycling process, which is induced during stress conditions, and has been shown to be required for the lifespan extension induced by decreased insulin/insulin-like growth factor-1 signaling (IIS) [11] or dietary restriction (DR) [12], [13]. We found that the extended lifespan of hyl-1;lagr-1 depends on autophagy, as knock down of atg-12 normalized longevity (Figure 1A, P = 0.3053). Depending on how dietary restriction is induced [14], DR-induced longevity depends on the transcription factors PHA-4/FOXA [13], [15] as well as the transcription factor SKN-1 [16]. Knock down of pha-4 and skn-1 decreased the lifespan of hyl-1;lagr-1 animals to that of pha-4(RNAi) and skn-1(RNAi) N2 animals (Figure 1B, P = 0.2369 and Figure 1D, P = 0.5476), implying that hyl-1;lagr-1 animals are dietary restricted. Mutations in eat-2, encoding a subunit of the nicotinic acetylcholine receptor, results in impaired pharyngeal pumping and defecation. eat-2 mutants are therefore often used as a genetic surrogate to study dietary induced lifespan extension [17]. Since knock down of eat-2 only increases lifespan in wild type animals and not in hyl-1;lagr-1 animals (Figure 1F) it supports the notion that hyl-1;lagr-1 animals are dietary restricted. Compromised IIS increases longevity in a FOXO/DAF-16 dependent manner [18] and interestingly, we found that knock down of daf-16 decreases the lifespan of hyl-1;lagr-1 beyond that of daf-16(RNAi) N2 animals (Figure 1C, P = 0.0002). Despite the fact that the lifespan extension of daf-2 animals depends on DAF-16 function, the increased level of autophagy in daf-2 animals has been shown to be independent of both DAF-16 and PHA-4, implying that autophagy on its own is not sufficient to extend lifespan and led to the suggestion that the role of DAF-16 is to orchestrate proper utilization of components released from autophagic events [15]. These observations prompted us to address whether knockdown of daf-2 affected hyl-1;lagr-1 longevity, and found that the lifespan of hyl-1;lagr-1 animals is further extended by daf-2 RNAi compared to both hyl-1;lagr-1 fed control bacteria and daf-2(RNAi) N2 animals (Figure 1E, P = 0.0002 and P<0.0001, respectively). These findings indicate that hyl-1;lagr-1 longevity is elicited independently of IIS and that hyl-1;lagr-1 animals are more susceptibility to functional loss of DAF-16. We also found that the life span extension of hyl-1;lagr-1 animals is independent of the AMP-dependent kinase AMPK, as knock down of aak-2, the catalytic subunit of AMPK [19], did not abolish the longevity phenotype of the dobbelt mutant (Figure 1G). Interestingly, we found that knock down of sphk-1 decreases hyl-1;lagr-1 lifespan to the same level as sphk-1(RNAi) N2 animals (Figure 1H, P = 0.8002), indicating that the turn-over of sphingolipids or specific sphingosine-1-phosphate(s) regulate the longevity response.
Figure 1. Knock-down of PHA-4, DAF-16, SKN-1, ATG-12, or SPHK-1 affect the extended longevity of hyl-1;lagr-1.
Cumulative survival curves of N2 and hyl-1;lagr-1 worms grown at 20°C subjected to either empty vector control bacteria (L4440) or the indicated RNAi from the early adult stage. (A) When subjected to atg-12 RNAi, the extended lifespan of hyl-1;lagr-1 is normalized to the extent of atg-12(RNAi) control animals, P = 0.3053. (B) When subjected to pha-4 RNAi, the extended lifespan of hyl-1;lagr-1 is normalized to the extent of pha-4(RNAi) control animals, P = 0.2369. (C) When subjected to daf-16 RNAi, the extended lifespan of hyl-1;lagr-1 is decreased beyond the extent of daf-16(RNAi) control animals, P = 0.0002. (D) When subjected to skn-1 RNAi, the extended lifespan of hyl-1;lagr-1 is normalized to the extent of skn-1(RNAi) control animals, P = 0.5476. (E) When subjected to daf-2 RNAi, hyl-1;lagr-1 lifespan is further extended compared to both hyl-1;lagr-1 control animals, P<0.0001, and daf-2(RNAi) control animals, P<0.0001. (F) When subjected to eat-2 RNAi, hyl-1;lagr-1 lifespan is decreased compared to hyl-1;lagr-1 control animals, P = 0.0002, while the lifespan of eat-2(RNAi) animals is extended compared to wild-type control animals, P<0.0001. (G) When subjected to aak-2 RNAi, hyl-1;lagr-1 lifespan is decreased compared to hyl-1;lagr-1 animals, P = 0.0009, while no lifespan effect is seen when comparing aak-2(RNAi) animals to wild-type control animals, P = 0.0975. (H) When subjected to sphk-1 RNAi, the extended lifespan of hyl-1;lagr-1 is normalized to the extent of sphk-1(RNAi) control animals, P = 0.8002. For additional details about these experiments, see Table 1.
Table 1. Adult lifespan of hyl-1;lagr-1 and N2 control worms subjected to empty vector control or the indicated RNAi at 20°C.
| Strain | Adult-only RNAi treatment | RNAi lifespana(days) | Number of animalsb (trials) | Control lifespanc (days) | Number of control animalsb (trials) | Lifespan change (%) | p-value vs. controld |
| N2 | pha-4 | 14/14.4 | 291/360 (3) | 19/17.6 | 293/360 (3) | −26.3 | <0.0001 |
| daf-16 | 18/16.2 | 280/360 (3) | 20/19.5 | 279/360 (3) | −10.0 | <0.0001 | |
| skn-1 | 22/21.2 | 156/192 (2) | 22/21.0 | 112/192 (2) | 0 | 0.8563 | |
| atg-12 | 22/22.2 | 135/192 (2) | 22/21.0 | 112/192 (2) | 0 | 0.5426 | |
| sphk-1 | 21/19.6 | 157/192 (2) | 22/21.0 | 112/192 (2) | −4.5 | 0.014 | |
| aak-2 | 21/20.2 | 143/192 (2) | 22/21.0 | 112/192 (2) | −4.5 | 0.0975 | |
| daf-2 | 28/26.4 | 67/96 (1) | 24/21.6 | 63/96 (1) | 27.3 | <0.0001 | |
| eat-2 | 23/22.1 | 243/360 (2) | 21/20.2 | 200/240 (2) | 9.5 | <0.0001 | |
| hyl-1;lagr-1 | pha-4 | 15/14.8 | 277/348 (3) | 23/20.6 | 239/351 (3) | −34.8 | <0.0001 |
| daf-16 | 15/15.0 | 304/360 (3) | 23/22.4 | 226/360 (3) | −34.8 | <0.0001 | |
| skn-1 | 21/21.0 | 146/192 (2) | 25/24.8 | 122/192 (2) | −16.0 | <0.0001 | |
| atg-12 | 22/24.8 | 138/192 (2) | 25/24.8 | 122/192 (2) | −12.0 | <0.0001 | |
| sphk-1 | 20/21.1 | 152/192 (2) | 25/24.8 | 122/192 (2) | −20.0 | <0.0001 | |
| aak-2 | 23/21.7 | 128/192 (2) | 25/24.8 | 122/192 (2) | −8.0 | 0.0009 | |
| daf-2 | 35/34.9 | 74/96 (1) | 26/25.2 | 47/96 (1) | 34.6 | 0.0089 | |
| eat-2 | 21/20.1 | 257/358 (2) | 23/22.6 | 159/240 (2) | −8.7 | 0.0002 | |
| hyl-1:lagr-1 vs. N2 | control | 23/20.6 | 239/351 (3) | 19/17.6 | 293/360 (3) | 21.1 | <0.0001 |
| pha-4 | 15/14.8 | 277/348 (3) | 14/14.4 | 291/360 (3) | 7.1 | 0.2369 | |
| control | 23/22.4 | 226/360 (3) | 20/19.5 | 279/360 (3) | 15.0 | <0.0001 | |
| daf-16 | 15/15.0 | 304/360 (3) | 18/16.2 | 280/360 (3) | −16.7 | 0.0002 | |
| control | 25/24.8 | 112/192 (2) | 22/21.0 | 122/192 (2) | 13.6 | <0.0001 | |
| skn-1 | 21/21.0 | 146/192 (2) | 22/21.2 | 156/192 (2) | −4.5 | 0.5476 | |
| atg-12 | 22/24.8 | 138/192 (2) | 22/22.2 | 135/192 (2) | 0 | 0.3053 | |
| sphk-1 | 20/21.1 | 152/192 (2) | 21/19.6 | 157/192 (2) | −4.8 | 0.8002 | |
| aak-2 | 23/21.7 | 128/192 (2) | 21/20.2 | 143/192 (2) | 9.5 | 0.0018 | |
| control | 26/25.2 | 47/96(1) | 24/21.6 | 63/96(1) | 8.3 | 0.0032 | |
| daf-2 | 35/34.9 | 74/96 (1) | 28/26.4 | 67/96 (1) | 25.0 | <0.0001 | |
| control | 23/22.6 | 159/240 (2) | 21/20.2 | 200/240 (2) | 9.5 | <0.0001 | |
| eat-2 | 21/20.1 | 257/358 (2) | 23/22.1 | 243/360 (2) | −8.7 | <0.0001 |
Median/mean RNAi lifespan of N2 and hyl-1;lagr-1 fed the specified RNAi-bacteria.
