Abstract
Objective
Breast milk is a major route of infant HIV infection, yet the majority of breast-fed, HIV-exposed infants escape infection by unknown mechanisms. This study aimed to investigate the role of HIV-specific breast milk cells in preventing infant HIV infection.
Design
A prospective study was designed to measure associations between maternal breast milk HIV-specific interferon-γ (IFN-γ) responses and infant HIV-1 detection at 1 month of age.
Methods
In a Kenyan cohort of HIV-infected mothers, blood and breastmilk HIV-gag IFN-γ ELISpot responses were measured. Logistic regression was used to measure associations between breast milk IFN-γ responses and infant HIV infection at 1 month of age.
Results
IFN-γ responses were detected in breast milk from 117 of 170 (69%) women. IFN-γ responses were associated with breast milk viral load, levels of macrophage inflammatory protein (MIP) 1α, MIP-1β, regulated upon activation, normal T-cell expressed, and secreted and stromal-cell derived factor 1 and subclinical mastitis. Univariate factors associated with infant HIV infection at 1 month postpartum included both detection and breadth of breast milk IFN-γ response (P =0.08, P =0.04, respectively), breast milk MIP-1β detection (P =0.05), and plasma (P =0.004) and breast milk (P =0.004) viral load. In multivariate analyses adjusting for breast milk viral load and MIP-1β, breast milk IFN-γ responses were associated with an approximately 70% reduction in infant HIV infection [adjusted odds ratio (aOR) 0.29, 95% confidence interval (CI) 0.092–0.91], and each additional peptide pool targeted was associated with an approximately 35% reduction in infant HIV (aOR 0.65, 95% CI 0.44–0.97).
Conclusion
These data show breast milk HIV-gag-specific IFN-γ cellular immune responses are prevalent and may contribute to protection from early HIV transmission. More broadly, these data suggest breast milk cellular responses are potentially influential in decreasing mother-to-child transmission of viruses.
Keywords: breastfeeding, breast milk cytotoxic T lymphocytes, cytokines, early postnatal transmission, infant, MIP-1β, pediatric, sub-Saharan Africa
Introduction
Vertical transmission of HIV accounts for a large percentage of new infections in many areas of the world. In the absence of antiretrovirals, breast milk transmission is estimated to account for 30–50% of infant HIV infections in Africa [1]. However, even without antiretrovirals for the prevention of mother-to-child HIV transmission (PMTCT), many breastfeeding infants of HIV-infected women remain uninfected, despite ongoing viral exposure [2]. Better understanding of the mechanism(s) of protection from vertical transmission may help in development of HIV vaccine.
Maternal antibodies in breast milk protect the infant against bacterial and viral pathogens. Breast milk also contains molecules and cells that mediate innate and adaptive immune responses including αβ and γδ T cells, chemokines, cytokines, monocytes/macrophages and B cells [3–11]. The quantity and function of breast milk HIV-specific CD8+ T cells have been characterized in small studies of HIV-infected women. These cells present in the breast milk of the majority of infected women predominantly display an effector memory phenotype and express mucosal homing receptors, demonstrate evidence of compartmentalization from peripheral blood, and recognize HIV-gag more frequently than env [9,10,12,13]. The biologic relevance of HIV-specific T cells in breast milk remains unclear; their presence may represent a marker of antigen exposure or could provide a protective mechanism against infection of the infant through cell-mediated lysis of virally infected cells present in breast milk [14]. To test the hypothesis that breast milk cellular immune responses in early milk may reduce vertical transmission of viruses, we evaluated HIV-gag-specific interferon-γ (IFN-γ) ELISpot responses in paired blood and breast milk specimens in a cohort of women enrolled in a perinatal HIV transmission cohort in Nairobi, Kenya, and examined the relationship between HIV-gag-specific IFN-γ responses and early postnatal HIV transmission.
