Abstract
Light signaling pathways and the circadian clock interact to help organisms synchronize physiological and developmental processes with periodic environmental cycles. The plant photoreceptors responsible for clock resetting have been characterized, but signaling components that link the photoreceptors to the clock remain to be identified. Here we describe a family of night light–inducible and clock-regulated genes (LNK) that play a key role linking light regulation of gene expression to the control of daily and seasonal rhythms in Arabidopsis thaliana. A genomewide transcriptome analysis revealed that most light-induced genes respond more strongly to light during the subjective day, which is consistent with the diurnal nature of most physiological processes in plants. However, a handful of genes, including the homologous genes LNK1 and LNK2, are more strongly induced by light in the middle of the night, when the clock is most responsive to this signal. Further analysis revealed that the morning phased LNK1 and LNK2 genes control circadian rhythms, photomorphogenic responses, and photoperiodic dependent flowering, most likely by regulating a subset of clock and flowering time genes in the afternoon. LNK1 and LNK2 themselves are directly repressed by members of the TIMING OF CAB1 EXPRESSION/PSEUDO RESPONSE REGULATOR family of core-clock genes in the afternoon and early night. Thus, LNK1 and LNK2 integrate early light signals with temporal information provided by core oscillator components to control the expression of afternoon genes, allowing plants to keep track of seasonal changes in day length.
The rotation of the earth around its own axis and its movement around the sun cause daily and seasonal oscillations in light intensity on our planet. The profound impact of these environmental changes on biological processes strongly contributed to the evolution of circadian clocks (1). Therefore, it is not surprising that circadian and light signaling networks are intimately connected. Indeed, although circadian rhythms normally persist in the absence of environmental cues with a period of ∼24 h, light/dark cycles entrain the clock and thereby ensure appropriate phasing of circadian rhythms in relation to changing sunrise and sunset throughout the year (2).
In plants, the effect of light on the clock is mediated by specific photoreceptors, such as phytochromes, cryptochromes, and members of the ZEITLUPE protein family (3–5). The plant circadian clock is mostly based on clock genes that mutually regulate each other expression (6), and some of these are acutely induced by phytochromes (7–9). Interestingly, cryptochromes and phytochromes are not essential for circadian oscillations in Arabidopsis plants (10–12), but circadian regulation of phototransduction pathways generates tight links between these two signaling networks (13). This phenomenon, known as gating, was originally described for the light-regulated activity of the promoter of the CHLOROPHYLL A/B BINDING PROTEIN II (CABII) gene (14). CABII expression is acutely induced by red light pulses, but the effectiveness of this treatment oscillates during a 24-h day, with maximal effects when photosynthetic activity is expected to be at its peak during the day and minimal effects during the night (14–16). Clock gating of light signaling is mediated, at least in part, by the clock gene EARLY FLOWERING 3 (15), which interacts directly with phytochrome B (17). Clock regulation of light signaling also influences physiological processes such stem elongation (18, 19), and the clock itself (15, 20). Indeed, in plants grown under light/dark cycles and then transferred to constant darkness brief light pulses are most effective in resetting the phase of circadian rhythms during the night rather than during the subjective day (i.e., the phase that would have been illuminated if the plants were kept under light/dark cycles) (20). This phenomenon is shared across kingdoms, suggesting that it is critical for the appropriate adjustment of circadian rhythms to the environment (21).
Despite the importance of the interactions between light and the circadian clock in the control of biological activities in plants, a comprehensive analysis of these interactions has been lacking. Light signaling and circadian networks operate primarily by transcriptional control (13, 22–24). To characterize these interactions in Arabidopsis, we evaluated the response of the Arabidopsis transcriptome to light pulses given at different times. A light pulse in the middle of the subjective day should modulate the expression of genes that contribute to maximizing process such as photosynthesis. In contrast, a light pulse in the middle of the night simulates either an earlier sunrise or a later sunset and may reveal genes involved in clock resetting and/or seasonal adjustment. Indeed, this analysis allowed us to identify a unique family of light and clock regulated morning genes. These genes control both the pace of circadian rhythms and the photoperiodic regulation of flowering time, apparently by promoting the expression of a subset of core-clock and clock-output genes in the afternoon.
Results
Light Treatments Are More Effective During the Subjective Day.
To investigate if and how time of day affects light regulation of gene expression at a global level, we used microarray analysis to evaluate the response of the Arabidopsis transcriptome to a 1-h light treatment given either in the middle of the subjective day or in the middle of the night (Fig. 1A). Many light-regulated genes showed a stronger response to a light pulse given during the subjective day compared with a similar treatment given during the night (Fig. 1B; Dataset S1). Among a total of 2,237 light-induced genes identified using a twofold change as cutoff, 1,537 responded at least twice as strongly to the light pulse given in the middle of the subjective day (Dataset S1A), and only 65 genes showed a stronger response during the night (Dataset S1B). Thus, almost 70% of light-induced genes behaved similarly to what had been reported for CABII (14). This group of day light–responsive genes was enriched in gene ontologies associated with metabolism, chloroplast components, responses to environmental stimuli, and responses to abiotic and biotic stress (Dataset S2A). The influence of time of day was less pronounced for light-repressed genes. Among a total of 1,672 light-repressed genes, only 607 responded at least twice as much during the subjective day compared with the night (Dataset S1C), and 78 showed the opposite response (Dataset S1D). The group more strongly repressed by light during the subjective day was mostly enriched in genes involved in amino acid catabolism (Dataset S2B), whereas those more responsive to light during the night were associated with hormonal regulation, among other processes (Dataset S2C).