Some animals were censored as they crawled of the plate, ruptured, or died as a “bag of worms”, however they are incorporated in the data set up until the day they were censored. The number of individual trials is in parentheses.
Median/mean control lifespan fed vector-only control bacteria.
P-values were determined using the Gehan-Breslow-Wilcoxon test using GraphPad Prism version 6.0 (GraphPad Software). The Bonferroni method was used to correct for multiple comparisons and P- values below 0.0125 are considered statistically significant equivalent to a significance level of 0.05 with four pair-wise comparisons. Cumulative statistics is shown in this table as experimental animals subjected to the same treatment behaved similarly between trials. Data shown in Figure 1.
Autophagy is Increased in hyl-1;lagr-1 in a DAF-16 and SKN-1 Dependent Manner
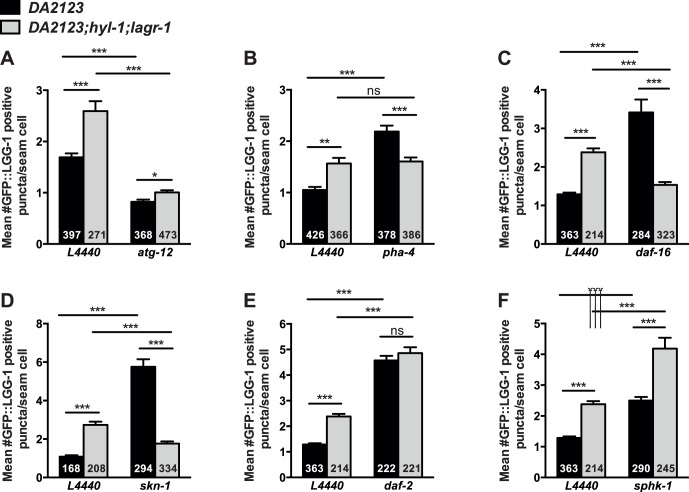
Since we identified ATG-12 function to be required for life span extension of hyl-1;lagr-1 animals we examined the level of autophagy in these animals. Autophagy is commonly addressed by examining the formation of preautophagosomal structures using transgenic strains expressing the C. elegans LC3-homolog LGG-1 tagged with GFP, which associates to the autophagosomal membrane [11]. In order to visualize the level of autophagy in hyl-1;lagr-1 animals, we introduced the hyl-1(ok976) and lagr-1(gk331) mutations into DA2123, a transgenic strain expressing GFP-tagged LGG-1. Under control conditions, hyl-1;lagr-1 animals have a consistently increased level of autophagy compared to wild type worms (Figure 2A and Figure S4). 3-Methyladenine inhibits class III phosphatidylinositol-3-kinases and is commonly used as a specific inhibitor of formation of autophagosomes [20]. We observed that 3-MA prevented the increase in the number of autophagosomes in hyl-1;lagr-1 animals (Figure S5). This and the observation that addition of concanamycin A (ConA), which blocks the lysosomal proton pump and thereby inhibits lysosome-dependent proteolysis [21], further increased the number of autophagosomes in both wild type and hyl-1;lagr-1 animals (Figure S5), suggest that autophagy is induced in response to functional loss of HYL-1 and LAGR-1. Consistently, knock down of atg-12 decreased the number of LGG-1::GFP positive puncta in both wild type and hyl-1;lagr-1 animals (Figure 2A). Knock down of pha-4 increased the number of LGG-1::GFP positive puncta in wild type worms, while hyl-1;lagr-1 animals were unresponsive to pha-4 knock down (Figure 2B), indicating the autophagy response in hyl-1;lagr-1 is independent of PHA-4. In contrast to this, we found that knock down of either daf-16 or skn-1 dramatically increased the level of autophagy in wild type, whereas daf-16 or skn-1 knock down in hyl-1;lagr-1 animals reduced the number of LGG-1::GFP positive puncta to the level found in wild type fed control RNAi (Figure 2C and D), indicating the autophagic response in hyl-1;lagr-1 depends on both DAF-16 and SKN-1. Despite that reduced IIS increases lifespan in both wild type and in hyl-1;lagr-1 animals (Figure 1E), knock down of daf-2 increased the level of autophagy to the same level in both wild type and in hyl-1;lagr-1 animals (Figure 2E). These observations and the fact that increased autophagy in daf-2 animals does not depend on DAF-16 [15] indicate that hyl-1;lagr-1 is somehow more susceptible to lack of DAF-16. Both ceramide and sphingosine-1-phosphate have been shown to be able to induce autophagy [22]. While the induction of autophagy by sphingosine-1-phosphate during starvation is associated with a moderate induction promoting cell survival, induction of autophagy by ceramide is associated with a more comprehensive autophagy response leading to cell death [23]. We found that knock down of sphk-1 increased the level of autophagy in both wild type and hyl-1;lagr-1 to similar extents (Figure 2F), implying that increased levels of ceramide or a subset of ceramide species or reduced sphingosine-1-phosphate levels can induce autophagy. Since sphk-1 knock down further increases the number of positive LGG-1::GFP puncta in hyl-1;lagr-1, induction of autophagy in hyl-1;lagr-1 animals does not depend on sphingosine-1-phosphate per se. It is therefore interesting that knock down of sphk-1 prevents the longevity extension of hyl-1;lagr-1 animals, indicating that altered ceramide or sphingosine-1-phosphate levels can have diverse regulatory properties.
Figure 2. Autophagy is increased in hyl-1;lagr-1 and the response mechanism differs from that of wild type.
LGG-1 is part of autophagosomal membranes and widely used as an indicator of autophagy in C. elegans. Bars represent the mean number of LGG-1::GFP-containing puncta per seem cell in non-starved wild type and hyl-1;lagr-1 worms grown at 20°C subjected to either empty vector control bacteria (L4440) or the indicated RNAi. The number in each bar indicates the total number of seam cells observed. (A) Knock-down of atg-12 lowers the level of autophagy in both wild type and hyl-1;lagr-1. (B) Knock-down of pha-4 does not change the increased level of autophagy in hyl-1;lagr-1 but increases autophagy in wild type. (C) Knock-down of daf-16 lowers the increased level of autophagy in hyl-1;lagr-1 but increases autophagy in wild type. (D) Knock-down of skn-1 lowers the increased level of autophagy in hyl-1;lagr-1 but increases autophagy in wild type. (E) Knock-down of daf-2 increases autophagy to the same extent in wild type and hyl-1;lagr-1. (F) Knock-down of sphk-1 increases the level of autophagy in hyl-1;lagr-1 beyond wild type level. Statistical analyses were performed by unpaired two-tailed t-test (with Welch’s correction if variances were significantly different) using GraphPad Prism version 6.0 (GraphPad Software). The Bonferroni method was used to correct for multiple comparisons and P values below 0.0125 were considered statistically significant equivalent to a significance level of 0.05. (*) P≤0.0125, (**) P≤0.001, and (***) P≤0.0001. N used for analysis is the total number of worms observed for each treatment (23–45 worms, two trials). Mean ± SEM is shown.