Participants and methods
Cohort
A perinatal HIV transmission cohort of HIV-infected women and their infants was studied from 1999 to 2005 in Nairobi, Kenya, as described elsewhere [15,16]. Women were recruited in pregnancy, provided written informed consent for participation and storage of specimens, and received zidovudine prophylaxis for PMTCT [17]. Clinically apparent mastitis was evaluated and recorded at each study visit. Breast milk and peripheral blood were collected 1 month postpartum. Infant blood was collected within 48 h of birth and at 1 month. This study was approved by Kenyatta National Hospital Ethics and Research Committee and University of Washington Institutional Review Board.
Specimen collection and preservation
Approximately 30 ml of breast milk was collected by manual expression. The cellular component was isolated from the supernatant and lipid layer by centrifugation (20 min, 710g). Breast milk supernatant was stored at −70°C and breast milk cells (BMCs) were washed in RPMI-1640 medium (Sigma, St Louis, Missouri, USA) and lymphocytes were counted on the basis of morphology and cryopreserved in freezing medium containing 10% dimethyl sulfoxide–90% fetal calf serum (FCS) (both from Sigma). Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient purification and cryopreserved as above.
Viral load measurements
The Gen-Probe (Gen-Probe Inc., San Diego, California, USA) assay was used to measure HIV RNA in blood plasma and breast milk supernatant as previously described [18,19].
Interferon γ ELISpot assays
The IFN-γ ELISpot assay was used for determination of HIV-gag-specific responses as previously described [12] with modifications. The technician performing the assays was blinded to HIV transmission status of the women. Cryopreserved PBMCs or BMCs were thawed, washed, and incubated for 4 h at 5% CO2, 37°C in RPMI 1640 medium supplemented with 10% FCS, and a 1% solution containing penicillin, streptomycin, and fungizone (R10; all from Sigma). Rested viable cells were added to duplicate wells, with mean concentrations 0.26 × 105 BMCs (±SD =0.38) and 1 × 105 PBMCs per well. Duplicate wells were stimulated with either peptide pools, 10 μg/ml phytohemagglutinin (PHA; Murex Biotech Limited, Dartford, UK) as a positive control, or R10 medium as negative control. Four pools of overlapping 15-mer peptides spanning clade A HIV-gag (provided by Dr Tomas Hanke, Oxford, UK) were used for antigen-specific stimulation (Supplemental Digital Content 1, http://links.lww.com/QAD/A257) [20,21]. Assays were considered valid if the PHA response was more than 100 spot-forming units (SFUs) and wells were visibly free from contamination. HIV-specific responses (HIVSFU) were defined as the mean SFU from peptide-stimulated wells minus the mean SFU of negative control wells. Positive responses were defined as HIVSFU per 106 cells at least 50 and peptide SFUs at least twice the negative control SFUs [22].
Infant HIV diagnosis
Infants were diagnosed with HIV infection as previously described [16]. Briefly, an infant was considered HIV infected if either HIV-gag DNA was detected from blood spotted onto filter papers by PCR [23] or HIV RNA was detected in plasma with the Gen-Probe HIV Viral Load Assay. Infection was considered peripartum if HIV DNA or RNA was undetectable in the birth specimen but detectable at 1 month.
Breast milk subclinical mastitis and chemokines
Subclinical mastitis, defined as sodium (Na+)-to-potassium (K+) ratio of more than 1 in whole breast milk [24], and breast milk concentration of macrophage inflammatory protein 1 alpha (MIP-1α), MIP-1β, regulated upon activation, normal T-cell expressed, and secreted (RANTES), and stromal-cell derived factor 1 (SDF-1) have been previously reported [25].
