Abstract
Background
Cystathionine β-lyase performs an essential role in the transsulfuration pathway by its primary reaction of forming homocysteine from cystathionine. Understanding how the Neurospora crassa met-2+ gene, which encodes cystathionine β-lyase, is regulated is important in determining the basis of the cellular control of transsulfuration. The aim of this study was to determine the nature of a potential regulatory connection of met-2+ to the Neurospora sulfur regulatory network.
Findings
The cystathionine β-lyase (met-2+) gene was cloned by the identification of a cosmid genomic clone capable of transforming a met-2 mutant to methionine prototrophy and subsequently characterized. The gene contains a single intron and encodes a protein of 457 amino acids with conserved residues predicted to be important for catalysis and pyridoxal-5′-phosphate co-factor binding. The expression of met-2+ in wild-type N. crassa increased 3.1-fold under sulfur-limiting growth conditions as compared to the transcript levels seen under high sulfur growth conditions (i.e., repressing conditions). In a Δcys-3 strain, met-2+ transcript levels were substantially reduced under either low- or high-sulfur growth conditions. In addition, the presence of CYS3 activator binding sites on the met-2+ promoter was demonstrated by gel mobility shift assays.
Conclusions
In this report, we demonstrate the sulfur-regulated expression of the met-2+ gene and confirm its connection to the N. crassa sulfur regulatory circuit by the reduced expression observed in a Δcys-3 mutant and the in vitro detection of CYS3 binding sites in the met-2+ promoter. The data further adds to our understanding of the regulatory dynamics of transsulfuration.
Keywords: Cystathionine β-lyase, met-2+, Transsulfuration, Sulfur gene regulation, CYS3 regulator, Neurospora crassa
Findings
Background
Cystathionine β-lyase (E.C. 4.4.1.8; also known as β-cystathionase) catalyzes the conversion of cystathionine to homocysteine, ammonia and pyruvate. Cystathionine β-lyase plays an important role in transsulfuration in that it allows for the utilization of the intracellular pool of cystathionine for the synthesis of homocysteine which serves as the immediate precursor to methionine. In combination with the action of the other transsulfuration and reverse transsulfuration reactions catalyzed by cystathionine γ-lyase, cystathionine γ-synthase, and cystathionine β-synthase (shown in Figure 1), this allows for either cysteine or methionine (via homocysteine) production from the intermediate cystathionine [1]. In yeast, secondary reactions of cystathionine β-lyase appear to involve aromatic thiol precursors (e.g., releasing 3-mecaptohexan-1-ol) [2]. Cystathionine β-lyase has been the subject of structural active-site characterization studies [3] and, due to its presence in bacteria and absence in humans, is a target enzyme for the development of novel antimicrobial agents [4]. Cystathionine β-lyase has also been overexpressed in higher plants in an attempt to enhance methionine biosynthesis [5].
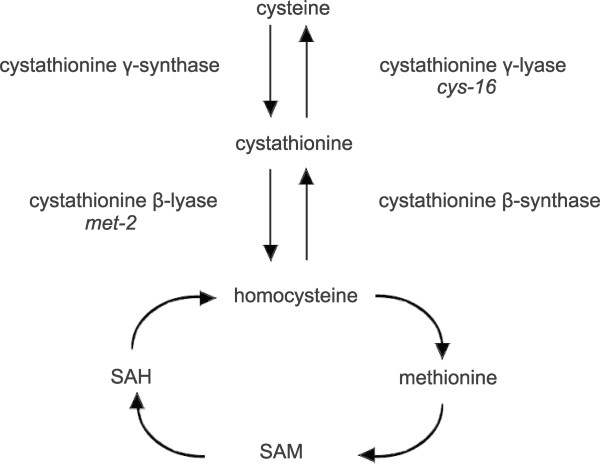
Figure 1.
Pathway of transsulfuration and methionine synthesis in Neurospora crassa. Cystathionine β-lyase, encoded by the met-2+ gene, is shown in its role in the conversion of cystathionine to homocysteine. Cystathionine γ-lyase (encoded by cys-16+), which converts cystathionine to cysteine, has been characterized previously [19]. Abbreviations: SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine.
