Abstract
Background and Aims
The evolution of seeds together with the mechanisms related to their dispersal into the environment represented a turning point in the evolution of plants. Seeds are produced by gymnosperms and angiosperms but only the latter have an ovary to be transformed into a fruit. Yet some gymnosperms produce fleshy structures attractive to animals, thus behaving like fruits from a functional point of view. The aim of this work is to increase our knowledge of possible mechanisms common to the development of both gymnosperm and angiosperm fruits.
Methods
B-sister genes from two gymnosperms (Ginkgo biloba and Taxus baccata) were isolated and studied. The Ginkgo gene was also functionally characterized by ectopically expressing it in tobacco.
Key Results
In Ginkgo the fleshy structure derives from the outer seed integument and the B-sister gene is involved in its growth. In Taxus the fleshy structure is formed de novo as an outgrowth of the ovule peduncle, and the B-sister gene is not involved in this growth. In transgenic tobacco the Ginkgo gene has a positive role in tissue growth and confirms its importance in ovule/seed development.
Conclusions
This study suggests that B-sister genes have a main function in ovule/seed development and a subsidiary role in the formation of fleshy fruit-like structures when the latter have an ovular origin, as occurs in Ginkgo. Thus, the ‘fruit function’ of B-sister genes is quite old, already being present in Gymnosperms as ancient as Ginkgoales, and is also present in Angiosperms where a B-sister gene has been shown to be involved in the formation of the Arabidopsis fruit.
Keywords: B-sister gene, fruit growth, fruit-like structure, Ginkgo biloba, MADS-box genes, Taxus baccata
INTRODUCTION
MADS-box genes are widespread in eukaryotes (Theissen et al., 1996). However, they are especially important in higher plants because the specification of the various floral components is under the control of different types of MADS-box genes that play homeotic roles in such a process (Ng and Yanofsky, 2001; Krizek and Fletcher, 2005; Theissen et al., 2000). Various tens of MADS-box genes can be found in the genome of the flowering plants (i.e. Angiosperms), while at present a lesser number of the same genes are present in Gymnosperms. In the latter plants the formation of the male and female strobili follows a model that, albeit simplified, is basically similar to the one operating in Angiosperms (Theissen and Melzer, 2007; Wang et al., 2010).
A particular type of MADS-box gene, named B-sister, was identified for the first time in the female cones of Gnetum gnemon (Becker et al., 2002). Subsequently, B-sister genes have been found and characterized also in some Angiosperms such as Arabidopsis (Nesi et al., 2002), Petunia (de Folter et al., 2006) and wheat (Yamada et al., 2009). In particular, in Angiosperm species it has been shown that B-sister genes are important for correct differentiation of the ovule/seed (Nesi et al., 2002; Kaufmann et al., 2005; de Folter et al., 2006; Yamada et al., 2009; Mizzotti et al., 2012) but also for the formation of the fruit in Arabidopsis (Prasad et al., 2010).
In fact, in Arabidopsis two different B-sister genes have been identified and studied: TRANSPARENT TESTA 16 (TT16) and GORDITA (GOA). In the case of TT16, Nesi et al. (2002) were able to show that the protein encoded by this gene is necessary for the correct development and pigmentation of the seed tegument. For GORDITA, the gene contributes to the early development of the ovule outer integument but seems especially important for the growth of fruits. In fact, in the loss-of-function gordita mutant the fruit appeared larger than those of the wild-type, while plants over-expressing this gene had smaller fruits than the wild-type, a finding that led the authors to postulate a role as a controller of fruit growth for GORDITA (Prasad et al., 2010; Prasad and Ambrose, 2010).
The GORDITA gene was also studied by other researchers who were able to demonstrate that it represents a recent duplication, and also that this duplication seems to be restricted to the Brassicaceae family of higher plants (Erdmann et al., 2010). GORDITA is not the only MADS-box gene involved in the development of fruits. For instance, C-class and SEPALLATA genes have also been demonstrated to play fundamental roles, particularly in the case of fleshy fruits. With regard to the latter, data are available relating to the development and ripening of fruits in tomato (Itkin et al., 2009; Vrebalov et al., 2009), peach (Tadiello et al., 2009), oil palm (Tranbarger et al., 2011), strawberry (Seymour et al., 2011), bilberry (Jaakola et al., 2010) and others.
As fruit proper derive from an ovary following a fertilization event, their development is normally studied in Angiosperms, the sole flower-producing plants. However, besides being the first taxon in which a B-sister gene was discovered, Gymnosperms represent an interesting group for the study of fruit evolution because in some species the seeds are accompanied by fleshy structures that may favour their dispersal through endozoochory, thus behaving like ‘fruits’ from a functional point of view. In particular, the Gymnosperm species Ginkgo biloba and Taxus baccata represent two different and interesting experimental models because the fleshy fruit-like structures produced by them have a different anatomical origin. In Ginkgo it is the outermost seed integument (sarcotesta) that grows and becomes fleshy, while in yew the fleshy aril that almost completely encloses the seed is formed de novo by an outgrowth of the peduncle at the basis of the ovule (Lovisetto et al., 2012).
In these two species it has recently been shown that various MADS-box genes belonging to similar subgroups are involved in the development of the fruit-like structures that surround their seeds (Lovisetto et al., 2012). The involvement of a common regulatory gene network in the development of fruit-like structures with different anatomical origin, in two species that are also quite distant from a phylogenetic point of view, made this finding of particular interest. As related MADS-box genes are known to be involved also in the development of the fleshy fruit of Angiosperms, the existence of a common molecular mechanism underlying the formation of the fleshy fruit habit was postulated, at least as far as the studied MADS-box genes are concerned (Lovisetto et al., 2012).