Statistical methods
All statistical analyses were performed using StataSE v11 (College Station, Texas, USA). Viral load, ELISpot HIVSFU, and chemokine levels were log10-transformed to normalize the data. Spearman’s correlation was used to test for independence between viral load and immune responses in breast milk. Fisher’s exact test, the independent t-test, and the Mann–Whitney U-test were used to compare proportions and continuous variables between women with and without breast milk responses and Spearman’s correlation was used to test for independence between correlates of breast milk responses and response magnitude. Logistic regression was used to build univariate and multivariate models of transmission. HIV-specific IFN-γ immune responses were modeled separately as dichotomized (detected/not detected) and continuous covariates (number of peptide pools recognized or response magnitude). Response magnitude was summarized for each subject as the sum of HIVSFU in all pools, the maximum HIVSFU of all pools, and the mean HIVSFU in all pools. For multivariate models, covariates were evaluated for co-linearity before inclusion. For all transmission analyses, the outcome of interest was infant infection at 1 month of age; thus, analyses were restricted to infants who were negative for HIV DNA and RNA within 48 h of birth. To enable comparability of odds ratios and confidence intervals, the sample size for both the univariate and multivariate analyses was restricted to infants with all covariates available (n =148). Final multivariate models were restricted to covariates that retained significance when they were adjusted individually for plasma or breast milk viral load. To evaluate effect of input cells per well, analyses were re-run with breast milk ELISpot assays restricted to those with a minimum cells per well of 0.1 × 105 and yielded similar results, indicating misclassification of ELISpot results from assays with low cell input numbers does not bias overall results (data not shown). We used the Holm Test to evaluate P values with nonindependent multiple comparisons [26]. All reported P values are two-tailed.
Results
Cohort characteristics
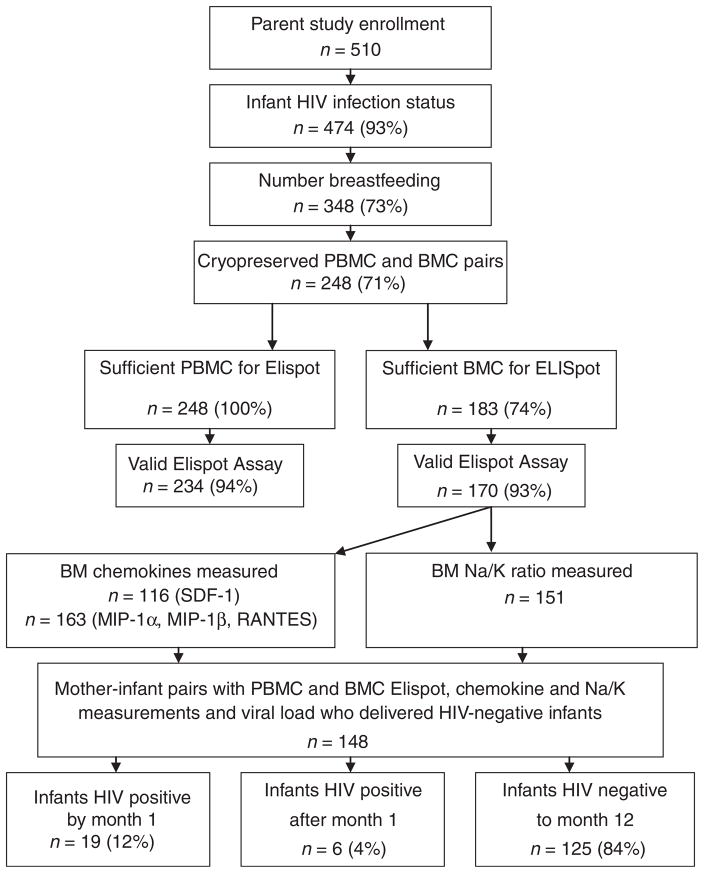
Detailed clinical characteristics of HIV-infected women enrolled in the perinatal cohort in Nairobi, Kenya, are described elsewhere [3,15]. The study enrolled 510 pregnant HIV-1 seropositive women, HIV-1 infection status was available on 474 infants (93%), and 348 (73%) women were breastfeeding their infants (Fig. 1). Paired PBMC and BMC specimens were cryopreserved from 248 (71%) of these women; however, sufficient cell numbers from breast milk were available from 183 (74%). Thus, 248 PBMC and 183 paired BMC ELISpot assays were evaluated. Transmission analyses were further restricted to the 148 women with BMC ELISpot, viral load, and chemokine data available who delivered HIV-negative infants. As in the parent cohort, the women were young, moderately immunosuppressed (median CD4 cell count 459 cells/μl) and had high HIV viral loads (mean 4.7 log10 copies/ml plasma). Cell-free HIV RNA was detected in breast milk of 170 of 180 (94%) of women at 1 month postpartum with a mean of 2.9 ±SD 1.1 log10 copies/ml (Table 1).
Fig. 1. Participant flow.