hyl-1;lagr-1 Animals Display Phenotypes Associated with Dietary-restricted Longevity
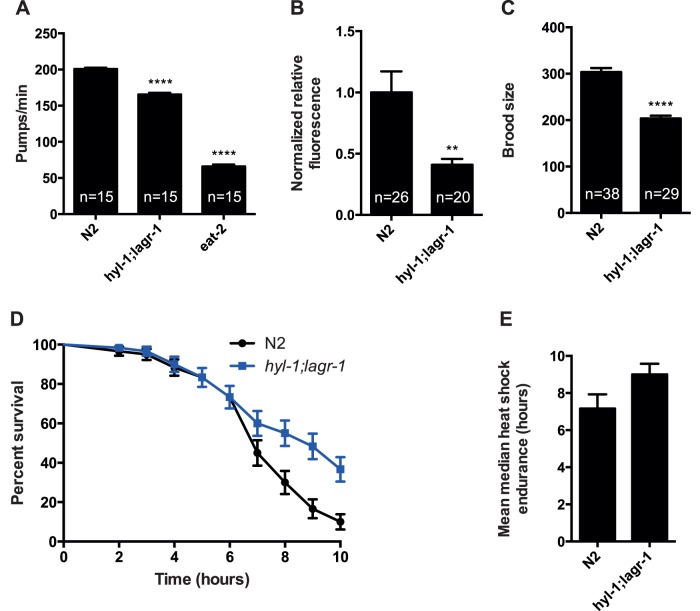
The above described observations imply that hyl-1;lagr-1 animals are dietary restricted. This prompted us to examine phenotypes, which are associated with dietary restriction. We found that hyl-1;lagr-1 animals had a decreased pumping rate of 17.4% compared to N2 (Figure 3A, P<0.0001), whereas eat-2 animals, which commonly are used as a genetic model for dietary restriction [17], displayed a 67.2% decrease in pumping rate compared to N2 (Figure 3A, P<0.0001). Consistently, we found that ingestion of bacteria mixed with fluorescent beads was reduced by 59% in hyl-1;lagr-1 animals compared to control (Figure 3B, P = 0.0026), but was unchanged in hyl-2 and in hyl-2;lagr-1 animals (Figure S6). Moreover, we found that the brood size of hyl-1;lagr-1 was 33% smaller than the brood size of N2 (Figure 3C, P<0.0001). Finally, increased thermotolerance has also been associated with dietary restriction [24] and consistently, we find that hyl-1;lagr-1 showed increased resistance to heat-shock at 37°C (Figure 3D, P = 0.0016 and Figure 3E,). Collectively, we interpret these observations as functional loss of HYL-1 and LAGR-1 renders C. elegans dietary restricted.
Figure 3. Phenotypes associated with lifespan extension and stress resistance are observed in hyl-1;lagr-1.
(A) Pumping rates of N2, hyl-1;lagr-1, and an eat-2 mutant. The latter displays a pronounced reduction in pumping rate and is commonly used as a genetic model for dietary restricted animals. Bars represent the mean number of pumps per minute. Compared to N2 displaying a mean pumping rate of 201±2 pumps/min, hyl-1;lagr-1 shows a 17.4% decrease with a mean pumping rate of 166±2, P<0.0001, while eat-2 shows a 67.2% decrease with a mean pumping rate of 66±3, P<0.0001. The data represents an mean ± SEM of 15 measurements in 5 worms of each genotype. (B) Quantification of fluorescent beads in the pharynx and the anterior part of the intestine following a feeding period of 30 minutes. Compared to N2, hyl-1;lagr-1 displays 59% less fluorescence, P = 0.0026. Mean ± SEM is shown, n indicates the number of worms. (C) Mean total brood size of N2 and hyl-1;lagr-1. Compared to N2 which displays a mean brood size of 304±9, hyl-1;lagr-1 shows a 33% decrease with a mean brood size of 204±6, P<0.0001. Mean ± SEM is shown, n = number of worms examined. (D) Survival curves of N2 and hyl-1;lagr-1 subjected to heat-shock at 37°C. Compared to N2, hyl-1;lagr-1 shows increased resistance, P = 0.0016. A total of 60 worms of each strain were assayed. Mean ± SD of 3 experiments is shown. N2-worms (6) and hyl-1;lagr-1 worms (22) were censored but are incorporated in the analysis until the time they were censored. (E) Bars represent mean median heat shock survival from the 3 experiments shown in D. Compared to N2 which has a median survival of 7 hours, hyl-1;lagr-1 displays a 29% increase in heat shock resistance with a median survival of 9 hours. Error bars represent ± SD.
Lipidomic Profiling Reveals Modified Sphingolipid Composition in hyl-1;lagr-1
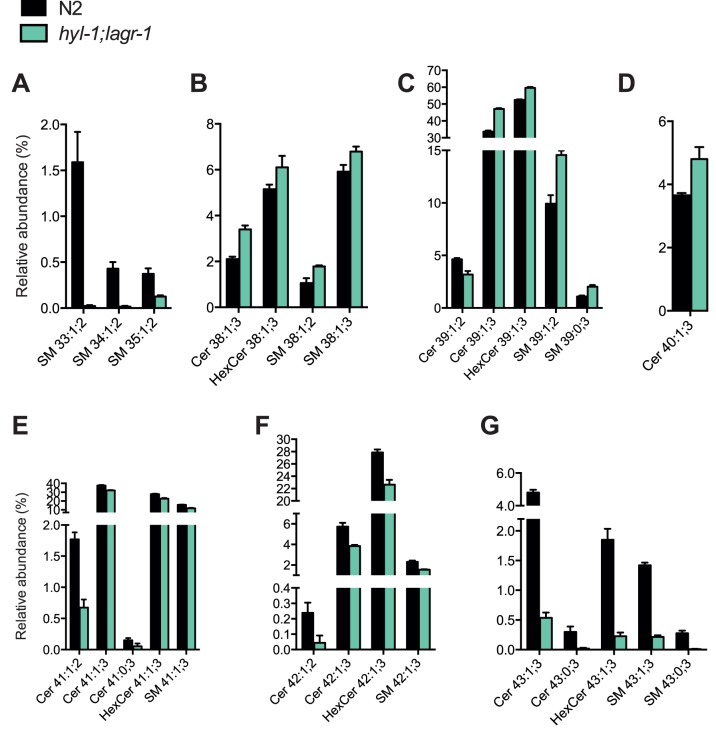
To address the underlying molecular mechanisms governing dietary restriction of hyl-1;lagr-1animals, we used LC/MS to examine the global changes in ceramides, glycosylceramides, and sphingomyelin in each of the ceramide synthase mutants (Figure 4, Figure S7, and Figure S8). Consistent with previous observations [10], we found that the presence of HYL-1 is required for the formation of sphingomyelin (SM) species containing C16-18 and C26 fatty acid residues (e.g. SM33:1;2, SM35:1;2 and 43:1;3) as these were drastically reduced in hyl-1 (Figure 4 and Figure S8C–D, J). We also found that HYL-2 has preference for incorporating C21-22 fatty acids (e.g. Cer/HexCer/SM 39:1;3) (Figure S8E–F) while HYL-1 appears to have a higher affinity towards incorporating C26 fatty acids primarily (e.g. Cer/HexCer/SM 43:1;3) (Figure S8J). The abundance of sphingolipids containing C22 fatty acids (e.g. Cer39:1;3 and HexCer39:1;3) was increased in hyl-1;lagr-1 animals (Figure 4C), while the levels of sphingolipids containing C24-26 fatty acids (Figure 4E–G) as well as sphingomyelin species containing C16-18 fatty acid moieties (SM 33–35:1;2) (Figure 4A) were much lower compared to wild type animals.
Figure 4. Lipidomic analysis reveals a modified sphingolipid composition in hyl-1;lagr-1.