In the present work we have investigated the presence of B-sister genes in the female reproductive structures of G. biloba and T. baccata. A B-sister gene has been evidenced in the ovules of both species, although only in Ginkgo does this gene appear to be particularly involved in the development of the fleshy fruit-like structure that surrounds the seed. The possible role played by the Gymnosperm B-sister genes in fruit growth has also been studied by means of over-expression experiments in tobacco plants. Given its involvement in the growth of the fruit-like structure, the sole B-sister gene from Ginkgo has been used. Interestingly, the involvement of this gene in the growth of the fruit has been confirmed also in tobacco.
METHODS
Plant material and RNA extraction
The Gymnosperm plant material came from the Botanic Garden of Padua. In yew the aril develops as a collar at the base of the ovule, which then becomes visible as a green leafy structure which grows thick and fleshy and starts gradually to develop a reddish colour (Supplementary Data Fig. S1). In Ginkgo initial samples consisted of whole ovules, and the fleshy sarcotesta could then be analysed separately at various development stages as indicated by dates in the figures, and a characterization of the various samples is shown in Supplementary Data Fig. S2. Total RNA was extracted from different tissues according to Chang et al. (1993). For each tissue the extraction was made using a pool of samples and was repeated at least twice. RNA yield and purity were checked by means of UV absorption spectra, whereas RNA integrity was ascertained by electrophoresis in agarose gel.
cDNA isolation, sequencing, phylogenetic analysis and protein analysis
Taxus baccata B-sister cDNA (TbBS) was obtained by RT-PCR using primers constructed following alignment of known sequences (Supplementary Data Table S1) and the template consisted of RNA extracted from young ovules. After amplifications, the fragments were cloned into a vector and then sequenced. DNA sequencing was performed at BMR Genomics (Padua). Sequence manipulations, analyses and alignments were performed using the ‘Lasergene’ software package (DNASTAR). The sequence was submitted to GenBank (accession number JX564539).
A Ginkgo biloba B-sister cDNA (GBM10) was identified from the GenBank nucleotide database (accession no. AB029472) with blastn alignments using GGM13 (accession no. AJ132219) as query.
To construct a phylogenetic tree, a set of MADS-box protein sequences downloaded from GenBank plus the sequences studied in this work was used. The GenBank accession numbers are: JX564539 (Taxus baccata TbBS); AJ132219 (Gnetum gnemon GGM13); AB029472 (Ginkgo biloba GBM10); AB035567 (Chara globularis CgMADS1); AF335242 (Petunia × ibrida FBP24); AJ307056 (Anthirrinum majus DEFH21); NM_203094 (Arabidopsis thaliana AGL32/ABS); NM_001198191 (Arabidopsis thaliana GORDITA/GOA/AGL63); AJ271208 (Zea mays ZMM17). The tree was constructed using the ‘MIK domain’, i.e. the MADS domain (60 amino acids) plus most of the I and K domains (90 amino acids) similarly to Winter et al. (1999), Becker and Theissen (2003) and Melzer et al. (2010). The amino acid sequences were aligned with the MUSCLE program and the obtained alignments were used to construct the tree with the MEGA5 program. The tree was constructed with the neighbour-joining method (Saitou and Nei, 1987) and evaluated by bootstrap analysis.
To compare the probability to form coiled-coils in the K domain, Taxus baccata TbBS, Ginkgo biloba GBM10, Arabidopsis thaliana TT16/ABS (accession no. NM_203094) and Arabidopsis thaliana GOA/AGL63 (accession no. NM_001198191) sequences were analysed. The Pfam database was used to identify the K domains (Punta et al., 2012); the database was available on line at http://pfam.sanger.ac.uk/ and these domains were used for coiled-coil prediction with the COILS program (Lupas et al., 1991) available on line at http://www.ch.embnet.org/software/COILS_form.html.
Analysis of gene expression
This analysis was performed by standard real time PCR. Six micrograms of total RNA was pre-treated with 2 U of DNase I (Promega). The first-strand cDNA was synthesized from 3 µg of the DNase I-treated RNA by means of the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA), using random hexamers as primers. The internal standards consisted of internal transcribed spacer (ITS) sequences specific for each species. Primers used for this analysis are available in Supplementary Data Table S1. PCR was carried out with the Gene Amp 7500 Sequence Detection System (Applied Biosystems). The obtained CT values were analysed by means of the Q-gene software by averaging three independently calculated normalized expression values for each sample. Expression values are given as the mean of the normalized expression values of the triplicates, calculated according to equation 2 of the Q-gene software (Muller et al., 2002).
Microscopy and in situ hybridization analysis
Tobacco leaves and fruits were observed without any treatment under low-pressure conditions by means of environmental scanning electron microscopy (ESEM) at the CUGAS facilities (University of Padua).
For histological analysis and for in situ hybridization, the samples were fixed with 2 % formaldehyde and 0·25 % glutharaldehyde in PBS buffer (pH 7·5). Clearing, infiltration, embedding and sectioning were performed according to Drews (1998). Paraffin was removed by washing twice with xylene. For histological observation the sections were stained with 0·1 % toluidine blue in water and extensively washed.
For in situ hybridization, pre-hybridization treatments and post-hybridization washes were performed according to Drews (1998), with minor modifications. PCR fragments of C domain were isolated, cloned and checked by sequencing. The primers used for the PCR are listed in Supplementary Data Table S2. The PCR products were cloned and the sequence validated. The digoxigenin (DIG)-labelled RNA sense and antisense probes were synthesized with the DIG RNA labelling Kit SP6/T7 (Roche Diagnostics, Germany), the immunological detection was performed with the anti-Digoxigenin-AP-Fab fragments antibody (Roche Diagnostics), and the SIGMA FAST BCIP/NBT tablets (Sigma, St Louis, MO, USA) were used as phosphatase substrate, all according to the manufacturers' instructions.