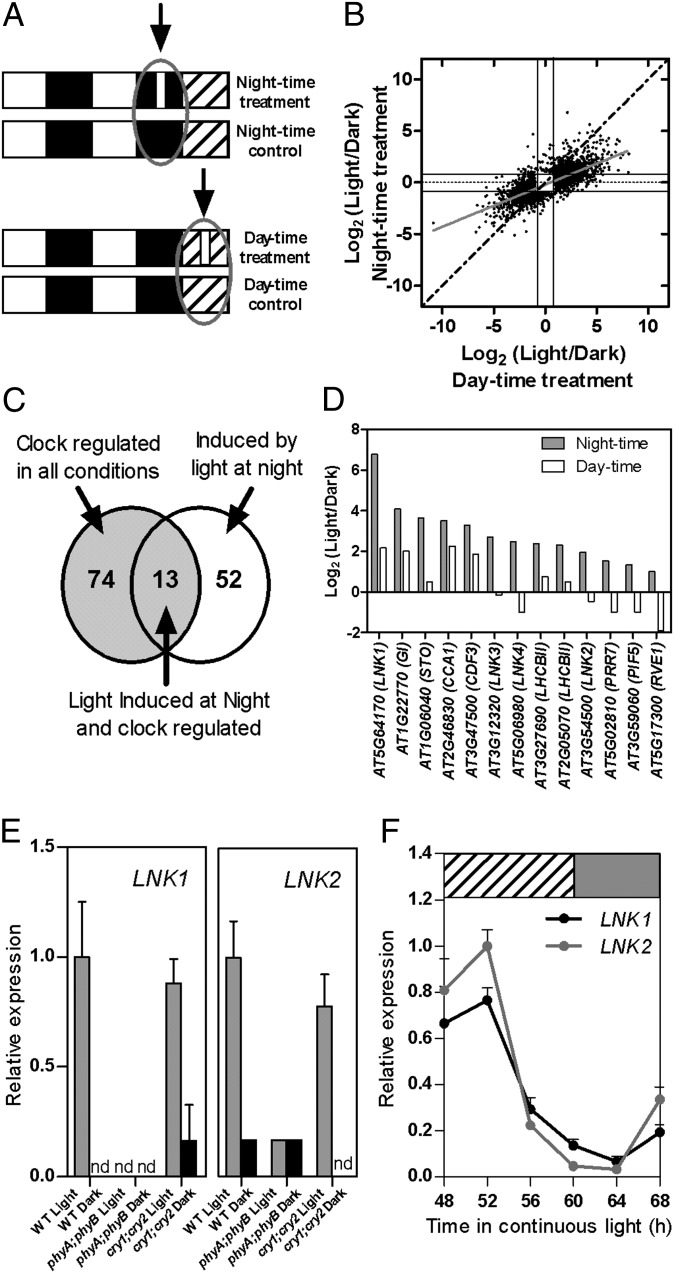
Fig. 1.
Genomewide analysis of light and clock interactions in the control of gene expression and identification of LNK genes. (A) Experimental design. Plants were grown under 12-h light/12-h dark cycles for 14 d and then exposed or not to a 1-h light pulse in the middle of the night or subjective day on the 15th day. (B) Comparative genomewide expression analysis of the effect of a light pulse given during subjective day time (x axis) vs. night time (y axis). (C) Overlap between 87 genes that are rhythmically expressed under multiple conditions (23) and 65 genes that showed a stronger induction by light during night time compared with subjective day time (Dataset S1). (D) Microarray data corresponding to the relative response of LNK genes to a 1-h light treatment given in the middle of the night or subjective day. (E) Relative expression levels of LNK1 and LNK2 measured by qRT-PCR. The analysis was conducted in WT, phyA;phyB, and cry;cry2 plants grown under 12-h light/12-h dark cycles and exposed or not to a 1-h light pulse in the middle of the night (n = 3). nd, not detectable. Data represent average + SEM. (F) Circadian expression of LNK1 and LNK2 genes. Expression was determined by qRT-PCR during the third day under free running conditions. n = 4. Data are average + SEM. White, dark, gray, and hatched boxes indicate day, night, subjective night, and subjective day, respectively.
Because plants were under starvation during the subjective day, the effect of light at this time of day could simply be the consequence of sucrose reaccumulation due to photosynthetic activity. However, no significant correlation was found between light induction of gene expression during the subjective day and changes in gene expression induced by sucrose or enhanced photosynthetic activity (Fig. S1). In contrast, a direct correlation was found for light-repressed genes (Fig. S1). Indeed, using quantitative RT-PCR (qRT-PCR), we found that light repression of two genes was unaffected in photoreceptor mutants, whereas light induction was significantly attenuated in phyA;phyB mutants and to a lesser extent in cry1;cry2 mutants (Fig. S2). In addition, the expression of these light-induced genes is not affected by sucrose or photosynthetic activity, whereas light-repressed genes were also repressed to some extent by sucrose or photosynthetic activity (Fig. S2). Thus, light induction of gene expression during the subjective day is mostly mediated by photomorphogenic photoreceptors, whereas repression is likely triggered by sucrose accumulation due to photosynthetic activity.