Relative abundance of detected sphingolipid species showing significant changes in hyl-1;lagr-1. Sphingolipids containing C24-26 fatty acids and sphingomyelin species containing C16-18 fatty acids are lowered in hyl-1;lagr-1, while sphingolipids containing C21-22 fatty acids acids are more abundant. C. elegans sphingolipids predominately contain C17 long-chain bases, thus making the fatty acid chain length readily deducible. The number of carbon atoms indicated is without head groups. (A) Sphingomyelins containing C16-18 fatty acids are significantly reduced in hyl-1;lagr-1. (B) All significantly changed sphingolipid species containing C21 fatty acids are more abundant in hyl-1;lagr-1. (C) All significantly changed sphingolipid species containing C22 fatty acids are more abundant in hyl-1;lagr-1. (D) The only significantly changed sphingolipid species containing C23 fatty acids is more abundant in hyl-1;lagr-1. (E) All significantly changed sphingolipid species containing C24 fatty acids are less abundant in hyl-1;lagr-1. (F) All significantly changed sphingolipid species containing C25 fatty acids are less abundant in hyl-1;lagr-1. (G) All significantly changed sphingolipid species containing C26 fatty acids are less abundant in hyl-1;lagr-1. Statistical analyses were performed by one way analysis of variance followed by Dunnett’s multiple comparisons test using GraphPad Prism version 6.0 (GraphPad Software). Results from two biological experiments are shown for each strain. Mean ± SD is shown. All species shown are at least significant different at the P<0.05 level. The entirety of detected sphingolipid species can be seen in Figure S7 and Figure S8.
The fact that loss of function of the different ceramide synthases differentially affect lifespan of C. elegans, prompted us to identify unique changes in the sphingolipidomes of each of these strains, which correlated with the observed lifespans. We found that the total level of sphingomyelin species containing C21-22 fatty acids (e.g. SM38:1;3 and SM39:1;3) was lower in hyl-2 and hyl-2;lagr-1 animals while increased in hyl-1;lagr-1(Figure S7C, Figure S8E–F). However, two sphingomyelin species containing C24 and C25 fatty acids (SM 41/42:1;3) was increased in hyl-2 and hyl-2;lagr-1 and lowered in hyl-1;lagr-1 (Figure S7C, Figure S8H–I). Interestingly, deletion of lagr-1 only had subtle effects on the ceramide and sphingolipid composition and level (Figure S8D–J). However, loss of LAGR-1 function in hyl-2 animals prevented sphingomyelins containing C16-C18 fatty acid moieties (SM 33–35:1;2) to increase as they do in hyl-2 animals (Figure S8C–D), arguing that LAGR-1 primarily contributes to the synthesis of long-chain fatty acid containing sphingomyelins.
These observations indicate that the SM species increased in hyl-1;lagr-1 and lowered in hyl-2 and hyl-2;lagr-1 may have a pro-longevity function while the SM species with the opposite distribution have pro-aging effects. These species can be seen in Figure S8K. Thus, we performed Principal Component Analysis to further address this model. The score plot of the sphingolipidomics data clustering showed a clear distinction between the three groups; “Decreased lifespan (mutant)”, “Increased lifespan (mutant)”, and “No change (ctrl N2)” (Figure S9). The corresponding loading plot revealed that Cer 41:1;3, HexCer 39:1;3, HexCer 41:1;3, SM 38:1;2, SM 39:0;2, SM 39:1;2, SM 41:1;3, SM 41:1;3 contributed the most to the score plot separation (Figure S9A–B). Among these species only the abundance of SM 38:1;2 and partly SM 39:1;2 and SM 41:1;3 (Figure S9C–E) are oppositely regulated taking all three longevity phenotypes into account, suggesting that the levels of these sphingolipid species may play a role in regulating longevity of C. elegans. We detected additional sphingolipid species differing between hyl-1 animals and hyl-1;lagr-1 animals (Figure S9B, D–E, Table S1). These observations may not directly account for the extended longevity of hyl-1;lagr-1 animals, however, it may indicate that specific sphingolipid species are involved in the longevity response. It also supports the notion that specific species within sphingolipid subgroups may have distinct functions and that these subgroups of lipid species cannot necessarily be viewed as units having the same properties and acting in the same pathways.
Discussion
The important role of ceramide synthases in organismal aging was first recognized when it was found that loss of the gene encoding the ceramide synthase Lag1 in the yeast Saccharomyces cerevisiae extended chronological lifespan [25], which is consistent with the recent observations that both genetic- and chemical inhibition of sphingolipid synthesis extend chronological lifespan of yeast [26]. Moreover, it has been shown that homologues of LAG1 from C. elegans not only complement Lag1 function in S. cerevisiae [7], [27], [28] but also differentially are required for normal longevity in C. elegans [8], [10]. In the present study we have further examined the role of ceramide synthases in aging in C. elegans and show that deletion of hyl-1(ok976) or lagr-1(gk331) does not affect lifespan (Figure S2), while deletion of hyl-2(ok1766) significantly shortens lifespan of C. elegans. Interestingly, we consistently find that deletion of both lagr-1 and hyl-2 further reduces lifespan, while deletion of both lagr-1 and hyl-1 significantly extends longevity compared to wild type C. elegans (Figure 1 and Figure S2). We did not succeed in generating hyl-1;hyl-2 animals, which is consistent with the observation that knock down of hyl-1 in hyl-2(ok1766) animals results in L1 arrest (data not shown). In contrast to our observations, Menuz et al. [10] found that loss of HYL-1 function (ok976) extends lifespan significantly, while deletion of hyl-2(ok1766) did not affect lifespan under normal conditions, but was required for survival under anoxic conditions [10]. Thus, our finding suggests that HYL-2 is not only required for survival at low oxygen tension, but also required for normal lifespan. We find that the extended lifespan of hyl-1;lagr-1 animals is independent of insulin signaling (Figure 1), but depends on ATG-12, a component of the autophagic machinery [29], and the three central transcription factors DAF-16, SKN-1, and partly PHA-4 (Figure 1), which previously have been shown to modulate aging under dietary restriction [14], [30]. Thus, loss of HYL-1 and LAGR-1 function may therefore induce a dietary restriction-like phenotype, which ultimately leads to extension of longevity. This scenario is supported by our observation that knock down of eat-2 does not further extend lifespan of hyl-1;lagr-1 animals (Figure 1), and by data showing that hyl-1;lagr-1 animals have decreased pumping rate, ingestion, and brood size, and increased resistance towards heat stress (Figure 3), which are all hallmarks of dietary restriction [31]. Furthermore, one of the best evolutionarily conserved cellular responses to dietary restriction is the activation of autophagy, a lysosomal degradation pathway in which the cell self-digests its own components to provide nutrients to maintain crucial cellular functions during fasting. Accordingly, the number of autophagosomes is significantly increased in hyl-1;lagr-1 animals. Interestingly, we find that knock down of atg-12, skn-1, and daf-16 in hyl-1;lagr-1 animals diminishes the number of GFP::LGG-1 positive puncti to wild type levels, while knock down of pha-4 does not increase the number of autophagosomes further as we observe in control animals (Figure 2). We also find that knock down of daf-2 expression increases the number of LGG-1::GFP positive puncti in both control and in hyl-1;lagr-1 animals. Thus, our results indicate that both lifespan extension and the increased number of autophagosomes in hyl-1;lagr-1 animals depends on a functional autophagy machinery as well as the transcription factors DAF-16, SKN-1, and partly PHA-4, and is independent of the insulin signaling pathway (Figure 2).
Our findings suggest that the lifespan extension of hyl-1;lagr-1 animals is not only mediated by a few genes, but rather a complex network of gene functions. This is in agreement with previous findings showing that the lifespan extension of eat-2 mutants, which commonly is used as a genetic surrogate for dietary restriction, depends on PHA-4 function [13], [14], while SKN-1 and PHA-4 both are required for lifespan extension by dietary restriction in liquid media [13], [16]. In contrast to these observations, the lifespan extension induced by dietary restriction on solid media depends on AAK-2 and DAF-16 [32].