Cohort numbers are based on available test results, focusing on breastfeeding women whose infants were HIV uninfected at birth. Chemokine and mastitis data are reported in [25]. BMCs, breast milk cells; PBMCs, peripheral blood mononuclear cells.
Table 1.
Characteristics of 183 women with paired breast milk and blood ELISpot assays 1 month postpartum.
| N (%)a | Mean (±SD) or median (IQR) | |
|---|---|---|
| Age (years) | 183 | 24 (21–27) |
| Parity | 182 (99) | 1 (1–2) |
| 32 weeks gestation (baseline) | ||
| CD4 cell count (cells/μl) | 177 (97) | 459 (313–618) |
| CD4 percentage | 177 (97) | 22 (17–29) |
| CD4: CD8 ratio | 177 (97) | 0.47 (0.33–0.71) |
| Plasma HIV RNA (log10 copies/ml) | 176 (96) | 4.7 (±0.80) |
| Month 1 postpartum | ||
| CD4 cell count (cells/μl) | 148 (81) | 562 (392–755) |
| CD4 percentage | 148 (81) | 24 (17–30) |
| CD4 : CD8 ratio | 148 (81) | 0.52 (0.30–0.73) |
| Plasma HIV RNA (log10 copies/ml) | 182 (99) | 4.7 (±0.92) |
| Breast milk HIV RNA (log10 copies/ml) | 180 (98)b | 2.9 (±1.1) |
Number of women with available data per test (%). Specimens and/or test results were not available for all women at all time points.
Number of women with detectable breast milk viral load =170 (94%).
Prevalence and magnitude of HIV-specific interferon γ responses in breast milk
The ELISpot assay was used to quantify HIV-specific IFN-γ production in response to stimulation with HIV-gag peptides. ELISpot assays that failed to meet validity criteria [14 (6%) PBMC and 13 (7%) BMC assays] were excluded from subsequent analyses. HIV-gag-specific responses were detected in breast milk from 117 of 170 (69%) women and in blood from 205 of 234 (88%). Of 170 women with valid paired blood and breast milk assays, 111 (66%) had concordant assays: 102 were concordant positive (92%) and nine were concordant negative (8%). Women with discordant assays were more likely to have a positive response in blood (44/59, 75%) than a positive response in breast milk (15/59, 25%; P =0.0002). Of the valid assays, the median background BMC response was 426 SFUs/106 cells [interquartile range (IQR) 166–1174 SFUs/106] and the median PHA response was 9575 SFUs/106 cells (IQR 3200–28823 SFUs/106). The median magnitude of positive breast milk responses to peptide pools 1–4 were 1400 HIVSFUs/106 cells (IQR 400–6314), 875 (IQR 350–2080), 825 (IQR 450–1680), and 933 cells (IQR 370–1600), respectively. These analyses are presented in detail in Supplemental Digital Contents 2 and 3, http://links.lww.com/QAD/A257.
