Abstract
Although population differences in gene expression have been established, the impact on differential gene expression studies in large populations is not well understood. We describe the effect of self-reported race on a gene expression study of lung function in asthma. We generated gene expression profiles for 254 young adults (205 non-Hispanic whites and 49 African Americans) with asthma on whom concurrent total RNA derived from peripheral blood CD4+ lymphocytes and lung function measurements were obtained. We identified four principal components that explained 62% of the variance in gene expression. The dominant principal component, which explained 29% of the total variance in gene expression, was strongly associated with self-identified race (P<10−16). The impact of these racial differences was observed when we performed differential gene expression analysis of lung function. Using multivariate linear models, we tested whether gene expression was associated with a quantitative measure of lung function: pre-bronchodilator forced expiratory volume in one second (FEV1). Though unadjusted linear models of FEV1 identified several genes strongly correlated with lung function, these correlations were due to racial differences in the distribution of both FEV1 and gene expression, and were no longer statistically significant following adjustment for self-identified race. These results suggest that self-identified race is a critical confounding covariate in epidemiologic studies of gene expression and that, similar to genetic studies, careful consideration of self-identified race in gene expression profiling studies is needed to avoid spurious association.
Keywords: ancestry, gene expression, population stratification, self-identified race
INTRODUCTION
High-throughput gene expression profiling provides an opportunity to investigate the molecular basis of disease susceptibility, and the feasibility of such studies in large clinical populations has been greatly enhanced by the improved affordability of expression microarrays. Though many early technical barriers to such studies, including issues regarding large sample processing and normalization, have been essentially resolved, it remains unclear to what extent population-specific differences in gene expression can interfere in the interpretation of differential expression studies. Several modestly powered studies have demonstrated population-specific differences in the expression of hundreds of genes, [Spielman et al., 2007; Storey et al., 2007; Zhang et al., 2008] with ongoing speculation regarding the relative contribution of genetic vs. environmental factors to these differences. In many instances, differential gene transcript abundance represents an intermediate stage between non-coding DNA sequence variation and susceptibility to complex diseases [Cheung et al., 2003], and suggests that between-population differences in gene expression may, in part, contribute to the observed ethnic differences across a spectrum of phenotypes, including susceptibility to infectious disease and other complex traits such as pharmacogenetic responses [Kirkpatrick and Dransfield, 2009; Ormerod et al., 2008; Van Dyke et al., 2009].
Common non-coding genetic variants have been mapped for a substantial proportion of human transcripts, with evidence of between-population allelic heterogeneity observed at many of these loci [Cheung and Spielman, 2009; Stranger et al., 2007]. Approximately 10–15% of total human DNA sequence variation can be attributed to between-population differences in ancestry that arose as a result of successive rounds of migration, genetic drift, and differential selective pressures [Barbujani et al., 1997; Romualdi et al., 2002; Rosenberg et al., 2002]. In genetic association studies, the presence of unrecognized subgroups of genetically distinct ancestry, so-called population substructure or population stratification, can result in spurious genotype-phenotype association in situations where the distributions of both the tested alleles and clinical phenotypes differ across subgroups. The potential negative impact of population substructure on genetic association studies has been well documented, motivating a variety of new statistical methodologies and study design strategies to address this problem, including careful matching of cases and controls according to genetic ancestry, measurement of and adjustment for population substructure [Patterson et al., 2006; Price et al., 2006], and family-based association testing methods [Lange et al., 2004].
In contrast, the impact of ancestry on interpretation of differential gene expression studies has been largely ignored. Previous studies of population differences in gene expression have been limited by relatively modest sample size, and have been performed exclusively in subsets of the Coriell HapMap lymphoblastoid cell lines. It has been suggested that some of the observed race-based differences in gene expression may have resulted from non-genetic technical confounders, thereby overestimating the impact of ancestry on gene expression [Akey et al., 2007]. We set out to explore these issues in a larger cohort of self-identified non-Hispanic white and African American subjects for whom gene expression profiles were generated from primary peripheral blood CD4+ lymphocytes. Herein, we confirm substantial differences in gene expression patterns between self-identified racial groups that cannot be explained by technical differences. Furthermore, we demonstrate that failure to account for these differences can introduce spurious evidence of differential gene expression with clinical outcomes, suggesting that self-reported racial differences in gene expression are a critical confounder of epidemiologic studies of gene expression.
METHODS
STUDY POPULATION AND SAMPLE COLLECTION
Study subjects were young adults ages 16–23 with mild-to-moderate persistent asthma who had previously participated in the Childhood Asthma Management Program (CAMP), a 4.5 year multicenter clinical trial established to investigate the long-term effects of inhaled anti-inflammatory medications for the treatment of asthma [1999]. Subjects were of self-reported non-Hispanic white or African American ancestry. Complete trial design, methodology, and the primary outcomes analysis of the CAMP study have been previously published [1999 [2000]. The clinical trial was followed by two consecutive 4-year observational studies: CAMP Continuation Studies 1 and 2 (CAMPCS/1 and CAMPCS/2) [Strunk et al., 2009]. The analysis presented here was limited to phenotype data, including a pre-bronchodilator spirometric measurement of lung function, as well as a concurrent blood draw for gene expression profiling studies, collected during the Year 3 or Year 4 CAMPCS/2 follow-up visit at four of the eight CAMP study centers (Baltimore, Boston, Denver, and St. Louis). Spirometry performance was required to meet American Thoracic Society criteria for acceptability and reproducibility [1995]. Approval was obtained from the Institutional Review Boards of Brigham and Women’s Hospital (Boston, MA) and each of the CAMP participating institutions. Informed consent was obtained from study participants if they were over the age of 18 years old. Otherwise, an informed consent was obtained from parents of participating children, and the child’s assent was obtained prior to study enrollment.