Neurospora crassa provides a useful model system to dissect the regulation and dynamics of the transsulfuration pathway [6-8]. Early work with met-2 mutants of N. crassa confirmed that they lacked cystathionine β-lyase activity [9]. In N. crassa, cystathionine β-lyase has been purified and enzymatic activity assayed under several growth conditions [9,10]. The most significant difference reported [10] was that the level of cystathionine β-lyase activity was reduced in wild-type by one-fourth over the reported baseline when grown with methionine supplemented at the highest level tested (i.e., 5 μmoles/ml) with little change noted at lower levels of supplementation. N. crassa microarray expression data collected at different stages of development demonstrate only modest changes in met-2+ expression during, for example, conidial germination or colony development (e.g., 1.4× higher early in germination versus 15 hr of growth) [11,12]. Limited data is available regarding the regulation of cystathionine β-lyase in other fungi. In Aspergillus nidulans, the cystathionine β-lyase gene is encoded by the metG locus and the gene has been cloned and characterized [13]. Analysis of the transcription of metG demonstrated constitutive expression with no apparent regulation by sulfur source [13]. Cystathionine β-lyase activity in A. nidulans was found to be repressed by methionine [14], but constitutively present in another study [15]. For Saccharomyes cerevisiae, comparable expression data for growth under different levels of sulfur supplementation is not available for cystathionine β-lyase, which is encoded by the STR3 gene [16]. Based on microarray expression data, STR3 expression does increase by 12-fold under fermentation stress response conditions [17]. In addition, the suggestion has been made that there is a role for Gto1 (omega-class glutathione transferase) in the redox regulation of S. cerevisiae cystathionine β-lyase [18].
An important question is whether there is a regulatory connection of the transsulfuration-related genes, including cystathionine β-lyase, to the N. crassa sulfur regulatory system which is made up of a set of trans-acting regulatory genes and a set of genes encoding a variety of sulfur-related enzymes and transporters. When N. crassa is cultured under sulfur-limiting conditions (i.e., derepressing conditions) then the coordinate expression of this set of sulfur-related genes occurs. The regulated genes include arylsulfatase, choline sulfatase, sulfate permease I and II, among others [6,8]. Recently, cystathionine γ-lyase has been added to the list of genes confirmed to be under control of this key regulatory circuit [19]. CYS3, a bZIP transcriptional activator, is the key regulator and necessary for regulation expression of these sulfur-related genes [6]. Based on binding-site studies, a consensus sequence for CYS3 binding has been determined [20].
We present here the cloning and regulatory analysis of the N. crassa met-2+ cystathionine β- lyase gene. This report demonstrates a new connection of cystathionine β-lyase to the CYS3-controlled regulatory network and provides us with important data for the continued dissection of the regulatory dynamics of transsulfuration.
Results and discussion
Sequence and characterization of the cystathionine β-lyase gene
The nucleotide sequence of the met-2+ gene along with flanking 5′ and 3′ regions is shown in Figure 2. The met-2+ gene, which has been mapped to chromosome IVR [21], was cloned by transforming the methionine-requiring met-2 mutant (FGSC # 4061) to prototrophy using chromosome IV-specific genomic clones from the Orbach/Sachs N. crassa library constructed in pMOCosX [22]. The cosmid vector pMOCosX carries a hygromycin resistance marker for selection [23] which allowed for less background growth following transformation during the screening process. A single clone from the library, designated G8F01 was found to transform met-2 to prototrophy. A subcloned 4.1 kb fragment capable of transforming the met-2 mutant was then sequenced for further study. The met-2+ gene was cloned and the sequence submitted as GenBank AF401237 prior to the completion of the N. crassa genomic sequence. Subsequently, the locus has been designated NCU07987 in the Broad Institute database [24]. The GenBank AF401237 sequence features as reported here are identical to those reported in NCU07987. The met-2+ gene has a single intron that is 63 nucleotides in length near the 3′ end of the gene as shown by cDNA and genomic sequence data. A putative transcriptional start, with the sequence TCATCACA, is located at −149 upstream of the initiator ATG. Putative binding sites for the CYS3 regulator, identified in experiments as described below, are included in Figure 2. The met-2+ gene encodes a polypeptide of 457 amino acids with important functional residues conserved as compared to other members within this group of pyridoxal-5′-phosphate dependent lyases [3,25,26]. In particular, conserved key residues include Tyr83, Arg87, Gly115, and Thr237 involved in cofactor binding; and Tyr139, Glu182, Asp213, Asn214, Lys238 (the residue forming the Schiff base linkage) and Arg399 involved in catalysis (Figure 2). The N. crassa cystathionine β-lyase shows substantial similarity to fungal (e.g., 69% identity to A. nidulans AABB03241 and 50% identity to S. cerevisiae NP_011331) and to plant (43% identity to Arabidopsis thaliana NP_850712) cystathionine β-lyases.