Correlates of breast milk HIV-specific interferon γ responses
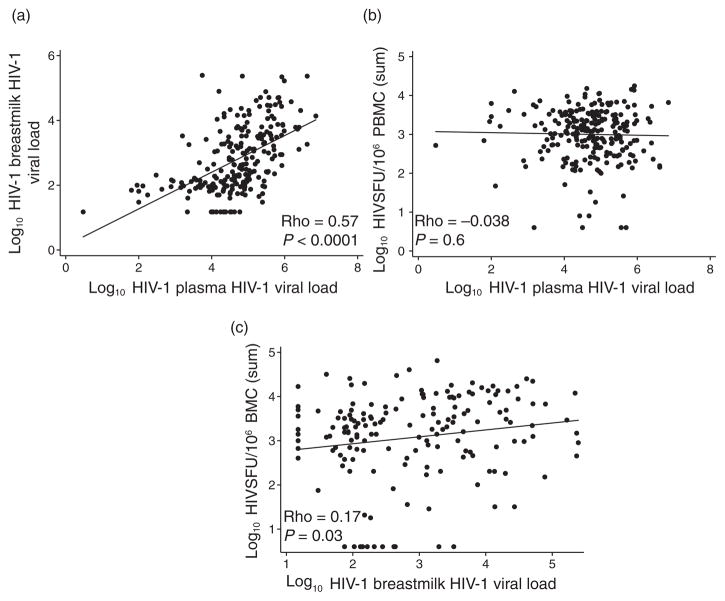
Breast milk and blood plasma viral loads were highly correlated, with viral load in breast milk approximately 2 log10 lower than that in plasma, as previously shown (Fig. 2a) [27]. We evaluated several potential correlates of breast milk HIV-specific IFN-γ responses including immunologic status, HIV plasma viral load, breast milk viral load, chemokines, and subclinical mastitis (Table 2). We did not observe a correlation between the magnitude of PBMC IFN-γ responses and plasma viral load (Fig. 2b and Table 2); however, breast milk HIV viral load was weakly associated with breast milk responses (Fig. 2c and Table 2). Breast milk MIP-1α, MIP-1β, RANTES, and SDF-1 levels were significantly correlated with the magnitude of breast milk IFN-γ responses (P <0.05 for each). Women with subclinical mastitis were more likely to have detectable breast milk responses (43%) compared with women without subclinical mastitis (22%, P =0.02), and the magnitude of the breast milk IFN-γ responses was weakly correlated with the Na: K ratio (ρ =0.24, P =0.003).
Fig. 2. HIV-specific interferon-γ responses and viral replication.
Scatter plots show the correlation between (a) plasma and breast milk HIV viral load, (b) plasma viral load and HIV-specific IFN-γ responses in peripheral blood mononuclear cells (PBMCs), and (c) breast milk viral load and HIV-specific IFN-γ responses in breast milk cells (BMCs). Correlation coefficients and P values were generated using Spearman’s correlation. IFN, interferon.
Table 2.
Correlates of breast milk interferon-γ responses.a
| IFN-γ responseb
|
IFN-γ-responseb magnitude | ||||
|---|---|---|---|---|---|
| n | Detected | Not detected | P | ||
|
|
|||||
| Immunologic/virologic | |||||
| CD4 cell count (cells/μl) | 139 | 556 (379–744) | 539 (408–717) | 0.9 | ρ =0.030, P =0.7 |
| CD4% | 139 | 23 (16–29) | 22 (18–30) | 0.9 | ρ =0.033, P =0.7 |
| CD8 (cells/μl) | 139 | 1108 (879–1465) | 1204 (797–1448) | 0.9 | ρ =0.046, P =0.6 |
| CD8% | 139 | 50 (41–56) | 45 (37–61) | 0.6 | ρ =0.081, P =0.3 |
| log10 plasma HIV RNA load | 169 | 4.7 (±0.92) | 4.8 (±0.96) | 0.5 | ρ =−0.0095, P =0.9 |
| log10 breast milk HIV RNA load | 167 | 3.0 (±1.1) | 2.7 (±0.91) | 0.08 | ρ =0.17, P =0.02 |
| Chemokine levels in breast milk (log10 pg/ml) | |||||
| MIP-1α | 162 | 1.4 (±0.39) | 1.3 (±0.37) | 0.07 | ρ =0.30, P =0.0001 |
| MIP-1β | 163 | 1.8 (±0.44) | 1.6(±0.48) | 0.02 | ρ =0.28, P =0.0003 |
| RANTES | 163 | 2.4 (±0.47) | 2.1 (±0.56) | 0.0005 | ρ =0.30, P =0.0001 |
| SDF-1 | 116 | 2.4 (±0.71) | 2.1(±0.72) | 0.07 | ρ =0.20, P =0.03 |
| Mastitis in the first month postpartum | |||||
| Clinical mastitis | 156 | 11 (12/109) | 8.5 (4/47) | 0.8 | c |
| Subclinical mastitisd | 151 | 43 (46/106) | 22 (10/45) | 0.02 | ρ =0.24, P =0.003e |
The values are given as mean (±SD), median (IQR), or percentage (n/N). IFN, interferon; IQR, interquartile range; MIP, macrophage inflammatory protein; RANTES, regulated upon activation, normal T-cell expressed, and secreted; SDF, stromal cell-derived factor.