PERIPHERAL BLOOD CD4+ LYMPHOCYTE EXPRESSION PROFILING
In total, 17 cc of blood was collected in BD Vacutainer CPT tubes (BD Diagnostics, Franklin Lakes, NJ) and placed on ice. Each clinical center followed a standardized protocol for the technical aspects of sample collection and CD4+ T cell isolation. Within 1 hr of collection, samples were centrifuged for 20 min at 1,700 RCF, followed by mononuclear cell layer isolation, and suspension in 10 ml of PBS. CD4+ lymphocytes were isolated using anti-CD4+ microbeads by positive column separation (Miltenyi Biotec, Auburn, CA) according to a previously published protocol [Jonuleit et al., 2000; Zorn et al., 2004]. Pilot studies in four subjects confirmed CD4+ lymphocyte yields of ~4 × 106 at ≥95% purity per collection using this technique. Total RNA was extracted using the RNeasy Mini Protocol (QIAGEN, Valencia, CA) [Chambers et al., 1999; Gonzalez et al., 1999; Gu et al., 2000]. Analysis using the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) confirmed average total RNA yields of 2 μg per collection, with no evidence of DNA contamination, minimal evidence of RNA degradation, and 28S:16S ratios of 2.0.
Genome-wide gene expression profiles were generated with Illumina HumanRef8 v2 BeadChip arrays (Illumina, San Diego, CA) using 100 ng of CD4+ total RNA from each sample and the Illumina BeadStation 500G according to the protocol. Briefly, total RNA was used to generate cDNA by reverse transcription, followed by biotin-labeled cRNA synthesis using the MessageAmp kit (Ambion, Austin, TX) [Pabon et al., 2001]. Labeled cRNA was combined with formamide and hybridization buffer, followed by overnight hybridization to HumanRef8Bead-Chip arrays. To avoid possible batch effects, chip batch and chip position (i.e. array positions A–H) were randomly assigned using a random number generator. Following randomization, we verified the effectiveness of this approach by assessing for stochastic differences in chip composition with respect to gender, clinic, race, and asthma severity, and observed essentially uniform distribution of these covariates, with no evidence of clustering by batch run or chip position. Following hybridization, chips were washed, blocked, stained with streptavidin-Cy3 dye, and scanned on the Illumina BeadArray scanner with images captured using the Illumina BeadScan software and processed with Illumina BeadStudio software (version 3.1.7). Raw expression intensities were processed using the lumi package [Du et al., 2008] with background adjustment with robust multi-array average convolution [Irizarry et al., 2003] of each array. Combined samples were normalized using variance stabilization and normalization [Huber et al., 2002; Lin et al., 2008]. The complete raw and normalized microarray data are available through the Gene Expression Omnibus of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE22324).
STATISTICAL ANALYSES
Two probe-filtering steps were applied prior to statistical testing. First, in order to avoid technical biases that can result from allele-specific differences in hybridization (i.e. SNPs residing within the probe can interfere with normal RNA-probe hybridization, suggesting between subject differences in expression when no such difference truly exists), we excluded from consideration the 1,659 probes that target genomic sequence known to harbor common sequence polymorphism (as reported in dbSNP) or that were not uniquely mappable to one genomic locus. Second, because only a fraction of all known transcripts are expressed in any given cell type, we excluded from consideration all probes mapping to transcripts that showed no evidence of dynamic expression in our CD4+ lymphocytes. We used the following criteria to define these uninformative probes: (i) those probes with low overall intensity (less than of 25% of samples with intensity of at least log2(100)); and (ii) those probes with low population variance across samples (less than twofold difference in interquartile range). By applying these filters, the final data set employed for all subsequent analyses included 10,294 probes corresponding to transcripts that map to autosomes.
We performed principal components analysis (PCA) using singular value decomposition (SVD) in the Bioconductor pcaMethods package [Stacklies et al., 2007]. Specifically, if X is the N × G matrix of expression values centered so that column means are zero, the SVD of X = UDVt, where V is a diagonal matrix of eigenvalues and U and V are orthogonal matrices. The decomposition is computed so that the elements of D are decreasing from the northwest corner. The columns of the matrix product XV are N-vectors, and the leftmost column is denoted the first principal component. This N-vector is a re-expression of transcript abundance data, and is regarded as a response variable in ANOVA or linear regression analyses; likewise for other columns of XV, which constitute lower order principal components. We then tested the identified principal components for association with known demographic, phenotypic, and environmental covariates using ANOVA for discrete variables and Spearman correlations for continuous variables.
Differential gene expression analysis was performed for 10,294 autosomal probes using the Significance Analysis of Microarray (SAM) test statistic as implemented in the siggenes package [Tusher et al., 2001]. An estimate of the number of differentially expressed genes between self-reported White (non-Hispanic) individuals and African Americans was derived using a conservative false-discovery rate (FDR) of 0.001 [Benjamini and Hochberg, 1995; Schwender, 2003]. Using the list of differentially expressed genes, we identified a subset of genes demonstrating consistent evidence of differential expression between self-identified racial groups from our analysis and in those that have been previously reported [Spielman et al., 2007; Storey et al., 2007; Zhang et al., 2008]. We then performed canonical pathway analysis using Ingenuity Pathway Analysis (Ingenuity Systems®, www.ingenuity.com) software on the list of differentially expressed genes that were common to the previously reported studies. To test the effect of self-identified race in an epidemiologic study of gene expression, we generated linear models using the limma package in Bioconductor [Smyth, 2004] to test for the association of gene expression with pre-bronchodilator forced expiratory volume in one second (FEV1), with and without covariate adjustment for self-identified race, age, gender, height, and height2. In order to determine the effect of principal components adjustment in gene expression studies, we tested the association of gene expression with pre-bronchodilator FEV1 by performing linear models with adjustment for PCs 1–4.