Figure 2.
Nucleotide sequence of the met-2+gene. The sequence is shown from 1066 nucleotides upstream of the translation start codon to 187 nucleotides downstream of the stop codon (indicated by an asterisk). The nucleotides are numbered relative to the initiator ATG codon. The predicted polypeptide sequence is given below the nucleotide sequence in single-letter code. The 5′ splice site, the 3′ splice site, and the internal lariat sequence within the intron are underlined. Putative CYS3-binding sites within the met-2+ promoter are bracketed and numbered 1 through 4. Conserved functional residues are highlighted within the predicted polypeptide sequence of 457 amino acids. Residues involved in cofactor binding are boxed; while residues involved in catalysis (including Lys238 involved in the Schiff base linkage) are circled.
Analysis of met-2+ gene expression
met-2+ transcript levels were initially assayed in wild-type N. crassa cultured on high and low levels of sulfur (i.e., repressing and derepressing conditions, respectively). Northern blots were prepared from isolated poly (A)+ mRNA and probed with the cloned met-2+ gene. Figure 3 shows a 1.7-kb message hybridizing to the met-2+ probe that was detectable under both sulfur repressing and derepressing conditions. The steady-state level of met-2+ transcript did, however, show a substantially higher level under sulfur derepressing conditions (3.1-fold more compared to the sulfur repressing condition based on phosphorimager quantitation of the data). All of the Northern blot experiments conducted used the constitutively expressed am+ gene, which encodes glutamate dehydrogenase, as a control probe for the blots to ensure that the bulk mRNA levels were comparable between samples. Given that in wild-type N. crassa the steady-state level of met-2+ transcript was substantially higher under sulfur derepressing conditions, an expression pattern similar to what has been observed for other components of the N. crassa sulfur regulatory circuit [6], the results suggest a possible role for CYS3 in the transcriptional control of met-2+. In particular, in relation to transsulfuration, met-2+ expression appears regulated in a similar fashion as with cys-16+ (which encodes cystathionine γ-lyase) [19]. The met-2+ levels observed in Figure 3 are directly comparable to those in our prior study with the cys-16+ locus [19]. The expression data suggests, at least in N. crassa, that the available pool of cystathionine is accessed by derepressing both cystathionine β-lyase (leading to homocysteine and subsequently methionine) and by cystathionine γ-lyase (leading to cysteine) when the organism is stressed by low sulfur availability (pathway shown in Figure 1). This is in marked contrast to the other similarly characterized fungal example, A. nidulans, where constitutive expression of cystathionine β-lyase has been shown [13].
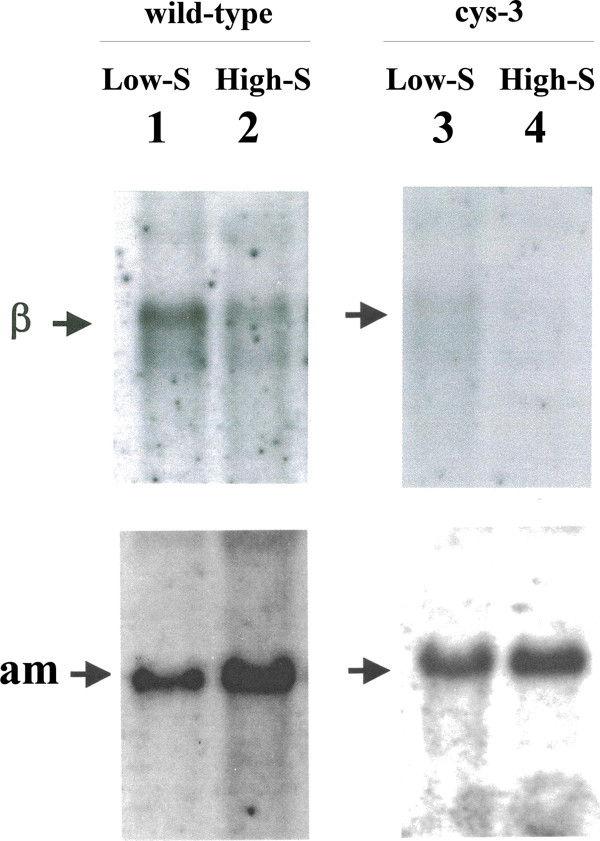
Figure 3.