Correlates were evaluated among 170 women with valid breast milk HIV-specific IFN-γ assays.
IFN-γ responses were evaluated as dichotomous (detected/not detected) and continuous (magnitude) variables. IFN-γ response magnitude is summarized by the median of the mean sum of pooled peptide responses; log10-transformed values were used in correlation analyses.
Not applicable.
Subclinical mastitis is defined as breast milk Na : K >1.
Correlation based on Na : K ratio.
Breast milk HIV-specific interferon γ responses and peripartum transmission
Univariate correlates of infant HIV infection included the number of gag peptide pools recognized in breast milk ELISpot [odds ratio (OR) 0.69, 95% CI 0.49–0.98], HIV viral load in breast milk and blood, and the detection of MIP-1β in breast milk (P <0.05 for each; Table 3). The magnitude of IFN-γ responses was not associated with infant HIV infection in either breast milk or blood when defined by sum of responses (P >0.05 for both), or alternatively defined by mean or maximum response (data not shown). There was a trend for reduced infection in infants born to women with any detectable breast milk IFN-γ response (OR 0.41, 95% CI 0.16–1.1). Detection of MIP-1β was associated with decreased odds of infant infection (OR 0.22, 95% CI 0.047–0.99). Because MIP-1α, RANTES, and SDF-1 were associated with breast milk IFN-γ responses, we also evaluated whether there was an association between these cytokines and infant HIV infection at month 1, and none were significant (data not shown).
Table 3.
Correlates of early breast milk HIV transmission.a
| OR for infant HIV infection (95% CI) | P | |
|---|---|---|
| Univariate model (n =148) | ||
| Breast milk HIV RNA load, 1 month postpartum | 2.0 (1.2–3.1) | 0.004b |
| Plasma HIV RNA load, 1 month postpartum | 2.7 (1.4–5.2) | 0.004b |
| HIV-gag response detected in breast milk | 0.41 (0.16–1.1) | 0.08 |
| HIV-gag response detected in blood | 1.1 (0.23–5.3) | 0.9 |
| No. of HIV-gag pools recognized in breast milk | 0.69 (0.49–0.98) | 0.04 |
| No. of HIV-gag pools recognized in blood | 0.74 (0.53–1.0) | 0.09 |
| Sum HIVSFU in breast milkc | 1.0 (0.62–1.6) | 0.97 |
| Sum HIVSFU in bloodc | 1.5 (0.68–3.4) | 0.3 |
| MIP-1β detected in breast milk | 0.22 (0.047–0.99) | 0.05 |
| Multivariate model 1 (n =148)d | ||
| HIV-gag response detected in breast milk | 0.29 (0.092–0.91) | 0.03 |
| MIP-1β detected in breast milk | 0.15 (0.024–0.95) | 0.04 |
| Breast milk HIV RNA load, 1 month postpartum | 2.5 (1.5–4.2) | 0.001 |
| Multivariate model 2 (n =148)d | ||
| No. of HIV-gag pools recognized in breast milk | 0.65 (0.44–0.97) | 0.03 |
| MIP-1β detected in breast milk | 0.13 (0.021–0.80) | 0.03 |
| Breast milk HIV RNA load, 1 month postpartum | 2.3 (1.4–3.8) | 0.001 |
95% CI, 95% confidence interval; HIVSFU, spot-forming unit; MIP, macrophage inflammatory protein; OR, odds ratio.
Analysis restricted to 148 breastfeeding mother infant pairs with viral load, ELISpot, and chemokine data available.
Univariate P values remain significant after Holm–Bonferroni test for multiple comparisons.
Similar estimates were obtained when alternatively using maximum and mean to summarize the magnitude of peptide pool responses.
Covariates that retained significance after adjustment for HIV breast milk or plasma viral load were included in the final multivariate models.