RESULTS
BASELINE CHARACTERISTICS
We generated Illumina HumanRef8 (v2) gene expression profiles for 254 young adults (205 self-identified non-Hispanic whites; 49 self-identified African Americans) with asthma on whom concurrent total RNA derived from peripheral blood CD4+ lymphocytes and measures of lung function were available. Characteristics of the subjects at the time of sample collection are shown in Table I. The age and gender distributions were similar between the self-identified non-Hispanic white and African American subjects. Furthermore, there was no significant difference in lung function (pre-bronchodilator FEV1 (% predicted), pre-bronchodilator FVC (% predicted), or pre-bronchodilator FEV1/FVC) or self-reported tobacco smoke exposure between the two groups. Asthma controller medication use was not significantly different between the two groups (P<0.05). Although African American subjects had higher Immunoglobulin E (IgE) levels (P = 0.02), other measures of asthma severity including the peripheral blood eosinophil level were not significantly different than non-Hispanic white subjects (P = 0.11).
TABLE I.
Baseline characteristics of subjects with CD4+ lymphocyte gene expression profiles in CAMP
| Variable | White (n = 205) | Black (n = 49) | P value |
|---|---|---|---|
| Male gender (n, %) | 123 (60%) | 28 (57%) | 0.74 |
| Age (years)a | 20.4 (2.2) | 20.6 (1.8) | 0.64 |
| Height (cm)a | 171.9 (9.1) | 170.5 (9.2) | 0.30 |
| Self-reported current smoking history (n, %) | 26 (13%) | 3 (6%) | 0.31 |
| Pre-bronchodilator FEV1 (% predicted) | 97.3 (12.2) | 94.4 (12.7) | 0.16 |
| Pre-bronchodilator FVC (% predicted) | 109.2 (10.8) | 106.9 (13.1) | 0.27 |
| Pre-bronchodilator FEV1/FVC | 77.4 (7.6) | 77.8 (8.4) | 0.74 |
| Inhaled corticosteroid use (n, %) | 49 (23%) | 11 (22%) | 0.85 |
| Long-acting bronchodilator use (n, %) | 27 (13%) | 6 (12%) | 0.83 |
| Total serum IgE level (IU/ml)b | 312.9 (134.7–781.4) | 581.7 (141–438.5) | 0.02 |
| Eosinophil count (cells/ml3)b | 212 (141–360.4) | 249 (141–438.5) | 0.11 |
FEV1, forced expiratory volume in one second; CAMP, Childhood Asthma Management Program; IgE, immunoglobulin E.
Mean (standard deviation).
Median (interquartile range).
PRINCIPAL COMPONENTS ANALYSIS
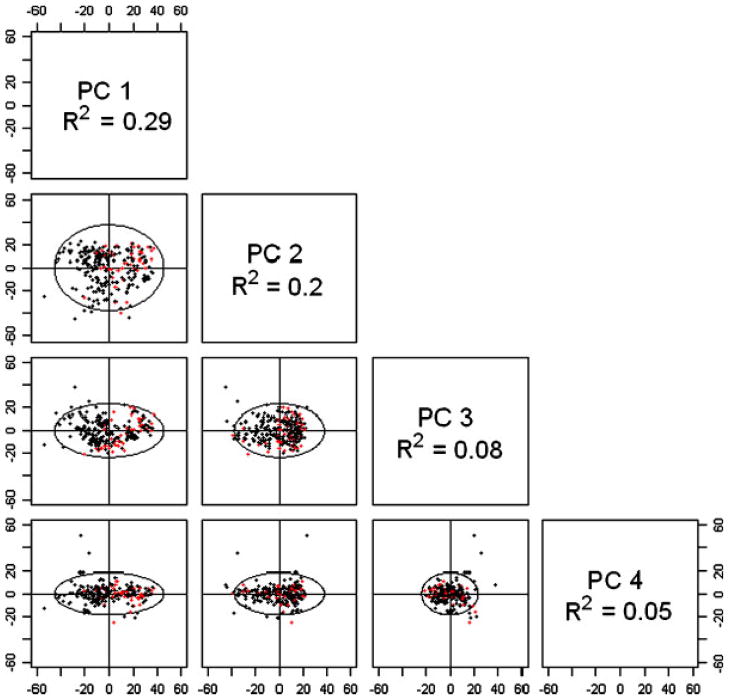
PCA was used to investigate the variance in gene expression across subjects in the study (Fig. 1). The first four principal components (PCS 1–4) explain a large proportion (62%) of the total variance in gene expression, the largest of which (PC1) explained 29%. Importantly, the distribution of PC1 loadings differed greatly by self-designated race, with positive PC1 values observed for most African American subjects and negative values for most self-reported white subjects (t-test P = 1.61 × 10−8). PC1 was also associated with study clinic, and weakly with a history of active smoking (Table II). Multivariable linear modeling confirmed self-identified race (not clinic) as the primary determinant of PC1, and clinic as the sole recognizable determinant of PC2 (data not shown). Though weaker associations were noted for gender with PC3 (P = 0.04), and IgE (P = 0.01) and FEV1 (P = 0.03) with PC4, we could not identify any measured covariate to explain the bulk of variation for these latter two principle components. Additional PCs (PC5–PC10) each explained less than 3% of the total variance in gene expression and were thus not considered further.
Fig. 1.
Non-Hispanic white subjects are shown in black, African American subjects are shown in red. First four principal components explain 62% of the variance in gene expression across the CAMP samples. CAMP, Childhood Asthma Management Program.