Night Light Is More Effective in Inducing the Expression of a Subset of Core-Clock Genes.
Clock entrainment is most sensitive to light pulses given during the night, a treatment that simulates seasonal changes in day length (20). Consistent with this, the subset of 65 genes responding at least twice as strongly to the night light treatment was significantly enriched in clock genes, a phenomenon that was specific for this particular class of light-regulated genes (Dataset S2D). Clock genes are also enriched among those with oscillations that are robust to different experimental conditions, such as continuous light, continuous darkness, short days, long days, temperature cycles, etc. (23). Thus, we reasoned that the list of genes that are more effectively induced by night light and also cycle under multiple conditions should contain new candidate clock regulators. Thirteen genes fulfilled both criteria, a 30-fold enrichment over expectation (P < 1 × 10−15, hypergeometric distribution; Fig. 1C). This group included the clock genes CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), PRR7, and GIGANTEA (GI), six genes involved in the control of stem elongation, flowering time or photosynthesis, as well as four genes that constitute a new family of plant specific proteins, which we named LNK1–4, for night light–inducible and clock-regulated genes 1–4 (Fig. 1D).
LNK1 (AT5G64170) and LNK2 (AT3G54500) are proteins of about 66 kDa, with 35% sequence similarity across their length. LNK3 (AT3G12320) and LNK4 (AT5G06980) proteins are smaller (each around 30 kDa), with 60% sequence similarity and with a third of conserved positions also shared with LNK1/LNK2 (Fig. S3). LNK homologs can be found throughout land plants, including nonvascular plants. LNK3 and LNK4 appear to be the result of a recent duplication event within the Brassicaceae (Fig. S4). Because LNK1 responded most strongly to the night light treatment (Dataset S1B), we focused on LNK1 and its closest homolog, LNK2. qRT-PCR analyses of WT and mutants indicated that these two genes are induced by a light pulse in the middle of the night via the phytochrome family of red/far-red light photoreceptors and that they are rhythmically expressed with maximum levels in the subjective morning (Fig. 1 E and F).
LNK1 and LNK2 Regulate Light Signaling and Biological Timing.
To determine whether LNK1 and LNK2 affect light- and clock-regulated developmental and physiological processes, several mutants with T-DNA insertions in these two genes were identified and characterized in detail (Fig. S5). An early developmental phenomenon under control of light and the circadian clock is the elongation of the hypocotyl, the embryonic stem. No significant differences in hypocotyl length were observed among WT plants and lnk1, lnk2, or lnk1;lnk2 mutants grown in complete darkness (Fig. S6A). In contrast, lnk1 mutants had longer hypocotyls than WT plants under continuous white light (Fig. 2A; Fig. S6B) or under continuous red light (Fig. S6C). lnk2 mutants also had longer hypocotyls than WT plants in red light (Fig. S6C), whereas the differences in hypocotyl length were not statistically significant under most other light conditions (Fig. 2A; Fig. S6). The lnk1;lnk2 double mutant had significantly longer hypocotyls than either single mutant or WT seedlings under continuous white light conditions, and the phenotype was stronger under red or white light than under blue light (Fig. 2A; Fig. S6). Taken together, these results indicate that LNK1 and LNK2 mediate light inhibition of hypocotyl elongation, in particular that triggered by the phytochrome family of red/far-red light photoreceptors.
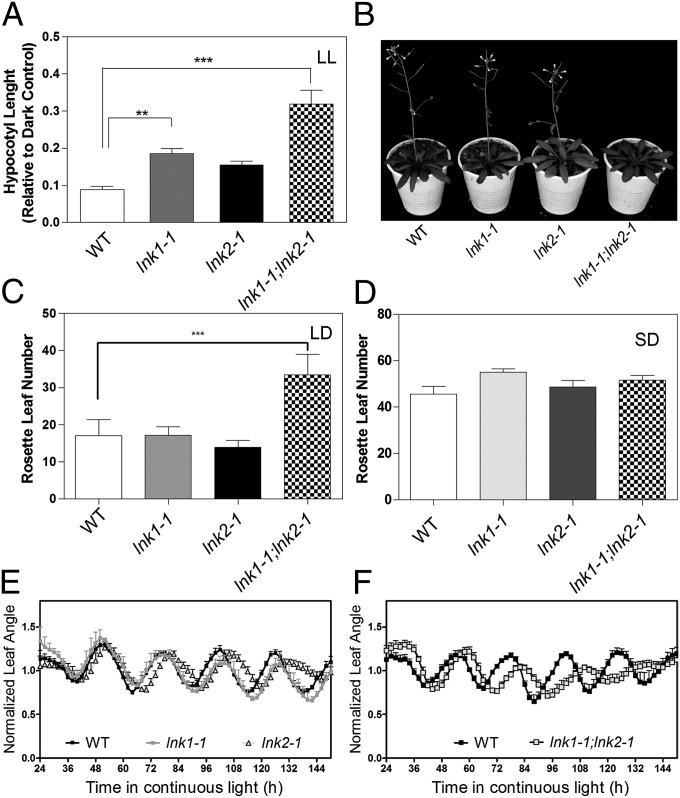
Fig. 2.