Analysis of met-2+expression. (Left panel) Northern hybridization analysis of poly (A)+ mRNA from wild-type N. crassa grown under low sulfur (or derepressing) (lane 1) and high sulfur (or repressing conditions) (lane 2). Northern blots were probed with 32P-labeled met-2+ DNA (designated β within figure) and am+ DNA (which served as a constitutively expressed control to allow for sample comparisons). These results are directly comparable to the prior cys-16+ study [19]. (Right panel) Northern analysis of poly (A)+ mRNA isolated from the ∆cys-3 (18–4) strain of N. crassa grown under low sulfur (or derepressing) (lane 3) or high sulfur (or repressing) (lane 4) conditions. Blots were probed as above.
In order to test whether met-2+ is subject to regulation by CYS3, Northern blot analysis was carried out using poly(A)+ mRNA isolated from a cys-3 mutant deletion mutant (Δcys-3) and probed with the met-2+ gene. For this case, the met-2+ transcript was detectable only at a very low level under either low- or high-sulfur culture conditions. In fact, the levels observed in Δcys-3 were slightly lower than in wild-type under repressing conditions (based on phosphorimager quantification). The reduction in met-2+ transcript level in the Δcys-3 background is typical of genes that have been demonstrated to be part of the N. crassa sulfur regulatory system in previous studies [6,8].
Gel mobility shift analysis of the met-2+ promoter
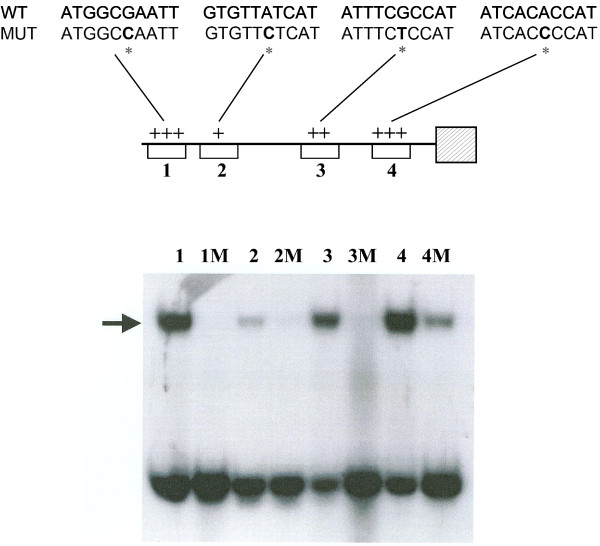
Based on in vitro binding and other studies the consensus binding site for CYS3 has been determined to be 5′ ATG GCGC CAT 3′ [20] . In order to analyze the met-2+ gene for the presence of CYS3 binding sites, the promoter region was subdivided into overlapping 100- to 200-bp segments by restriction endonuclease digestion and examined for gel mobility shifts with CYS3 extract (data not shown). The fragments demonstrating mobility shifts were progressively narrowed down to 24 bp segments that were still able to bind CYS3. Figure 4 shows that among the group of defined segments, CYS3 binding was strongest to site 4 (at −138 to −147) which had a sequence of 5′ ATC ACAC CAT 3′ which corresponded to the closest match to the CYS3 consensus binding site found in the met-2+ promoter. Previous studies have shown that the core of the consensus binding site commonly contains RYRY variations as in 5′ ATG RYRY CAT 3′) [8,20]. The weakest binding was observed at site 2 (−916 to −925) which had the greatest divergence from the consensus binding site (i.e., 5′ GTG TTAT CAT 3′). To confirm the specificity of CYS3 binding, the sixth base position within the 10 bp core sequence was mutated (from purine to pyrimidine), and the effect on gel mobility shifts determined. Figure 4 shows that the mutated segments showed no or reduced binding of CYS3and, therefore, demonstrate the specificity of the binding site assay.