TABLE II.
Relationship between principal components of gene expression and relevant demographic and clinical characteristics in the CAMP cohort
| Covariate | Association P-value
|
|||
|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC4 | |
| Discrete variablesa | ||||
| Race | 1.61 × 10−8 | 0.03 | 0.40 | 0.60 |
| Gender | 0.24 | 0.11 | 0.04 | 0.48 |
| Clinic | 1.1 × 10−6 | 2 × 10−16 | 0.07 | 0.02 |
| Active smoking history | 0.007 | 0.61 | 0.08 | 0.88 |
| Asthma medication use | ||||
| Inhaled corticosteroids | 0.71 | 0.05 | 0.77 | 0.27 |
| Long-acting beta agonists | 0.50 | 0.98 | 0.60 | 0.64 |
| Continuous variables | ||||
| Age | 0.96 | 0.62 | 0.76 | 0.88 |
| Height | 0.21 | 0.97 | 0.32 | 0.32 |
| Pre-bronchodilator FEV1 | 0.71 | 0.51 | 0.07 | 0.03 |
| Immunoglobulin E (IgE level) | 0.22 | 0.74 | 0.57 | 0.01 |
FEV1, forced expiratory volume in one second; IgE, immunoglobulin E.
Discrete variables tested by two-sided t-test or ANOVA. Continuous variables tested by Spearman’s correlation.
Given the noted association of principle components 1, 2, and 4 with study clinic, we repeated the PCA, stratified by study clinic, to address whether the differences in gene expression by self-reported race were stable when accounting for possible site-specific differences in sampling (Online Supplemental Table EI). Despite the smaller sample sizes of these stratified analyses, persistent differential expression by self-identified race was noted in all four centers, suggesting that the effect of self-reported race on gene expression are independent of the study site and are not a function of study design.
DIFFERENTIAL EXPRESSION BY RACE
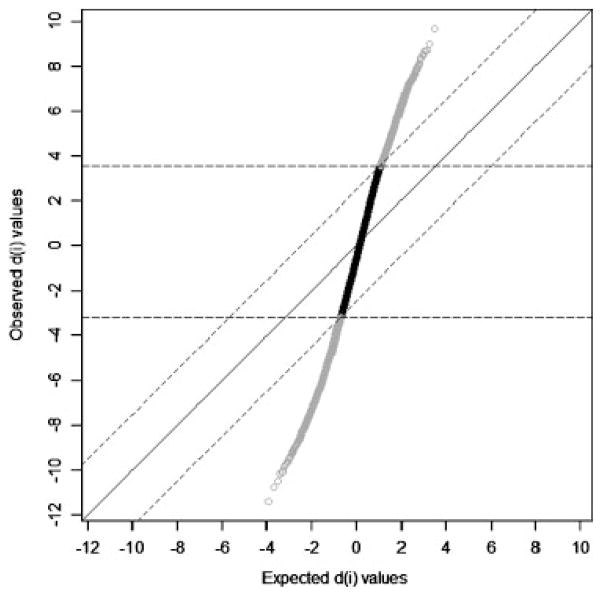
Given our findings of self-designated race as a major determinant of global gene expression patterns in the CAMP data set, we performed differential gene expression analysis across the two self-identified racial groups (Fig. 2). At a conservative FDR of 0.001, we found evidence for differential gene expression between self-identified non-Hispanic white and African American subjects of 3,743 genes (36%)—a similar proportion to that observed by Zhang et al. [2008] (Online Supplement Table EII). When comparing this gene set with the prior studies [Spielman et al., 2007; Storey et al., 2007; Zhang et al., 2008], we identified between 297 and 871 genes that were differentially expressed in both our data and in at least one of the previous studies. A total of 105 genes demonstrated consistent evidence of differential expression between racial groups in all previous studies, several of which have been implicated in racial differences in disease susceptibility. Examples include CREB1 in major depression [Dong et al., 2009] and SMARCA4 in breast cancer risk [Haiman et al., 2009]. We note that many of the genes differentially expressed by self-designated race clustered in specific gene networks and pathways, including several overlapping immune-response pathways, apoptosis signaling, and hormonal and metabolic pathways (Table III).
Fig. 2.
SAM plot demonstrating differential expression of 3,743 genes between non-Hispanic white and African American subjects in CAMP. The genes shown in grey (n = 3,743) are differentially expressed at a FDR = 0.001. CAMP, Childhood Asthma Management Program; SAM, significance analysis of microarray.
TABLE III.
IPA-defined pathways enriched for genes with differential expression by race
| Ingenuity canonical pathwaya | −log10P value | Molecules |
|---|---|---|
| PI3K/AKT signaling | 4.27 | JAK1, GYS1, PPP2R4, SOS1, YWHAZ, ITGA5, IKBKE, NFKB2, MAPK7, NFKBIB, CTNNB1, ITGA4 |
| Role of PKR in interferon induction and antiviral response | 4.05 | TRAF2, TRAF3, TNFRSF1A, IKBKE, NFKB2, CASP8, NFKBIB |
| Apoptosis signaling | 4.00 | ENDOG, TNFRSF1A, IKBKE, NFKB2, MAPK7, CAPN7, RPS6KA1, CASP8, NFKBIB, CAPN3 |
| Role of RIG1-like receptors in antiviral innate immunity | 3.84 | DHX58, TRAF2, TRAF3, IKBKE, NFKB2, CASP8, NFKBIB |
| CD40 signaling | 3.63 | TRAF2, TRAF3, CD40, IKBKE, NFKB2, MAPK7, NFKBIB, MAP2K5 |
| Death receptor signaling | 3.02 | DAXX, TRAF2, TNFRSF1A, IKBKE, NFKB2, CASP8, NFKBIB |
| Induction of apoptosis by HIV1 | 2.98 | DAXX, TRAF2, TNFRSF1A, IKBKE, NFKB2, CASP8, NFKBIB |
| 4-1BB signaling in Tlymphocytes | 2.93 | TRAF2, IKBKE, NFKB2, MAPK7, NFKBIB |
| FAK signaling | 2.70 | PAK4, SOS1, ITGA5, MAPK7, CAPN7, ITGA4, CAPN3, GIT2 |
| CD27 signaling in lymphocytes | 2.65 | TRAF2, IKBKE, NFKB2, CASP8, NFKBIB, MAP2K5 |
Top 10 ingenuity pathways are shown.