Physiological characterization of LNK1 and LNK2. (A) Hypocotyl length of WT, lnk1, lnk2, and lnk1;lnk2 mutant seedlings grown under continuous white light (LL) (n = 6 replicates of 10 seedlings each). (B) LNK1 and LNK2 control the floral transition in plants grown under LD (16-h light/8-h dark) conditions. (C and D) Flowering time measured as the number of rosette leaves at bolting in LD (C) and SD (D) conditions (8-h light/16-h dark). ANOVA followed by a Tukey´s multiple comparison test was used to evaluate the statistical significance of differences observed between genotypes. Error bars indicate +SEM (***P < 0.001, *P < 0.05). (E and F) Circadian rhythms of leaf movement in continuous light (n = 7). Plants were grown under LD cycles and then transferred to constant light and temperature conditions. Error bars indicate +SEM. Open and hatched boxes indicate subjective day and subjective night, respectively.
Another physiological process that depends on the interactions between light signaling and the circadian clock is photoperiod-dependent flowering (25). lnk1;lnk2 double mutant flowered later than WT plants or lnk1 or lnk2 single mutants under long days (LD; 16-h light/8-h dark; Fig. 2 B and C). Under short days (SD; 8-h light/16-h dark), no delay in flowering was observed (Fig. 2D), confirming that LNK1 and LNK2 are indeed only required for long day–dependent acceleration of flowering rather than the transition to flowering per se.
To observe circadian behavior directly, we monitored the circadian rhythm of leaf movement in WT plants and lnk1, lnk2, and lnk1;lnk2 mutants by time lapse photography. Leaf movement of lnk2 mutants had a longer circadian period than WT or single lnk1 mutant plants (Fig. 2E; Fig. S6F), and the lnk1;lnk2 double mutant was even more strongly affected (Fig. 2F; Fig. S6F). Similar photomorphogenic and circadian phenotypes were observed in additional mutant alleles of LNK1 and LNK2 (Fig. S6), confirming that these two genes play important and partially redundant roles controlling light- and clock-regulated processes in Arabidopsis.
LNK1 and LNK2 Activate Clock-Controlled Genes with Afternoon Peak.
LNK proteins lack known functional domains, but LNK1:YFP localized mostly to the nucleus in Arabidopsis thaliana hypocotyl cells, suggesting a role in the regulation of gene expression (Fig. 3A). To identify genes controlled by LNK1 and LNK2, we compared the transcriptome of WT and lnk1;lnk2 mutant plants using RNA sequencing (RNA-seq). In plants grown under constant light and temperature, we found 806 genes differentially expressed using a false discover rate (FDR)-adjusted P < 0.01 as a cutoff (Dataset S3A). Genes down-regulated in lnk1;lnk2 mutants were significantly enriched for genes that peak in LD at Zeitgeber time 10 (ZT10), i.e., 10 h after lights on. Up-regulated genes were slightly enriched for genes that peak late at night (Fig. 3B).
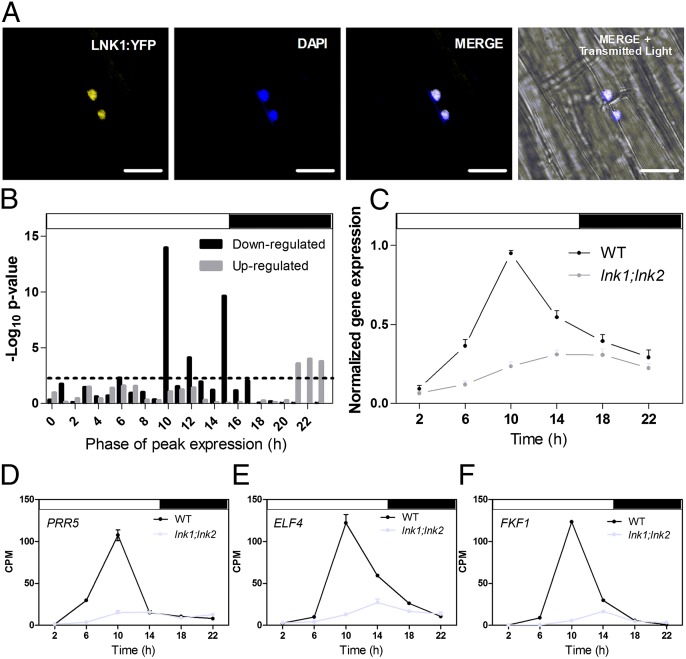
Fig. 3.
LNK1, a nuclear protein, positively regulates expression of circadian genes with an afternoon phase. (A) LNK:YFP detection by confocal microscopy in the hypocotyl of seedlings expressing 35S:LNK:YFP in a WT background is shown on the left (first panel). Fluorescence following staining with DAPI is shown in blue (second panel). The merged image (white) and the image with the transmitted light channel are also shown (third and fourth panels, respectively). (Scale bar, 20 µm.) (B) Phase enrichment of circadian-regulated genes whose expression was down- or up-regulated in lnk1;lnk2 mutant compared with WT plants, according to RNA-seq data of plants grown under continuous light conditions. The phase overrepresentation analysis was conducted with Phaser (http://phaser.mocklerlab.org/) and was based on the phases of gene expression estimated from data obtained using WT plants grown under LD conditions (23). Dashed line corresponds to P = 0.01. (C) Average normalized expression of 36 genes from the cluster with the largest number of genes whose expression was altered in lnk1:lnk2 mutants compared with WT plants grown under LD conditions. Normalized expression of PRR5 (D), ELF4 (E), and FKF1 (F), three genes present in the cluster shown in C. (C–F) Plants grown under LD cycles were sampled every 4 h, starting 2 h after lights on. n = 3, Error bars indicate + SEM.