Transformation of tobacco
The ginkgo GBM10 cDNA was cloned into the pBin-AR vector (Hoefgen and Willmitzer, 1988). The resulting binary plasmid was inserted in Agrobacterium tumefaciens (strain LBA4404) cells that were used to transform tobacco according to Fisher and Guiltinan (1995). Kanamycin-resistant plants have been confirmed for the presence of the transgene by means of PCR.
RESULTS
cDNAs coding for B-sister transcription factors were obtained for both Ginkgo and yew although with different approaches. By considering that these genes are important for ovule formation (Becker et al., 2002; Nesi et al., 2002; de Folter et al., 2006), in the case of yew RNA extracted from young ovules was used as starting material to clone a cDNA that was named TbBS (Taxus baccata B-sister). Regarding Ginkgo, many sequences coding for MADS-box genes are present in public databases so an in silico search was made and a B-sister encoding full-length cDNA (GBM10) was singled out.
The two sequences were used together with other known Gymnosperm and Angiosperm B-sister sequences to construct a phylogenetic tree (Fig. 1). As expected, the Gymnosperm B-sister sequences formed a separate clade while, within the Angiosperm sequences, the monocotyledonous maize appeared separated from the dicotyledonous species.
Fig. 1.
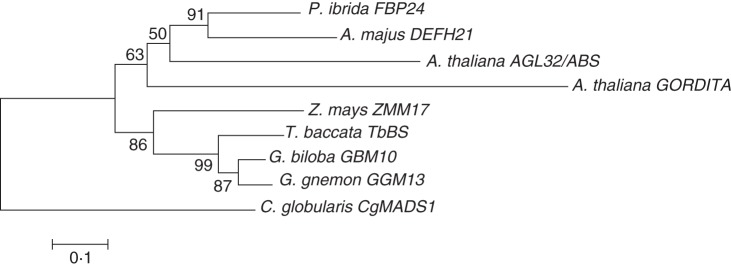
Phylogenetic tree showing the relationships between Gymnosperm and Angiosperm B-sister genes. Gymnosperm sequences form a group while Angiosperm sequences show a separation between monocotyledons and dicotyledons.
In very young ovules, expression of the two genes was analysed by in situ hybridization experiments. As soon as the structures destined to become fleshy could be isolated, gene expression was analysed by real-time PCR.
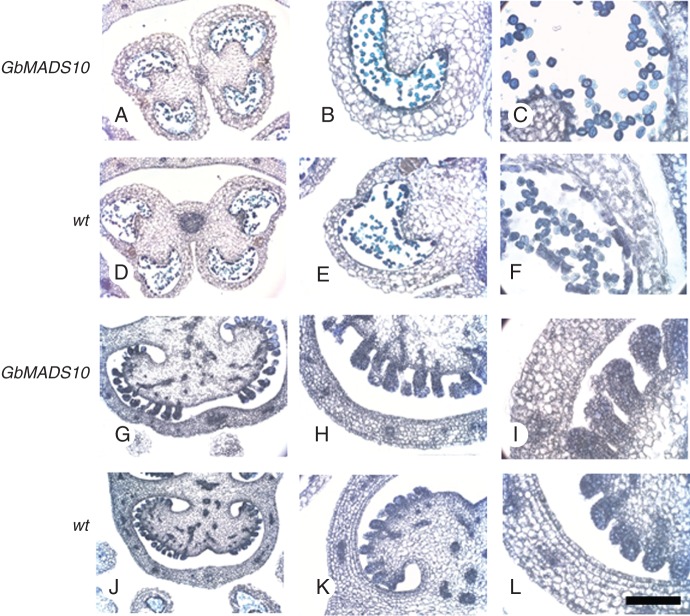
In the young reproductive structures of yew, when the ring-shaped aril primordium was just detectable as a very reduced structure protruding from the basal part of the ovule, the B-sister gene was weakly and uniformly expressed throughout the whole ovule, in the emerging aril primordium and in the distal portion of the peduncle (Fig. 2A, B). A similar weak and uniform expression level was detectable later in development, when at the base of the young ovule the growing aril primordium was more clearly visible (Fig. 2D). In Ginkgo a much stronger B-sister expression was detectable in the young ovules and in the distal part of the receptacle (Fig. 2F); a strong level of expression was maintained throughout the ovule also in later stages of development, when a layered organization of the integument had clearly developed, and both the inner and the outer integuments could be distinguished (Fig. 2H).
Fig. 2.

Expression of B-sister analysed by in situ hybridization in young Taxus baccata (A–E) and Ginkgo biloba (F–I) female reproductive structures. S, sense probe; α, antisense probe; oi, outer integument; ii, inner integument. Asterisks: young aril primordium.
Real-time PCR analyses showed that no transcripts of the B-sister genes could be detected in either leaves or male strobili of both Ginkgo and yew, while expression of the gene was observed in whole ovules of both species (Fig. 3A, B), thus confirming the in situ hybridization data. However, in Ginkgo the GBM10 gene was generally expressed at much higher levels than in yew and continued to be expressed during growth of the fleshy fruit-like structure. In particular, in the pulp separated from the developing stone the transcript level increased steadily to a maximum in the 23/5 sample where it reached levels of expression about fourfold greater than those present in the young ovules, and gradually decreased afterwards so that in the 4/8 sample they were almost undetectable, and no signal was visible in the ripening pulp (Fig. 3A).