THE EFFECT OF RACE IN EPIDEMIOLOGIC ANALYSES OF GENE EXPRESSION
Given the extensive influence of self-identified race on gene expression, we explored the potential impact of these differences on the context of epidemiologic studies of clinical phenotypes, by modeling the relationship between peripheral blood CD4+ lymphocyte gene expression and FEV1, a spirometric measure of lung function (Table IV). Whereas unadjusted linear models identified 624 genes significantly associated with FEV1 (P<0.05 after correction for multiple comparisons), inclusion of self-identified race as a covariate resulted in a substantial drop–(95%)—in the number of detected genes. All the genes that dropped out with this adjustment were among those differentially expressed by self-identified race (Online Supplementary Table EII). Though further adjustment for other covariates known to be associated with lung function led to additional gene drop out (with no gene identified in the final model), the covariate with the single greatest impact on the analysis was self-identified race (Table IV). Linear models of pre-bronchodilator FEV1 percent predicted, which incorporates adjustment for age, gender, race, height, and height2, were used to confirm these findings and demonstrated no significant associations after these adjustments. Of note, when modeling FEV1 with principal components 1–4 included as covariates several genes were identified that were significantly associated with FEV1 (Table V). Similar results were obtained without adjustment for PC4, despite the observed correlation of PC4 with FEV1 (results not shown).
TABLE IV.
Effect of adjusting for race in an epidemiologic study of lung function
| Covariate adjustment | Number of significant genes (P<0.05 after correction for multiple comparisons) | Most significant gene
|
||
|---|---|---|---|---|
| Symbol | Unadjusted P-value | Adjusted P-value | ||
| Unadjusted | 624 | NRCAM | 7.07 × 10−8 | 0.0003 |
| Race | 31 | PRSS16 | 3.22 × 10−7 | 0.003 |
| Age, gender, race, height, height2 | 0 | XTP3TPA | 0.0005 | 0.97 |
TABLE V.
Effect of adjusting for principal components in an epidemiologic study of lung functiona
| HUGO gene | LOG fold change | P-value | Adjusted P-value |
|---|---|---|---|
| MMEL1 | 0.66 | 1.39 × 10−8 | 0.0001 |
| C16orf74 | 0.10 | 2.01 × 10−8 | 0.0001 |
| NRCAM | 0.08 | 1.51 × 10−6 | 0.005 |
| E2F3 | 0.10 | 2.05 × 10−6 | 0.005 |
| RSU1 | 0.07 | 5.18 × 10−6 | 0.01 |
| PARP3 | 0.06 | 8.5 × 10−6 | 0.01 |
| TSPYL5 | 0.05 | 1.70 × 10−5 | 0.02 |
| CD300C | 0.08 | 1.85 × 10−5 | 0.02 |
| SNX19 | 0.06 | 2.31 × 10−5 | 0.03 |
| PRSS16 | −0.04 | 2.69 × 10−5 | 0.03 |
Linear models adjusted for PCs 1–4. This table is restricted to results that are significant following the adjustment for multiple comparisons.
DISCUSSION
We explored the impact of self-identified race on epidemiological gene expression studies using microarray and spirometric data from a cohort of self-reported non-Hispanic white and African American asthmatics. In this study, we not only confirmed differences in gene expression due to self-identified race, but also demonstrated how these differences can confound epidemiologic studies. These results suggest that similar to genetic association studies, self-identified race is a critical confounder of epidemiologic studies of gene expression and must be considered and accounted for in population-based gene expression studies.
Self-reported racial designation does not arise through biological processes, but rather reflects a social construct derived from geographic ancestry, social dynamics, and historical mating patterns. Nevertheless, genetic divergence in allele frequency distributions and linkage disequilibrium patterns between racial groups is well documented [Gabriel et al., 2002; Stephens et al., 2001]. Though these genetic differences likely explain a proportion of the observed racial differences in disease susceptibility and drug response [Burchard et al., 2003], non-genetic differences such as epigenetic factors, environmental exposures, and cultural differences play significant roles as well. Similarly, though race-specific differences in gene expression due to between-population differences in allele frequency distribution have been documented [Cheung and Spielman, 2009; Stranger et al., 2007; Zhang et al., 2008], evidence for a larger non-genetic influence is also clear. While the source of these differences, whether epigenetic, environmental, or behavioral, remains unclear, a better understanding of these differences and how they influence studies of gene expression in health and disease is needed.