To learn more about LNK1/LNK2 target genes, we used RNA-seq to characterize the daily transcriptome of LD-grown WT and lnk1;lnk2 mutant plants. Using stringent criteria aimed at identifying genes with altered overall mRNA levels, and not simply changed temporal patterns of expression, we identified 387 genes that differed between WT and lnk1;lnk2 mutant plants (Dataset S3B). A cluster analysis revealed that most of the genes down-regulated in lnk1;lnk2 mutant oscillated in WT plants with peak expression in the afternoon or early night (Fig. S7), with the largest cluster peaking at ZT10 (Fig. 3C), providing independent support for the initial phase enrichment analysis, which had suggested that LNK1/LNK2 activity is maximal in the afternoon (Fig. 3B).
To identify genes likely responsible for the phenotypic defects in lnk1;lnk2 mutants, we focused our analysis on the 101 down-regulated and the 31 up-regulated genes in both RNA-seq data sets (Dataset S3C). Down-regulated genes included two core-clock genes, PRR5 (Fig. 3D) and EARLY FLOWERING 4 (ELF4; Fig. 3E), which were present in the cluster of genes with peak expression in WT plants at ZT10 (Fig. 3C) and might be primary targets of LNK1/LNK2 activity. Other clock and light signaling genes were also misregulated in lnk1;lnk2 mutants (Dataset S3C). However, these genes were affected more subtly, suggesting that they might be secondary targets of LNK1/LNK2 activity. Down-regulated genes also included the flowering time genes FLAVIN-BINDING KELCH REPEAT F-BOX 1 (FKF1; Fig. 3F), which was also present in the cluster of genes with peak expression at ZT10 (Fig. 3C), as well as FLOWERING LOCUS T (FT) and SUPPRESOR OF CONSTANS OVEXPRESSION 1 (SOC1) (Fig. S8). All three genes are positive regulators of flowering time, with FKF1 acting upstream of FT and SOC1 (26). Therefore, the late flowering of lnk1;lnk2 mutants under LD is likely due, at least in part, to reduced FKF1 expression, which in turn leads to reduced FT and SOC1 mRNA levels (Fig. S8). FT expression is controlled by the transcription factor CONSTANS (CO), whose transcript and protein levels are independently regulated by FKF1 (26, 27). CO transcript levels were only slightly reduced in lnk1;lnk2 mutants throughout the afternoon of a long day (Fig. S8), suggesting that the strong down-regulation of FT mRNA levels in the lnk1;lnk2 mutants might result from an effect of FKF1 on CO protein (27).
PRR5 Expression Is Severely Affected in lnk1;lnk2 Mutants Under Free-Running Conditions.
To investigate the effect of LNK1 and LNK2 on the central clock in more detail, we analyzed the expression of clock components in plants that had been entrained under 12-h light/12-h dark cycles at 22 °C and were then transferred to constant light and temperature (i.e., free-running) conditions. The plant circadian clock is based on interlocking transcriptional feedback loops in which the morning clock factors CCA1 and LATE ELONGATED HYPOCOTYL (LHY) repress the expression of evening clock genes such as TOC1/PRR1 (28). In addition, CCA1 and LHY also promote the expression of PRR9 and PRR7 (29), which, sequentially with PRR5 and TOC1/PRR1, repress CCA1 and LHY expression throughout the remaining of the day and early night (30–32).