Fig. 3.
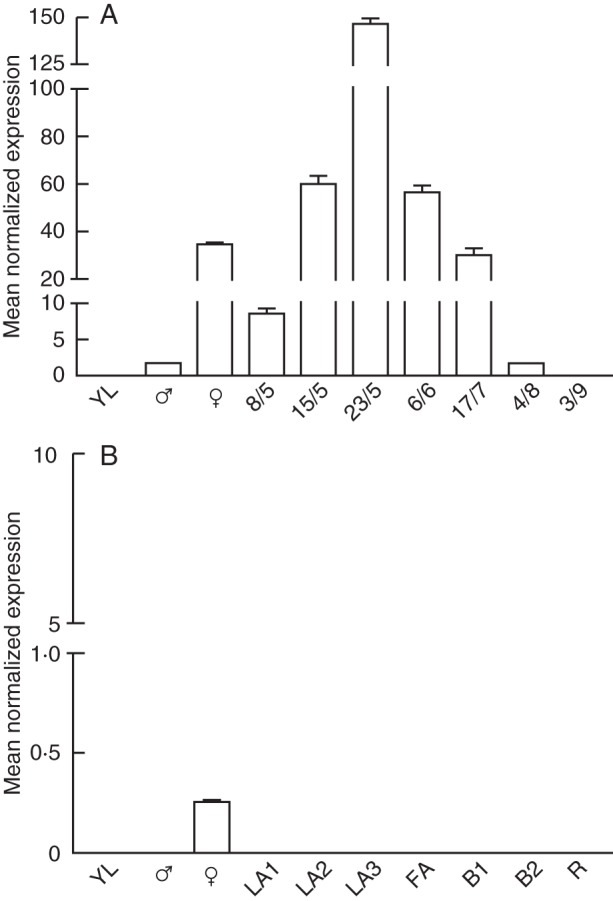
Relative expression profiles of G. biloba (A) and T. baccata (B) B-sister MADS-box genes. Ginkgo biloba samples: YL, young leaf; ♂, male strobili; ♀, young ovules; 8/5, 15/5, 23/5, 6/6, 17/7, 4/8, 3/9, sampling dates (2010 season) of pulps at different developmental stages. Taxus baccata samples: YL, young leaf; ♂ male strobili; ♀ young ovules; LA1, LA2, LA3, leafy arils at different stages of development; FA, fleshy green arils; B1, breaker-1 arils; B2, breaker-2 arils; R, red ripe arils. Values (means of the normalized expression) have been obtained by real-time PCR analyses. Bars are the standard deviations from the means.
As regards yew, the expression was analysed in arils at increasing stages of growth, two of them still occurring below the ovule bracts and four being visible outside the bracts and ranging from green leafy arils (LA3) to the fleshy red ripe arils. In both the very young arils masked by the ovule bracts (LA1 and LA2, Fig. 3B ) and in all the subsequent stages of aril development the TbBS transcripts were undetectable.
From the expression data it appeared that only the Ginkgo B-sister gene was significantly involved in the growth of the fruit-like structure. Therefore, the role of the sole Ginkgo B-sister gene was further appraised by over-expressing it in tobacco. The transformation experiments yielded 25 clones that were checked by PCR for the presence of the transgene (data not shown). Twelve transgenic clones were further grown in pots until flowering.
The most evident phenotype was a reduction in size of the transgenic seedlings compared with wild-type seedlings (Fig. 4A) and, in a few cases, also a loss of apical dominance with growth of lateral branches was observed (not shown). Transgenic leaves were generally smaller than wild-type leaves and, in a few cases, they also had a curled appearance (Fig. 4B, C). The epidermal cells of the small transgenic leaves were examined by means of ESEM and they appeared to be much larger than the corresponding wild-type cells (Fig. 4D, E).
Fig. 4.
Phenotypic analysis of 35S::GBM10 plants and comparison with wild-types seedlings. In (A) both wild-type (WT) and 35S::GBM10 plants of the same age are shown. Details of wild-type (F) and transgenic (G) shoot apices. In (B) and (C) leaves from the same internode of WT and 35S::GBM10 plants at the same age are visible. Panels (D, transgenic) and (E, WT) show ESEM images of leaf adaxial surfaces (scale bar 100 μm for both images).
The transgenic plants started to flower at a very reduced size (Fig. 4G, F), although the flowers had no apparent macroscopic defect in the transgenic seedlings. Anthers and ovaries of both transgenic and wild-type mature flowers were analysed by light microscopy using the same enlargement. To analyse flowers at comparable developmental stages, only flowers whose anthers contained young pollen grains and a largely degenerated tapetum were analysed (Fig. 5C, F). The anthers of the B-sister over-expressing flowers had pollen sacs whose enclosure looked much thicker than the corresponding wild-type one (Fig. 5A, D). The increased thickness appeared to be due mainly to the presence of radially enlarged cells, while no apparent difference was detectable in the number of cell layers (from three to four in the outer portion of the anthers; Fig. 5B, E). Furthermore, because all the analysed flowers were at comparable stages of development (stage 3–4 according to Koltunov et al., 1990), it was of particular interest that in the transgenic young ovaries the ovules appeared much larger than the corresponding wild-type ovules (Fig. 5G–I, J–L).
Fig. 5.
Anthers (A–F) and ovaries (G–L) from tobacco plants overexpressing GBM10 (A–C, G–I) and from wild-type plants (D–F, J–L). Scale bars: (A, D, G, J) = 400 µm; (B, E, H, K) = 200 µm; (C, F, I, L) = 100 µm.