Though population differences in gene expression have been estimated in four prior reports [Spielman et al., 2007; Storey et al., 2007; Stranger et al., 2007; Zhang et al., 2008], our observations complement and extend these initial works in several important aspects. Foremost among these is that all four prior studies were performed in subsets of the HapMap lymphoblastoid cell lines, and as such cannot be considered truly independent from one another. Moreover, because the HapMap cell lines of Western European ancestry (CEU) were established many years before either the Asian (CHB and JPT) or Yoruba (YRI) samples, the validity of between-population comparisons using these samples is questionable [Akey et al., 2007]. In contrast, our studies were performed using RNA derived from freshly collected primary cells (circulating peripheral blood CD4+ lymphocytes), processed in uniform fashion irrespective of self-reported racial designation. Furthermore, our micro-array studies included sample randomization during allocation of chip and array position. We also filtered our data set so as not to consider transcripts interrogated by probes that target polymorphic sequence, thus limiting spurious association due to population differences in allele frequencies of SNP underlying probes. As such, we are assured that our observation of abundant differential expression between self-identified racial groups are not influenced by either batch effects resulting from differential sample processing, or due to potential race-specific differences in the lymphocyte immortalization process in response to Epstein-Barr virus infection.
We observed greater evidence of expression differences due to self-reported race in our cohort compared to the prior studies, both in terms of proportion (36% vs. 5–29%) and absolute number (3,743 vs. 464–1,320 genes). Several factors, both statistical and technical, may explain these differences. With respect to statistical considerations, we note that we studied a substantially larger number of subjects (on average two to three times greater than prior reports); the greater number of detected genes in the current study likely reflects the enhanced statistical power to detect these differences. Second, our focus on a primary cell type (vs. immortalized cell lines in prior studies) may have improved our ability to detect population-specific differences, in several ways. First, the technical biases that may have been introduced through the process of cell immortalization (which can bias both towards and away from the null) are not a factor in primary lines. Moreover, because the current study examined RNA that was isolated from primary cells within hours of blood draw, the observed race-specific differences in gene expression patterns in these samples reflects both the genetic differences and the contemporary environmental differences that exist between the self-identified racial groups. It is likely that the race-specific differences in expression were attenuated in the prior studies as a consequence of the immortalization process and because these cells were stored under fairly uniform (in vitro) environmental conditions. Thus, genes that show differential patterns of expression by racial group due to race-specific environmental differences would be underrepresented in prior studies.
Could our results be influenced by our exclusive focus on asthmatics such that the observed differences are a consequence of racial differences in asthma severity? In order for asthma status to have significantly confounded our results, two conditions would be required: (i) significant racial differences in asthma severity in our cohort, and (ii) significant differences in gene expression patterns by asthma severity. Our data suggest that neither condition is prominent here. First, as described above, the observed principle components of gene expression correlated most strongly with demographic features (including self-identified race) but not measures of asthma severity (such as lung function or current asthma medication usage). Second, with the exception of total serum IgE levels, there were no significant differences between racial groups in their baseline characteristics, including several markers of asthma severity, such as lung function and blood eosinophil counts (Table I). Though mean total serum IgE levels were higher in African Americans as compared to non-Hispanic whites (P = 0.02), there was no correlation between total serum IgE level and the first three principal components, which explain in total 57% of variation in gene expression (see Table II). Moreover, we note that a correlation analysis of total gene expression with total serum IgE levels identified only one transcript (IL17RB) that was significantly correlated with total serum IgE levels (data not shown). This correlation was observed in all subjects regardless of self-identified race. Moreover, IL17RB was not differentially expressed by self-designated race in the current analysis here. Together, these data suggest that neither IgE level nor other measures of asthma severity are confounders of the racial differences in gene expression described herein.
The confirmation of widespread differences in gene expression between self-identified racial groups has several important implications for efforts aimed at identifying the underlying molecular causes of disease. First, as suggested in previous studies, some of these differences may offer insights into the mechanisms underlying population differences in disease susceptibility. For instance, racial differences in chemotherapeutic response and cancer survival rates [Caudle et al.; Polite et al., 2008] may in part be explained by the observed differences in expression of genes belonging to the phosphatidylinositol-3 kinase pathway, which has been implicated in the regulation of cell survival regulation, cell cycle progression, and cell growth. Modulation of the PI3k/Akt signaling pathway can result in failure to activate the apoptotic pathway and has been proposed as a mechanism of drug resistance [Fresno Vara et al., 2004]. We found this pathway to be significantly enriched for genes differentially expressed by self-identified racial designation. Similar findings of genes belonging to diabetes signaling pathways may in part explain the difference in prevalence rates and natural history of this disease between Caucasians and individuals of African descent [Fukushima et al., 2010], while the numerous differences observed in immune-related pathways could in part explain the racial differences in humoral and cellular responses to infection [de la et al., 2007; Donlin et al., 2010; Gale et al., 1998].
The observed between-population differences in expression also have implications for epidemiologic studies, as illustrated in our analysis of lung function, where the relationship between gene expression and spirometric measurements was confounded by self-reported race. Racial designation is an important determinant of lung size, together with age and anthropomorphic measurements, and adjustment of spirometric measures for self-identified race is critical for reliable interpretation of epidemiological studies. Thus, though our illustration of the impact of self-identified race on gene expression studies of lung function may represent somewhat of a “straw man,” it nonetheless makes the point that failure to account for population ancestry can have deleterious consequences on the ability to reliably detect truly differentially expressed genes in population-based studies.
Similar to genome-wide genetic association studies where principal components adjustment is used to address population structure, our results suggest that principal components may also be used in epidemiologic studies of gene expression. Addition of principal components as covariates in gene expression studies allows for the adjustment of both known confounders (ie technical confounders like batch) and unmeasured confounders including genetic ancestry when genotype data is unavailable. Using PCs to model the association of gene expression with pre-bronchodilator FEV1, we were able to identify several biologically plausible candidate genes for further investigation. Of the 10 genes identified to be associated with FEV1, three have been associated with smoking-related lung disease or lung cancer. For example, E2F3 is a transcription factor that has an established role in controlling the cell cycle. In addition, overexpression of E2F3 has been implicated the pathogenesis of small cell lung cancer [Cooper et al., 2006]. TSPLY5 has been shown to be differentially expressed in squamous cell lung cancer [Vachani et al., 2007]. Furthermore, PARP3, which is involved with DNA repair, is downregulated in non-small cell lung cancers that reactivate telomerase decreasing telomere lengths in these subjects [Frias et al., 2008]. In addition to its role in lung cancer, telomere length is lower in patients with chronic obstructive pulmonary disease (COPD) [Savale et al., 2009]. Both lung cancer and COPD are smoking-related lung diseases, which can be associated with airflow obstruction and low lung function in adults. While these genes are interesting candidates for investigation for their role in impaired lung function, it is important to recognize that none of these genes would have been identified using traditional modeling approaches. Our results suggest that a more comprehensive analysis of the use of principal component adjustment of gene expression studies is warranted.