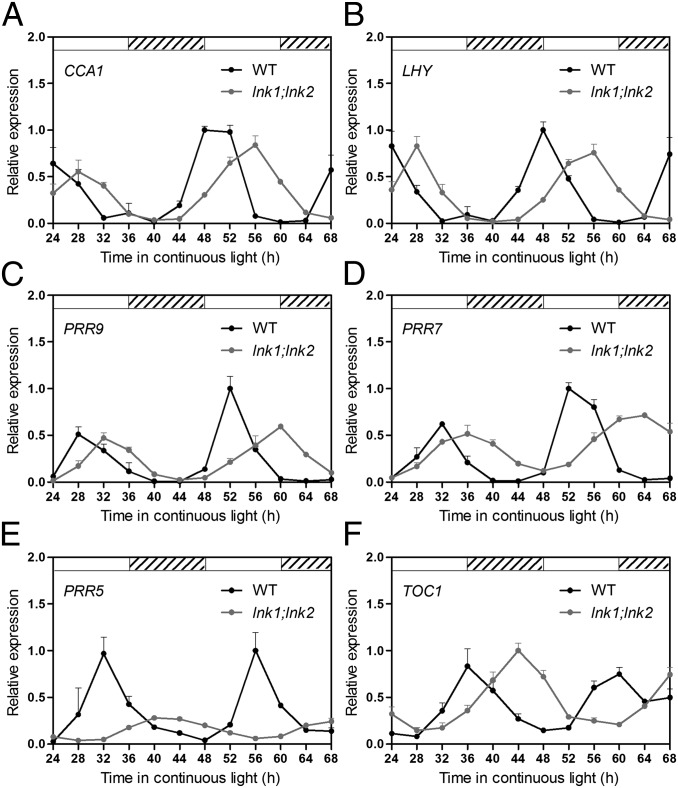
We observed a substantial delay in the phase of CCA1, LHY, PRR9, and PRR7 expression during the second day in continuous light. The delay increased to 8 h on the third day, consistent with a lengthening of circadian period by ∼2.5 h in the lnk1;lnk2 mutant compared with WT plants (Fig. 4 A–D; Fig. S6F). Despite the strong effect of LNK1 and LNK2 on the period and/or phase of circadian oscillations, the overall mRNA levels of morning and early afternoon clock components were largely unaffected in lnk1;lnk2 mutants. In contrast, significant down-regulation coupled to a much longer delay in the phase of expression, i.e., close to 12 h on the third day, was observed for PRR5, which is normally expressed in the afternoon (Fig. 4E). A similar phase delay, but lacking differences in the overall mRNA levels, was observed for the TOC1/PRR1 gene that is expressed slightly after PRR5 (Fig. 4F). A strong delay in the timing of TOC1 expression, coupled with a slight reduction in overall levels, was observed for this clock gene under LD conditions (Fig. S9). Taken together, these results suggest that LNK1 and LNK2 act initially as transcriptional activators, controlling the levels and timing of expression of a subset of genes with peak expression in the afternoon, such as PRR5 (Figs. 3D and 4E), ELF4 (Fig. 3E), TOC1 (Fig. S9), and FKF1 (Fig. 3F), which later affect the rhythmic expression of other core-clock and clock-output genes.
Fig. 4.
LNK1 and LNK2 are necessary for the proper function of the circadian clock. CCA1 (A), LHY (B), PRR9 (C), PRR7 (D), PRR5 (E), and TOC1 (F) mRNA expression measured by qRT-PCR in plants grown under 12-h light/12-h dark cycles and then transferred to continuous light. Values are expressed relative to PP2A and normalized to the maximum value of each gene. Data represent average +SEM (n = 4). Open and hatched boxes indicate subjective day and subjective night periods, respectively.
LNK1 and LNK2 Are Repressed by Members of the TOC1/PRR1 Family of Clock Genes.
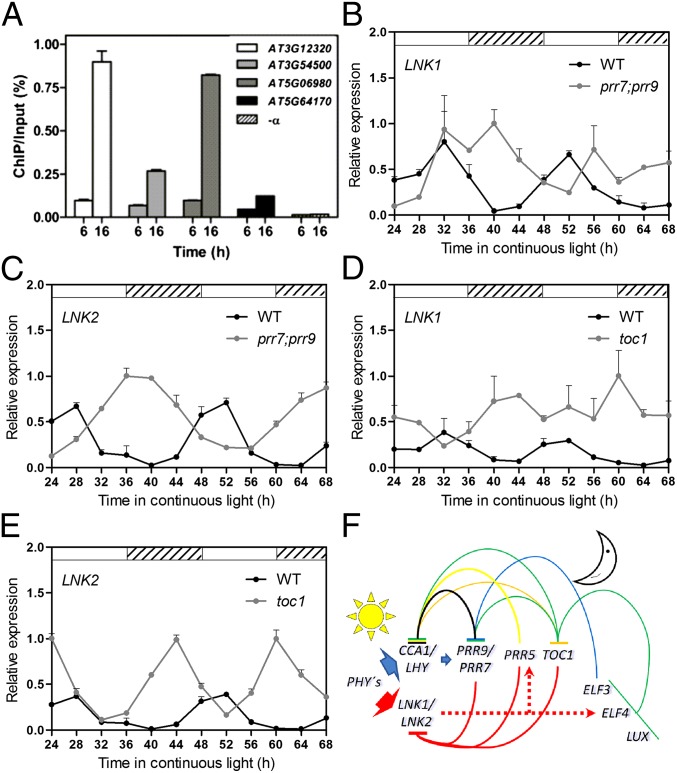
Many clock-regulated genes with peak expression in the morning are repressed throughout the day and during the early night by members of the TOC1/PRR1 family of clock proteins. To determine whether the LNKs were regulated by members of this protein family, we reexamined data describing TOC1/PRR1 and PRR5 binding sites in the Arabidopsis genome using ChIP followed by sequencing (ChIP-seq) (31, 33). Indeed, we found that the regulatory region of LNK3 was directly bound by TOC1/PRR1 (31). ChIP followed by qPCR not only confirmed this, but also revealed that TOC1/PRR1 binds directly to the regulatory regions of LNK1, LNK2, and LNK4 (Fig. 5A). Furthermore, PRR5, PRR7, and PRR9 were also found to bind directly to the regulatory regions of LNK1–4 (33).
Fig. 5.