Macroscopic anomalies started to become evident during subsequent development of the fruit. In particular, the very short transgenic seedlings yielded fruits that were very small and had a wrinkled surface compared with wild-type seedlings (Fig. 6A, D). These fruits were apparently unable to bear seeds and appeared empty at maturity (Fig. 6E). An ESEM analysis revealed massive abortion, as judged by the numerous remnants of undeveloped ovules/seeds compared with the wild-type (Fig. 6F and 6C, respectively).
Fig. 6.
Phenotypic analyses of 35S::GBM10 fruits. In (A), (B), (D) and (E) different stages of fruit development of wild-type (A, B) and 35S::GBM10 (D, E) tobacco plants are shown. Panels (C) and (F) are two ESEM images showing ovules from wild-type (C) and 35S::GBM10 (F) tobacco plants (scale bar = 1 mm).
To understand whether the reduced fertility was due to defects in either the male or the female parts, cross-pollination experiments were carried out between transgenic and wild-type seedlings. Moreover, the viability of the transgenic pollen was assayed as indicated by Rodriguez-Riano and Dafni (2000), Khatum and Flowers (1995) and Norton (1996). For this, about 65 % of the transgenic pollen appeared viable compared with the 90 % of the wild-type pollen (Supplementary Data Fig. S3) so the transgenic flowers had more than enough viable pollen for fertilization of the ovules. As regards the cross-pollination experiments, in both cases they yielded fruits of quasi normal size that contained comparable amounts of seeds (Fig. 7A) whose dimensions were comparable to those of the wild-type seeds (Fig. 7B). The vitality of the above seeds was assayed by germination experiments: we found 80 % germination in the case of seeds derived by a cross between wild-type pollen and transgenic pistils (Fig. 8A, B), while 88 % germination was obtained for seeds derived as a result of crosses between transgenic pollen and wild-type pistils (Fig. 8C, D).
Fig. 7.

Seeds obtained from cross-pollination experiments. The table in (A) shows the average number of seeds per fruit. For each cross, the seeds of five different capsules were counted. The values are given with the standard deviation. Seeds obtained from the various cross-pollination experiments are shown in (B). Scale bar = 1000 µm.
Fig. 8.
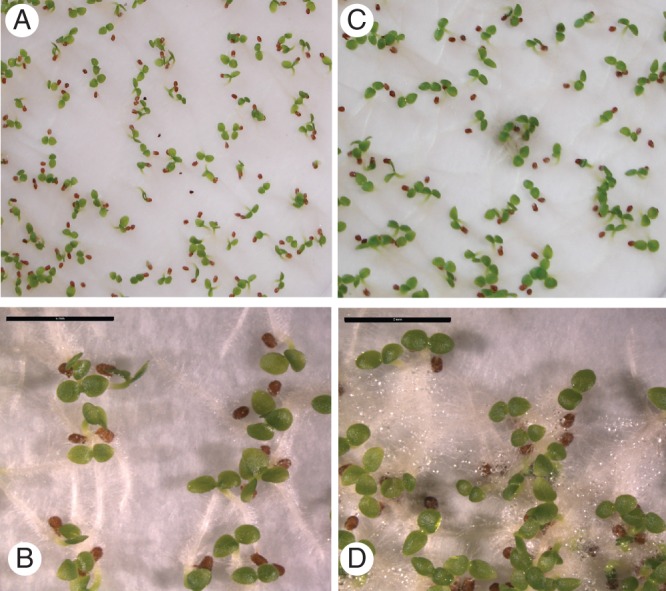
Germination experiments. (A, B) Germinating seeds obtained from cross-pollination using wild-type pollen to hand-pollinated transgenic pistils. (C, D) Germinating seeds obtained from cross-pollination using transgenic pollen to hand-pollinated wild-type pistils (scale bar = 5 mm).
DISCUSSION
Thanks to the development of the seed, Spermatophytes (i.e. seed plants) experienced great evolutionary success because this new structure increased enormously their capacity to spread to new habitats (Taylor et al., 2009). Given its importance, the process of seed dispersal went through a series of specializations aimed at improving its efficiency. Thus, a series of structures evolved, the pinnacle today being the differentiation of the Angiosperm typical fruit.
Among the various Angiosperm fruits, the fleshy fruits represent a specialized typology because, by being sought by frugivorous animals, they exploit the animals' mobility to have their seeds dispersed over great distances through the animal faeces. Fleshy structures that attract frugivorous animals and behave like fruits from a functional point of view are actually present also in various Gymnosperm species, and the existence of a common molecular regulatory network, formed by a number of MADS-box genes, has recently been shown to be active during the differentiation and ripening of fleshy structures in both Gymnosperms and Angiosperms (Lovisetto et al., 2012).
In Arabidopsis the ABS/TT16 MADS-box gene participates in correct development of the seed (Nesi et al., 2002). This gene belongs to the B-sister group and in Arabidopsis a second copy, named GORDITA, has recently been identified that seems to be present in the sole Brassicaceae family. GORDITA has been postulated to have acquired de novo a ‘fruit’ function (Erdmann et al., 2010) while ABS/TT16 seems to have maintained the ‘seed’ function (Nesi et al., 2002). Given that, apart from the Brassicaceae, one copy of B-sister is present both in Gymnosperms and in Angiosperms, which general function might be ascribed to this common gene?
As regards the two Gymnosperm species studied here, the gene is not expressed in either leaves or male strobili while it is expressed in young ovules, thus confirming the ovule specificity of the B-sister genes (Becker et al., 2002; de Folter et al., 2006). In particular, in Ginkgo in situ hybridization analysis has shown that expression tends to increase at the level of the inner integument, i.e. the tissue that will later become the sclerotesta and will thus have an important role in protection of the future embryo. Note that in Arabidopsis the ABS/TT16 gene is especially expressed in the innermost layer of the internal integument, known as the endothelium (Mizzotti et al., 2012). Thus, it appears that the ovule/seed function of the B-sister genes is very ancient and has been maintained in the course of plant evolution. Also in yew the B-sister gene is expressed at the level of ovule and integument, further confirming the role played by these genes in the development of seeds.