Several limitations to this study must be discussed. While we demonstrate that the effect of gene expression on lung function is confounded by the effects of self-designated race, we do recognize that several of the genes that are differentially expressed between populations may contribute to fundamental differences in lung function. Thus, adjustments for self-identified race may in fact result in some false-negative results as well. Finally, our gene expression analysis of lung function was performed in peripheral blood CD4+ lymphocytes obtained from asthmatic individuals. It is therefore possible that self-identified race would not dominate the analysis if perhaps a tissue more proximal to the phenotype (ie airway epithelial cells) were studied. Nonetheless, consideration of population ancestry would remain necessary in these studies.
In summary, this study demonstrates that racial differences exist in gene expression, and shows that self-identified race may be an important confounding factor in studies of gene expression that examine people of diverse racial backgrounds. Furthermore, population differences in gene expression are widespread, apparently affecting a considerably larger number of genes than previously suspected. We have demonstrated the impact of these differences on epidemiological gene expression studies of disease and show how principal components adjustment may be beneficial in these studies. In the field of genetics, several well-developed statistical approaches are available to address the confounding effects of ancestry, including stratified analysis, family-based tests, and genomic control strategies [Devlin et al., 2001; Horvath et al., 2001; Patterson et al., 2006; Price et al., 2006]. Our observations support the need for the development of similar strategies for gene expression studies.
Supplementary Material
Acknowledgments
We thank all subjects for their ongoing participation in this study. We acknowledge the CAMP investigators and research team, supported by NHLBI, for collection of CAMP Genetic Ancillary Study data. Special thanks to Anne Plunkett, Teresa Concordia, Debbie Bull, Denise Rodgers, and D. Sundstrom for their assistance with sample collection; to Huiqing Yin-DeClue, Ph.D., Michael McLane and Chris Allaire for their assistance with T-cell isolations and RNA preparation; and to Ankur Patel for his assistance in running the microarrays. All work on data collected from the CAMP Genetic Ancillary Study was conducted at the Channing Laboratory of the Brigham and Women’s Hospital under appropriate CAMP policies and human subject protections. This work is supported by grant R01 HL086601 and RC2 HL101543 from the National Heart, Lung and Blood Institute, National Institutes of Health (NIH/NHLBI). The CAMP Genetics Ancillary Study is supported by U01 HL075419, U01 HL65899, P01 HL083069 and T32 HL07427 from the NIH/NHLBI, and through the Colorado CTSA grant 1 UL1 RR025780 from the National Institutes of Health (NIH) and National Center for Research Resources (NCRR).
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- American Thoracic Society. Standardization of spirometry, 1994 update. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- Akey JM, Biswas S, Leek JT, Storey JD. On the design and analysis of gene expression studies in human populations. Nat Genet. 2007;39:807–808. doi: 10.1038/ng0707-807. author reply 808–809. [DOI] [PubMed] [Google Scholar]
- Barbujani G, Magagni A, Minch E, Cavalli-Sforza LL. An apportionment of human DNA diversity. Proc Natl Acad Sci USA. 1997;94:4516–4519. doi: 10.1073/pnas.94.9.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Perez-Stable EJ, Sheppard D, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348:1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- Caudle AS, Gonzalez-Angulo AM, Hunt KK, Liu P, Pusztai L, Symmans WF, Kuerer HM, Mittendorf EA, Hortobagyi GN, Meric–Bernstam F. Predictors of tumor progression during neo-adjuvant chemotherapy in breast cancer. J Clin Oncol. 28:1821–1828. doi: 10.1200/JCO.2009.25.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan JS, Bittner A, Frueh K, Jackson MR, Peterson PA, Erlander MG, Ghazal P. DNA microarrays of the complex human cyto-megalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73:5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet. 2009;10:595–604. doi: 10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- Cooper CS, Nicholson AG, Foster C, Dodson A, Edwards S, Fletcher A, Roe T, Clark J, Joshi A, Norman A, et al. Nuclear over-expression of the E2F3 transcription factor in human lung cancer. Lung Cancer. 2006;54:155–162. doi: 10.1016/j.lungcan.2006.07.005. [DOI] [PubMed] [Google Scholar]
- de la CSB, Kouri G, Guzman MG. Race: a risk factor for dengue hemorrhagic fever. Arch Virol. 2007;152:533–542. doi: 10.1007/s00705-006-0869-x. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- Dong C, Wong ML, Licinio J. Sequence variations of ABCB1, SLC6A2, SLC6A3, SLC6A4, CREB1, CRHR1 and NTRK2: association with major depression and antidepressant response in Mexican-Americans. Mol Psychiatry. 2009;14:1105–1118. doi: 10.1038/mp.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin MJ, Cannon NA, Aurora R, Li J, Wahed AS, Di Bisceglie AM, Tavis JE. Contribution of genome-wide HCV genetic differences to outcome of interferon-based therapy in Caucasian American and African American patients. PLoS One. 2010;5:e9032. doi: 10.1371/journal.pone.0009032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzalez-Baron M. PI3K/Akt signaling pathway and cancer. Cancer Treat Rev. 2004;30:193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Frias C, Garcia-Aranda C, De Juan C, Moran A, Ortega P, Gomez A, Hernando F, Lopez-Asenjo JA, Torres AJ, Benito M, et al. Telomere shortening is associated with poor prognosis and telomerase activity correlates with DNA repair impairment in non-small cell lung cancer. Lung Cancer. 