Consistent with previous findings our lipidomic profiling supports the notion that HYL-1 promotes synthesis of ceramides containing very-long acyl-chains, while HYL-2 confers synthesis of ceramide species with shorter fatty acid moieties (Figure S8) [10]. Notably, we find that deletion of lagr-1 has only subtle effects on the level and molecular composition of sphingolipids, independent of the presence of functional hyl-1 and hyl-2 (Figure S8A–K). Despite its minor contribution to the overall molecular sphingolipid species composition, LAGR-1 contributes to C. elegans longevity as hyl-1;lagr-1 animals live significantly longer while hyl-1 and lagr-1 animals do not (Figure S2). This and the fact that HYL-1, HYL-2, and LAGR-1 are expressed in different tissues (Figure S10), suggest that impaired synthesis of specific ceramide species in a limited number of cells can induce a DR-like response. Our observation that ingestion is severely reduced in hyl-1;lagr-1 animals (Figure 3) and the fact that impaired synthesis of glycosphingolipids by ceramide glucosyltransferases in a subset of cells in the digestive tract impairs feeding [9] lend credence to the notion that loss of specific ceramides and sphingolipids are required for specific cellular functions in a subset of tissues. Principal component analyses of the sphingolipidomic data (Figure S9) indicate that the abundance of specific sphingolipids like SM 38:1;2, 39:1;2, 39:1;3, 41:1;3, and HexCer 39:1;3 could contribute to the diminished lifespan of hyl-2 animals, while the level of SM 38:1;2 could contribute to the lifespan extension of hyl-1;lagr-1 animals.
Our results show that primarily HYL-1 and HYL-2 functions contribute to the overall abundance and molecular composition of ceramide and other sphingolipids in C. elegans. Despite the subtle contribution of LAGR-1 to the synthesis of these lipids, deletion of lagr-1 further impairs longevity of hyl-2 animals, while deletion of lagr-1 extends the lifespan of hyl-1 animals. The observation that knock down of sphk-1 expression decreases hyl-1;lagr-1 lifespan to wild type level but increases the number of autophagosomes in hyl-1;lagr-1 animals, suggests that the longevity response to sphk-1 knock down is induced by mechanisms, which do not involve autophagy. Interestingly, although our observations supports the notion that autophagy supports cell survival, recent studies have shown that autophagy can promote cell death under certain conditions (reviewed in [33]). Accordingly, it has recently been found that chemical inhibition of sphingosine kinase 2 induces the formation of autophagosomes while promoting nonapoptotic cell death in human kidney carcinoma cells [34].
While the present work shows that synthesis and turn-over of sphingolipids and sphingosine-1-phosphate(s) can regulate the longevity and autophagy, the genetic tractability of C. elegans and the mutant strains we have obtained so far provide an excellent framework to further delineate how specific ceramide and sphingolipid species regulate organismal longevity and autophagy.
Materials and Methods
Strains
Standard procedures were used for culturing [35]. Single mutants: RB1036: hyl-1(ok976) IV, RB1498: hyl-2(ok1766) X, VC765: lagr-1(gk331) I, DA1116: eat-2(ad1116) II. Double mutants: FE0007: hyl-1(ok976) IV;lagr-1(gk331) I, FE0008: hyl-2(ok1766) X; lagr-1 (gk331) I. Transgenic strains: DA2123: adIS2122[lgg-1::lgg-1-GFP; rol-6(su1006)] (kind gift from Malene Hansen, Sanford-Burnham Medical Research Institute, La Jolla, California), FE0015: hyl-1(ok976) IV;lagr-1(gk331) I; adIS2122[lgg-1::lgg-1-GFP; rol-6(su1006)], BC10421: dpy-5(e907) I; sEx10421[rCesC09G4.1::GFP+pCeh361], VB2360: svEX798-800 [hyl-2::GFP;rol-6 (su1006)], BC10482: dpy-5(e907) I; sEX10482[lagr-1::GFP]. All single mutant strains were obtained from the Caenorhabditis Genetics Centre (CGC) and unless otherwise noted generation of the remaining strains listed were conducted in our lab. Generating a hyl-1;hyl-2 strain was also attempted but worms homozygous for both mutations were not attained and we later confirmed by RNAi that worms lacking both HYL-1 and HYL-2 arrest development and die. The three single mutant strains were out-crossed seven times to N2 Bristol wild type worms.
Confocal Microscopy
Transgenic L4 worms expressing GFP fusions were mounted in M9 buffer containing Tetramisol (10 µM) (Sigma-Aldrich, St. Louis, Montana, USA) on a 2% agar pad on a microscope slide and covered with a coverslip and analyzed by confocal microscopy on an LSM 510 META microscope (Carl Zeiss MicroImaging Inc., Germany). Primary image analysis was performed using LSM Image Browser (Carl Zeiss MicroImaging Inc., Germany).
RNAi
All clones were from the Julie Ahringer RNAi library [36]. RNAi by feeding was performed according to Ahringer [37].
Lifespan Analysis
All lifespan analyses were conducted at 20°C on worms non-starved for at least two generations. For the RNAi lifespan assays, 3-day-old synchronized worms were transferred to gene-specific RNAi bacterial plates. The worms were counted and moved to new plates every second day during the reproductive period and afterwards only moved to new plates every forth day. Lifespan is defined as the time from day one of adulthood to the time when they were scored as dead (i.e. no longer responded to gentle prodding with a platinum wire). Worms that “exploded”, were bagged, or went missing were censored the day the event was observed. For the OP50 lifespan assays (Figure S2), synchronized L4 worms were transferred to plates containing 100 µM FUDR (5-fluoro-2′-deoxyuridine, 50503 Sigma) to prevent progeny from developing and counted every 1–2 days. Lifespan is defined as the time from when the worms were placed on FUDR plates to the time when they were scored as dead.
Autophagy Assay
The level of autophagy was investigated using an LGG-1::GFP translational reporter [11]. GFP positive puncta/seem cell were counted in L4 transgenic worms at 1000× magnification using a Leica DMI 6000 B microscope. Counting puncta is usually performed in L3 animals as autofluorescence increases with age and obscures LGG-1::GFP visualization. We investigated the level of autophagy in young L4 worms in order to age-match the assay with the feeding and pumping rate assays. All animals were kept at 20°C and assayed by the same experimenter. The number of autophagosomes per seem cell was averaged for each worm and this average was used for calculating the mean number of LGG-1::GFP containing puncta/seem cell.
Feeding Assay
NGM plates were seeded with a 500∶1(vol:vol) blend of OP50 bacteria and Fluoresbrites Multifluorescent microspheres, 0.2 µm (Polyscience, Inc.) 22 hours prior to the assay. L4 larvae of N2 and hyl-1;lagr-1 were allowed to feed for 30 min on the bead-plates after which they were transferred to eppendorf tubes with 1 ml of 10 mM levamisole (Sigma-Aldrich, St. Louis, Montana, USA) and centrifuged for 2 min at 2000 g, room temperature. The supernatant was removed and ∼30 µl of resuspended worms were transferred to a freshly made agarose pad (2% agarose in M9). The amounts of ingested beads were photographed at 1000× using a Leica DMI 6000 B microscope and quantified using NIH ImageJ freeware [38]. All images were acquired using identical settings and exposure time and the presented images, are representative of general observations.
Pumping Rate Assay
Five non-starved L4 worms of each strain were transferred to NGM plates seeded with OP50 and filmed three times 15 consecutive seconds using a Leica DMI 6000 B microscope with Infinity Capture Software. Pumping rates were counted by playing the recordings at one fourth of the normal speed and mean pumping rate calculated from the fifteen measurements of each genotype.
Brood Size Assay
Brood size assays were performed at 20°C by picking late L4s to fresh NGM plates and transferring them to new plates every 24 hours for 4–5 days. After this worms were left on the same plates for 3 days. Worms that “exploded”, were bagged, or went missing were omitted from the analysis. All progeny plates were incubated at 20°C for about two days before the number of developed progeny was scored.
Heat Shock Assay
Synchronized L4 worms grown at 20°C were picked to fresh plates and subjected to heat shock at 35°C for one hour after which the worms were counted. Worms were scored dead when they no longer responded to gentle prodding with a platinum wire. The worms were continuously subjected to heat stress one hour followed by scoring.