In the case of GORDITA, Erdmann et al. (2010) showed that the gene represents a recent duplication and concluded that its ‘fruit’ function represents a neo-functionalization that is likewise recent. The expression data of B-sister in yew appear in agreement with the above idea as the gene transcripts are practically undetectable in the fruit-like aril throughout its growth.
The situation in the more ancient Ginkgo is quite different where, besides being generally present at much higher levels than in yew, the GBM10 gene transcripts show an increase that parallels the increase in size of the pulp, with levels of expression in the fast growing fleshy structure becoming about four-fold greater than those present in the whole young ovules. Thus, the pattern of increasing expression indicates clearly that in Ginkgo the B-sister gene must play a role in the growth of the fleshy tissue surrounding the seed. The results of the ectopic expression of this gene in tobacco confirms that it is involved in the formation of fruits, and that its role is a positive one.
The over-expression of GORDITA in Arabidopsis led to the production of smaller seedlings, siliques and leaves. In particular, the epidermal cells of these leaves were smaller than the wild-type leaves, and therefore the hypothesis was made that GORDITA negatively controls growth, and this was in agreement with the larger siliques produced by the loss-of-function gordita mutant (Prasad et al., 2010).
Similarly to what observed in Arabidopsis, in tobacco the plants over-expressing the Ginkgo B-sister gene were shorter and yielded fruits that were smaller than those of the wild-type, and had leaves with reduced size. However, in this case, the smaller leaf size accompanied epidermal cells that were much larger than the corresponding wild-type cells. Moreover, larger cells were found also at the level of the tissue enclosing the pollen sacs, and transgenic young ovules in flowers at comparable stages of development (stage 3–4 according to Koltunov et al., 1990) also appeared much larger than the corresponding wild-type ovules. The latter findings are in contrast to those in Arabidopsis for GORDITA, and we have no explanation for this discrepancy. However, given the increasing gene expression that accompanies the growth of the Ginkgo fruit-like structure, it appears that the B-sister gene may play a positive role in fruit development. The wrinkled appearance of the small capsules observed in the transgenic tobacco might be the consequence of the massive ovule/seed abortion that would thus be unable to support normal growth of the fruit.
It appears that the Ginkgo B-sister gene must have characteristics differentiating it from GORDITA in spite of their common involvement in the formation of fruits. This idea seems to be supported by the fact that in a few cases the tobacco plants over-expressing the Ginkgo GBM10 gene produced curly leaves, a characteristic that was found in Arabidopsis over-expressing ABS/TT16, the other B-sister gene of this species.
In contrast to wild-type fruits that at maturity appeared packed with seeds, the small and wrinkled tobacco capsules appeared empty. ESEM analysis showed that in the capsules of the transgenic plants the apparently empty area was actually full of aborted ovules still attached to the placenta. Hence, over-expression of the Ginkgo GBM10 gene appears to have had a dramatic effect on the development of ovules/seeds.
The Arabidopsis B-sister (ABS/TT16) loss-of-function mutant yielded seeds that were normally viable, albeit with defects in the production of tegumental pro-anthocyanidins (Nesi et al., 2002; Mizzotti et al., 2012). By contrast, the double mutant seedstick/abs showed a dramatic decrease in the amount of viable seeds produced. This indicated that the interaction between the two MADS-box genes SEEDSTICK and ABS/TT16 is necessary for correct seed development (Mizzotti et al., 2012).
Kaufmann et al. (2005) obtained high levels of infertility in Arabidopsis plants over-expressing the ABS/TT16 gene. Also, de Folter et al. (2006) obtained a dramatic reduction in the number of seeds in Petunia, but in this case the reduction was obtained through silencing of the B-sister (FBP24) gene, probably by a co-suppression effect. Finally, Deng et al. (2012) found that by silencing the ABS/TT16 gene in Brassica napus through RNA interference, the transgenic plants produced shorter siliques with fewer seeds, and the latter result was ascribed to defects in pollen tube guidance. In tobacco, the observed infertility cannot be explained in terms of problems with pollen tube guidance because in our cross-pollination experiments both types of pollination yielded comparable seed production and germinability. Whatever the method used, it appears that a disequilibrium in the dosage of B-sister products may lead to a decrease in fertility.
In Arabidopsis (Kaufmann et al., 2005) and in Petunia (de Folter et al., 2006) it has been shown that the B-sister proteins can make higher-order complexes with MADS-box proteins belonging to other classes: Bs-D-E and Bs-C-E. Moreover, by means of FRET-SLIM analyses, it has been shown in Petunia that the complex Bs-D-E occurs in planta (Nougalli Tonaco et al., 2006). Kaufmann et al. (2005) have suggested a model to explain the defects observed by them in Arabidopsis seedlings over-expressing the ABS/TT16 gene. This model is based on the possibility of the B-sister proteins making higher-order complexes with other MADS-box proteins, and implies that modifications of the B-sister gene expression may lead to modifications of ‘dosage- and affinity-dependent titration of the protein complexes’ involved in the development of floral organs (Kaufmann et al., 2005).