Figure 4.
Gel mobility-shift analysis of CYS3 binding sites within the met-2 + promoter. Lanes: 1, CYS3-binding site 1 (nt −1017 to −1026); 1 M, mutated CYS3-binding site 1; 2, CYS3-binding site 2 (nt −916 to −925); 2 M, mutated CYS3-binding site 2; 3, CYS3-binding site 3 (nt −413 to −422); 3 M, mutated CYS3-binding site 3; 4, CYS3-binding site 4 (nt −138 to −147); 4 M, mutated CYS3-binding site 4. Each CYS3 binding site was mutated with a single point mutation to determine the specificity of CYS binding.. The arrow indicates DNA fragments exhibiting reduced electrophoretic mobility due to protein-DNA interactions. The schematic of the met-2+ promoter (top) depicts the affinity of CYS3 for each binding site (+, weakest affinity; +++ strongest affinity) and includes the corresponding binding site nucleotide sequences for both wild type (WT) and mutated (MUT).
Conclusions
The expression of the N. crassa met-2+ gene, which encodes cystathionine β-lyase, was found to increase in level under culture conditions of sulfur limitation. Further, the reduced in vivo expression of met-2+ seen in a Δcys-3 background along with the in vitro detection of CYS3 binding sites within the met-2+ promoter supports a role for CYS3-mediated regulation of this gene. The data add to the continuing analysis of the regulatory dynamics of transsulfuration in this model system.
Methods
Strains, plasmids and culture conditions
The met-2 mutant strain FGSC#4061 and Orbach/Sachs N. crassa genomic library [22] that was constructed with the cosmid pMOCosX [23] were obtained from the Fungal Genetics Stock Center (University of Missouri-Kansas City). The ∆cys-3 (18–4) deletion strain was constructed as described previously [27]. The strain 74OR23-1a was used as wild-type for these experiments. Vogel minimal medium [28], with supplements as required, was used. Sulfur repression and derepression experiments were carried out as described previously by culturing mycelia at 25°C on Vogel-minus-sulfur medium plus high sulfur (5.0 mM methionine) and low-sulfur medium (0.25 mM methionine) medium, respectively [29].
Gene cloning and sequencing
The N. crassa cosmid library was subdivided into chromosome IV specific clones which were then used in individual transformations of met-2 with selection for methionine prototrophy and hygromycin resistance. A single clone designated G8F01 was found that was capable of transforming met-2. Subsequent met-2 transformation tests identified a 4.1 kb fragment which contained the met-2+ gene. Following subcloning into pSPORT, the segment was subjected to automated sequencing (Cleveland Genomics; Cleveland, OH) by primer walking. The genomic sequence was submitted to GenBank as AF401237. The met-2+ gene has been assigned the locus designation NCU07987 in the Broad Institute Neurospora crassa database [24].
Northern analysis
Total RNA was isolated by the phenol extraction procedure described previously [29]. Following sodium acetate washes, poly(A)+ mRNA was isolated by oligo(dT) cellulose chromatography. Oligolabeling of DNA fragments was used to prepare 32P-labeled probes and used to probe Northern blots prepared as outlined elsewhere [29]. Quantitation of Northern blots was accomplished by using a Molecular Dynamics Phosphorimager.