RNA Isolation, cDNA Synthesis, and Quantitative Real-time PCR
Total RNA was extracted from four independent synchronized worm populations of each strain as described (51). cDNA was synthesized from 500 ng total RNA as previously described (52). Quantitative real-time PCR (qRT-PCR) was performed on an ABI PRISM 7700 RealTime PCR-machine (Applied Biosystems, Carlsbad, California, USA) or Stratagene MXPro 3000 (Agilent Technologies, Santa Clara, California, USA) using 2× SYBR Green JumpStart™ Taq ReadyMix™ and Sigma Reference Dye (Sigma-Aldrich, St. Louis, Montana, USA) as described by the manufacturer. PCR reactions were performed in 25 µl reactions containing 1.5 µl diluted cDNA. Reactions were incubated at 95°C for 2 min followed by 40 cycles of 95°C for 15 s, 60°C for 45 s and 72°C for 45 s. All reactions were performed in duplicates and normalized to the level of the tbb-2 gene (encoding the C. elegans orthologue of the human β-tubulin). Primers for qRT-PCR were designed using Primer Express version 2.0 (Applied Biosystems, Carlsbad, California, USA). Primer sequences can be obtained upon request. Statistical analyses were performed by unpaired two-tailed t-test using GraphPad Prism version 6.0 (GraphPad Software).
Lipid Extraction
Synchronized L4 worms were washed twice with 10 mL 0.9% NaCl and incubated end-over-end for 15 min to ensure intestinal emptying. Worms were washed trice with 10 mL 150 mM ammonium acetate in H2O. All centrifugation steps were carried out at 20°C, 1000 rpm, 1 min. Liquid was aspirated to 600 µL and 550 µl were transferred to an eppendorf tube. Samples were flash frozen in liquid nitrogen and stored at −80°C. Lipid extraction was carried out at 4°C. Approximately 1000 worms were aliquotted (∼100 µl) mixed with 100 µl 150 mM ammonium acetate before adding 990 µl chloroform/methanol (2∶1). Samples were vortexed at 4°C for 45 min and centrifuged 2 min (2000 g, 4°C). The lower organic phase was transferred to a new tube and subjected to vacuum evaporation. Lipid extracts were dissolved in 100 µl chloroform/methanol (2∶1).
Lipid Analysis by Mass Spectrometry
Lipid extracts were analyzed by normal-phase liquid chromatography using a PVA-SIL column (YMC Europe GmbH) interfaced with a nanoflow ion source Triversa NanoMate (Advion Biosciences, Inc.) and a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Cer, HexCer and SM species were monitored in negative ion mode by recording FT MS analysis using a target mass resolution of 100,000. Lipid species were quantified by extracting their peak intensities and using ALEX software as previously described [39]. The relative abundance of lipid species was determined by normalizing the intensity of monitored sphingolipid species to the total intensity of all monitored lipid species. Sphingolipid species was annotated according to previous reports [39]–[41].
Statistical Analyses
All statistical analyses were performed using GraphPad Prism version 6.0 (GraphPad Software), except for the Principal Component Analysis (PCA), which was performed using Markerview software (AB SCIEX).
Lifespan analysis
Statistical analyses were performed by Gehan-Breslow-Wilcoxon tests. The Bonferroni method was used to correct for multiple comparisons and P-values equivalent to a significance level of 0.05 were considered statistically significant. Worms that “exploded”, were bagged, or went missing were censored the day the event was observed. Further statistical lifespan details can be found in Table 1 and Figure S2.
Autophagy assay
Statistical analysis was performed by unpaired two-tailed t-test (with Welch’s correction if variances were significantly different). The Bonferroni method was used to correct for multiple comparisons and P-values equivalent to a significance level of 0.05 were considered statistically significant. N used for analysis is the total number of worms observed for each treatment.
Feeding assay and quantitative real-time PCR
Statistical analyses were performed by unpaired two-tailed t-test (with Welch’s correction if variances were significantly different).
Pumping rate and brood size assays
Statistical analyses were performed by one way analysis of variance followed by Dunnett’s multiple comparisons test.
Heat shock assay
Statistical analysis was performed by Log-rank (Mantel-Cox) tests. Some animals were censored as they crawled of the plate or ruptured, however they are incorporated in the data set up until the time point they were censored.
Lipid analysis by mass spectrometry
Statistical analyses were performed by one way analysis of variance followed by Dunnett’s multiple comparisons test and P-values below 0.05 were considered statistically significant. Results are from two biological experiments.
Principal component analysis
Supervised principal component analysis was performed with the following preprocessing parameters; Weighting = none, Scaling = Pareto. We excluded species which were not significantly altered between at least two of the three lifespan categories: “No change”, “Increased lifespan”, and “Decreased Lifespan”. P-values from the t-test of hyl-1 and hyl-1;lagr-1 (Table S1) have been corrected for multiple comparisons and accordingly, P-values below 0.05 are considered statistically significant.
Supporting Information
Structure examples of selected sphingosine-, ceramide-, sphingomyelin- and glucosyl ceramide species from C. elegans .
(PDF)
Lifespan analyses of all ceramide synthase mutants. Cumulative lifespan analyses of N2, hyl-1, hyl-2, lagr-1, hyl-1;lagr-1, and hyl-2;lagr-1 grown on OP50 at 20°C. (A) hyl-1 displays no change in lifespan compared to N2 (P = 0.3592). (B) hyl-2 displays a shortened lifespan compared to N2 (P<0.0001). (C) lagr-1 displays no change in lifespan compared to N2 (P = 0.1651). (D) hyl-1;lagr-1 displays an extended lifespan compared to N2 (P<0.0001). (E) hyl-2;lagr-1 displays shortened lifespan compared to N2 (P<0.0001). (F) Table summarizing the depicted lifespan analyses. aSome animals were censored as they crawled of the plate, ruptured, or died as a “bag of worms”, however they are incorporated in the data set up until the day they were censored. The number of individual trials is in parentheses. Statistical analyses were performed by Gehan-Breslow-Wilcoxon tests using GraphPad Prism version 6.0 (GraphPad Software). bThe Bonferroni method was used to correct for multiple comparisons and P-values below 0.01 are considered statistically significant equivalent to a significance level of 0.05. Cumulative statistics is shown in this table as experimental animals subjected to the same treatment behaved similarly between trials.
(PDF)
Expression levels of longevity and autophagy genes in wild type N2 and in hyl-1;lagr-1 animals. Total RNA was harvested from N2 worms at the L4 stage. The expression level of the indicated genes was quantified using qRT-PCR, and normalized to tbb-2 mRNA and shown in arbitrary units (a. u.). Mean ± SEM is shown, number of independent experiments ranged from 3 to 9. (*) P≤0.05. Only the expression of pha-4 and atg-12 in hyl-1;lagr-1 animals is significantly changed relatively to wild type animals.
(PDF)
LGG-1::GFP-positive puncta in hypodermal seam cells of wild type and hyl-1;lagr-1 animals. Representative micrographs of young L4 N2 (DA2123) or hyl-1;lagr-1 larvae expressing GFP-tagged LGG-1 in hypodermal seam cells. A yellow arrow head indicates a hypodermal seam cell, while blue arrow heads indicate LGG-1::GFP positive puncti. Using fluorescence microscopy, LGG-1::GFP positive puncti were counted in 3–10 seam cells were counted in each of 23–45 animals and averaged. Scale bar: 20 µm.
(PDF)
Effect of 3-methyladenine and Concanamycin A on autophagy. Transgenic animals expressing LGG-1::GFP were treated with 3-MA (1 mM) or Concanamycin A (50 nM) for 24 hours after they reaching the young L4 larval stage. Bars represent mean number of LGG-1::GFP-containing puncta per seem cell in non-starved wild type and hyl-1;lagr-1 worms grown at 20°C. The number in each bar indicates the total number of seam cells observed. N used for analysis is the total number of worms observed for each treatment (the number of worms examined ranged from 12 to 21). Mean ± SEM is shown. (**) P≤0.01 and (****) P≤0.0001.