2008;60:416–425. doi: 10.1016/j.lungcan.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Fukushima A, Loh K, Galic S, Fam B, Shields B, Wiede F, Tremblay ML, Watt MJ, Andrikopoulos S, Tiganis T. TCPTP attenuates STAT3 and insulin signaling in the liver to regulate gluconeogenesis. Diabetes. 2010;59:1906–1914. doi: 10.2337/db09-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gale MJ, Jr, Korth MJ, Katze MG. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin Diagn Virol. 1998;10:157–162. doi: 10.1016/s0928-0197(98)00034-8. [DOI] [PubMed] [Google Scholar]
- Gonzalez P, Zigler JS, Jr, Epstein DL, Borras T. Identification and isolation of differentially expressed genes from very small tissue samples. Biotechniques. 1999;26:884–886. 888–892. doi: 10.2144/99265st01. [DOI] [PubMed] [Google Scholar]
- Gu L, Tseng S, Horner RM, Tam C, Loda M, Rollins BJ. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407–411. doi: 10.1038/35006097. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Garcia RR, Hsu C, Xia L, Ha H, Sheng X, Le Marchand L, Kolonel LN, Henderson BE, Stallcup MR, et al. Screening and association testing of common coding variation in steroid hormone receptor co-activator and co-repressor genes in relation to breast cancer risk: the Multiethnic Cohort. BMC Cancer. 2009;9:43. doi: 10.1186/1471-2407-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jonuleit H, Schmitt E, Schuler G, Knop J, Enk AH. Induction of interleukin 10-producing, nonproliferating CD4(+) T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J Exp Med. 2000;192:1213–1222. doi: 10.1084/jem.192.9.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick P, Dransfield MT. Racial and sex differences in chronic obstructive pulmonary disease susceptibility, diagnosis, and treatment. Curr Opin Pulm Med. 2009;15:100–104. doi: 10.1097/MCP.0b013e3283232825. [DOI] [PubMed] [Google Scholar]
- Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormerod S, McDowell SE, Coleman JJ, Ferner RE. Ethnic differences in the risks of adverse reactions to drugs used in the treatment of psychoses and depression: a systematic review and meta-analysis. Drug Saf. 2008;31:597–607. doi: 10.2165/00002018-200831070-00005. [DOI] [PubMed] [Google Scholar]
- Pabon C, Modrusan Z, Ruvolo MV, Coleman IM, Daniel S, Yue H, Arnold LJ., Jr Optimized T7 amplification system for microarray analysis. Biotechniques. 2001;31:874–879. doi: 10.2144/01314mt05. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polite BN, Cirrincione C, Fleming GF, Berry DA, Seidman A, Muss H, Norton L, Shapiro C, Bakri K, Marcom K, et al. Racial differences in clinical outcomes from metastatic breast cancer: a pooled analysis of CALGB 9342 and 9840—Cancer and Leukemia Group B. J Clin Oncol. 2008;26:2659–2665. doi: 10.1200/JCO.2007.13.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Romualdi C, Balding D, Nasidze IS, Risch G, Robichaux M, Sherry ST, Stoneking M, Batzer MA, Barbujani G. Patterns of human diversity, within and among continents, inferred from biallelic DNA polymorphisms. Genome Res. 2002;12:602–612. doi: 10.1101/gr.214902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Savale L, Chaouat A, Bastuji-Garin S, Marcos E, Boyer L, Maitre B, Sarni M, Housset B, Weitzenblum E, Matrat M, et al. Shortened telomeres in circulating leukocytes of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2009;179:566–571. doi: 10.1164/rccm.200809-1398OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender H. Assessing the False Discovery Rate in a Statistical Analysis of Gene Expression Data. Dortmund: University of Dortmund; 2003. [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods—a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23:1164–1167. doi: 10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH, et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM. Gene-expression variation within and among human populations. Am J Hum Genet. 2007;80:502–509. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk RC, Sternberg AL, Szefler SJ, Zeiger RS, Bender B, Tonascia J. Long-term budesonide or nedocromil treatment, once discontinued, does not alter the course of mild to moderate asthma in children and adolescents. J Pediatr. 2009;154:682–687. doi: 10.1016/j.jpeds.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med. 2000;343:1054–1063. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachani A, Nebozhyn M, Singhal S, Alila L, Wakeam E, Muschel R, Powell CA, Gaffney P, Singh B, Brose MS, et al. A 10-gene classifier for distinguishing head and neck squamous cell carcinoma and lung squamous cell carcinoma. Clin Cancer Res. 2007;13:2905–2915. doi: 10.1158/1078-0432.CCR-06-1670. [DOI] [PubMed] [Google Scholar]
- Van Dyke AL, Cote ML, Wenzlaff AS, Land S, Schwartz AG. Cytokine SNPs: comparison of allele frequencies by race and implications for future studies. Cytokine. 2009;46:236–244. doi: 10.1016/j.cyto.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn E, Miklos DB, Floyd BH, Mattes-Ritz A, Guo L, Soiffer RJ, Antin JH, Ritz J. Minor histocompatibility antigen DBY elicits a coordinated B and T cell response after allogeneic stem cell transplantation. J Exp Med. 2004;199:1133–1142. doi: 10.1084/jem.20031560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.