Gel mobility shifts
Initially, the met-2+ promoter was subjected to restriction endonuclease digestion to generate a series of overlapping fragments tested in gel mobility shift assays. Subsequent identification of fragments that bound CYS3 allowed for the synthesis of short segments (24 bp) that retained binding activity. The 24 bp oligonucleotides representing the binding sites to be further tested were synthesized with an Applied Biosystems 391EP synthesizer, labeled using [ϒ-32P]ATP, annealed and gel purified as described previously [20,30]. Four oligonucleotides (and complementary strands) representing putative CYS3 binding sites on the met-2+ promoter were analyzed: Site 1 [ 5′ GAAAAGGATGGCGAATTTTAGTGA 3′], Site 2 [ 5′ GGTCTAGGTGTTATCATCTGGTGG 3′], Site 3 [5′GGCCCTGATTTCGCCATTTTCTTT 3′], and Site 4 [5′ TTGACTCATCACACCATCGGCCTC 3′]. Mutated CYS3 binding sites had a purine to pyrimidine substitution at the sixth position of the 10 bp core of the CYS3 consensus binding site: Site 1 M [ 5′ GAAAAGGATGGCCAATTTTAGTGA 3′], Site 2 M [5′ GGTCTAGGTGTTCTCATCTGGTGG 3′], Site 3 M [5′ GGCCCTGATTTCTCCATTTTCTTT 3′], and Site 4 M [5′ TTGACTCATCACCCCATCGGCCTC 3′]. E. coli produced CYS3 protein [20] was used for gel shift assays as we have described [20,30]. A Molecular Dynamics Phosphorimager was used to quantify the gel shift binding assays.
Availability of supporting data
The sequence data supporting the results of this article is available in the GenBank respository [AF401237, http://www.ncbi.nlm.nih.gov/nuccore/AF401237].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BR carried out the majority of experiments. JP and BR designed the experiments and prepared the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Brad S Reveal, Email: brad.reveal@nih.gov.
John V Paietta, Email: john.paietta@wright.edu.
Acknowledgements
The work was supported by a Medical Innovations Grant from the Wright State University Boonshoft School of Medicine.
References
- Flavin M. Transsulfuration reactions: an introduction. Methods Enzymol. 1971;17B:416–417. [Google Scholar]
- Holt S, Corente AG, Williams SJ, Capone DL, Jitjaroen W, Menz IR, Curtin C, Anderson PA. Engineering Saccharomyces cerevisiae to release 3-mercaptohexan-1-ol during fermentation through overexpression of an S. cerevisiae gene, STR3, for improvement of wine aroma. Appl Environ Microbiol. 2011;77:3626. doi: 10.1128/AEM.03009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken SM, Lodha PH, Morneau DJK. The enzymes of the transsulfuration pathways: active-site characterizations. Biochim Biophys Acta. 1814;2011:1511–1517. doi: 10.1016/j.bbapap.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Ejim LJ, Blanchard JE, Koteva KP, Sumerfield R, Elowe NH, Chechetto JD, Brown D, Junop MS, Wright GD. Inhibitors of bacterial cystathionine beta-lyase: leads for new antimicrobial agents and probes of enzyme structure and function. J Med Chem. 2007;22:755–764. doi: 10.1021/jm061132r. [DOI] [PubMed] [Google Scholar]
- Maimann S, Hoefgen R, Hesse H. Enhanced cystathionine beta-lyase activity in transgenic potato plants does not force metabolite flow towards methionine. Planta. 2001;214:163–170. doi: 10.1007/s004250100651. [DOI] [PubMed] [Google Scholar]
- Paietta JV. In: Cellular and molecular biology of filamentous fungi. Borkovich KA, Ebbole DJ, editor. Washington, DC: American Society for Microbiology; 2010. Sulfur, phosphorus and iron metabolism; pp. 359–375. [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pederson D, Paietta JV, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff C, Galagan JE, Dunlap JC, Loros J, Catheside D, Inoue H, Aramayo R, Polymenis M, Selker EU, Sachs MS, Marzluf GA, Paulsen I, Davis R, Ebbole DJ. et al. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta JV. In: The mycota. III. Biochemistry and molecular biology. 2. Brambl R, Marzluf GA, editor. Berlin: Sprinter-Verlag; 2004. Regulation of sulfur metabolism in mycelial fungi; pp. 369–383. [Google Scholar]
- Flavin M, Slaughter C. Cystathionine cleavage enzymes of Neurospora. J Biol Chem. 1964;239:2212–2219. [PubMed] [Google Scholar]