(PDF)
Ingestion of fluorescent beads in hyl-2 and in hyl-2;lagr-1 animals. Quantification of fluorescent beads in the pharynx and the anterior part of the intestine following a feeding period of 30 minutes. Fluorescence intensities were normalized to the level in wild type animals. Mean ± SEM is shown, n = number of worms analyzed.
(PDF)
Overview of all Cer, HexCer, and SM species detected in hyl-1;lagr-1 . (A) Top: Relative abundance of all Cer species detected in hyl-1;lagr-1. Bottom: Zoom of low abundance Cer species detected in hyl-1;lagr-1. (B) Relative levels of all HexCer species detected in hyl-1;lagr-1. (C) Top: Relative abundance of all SM species detected in hyl-1;lagr-1. Bottom: Zoom of low abundance SM species detected in hyl-1;lagr-1.
(PDF)
Overview of all Cer, HexCer, and SM species detected in the five different ceramide synthase mutants. (A) Top: abundance levels of all Cer species detected in ceramide synthase mutants. Bottom: Zoom of low abundance Cer species detected in ceramide synthase mutants. (B) Top: Relative levels of all HexCer species detected in ceramide synthase mutants. Bottom: Zoom of low abundance HexCer species detected in ceramide synthase mutants. (C) Top: Relative levels of all SM species detected in ceramide synthase mutants. Bottom: Zoom of low abundance SM species detected in ceramide synthase mutants. (D) Relative levels of all C33, C34, and C35 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (E) Relative levels of all C38 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (F) Relative levels of all C39 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (G) Relative abundance of all C40 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (H) Relative levels of all C41 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (I) Relative levels of all C42 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (J) Relative levels of all C43 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (K) Relative levels of all species significantly altered according to lifespan changes (e.g. Oppositely regulated in long-lived and short-lived strains). Statistical analyses were performed by one way analysis of variance followed by Dunnett’s multiple comparisons test using GraphPad Prism version 6.0 (GraphPad Software). Two technical replicates of two biological replicates were analysed for each strain. (*) P≤0.05, (**) P≤0.001, and (***) P≤0.0001. Mean ± SD is shown.
(PDF)
Multivariate analysis by principal component analysis of sphingolipidomic data segregated according to lifespan changes. (A) Score plot projecting the first (D1) and second (D2) principal components show a clear separation of the strains when grouped according to the following lifespan changes: “Decreased lifespan ”, “Increased lifespan ”, and “No change ”. D1 and D2 account for 100% of the total sample variance. (B) Loading plot depicting the sphingolipid species contributing the most to the total sample variance. Red stars denotes the species significantly altered when comparing hyl-1 (no lifespan change) to hyl-1;lagr-1(increased lifespan). (C) Relative abundance of the four species contributing the most to the score plot separation. (D and E) The 5 species which contribute most to the separation and are significantly altered in hyl-1;lagr-1 compared to hyl-1 (denoted by red stars in B).
(PDF)
Expression patterns of the ceramide synthase genes at the L4 stage. (A) HYL-1 shows expression in the body wall muscles, the pharyngeal muscles PM3 and PM5, and unidentified cells in the pharynx. (B) HYL-2 shows expression in the body wall muscles and the nervous system. (C) LAGR-1 shows expression in the pharyngeal muscles PM3-5 and in pharyngeal nerves. Scale bar: 80 µm.
(PDF)
P-values from the t-test of hyl-1 and hyl-1;lagr-1 of the Principal Component Analysis.
(XLSX)
Acknowledgments
We gratefully appreciate Connie Gram for technical assistance and Elena Sokol for assisting the Principal Component Analysis.
Funding Statement
The present work was supported by funds from The Danish Research Council for Independent Research (grant numbers 23363 (NJF), 09-072484 (CSE)), The Novo-Nordisk Foundation (grant number 13514 (NJF)), and Lundbeckfonden (95-310-13591 (CSE)). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bartke N, Hannun YA (2009) Bioactive sphingolipids: metabolism and function. J Lipid Res 50 Suppl: S91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pitson SM (2011) Regulation of sphingosine kinase and sphingolipid signaling. Trends Biochem Sci 36: 97–107. [DOI] [PubMed] [Google Scholar]
- 3. Hannun YA, Obeid LM (2011) Many ceramides. The Journal of biological chemistry 286: 27855–27862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stiban J, Tidhar R, Futerman AH (2010) Ceramide synthases: roles in cell physiology and signaling. Adv Exp Med Biol 688: 60–71. [DOI] [PubMed] [Google Scholar]
- 5. Mullen TD, Spassieva S, Jenkins RW, Kitatani K, Bielawski J, et al. (2011) Selective knockdown of ceramide synthases reveals complex interregulation of sphingolipid metabolism. J Lipid Res 52: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spassieva S, Seo JG, Jiang JC, Bielawski J, Alvarez-Vasquez F, et al. (2006) Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. The Journal of biological chemistry 281: 33931–33938. [DOI] [PubMed] [Google Scholar]
- 7. Tedesco P, Jiang J, Wang J, Jazwinski SM, Johnson TE (2008) Genetic analysis of hyl-1, the C. elegans homolog of LAG1/LASS1. Age (Dordr) 30: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deng X, Yin X, Allan R, Lu DD, Maurer CW, et al. (2008) Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans . Science 322: 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marza E, Simonsen KT, Faergeman NJ, Lesa GM (2009) Expression of ceramide glucosyltransferases, which are essential for glycosphingolipid synthesis, is only required in a small subset of C. elegans cells. J Cell Sci 122: 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Menuz V, Howell KS, Gentina S, Epstein S, Riezman I, et al. (2009) Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science 324: 381–384. [DOI] [PubMed] [Google Scholar]
- 11. Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, et al. (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans . Science 301: 1387–1391. [DOI] [PubMed] [Google Scholar]
- 12. Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, et al. (2008) A role for autophagy in the extension of lifespan by dietary restriction in C. elegans . PLoS Genet 4: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A (2007) PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans . Nature 447: 550–555. [DOI] [PubMed] [Google Scholar]
- 14. Greer EL, Brunet A (2009) Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans . Aging Cell 8: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, et al. (2008) A role for autophagy in the extension of lifespan by dietary restriction in C. elegans . PLoS genetics 4: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bishop NA, Guarente L (2007) Two neurons mediate diet-restriction-induced longevity in C. elegans . Nature 447: 545–549. [DOI] [PubMed] [Google Scholar]
- 17. Lakowski B, Hekimi S (1998) The genetics of caloric restriction in Caenorhabditis elegans . Proc Natl Acad Sci U S A 95: 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- 19. Curtis R, O'Connor G, DiStefano PS (2006) Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell 5: 119–126. [DOI] [PubMed] [Google Scholar]
- 20. Seglen PO, Gordon PB (1982) 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A 79: 1889–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mordier S, Deval C, Bechet D, Tassa A, Ferrara M (2000) Leucine limitation induces autophagy and activation of lysosome-dependent proteolysis in C2C12 myotubes through a mammalian target of rapamycin-independent signaling pathway. J Biol Chem 275: 29900–29906. [DOI] [PubMed] [Google Scholar]
- 22. Lavieu G, Scarlatti F, Sala G, Levade T, Ghidoni R, et al. (2007) Is autophagy the key mechanism by which the sphingolipid rheostat controls the cell fate decision? Autophagy 3: 45–47. [DOI] [PubMed] [Google Scholar]
- 23. Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, et al. (2006) Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem 281: 8518–8527. [DOI] [PubMed] [Google Scholar]
- 24. Munoz MJ (2003) Longevity and heat stress regulation in Caenorhabditis elegans . Mech Ageing Dev 124: 43–48. [DOI] [PubMed] [Google Scholar]
- 25. D'Mello N P, Childress AM, Franklin DS, Kale SP, Pinswasdi C, et al. (1994) Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J Biol Chem 269: 15451–15459. [PubMed] [Google Scholar]
- 26. Huang X, Liu J, Dickson RC (2012) Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet 8: e1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang JC, Kirchman PA, Zagulski M, Hunt J, Jazwinski SM (1998) Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res 8: 1259–1272. [DOI] [PubMed] [Google Scholar]
- 28. Guillas I, Jiang JC, Vionnet C, Roubaty C, Uldry D, et al. (2003) Human homologues of LAG1 reconstitute Acyl-CoA-dependent ceramide synthesis in yeast. J Biol Chem 278: 37083–37091. [DOI] [PubMed] [Google Scholar]
- 29. Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, et al. (2007) Autophagy regulates ageing in C. elegans . Autophagy 3: 93–95. [DOI] [PubMed] [Google Scholar]
- 30. Lapierre LR, Hansen M (2012) Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol Metab 23: 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mair W, Dillin A (2008) Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem 77: 727–754. [DOI] [PubMed] [Google Scholar]
- 32. Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, et al. (2007) An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans . Curr Biol 17: 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ponnusamy S, Meyers-Needham M, Senkal CE, Saddoughi SA, Sentelle D, et al. (2010) Sphingolipids and cancer: ceramide and sphingosine-1-phosphate in the regulation of cell death and drug resistance. Future Oncol 6: 1603–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beljanski V, Knaak C, Smith CD (2010) A novel sphingosine kinase inhibitor induces autophagy in tumor cells. J Pharmacol Exp Ther 333: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brenner S (1974) The genetics of Caenorhabditis elegans . Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- 37. Kamath RS, Ahringer J (2003) Genome-wide RNAi screening in Caenorhabditis elegans . Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- 38. Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, et al. (2009) Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A 106: 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, et al. (2011) Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci U S A 108: 1903–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, et al. (2012) Flexibility of a eukaryotic lipidome–insights from yeast lipidomics. PLoS One 7: e35063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure examples of selected sphingosine-, ceramide-, sphingomyelin- and glucosyl ceramide species from C. elegans .