LNK1 and LNK2 are repressed by the TOC1/PRR1 family of circadian clock components. (A) TOC1 binds to LNK1–4 gene promoters. ChIP-qPCR assays were conducted using TOC1. Minigene (TMG) seedlings grown under 12-h light/12-h dark cycles. Samples were collected at ZT 6 and ZT16 in the light and dark, respectively. (B–E) LNK1 (B and D) and LNK2 (C and E) expression measured by qRT-PCR in continuous light relative to PP2A (n = 4). Plants were grown under 12-h light/12-h dark cycles and then transferred to continuous light. Error bars indicate +SEM. Open and hatched boxes indicate subjective day and subjective night, respectively. (F) Model showing the proposed function of LNK1 and LNK2 in the circadian clock. Light regulates LNK1 and LNK2 expression in the morning, which then act to promote, directly or indirectly (dashed line), the expression of a subset of afternoon genes, including the core clock genes PRR5 and ELF4. During the afternoon and early evening, PRR9, PRR7, PRR5, and TOC1 bind to the LNK promoters blocking their expression.
To evaluate the functional consequence of the binding of these factors to LNK1–4 promoters, we compared the expression patterns of LNK1 and LNK2 in WT, toc1, or prr9;prr7 mutant plants, entrained under light/dark cycles and then transferred to constant light conditions. Not only did we observe progressively larger delays in the phase of the circadian oscillations of LNK1 and LNK2, but their mRNA levels were increased in the prr9;prr7 double mutant at the trough of the circadian oscillations (Fig. 5 B and C). A larger overall increase in LNK1 and LNK2 mRNA levels, coupled to progressive phase advances, was also observed in the short period mutant toc1 over the entire time course (Fig. 5 D and E), indicating that TOC1 is a direct repressor of these genes.
Discussion
Light and the circadian clock interact to regulate many biological processes in plants, such as flowering time (25) and stem growth (18, 19). In addition, this interaction is also required for robust functioning of the circadian clock itself (15, 20). Our genomewide analysis revealed that these physiological interactions are mirrored by global interactions at the transcriptional level. In particular, we found that 70% of light-induced genes responded more strongly to a light pulse during the subjective day than during the night, likely optimizing the energy spent on light-dependent biological processes that have maximal activity at midday, when light intensity is at its peak under natural conditions. At the same time, a light stimulus during the night preferentially promoted the expression of certain key clock components, consistent with the general observation that light present at the beginning or end of the photoperiod adjusts the circadian clock to seasonal changes in day length (2, 21).
The characterization of genes that are preferentially induced by light at night and that are also rhythmic across multiple conditions led to the identification of LNK genes, a partially redundant family of plant-specific genes that control photomorphogenic and photoperiodic responses, as well as circadian rhythms. LNK1 and LNK2 are regulated by the phytochrome photoreceptors and predominantly affect photomorphogenic responses to red light, pointing to an important role in phytochrome signaling. Additionally, LNKs are expressed rhythmically with peak expression in the morning or at noon, likely due to their repression by members of the TOC1/PRR1 family of core clock regulators during the afternoon and early night. Thus, LNK1 and LNK2 link phytochrome and circadian signaling to regulate many physiological processes, including time keeping by the clock itself.
A comparison of LNK genes with other light-induced clock genes or regulators is informative. Like LNK genes, CCA1 and LHY are light-induced genes whose mRNAs reach peak levels in the early morning (9, 34). Mutations in CCA1 and LHY, however, shorten the period of circadian rhythms, whereas mutations in LNK1 and LNK2 lengthen it (6). GI is a light-induced clock regulator, and loss-of-function mutations in this gene lengthen circadian period but, in contrast to LNK1 and LNK2, GI is expressed with peak levels in the late afternoon (35, 36). Finally, PRR9 and PRR7 are similar to LNK1 and LNK2 in that they are expressed during the morning and early afternoon, are induced by light, and decrease period length and promote flowering (29). Different from LNK2 and LNK2, which at least under constant light do not seem to be required for normal CCA1 and LHY expression (Fig. 4), PRR9 and PRR7 are repressors of CCA1 and LHY (29). Thus, LNK1 and LNK2 act differently from previously described light-induced clock genes or regulators.
LNK1 and LNK2 are plant-specific proteins without recognizable functional domains. This is reminiscent of the clock components ELF3 and ELF4, which only very recently were shown to participate in an evening phased protein complex that represses the expression of a subset of morning genes, such as PRR9 (19, 37–39). The precise mechanism through which LNK1 and LNK2 affect the pace of the clock is uncertain. They activate the expression of afternoon/early evening genes, including PRR5 and ELF4, but the long period phenotype is unlikely to be simply the result of reduced expression of these two genes. If that was the case, lnk1;lnk2 mutants should be either short period or arrhythmic, such as prr5 or elf4 mutants, respectively. Thus, the long period phenotype may result from delayed activation of afternoon/early evening genes rather than simply from, or in addition to, reduced levels of these genes. In summary, our work supports a model in which light perceived through phytochromes activates the expression of the LNKs, as well as that of the CCA1 and LHY (9), in the early morning. CCA1 and LHY then promote the expression of PRR9 and PRR7 (29), whereas LNK1 and LNK2 act later during the day to activate clock genes with peak expression in the afternoon, such as PRR5 and ELF4. Simultaneously, members of the TOC1/PRR family repress these morning genes throughout the afternoon and beginning of the night (30–32, 40). Finally, the progressive reduction in TOC1/PRR levels leave CCA1, LHY, and the LNK genes poised to respond again to light signals that reset the clock every morning (Fig. 5F).
Materials and Methods
Plant Material.