As MADS-box genes belonging to C-, D- and E-class are expressed in young ovules and during seed formation (Colombo et al., 1995; Ferrario et al., 2003; Pinyopich et al., 2003; de Folter et al., 2006), it may be hypothesized that any changes in the levels of the B-sister gene transcripts, either by over-expression or by silencing, may disrupt the ability to correctly form the higher-order complexes necessary for the development of ovule to seed, and this might explain the reduced fertility observed in tobacco plants over-expressing the Ginkgo B-sister gene following auto-pollination but not in the case of cross-pollination in which one of the parents is a wild-type plant.
The results of the over-expression experiments indicate that the Ginkgo B-sister gene includes characteristics that in Arabidopsis are divided between ABS/TT16 and GORDITA. In particular, GORDITA plays a role in the development of the seed outer integument (Prasad et al., 2010) and in Ginkgo the gene is involved in the formation of the fruit-like structure that is a modified seed outer integument. Ginkgoales are very ancient plants (Zhou, 2009) and therefore it is plausible that the Ginkgo B-sister gene may represent a more ancestral form than the Arabidopsis ones, and accordingly the separation of function observed in Arabidopsis might represents a process of sub-functionalization.
Erdmann et al. (2010) identified interesting characteristics that differentiated ABS/TT16 from GORDITA. In its K-domain the ABS/TT16 protein presents the three characteristic strings of heptads (named K1, K2 and K3) that are probably involved in the formation of the coiled coils necessary for interactions with other MADS-box proteins (Fan et al., 1997; Yang and Jack, 2004). By contrast, in the K-domain of the GORDITA protein only one (K1) is present with high probability to form coiled coils. The same analysis performed by us on the K-domain of the B-sister proteins of Ginkgo and yew showed that both proteins contain the three sequences K1, K2 and K3 with high probabilities of forming the coiled coils that are characteristics of ABS/TT16 (Supplementary Data Fig. S4). This finding confirms that in Arabidopsis ABS/TT16 represents the more ancient gene form, while GORDITA is a recent duplication, as suggested by Erdmann et al. (2010).
Recently, Lovisetto et al. (2012) showed that in G. biloba and T. baccata, two Gymnosperm species that produce fleshy fruit-like structures with different anatomical origins, MADS-box genes of the same types were involved in their development. In the present work it has been shown that, besides the above common molecular networks, particular situations can be found that might be linked to the anatomical specificity of the tissues that will become fleshy. In Ginkgo, where it is the outer ovule integument that becomes fleshy, the ovule-specific B-sister gene participates in the growth of the fleshy fruit-like structure that surrounds the seed. By contrast, in yew, where the aril is formed de novo at the base of the ovule as an outgrowth of the peduncle, the ovule-specific B-sister gene does not seem to be particularly involved in the development of the fleshy aril. Therefore, the primordial gymnosperm B-sister gene appears to be ovule-specific and seems to have both a general ‘ovule/seed’ function and a more particular ‘fruit’ function, the latter limited to those cases where the ‘fruit’ represents a modified part of the ovule as occurs in Ginkgo.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
All the gymnosperm plant material was supplied by the Botanic Garden of the Padua University. A. Pavanello is thanked for skillful technical help. This work was supported by a grant from Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR), Italy, to G.C.
LITERATURE CITED
- Becker A, Theissen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetic Evolution. 2003;29:464–489. doi: 10.1016/s1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Becker A, Kaufmann K, Freialdenhoven A, et al. A novel MADS-box gene subfamily with a sister-group relationship to class B floral homeotic genes. Molecular Genetics and Genomics. 2002;266:942–950. doi: 10.1007/s00438-001-0615-8. [DOI] [PubMed] [Google Scholar]
- Chang S, Puyear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Colombo L, Franken J, Koetje E, et al. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell. 1995;7:1859–1868. doi: 10.1105/tpc.7.11.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Shchennicova AV, Franken J, et al. A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant Journal. 2006;47:934–946. doi: 10.1111/j.1365-313X.2006.02846.x. [DOI] [PubMed] [Google Scholar]
- Deng W, Chen G, Peng F, Truksa M, Snyder CL, Weselake RJ. Transparent Testa16 plays multiple roles in plant development and is involved in lipid synthesis and embryo development in Brassica napus. Plant Physiology. 2012;160:978–989. doi: 10.1104/pp.112.198713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN. In situ hybridization. In: Martinez-Zapater JM, Salinas J, editors. Arabidopsis protocols. Totowa, NJ: Humana Press; 1998. pp. 353–371. [Google Scholar]
- Erdmann R, Gramzow L, Melzer R, Theissen G, Becker A. GORDITA (AGL63) is a young paralog of the Arabidopsis thaliana Bsister MADS box gene ABS (TT16) that has undergone neofunctionalization. Plant Journal. 2010;63:914–924. doi: 10.1111/j.1365-313X.2010.04290.x. [DOI] [PubMed] [Google Scholar]
- Fan HY, Hu Y, Tudor M, Ma H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant Journal. 1997;12:999–1010. doi: 10.1046/j.1365-313x.1997.12050999.x. [DOI] [PubMed] [Google Scholar]
- Ferrario S, Immink RG, Shchennikova A, Busscher-Lange J, Angenent GC. The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant cell. 2003;15:914–925. doi: 10.1105/tpc.010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DK, Guiltinan MJ. Rapid, efficient production of homozygous transgenic tobacco plants with Agrobacterium tumefaciens: a seed-to-seed protocol. Plant Molecular Biology Reporter. 1995;13:278–289. [Google Scholar]
- Hoefgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Research. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant Journal. 2009;60:1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- Jaakola L, Poole M, Jones MO, et al. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiology. 2010;153:1619–1629. doi: 10.1104/pp.110.158279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Anfang N, Saedler H, Theissen G. Mutant analysis, protein–protein intereactions and subcellular localization of the Arabidopsis Bsister (ABS) protein. Molecular Genetics Genomics. 2005;274:103–118. doi: 10.1007/s00438-005-0010-y. [DOI] [PubMed] [Google Scholar]
- Khatum S, Flowers TJ. The estimation of pollen viability in rice. Journal of Experimental Botany. 1995;46:151–154. [Google Scholar]
- Koltunov AM, Truettner J, Cox KH, Wallroth M, Goldberg R. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell. 1990;2:1201–1224. doi: 10.1105/tpc.2.12.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. Molecular mechanism of flower development: an armchair guide. Nature Reviews Genetics. 2005;6:688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- Lovisetto A, Guzzo F, Tadiello A, Toffali K, Favretto A, Casadoro G. Molecular analyses of MADS-box genes trace back to Gymnosperms the invention of fleshy fruits. Molecular Biology Evolution. 2012;29:409–419. doi: 10.1093/molbev/msr244. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Melzer R, Wang YQ, Theissen G. The naked and the dead: the ABCs of gymnosperm reproduction and the origin of the angiosperm flower. Seminars in Cell and Developmental Biology. 2010;21:118–128. doi: 10.1016/j.semcdb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Mizzotti C, Mendes MA, Caporali E, et al. The MADS box genes SEEDSTICK and ARABIDOPSIS Bsister play a maternal role in fertilization and seed development. Plant Journal. 2012;70:409–420. doi: 10.1111/j.1365-313X.2011.04878.x. [DOI] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative realtime RT-PCR. Biotechniques. 2002;32:1372–1379. [PubMed] [Google Scholar]
- Nesi N, Debeauion I, Jond C, et al. The TRANSPARENT TESTA 16 locus encodes the Arabidopsis bsister MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002;14:2463–2479. doi: 10.1105/tpc.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky F. Function and evolution of the plant MADS-box gene family. Nature Reviews Genetics. 2001;2:186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- Norton RB. Testing of plum pollen viability with tetrazolium salts. Journal of the American Society for Horticultural Science. 1966;89:132–134. [Google Scholar]
- Nougalli Tonaco IA, Borst JW, de Vries SC, Angenent GC, Immink RG. In vivo imaging of MADS-box transcription factor interactions. Journal of Experimental Botany. 2006;57:33–42. doi: 10.1093/jxb/erj011. [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, et al. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- Prasad K, Ambrose BA. Control of fruit size by an Arabidopsis B-sister MADS-box gene. Plant Signaling and Behavior. 2010;5:899–902. doi: 10.4161/psb.5.7.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Zhang X, Tobón E, Ambrose BA. The Arabidopsis B-sister MADS-box protein, GORDITA, represses fruit growth and contributes to integument development. Plant Journal. 2010;62:203–214. doi: 10.1111/j.1365-313X.2010.04139.x. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Research. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Riano T, Dafni A. A new procedure to assess pollen viability. Sexual Plant Reproduction. 2000;12:241–244. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Seymour GB, Ryder CD, Cevik V, et al. A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria×ananassa Duch.) fruit, a non-climacteric tissue. Journal of Experimental Botany. 2011;62:1179–1188. doi: 10.1093/jxb/erq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadiello A, Pavanello A, Zanin D, et al. A PLENA-like gene of peach is involved in carpel formation and subsequent transformation into a fleshy fruit. Journal of Experimental Botany. 2009;60:651–661. doi: 10.1093/jxb/ern313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TN, Taylor EL, Krings M. Paleobotany. The biology and evolution of fossil plants. 2nd edn. New York: Academic Press; 2009. [Google Scholar]
- Theissen G, Melzer R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Annals of Botany. 2007;100:603–619. doi: 10.1093/aob/mcm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Kim JT, Saedler H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of Eukaryotes. Journal of Molecular Evolution. 1996;43:484–516. doi: 10.1007/BF02337521. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, et al. A short history of MADS-box genes in plants. Plant Molecular Biology. 2000;42:115–149. [PubMed] [Google Scholar]
- Tranbarger TJ, Dussert S, Joët T, et al. Regulatory mechanisms underlying oil palm fruit mesocarp maturation, ripening, and functional specialization in lipid and carotenoid metabolism. Plant Physiology. 2011;156:564–584. doi: 10.1104/pp.111.175141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, et al. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell. 2009;21:3041–3062. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Melzer R, Theissen G. Molecular interactions of orthologues of floral homeotic proteins from the gymnosperm Gnetum gnemon provide a clue to the evolutionary origin of ‘floral quartets. Plant Journal. 2010;64:177–190. doi: 10.1111/j.1365-313X.2010.04325.x. [DOI] [PubMed] [Google Scholar]
- Winter KU, Becker A, Münster T, Kim JT, Saedler H, Theissen G. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proceedings of the National Academy of Sciences USA. 1999;96:7342–7347. doi: 10.1073/pnas.96.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Saraike T, Shitsukawa N, Hirabayashi C, Takumi S, Murai K. Class D and B(sister) MADS-box genes are associated with ectopic ovule formation in the pistil-like stamens of alloplasmic wheat (Triticum aestivum L.) Plant Molecular Biology. 2009;71:1–14. doi: 10.1007/s11103-009-9504-z. [DOI] [PubMed] [Google Scholar]
- Yang Y, Jack T. Defining subdomains of the K domain important for protein–protein interactions of plant MADS proteins. Plant Molecular Biology. 2004;55:45–59. doi: 10.1007/s11103-004-0416-7. [DOI] [PubMed] [Google Scholar]
- Zhou ZY. An overview of fossil Ginkgoales. Palaeoworld. 2009;18:1–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.