(PDF)
Lifespan analyses of all ceramide synthase mutants. Cumulative lifespan analyses of N2, hyl-1, hyl-2, lagr-1, hyl-1;lagr-1, and hyl-2;lagr-1 grown on OP50 at 20°C. (A) hyl-1 displays no change in lifespan compared to N2 (P = 0.3592). (B) hyl-2 displays a shortened lifespan compared to N2 (P<0.0001). (C) lagr-1 displays no change in lifespan compared to N2 (P = 0.1651). (D) hyl-1;lagr-1 displays an extended lifespan compared to N2 (P<0.0001). (E) hyl-2;lagr-1 displays shortened lifespan compared to N2 (P<0.0001). (F) Table summarizing the depicted lifespan analyses. aSome animals were censored as they crawled of the plate, ruptured, or died as a “bag of worms”, however they are incorporated in the data set up until the day they were censored. The number of individual trials is in parentheses. Statistical analyses were performed by Gehan-Breslow-Wilcoxon tests using GraphPad Prism version 6.0 (GraphPad Software). bThe Bonferroni method was used to correct for multiple comparisons and P-values below 0.01 are considered statistically significant equivalent to a significance level of 0.05. Cumulative statistics is shown in this table as experimental animals subjected to the same treatment behaved similarly between trials.
(PDF)
Expression levels of longevity and autophagy genes in wild type N2 and in hyl-1;lagr-1 animals. Total RNA was harvested from N2 worms at the L4 stage. The expression level of the indicated genes was quantified using qRT-PCR, and normalized to tbb-2 mRNA and shown in arbitrary units (a. u.). Mean ± SEM is shown, number of independent experiments ranged from 3 to 9. (*) P≤0.05. Only the expression of pha-4 and atg-12 in hyl-1;lagr-1 animals is significantly changed relatively to wild type animals.
(PDF)
LGG-1::GFP-positive puncta in hypodermal seam cells of wild type and hyl-1;lagr-1 animals. Representative micrographs of young L4 N2 (DA2123) or hyl-1;lagr-1 larvae expressing GFP-tagged LGG-1 in hypodermal seam cells. A yellow arrow head indicates a hypodermal seam cell, while blue arrow heads indicate LGG-1::GFP positive puncti. Using fluorescence microscopy, LGG-1::GFP positive puncti were counted in 3–10 seam cells were counted in each of 23–45 animals and averaged. Scale bar: 20 µm.
(PDF)
Effect of 3-methyladenine and Concanamycin A on autophagy. Transgenic animals expressing LGG-1::GFP were treated with 3-MA (1 mM) or Concanamycin A (50 nM) for 24 hours after they reaching the young L4 larval stage. Bars represent mean number of LGG-1::GFP-containing puncta per seem cell in non-starved wild type and hyl-1;lagr-1 worms grown at 20°C. The number in each bar indicates the total number of seam cells observed. N used for analysis is the total number of worms observed for each treatment (the number of worms examined ranged from 12 to 21). Mean ± SEM is shown. (**) P≤0.01 and (****) P≤0.0001.
(PDF)
Ingestion of fluorescent beads in hyl-2 and in hyl-2;lagr-1 animals. Quantification of fluorescent beads in the pharynx and the anterior part of the intestine following a feeding period of 30 minutes. Fluorescence intensities were normalized to the level in wild type animals. Mean ± SEM is shown, n = number of worms analyzed.
(PDF)
Overview of all Cer, HexCer, and SM species detected in hyl-1;lagr-1 . (A) Top: Relative abundance of all Cer species detected in hyl-1;lagr-1. Bottom: Zoom of low abundance Cer species detected in hyl-1;lagr-1. (B) Relative levels of all HexCer species detected in hyl-1;lagr-1. (C) Top: Relative abundance of all SM species detected in hyl-1;lagr-1. Bottom: Zoom of low abundance SM species detected in hyl-1;lagr-1.
(PDF)
Overview of all Cer, HexCer, and SM species detected in the five different ceramide synthase mutants. (A) Top: abundance levels of all Cer species detected in ceramide synthase mutants. Bottom: Zoom of low abundance Cer species detected in ceramide synthase mutants. (B) Top: Relative levels of all HexCer species detected in ceramide synthase mutants. Bottom: Zoom of low abundance HexCer species detected in ceramide synthase mutants. (C) Top: Relative levels of all SM species detected in ceramide synthase mutants. Bottom: Zoom of low abundance SM species detected in ceramide synthase mutants. (D) Relative levels of all C33, C34, and C35 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (E) Relative levels of all C38 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (F) Relative levels of all C39 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (G) Relative abundance of all C40 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (H) Relative levels of all C41 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (I) Relative levels of all C42 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (J) Relative levels of all C43 sphingolipid species detected to have significantly different levels in one or more ceramide synthase mutant compared to N2. (K) Relative levels of all species significantly altered according to lifespan changes (e.g. Oppositely regulated in long-lived and short-lived strains). Statistical analyses were performed by one way analysis of variance followed by Dunnett’s multiple comparisons test using GraphPad Prism version 6.0 (GraphPad Software). Two technical replicates of two biological replicates were analysed for each strain. (*) P≤0.05, (**) P≤0.001, and (***) P≤0.0001. Mean ± SD is shown.
(PDF)
Multivariate analysis by principal component analysis of sphingolipidomic data segregated according to lifespan changes. (A) Score plot projecting the first (D1) and second (D2) principal components show a clear separation of the strains when grouped according to the following lifespan changes: “Decreased lifespan ”, “Increased lifespan ”, and “No change ”. D1 and D2 account for 100% of the total sample variance. (B) Loading plot depicting the sphingolipid species contributing the most to the total sample variance. Red stars denotes the species significantly altered when comparing hyl-1 (no lifespan change) to hyl-1;lagr-1(increased lifespan). (C) Relative abundance of the four species contributing the most to the score plot separation. (D and E) The 5 species which contribute most to the separation and are significantly altered in hyl-1;lagr-1 compared to hyl-1 (denoted by red stars in B).
(PDF)
Expression patterns of the ceramide synthase genes at the L4 stage. (A) HYL-1 shows expression in the body wall muscles, the pharyngeal muscles PM3 and PM5, and unidentified cells in the pharynx. (B) HYL-2 shows expression in the body wall muscles and the nervous system. (C) LAGR-1 shows expression in the pharyngeal muscles PM3-5 and in pharyngeal nerves. Scale bar: 80 µm.
(PDF)
P-values from the t-test of hyl-1 and hyl-1;lagr-1 of the Principal Component Analysis.
(XLSX)