All of the Arabidopsis lines used in this study were Columbia ecotype. lnk1-1 (SALK_024353), lnk1-2 (SALK_063322), lnk1-3 (GK_044A09), lnk2-1 (GK_484F07), lnk2-2 (SALK_116103), and lnk2-3 (SALK_141609) mutants were obtained from the Arabidopsis Biological Research Center (ABRC) and the Gabi Kat T-DNA insertion collections. The lnk1;lnk2 double mutant was obtained by crossing the simple mutants lnk1-1 and lnk2-1. The clock and photoreceptor mutants used in this study were prr7-3;prr9-1, toc1-101, phyA-211;phyB-9, and cry1-b104;cry2-1.
Growth Conditions.
For flowering time experiments, the plants were grown on soil at 22 °C under long days (LD; 16-h light/8-h dark cycles; 70 µmol⋅m−2⋅s−1 of white light), short day (SD; 16-h light/8-h dark cycles; 140 µmol⋅m−2⋅s−1 of white light), or continuous light (LL; 50 µmol⋅m−2⋅s−1 of white light), depending on the experiment.
Physiological Measurements.
Detailed information is in SI Materials and Methods.
Subcellular Localization of LNK1.
Detailed information is in SI Materials and Methods.
qRT-PCR, Microarray, and RNA-Seq Analysis.
Detailed information is in SI Materials and Methods.
ChIP Analysis.
Detailed information is in SI Materials and Methods.
Protein Sequence Alignment and Phylogenetic Analysis.
Detailed information is in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Leonardo Storani, Esteban Beckwith, and Santiago Mora García for technical assistance and valuable discussions. This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica, the International Centre for Genetic Engineering and Biotechnology, and The Howard Hughes Medical Institute (to M.J.Y.) and from the Max Planck Institute (to D.W.). Work in the laboratory of P.M. was supported by grants from the Ramón Areces Foundation, the Spanish Ministry of Science and Innovation, European Heads of Research Councils, and the European Science Foundation through the European Young Investigator award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE43865 and GSE46621).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302170110/-/DCSupplemental.
References
- 1.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol. 2009;7(3):e62. doi: 10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pittendrigh CS, Minis DH. The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am Nat. 1964;98(902):261–294. [Google Scholar]
- 3.Kim WY, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449(7160):356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 4.Más P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408(6809):207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- 5.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282(5393):1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 6.Nagel DH, Kay SA. Complexity in the wiring and regulation of plant circadian networks. Curr Biol. 2012;22(16):R648–R657. doi: 10.1016/j.cub.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khanna R, Kikis EA, Quail PH. EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol. 2003;133(4):1530–1538. doi: 10.1104/pp.103.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makino S, Matsushika A, Kojima M, Oda Y, Mizuno T. Light response of the circadian waves of the APRR1/TOC1 quintet: When does the quintet start singing rhythmically in Arabidopsis? Plant Cell Physiol. 2001;42(3):334–339. doi: 10.1093/pcp/pce036. [DOI] [PubMed] [Google Scholar]
- 9.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93(7):1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 10.Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12(12):2499–2510. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdán PD. Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA. 2010;107(10):4776–4781. doi: 10.1073/pnas.0910446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanovsky MJ, Mazzella MA, Casal JJ. A quadruple photoreceptor mutant still keeps track of time. Curr Biol. 2000;10(16):1013–1015. doi: 10.1016/s0960-9822(00)00651-5. [DOI] [PubMed] [Google Scholar]
- 13.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290(5499):2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 14.Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA. 1996;93(26):15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature. 2000;408(6813):716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- 16.Hall A, et al. The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell. 2003;15(11):2719–2729. doi: 10.1105/tpc.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13(6):1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448(7151):358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 19.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475(7356):398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Covington MF, et al. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001;13(6):1305–1315. doi: 10.1105/tpc.13.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CH. Forty years of PRCs—what have we learned? Chronobiol Int. 1999;16(6):711–743. doi: 10.3109/07420529909016940. [DOI] [PubMed] [Google Scholar]
- 22.Casal JJ, Yanovsky MJ. Regulation of gene expression by light. Int J Dev Biol. 2005;49(5-6):501–511. doi: 10.1387/ijdb.051973jc. [DOI] [PubMed] [Google Scholar]
- 23.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4(2):e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malapeira J, Khaitova LC, Mas P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc Natl Acad Sci USA. 2012;109(52):21540–21545. doi: 10.1073/pnas.1217022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanovsky MJ, Kay SA. Living by the calendar: How plants know when to flower. Nat Rev Mol Cell Biol. 2003;4(4):265–275. doi: 10.1038/nrm1077. [DOI] [PubMed] [Google Scholar]
- 26.Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 27.Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336(6084):1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alabadí D, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293(5531):880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 29.Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15(1):47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 30.Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA. 2012;109(8):3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336(6077):75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 32.Nakamichi N, et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22(3):594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamichi N, et al. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA. 2012;109(42):17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaffer R, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93(7):1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 35.Fowler S, et al. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18(17):4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park DH, et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285(5433):1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 37.Dixon LE, et al. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol. 2011;21(2):120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helfer A, et al. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol. 2011;21(2):126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrero E, et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell. 2012;24(2):428–443. doi: 10.1105/tpc.111.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pokhilko A, et al. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol. 2012;8:574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.