- Flavin M, Slaughter C. The derepression and function of enzymes of reverse transsulfuration in Neurospora. Biochim Biophys Acta. 1967;132:406–411. doi: 10.1016/0005-2744(67)90159-3. [DOI] [PubMed] [Google Scholar]
- Kasuga T, Townsend JP, Tian C, Gilbert LB, Mannhaupt G, Taylor JW, Glass NL. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 2005;33:6469–6485. doi: 10.1093/nar/gki953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Li J, Glass NL. Exploring the bZIP transcription factor regulatory network in Neurospora crassa. Microbiology. 2011;157:747–759. doi: 10.1099/mic.0.045468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienko M, Paszewski A. The metG gene of Aspergillus nidulans encoding cystathionine β-lyase: cloning and analysis. Curr Genet. 1999;35:638–646. doi: 10.1007/s002940050463. [DOI] [PubMed] [Google Scholar]
- Paszewski A, Grabski J. Studies on β-cysthionase and O-acetylhomoserine sulfhydrylase as the enzymes of alternative methionine biosynthetic pathways in Aspergillus nidulans. Acta Biochim Pol. 1973;20:159–168. [PubMed] [Google Scholar]
- Natorff R, Balinska M, Paszewski A. At least four regulatory genes control sulphur metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1993;238:185–192. doi: 10.1007/BF00279546. [DOI] [PubMed] [Google Scholar]
- Hansen J, Johannesen F. Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:535–542. doi: 10.1007/s004380051199. [DOI] [PubMed] [Google Scholar]
- Marks VD, Ho Sui SJ, Erasmus D, van der Merwe GK, Brumm J, Wasserman WW, Bryan J, van Vuuren HJJ. Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res. 2008;8:35–52. doi: 10.1111/j.1567-1364.2007.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto L, Garcera A, Jansson K, Sunnerhagen P, Herrero E. A peroxisomal glutathione transferase of Saccharomyces cerevisiae is functionally related to sulfur amino acid metabolism. Eukaryot Cell. 2006;5:1748–1759. doi: 10.1128/EC.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveal BS, Paietta JV. Analysis of the sulfur-regulated control of the cystathionine γ-lyase gene of Neurospora crassa. BMC Res Notes. 2012;5:339. doi: 10.1186/1756-0500-5-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta JV. DNA-binding specificity of the CYS3 transcription factor of Neurospora crassa defined by binding-site selection. Fungal Genet Biol. 2008;45:1166–1171. doi: 10.1016/j.fgb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Perkins DD, Radford A, Newmeyer D, Mjorkman M. Chromosomal loci of Neurospora crassa. Microbiol Rev. 1982;46:426–570. doi: 10.1128/mr.46.4.426-570.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fungal Genetics Stock C. The Orbach/Sachs cosmid library of N. crassa DNA sequences (pMOCosX) Fungal Genetics Newsl. 1991;38:97. [Google Scholar]
- Orbach MJ. A cosmid with a HygR marker for fungal library construction and screening. Gene. 1994;150:159–162. doi: 10.1016/0378-1119(94)90877-X. [DOI] [PubMed] [Google Scholar]
- Broad Institute Neurospora crassa database. http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html.
- Messerschmidt A, Pohlenz H, Laber B, Huber R, Clausen T. Crystal structure of the pyridoxal-5′-phosphate dependent cystathionine β-lyase from Eschericia coli. J Mol boil. 1996;262:202–224. doi: 10.1006/jmbi.1996.0508. [DOI] [PubMed] [Google Scholar]
- Alexander FW, Sandmeier PK, Christen P. Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes. Eur J Biochem. 1994;219:953–960. doi: 10.1111/j.1432-1033.1994.tb18577.x. [DOI] [PubMed] [Google Scholar]
- Paietta JV. Production of the CYS3 regulator, a bZIP DNA-binding protein, is sufficient to induce sulfur gene expression in Neurospora crassa. Mol Cell Biol. 1992;12:1568–1577. doi: 10.1128/mcb.12.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH, DeSerres EF. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17A:79–143. [Google Scholar]
- Paietta JV. Molecular cloning and regulatory analysis of the arylsulfatase structural gene of Neurospora crassa. Mol Cell Biol. 1989;9:3630–3637. doi: 10.1128/mcb.9.9.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Paietta JV. An additional role for the F-box motif: gene regulation within the Neurospora crassa gene control network. Proc Natl Acad Sci USA. 1998;95:2417–2422. doi: 10.1073/pnas.95.5.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]