We evaluated the effect of breast milk IFN-γ responses on peripartum transmission in multivariate analyses adjusting for factors associated with transmission. We constructed two complementary models that included detection of MIP-1β and breast milk viral load but differed in terms of categorization of breast milk IFN-γ responses: model 1 included detection of any breast milk IFN-γ response, whereas model 2 included a number of gag pools recognized in breast milk. In model 1, detection of a breast milk IFN-γ response was associated with a 71% decrease in infant infection (OR 0.29, 95% CI 0.092–0.91). In model 2, infant infections were decreased by approximately 35% for each additional peptide pool recognized (OR 0.65, 95% CI 0.44–0.97). Breast milk HIV RNA viral load and MIP-1β detection remained significantly associated with infant infection in the multivariate models. Results were similar when alternatively adjusting for plasma viral load (data not shown).
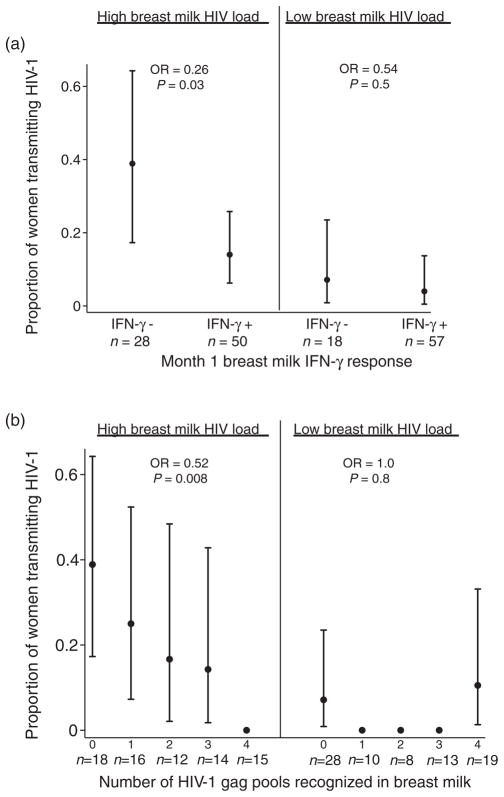
To test for effect of modification of breast milk IFN-γ responses by HIV viral load, we examined transmission data stratified by breast milk viral load above and below the median (Fig. 3a). Among women with high breast milk HIV viral load, those with detectable IFN-γ responses in breast milk were at approximately 74% lower odds of transmitting HIV compared with those who lacked detectable responses (OR 0.26, 95% CI 0.077–0.86; P =0.03). This effect was not significant in the low viral load stratum (OR 0.54, 95% CI 0.072–4.1; P =0.5), which experienced fewer transmissions. Similarly, in women with high breast milk viral load, each additional peptide pool recognized was associated with an approximately 48% reduction in the odds of infant infection (OR 0.52, 95% CI 0.32–0.84; P =0.008). The number of pools recognized was not significantly associated with infant infection in the low breast milk viral load stratum (OR 1.0, 95% CI 0.58–2.0; P =0.8) (Fig. 3b).
Fig. 3. Stratified analysis of early peripartum transmission and breast milk HIV-specific interferon-γ responses.
Markers show the proportion of infants with HIV infection at 1 month of age, stratified by breast milk viral load and (a) HIV-gag-specific response detected in breast milk and (b) the number of peptide pools eliciting a positive HIV-gag-specific response in breast milk. Bars show exact 95% confidence intervals calculated from the binomial distribution. The number of mother–child pairs is shown for each category (n). No confidence intervals are shown for categories where no transmission occurred. P values are shown for logistic regression within strata.
Definitive diagnosis of late in-utero, intrapartum, and early breastfeeding transmission was not possible in this cohort. However, we can estimate an intrapartum transmission rate based on the number of infants who became infected after birth whose mothers were not breastfeeding (7/102, 6.9%). If we apply this estimate obtained to our transmission analysis of 148 mother–infant pairs, we could predict that 10 intrapartum transmissions may be misclassified as early breast milk transmissions. Assuming the misclassification of transmission route is distributed equally among women who had and who lacked breast milk IFN-γ responses, we evaluated the associations, which remained significant (data not shown).
Lastly, to address the possibility that ingested maternal breast milk cells could persist in the infant and reduce risk of infection, we compared HIV-specific IFN-γ responses in children born to women with detectable and undetectable breast milk responses. Among HIV-exposed uninfected infants, there was no significant difference in frequency of responses in children born to women with detectable breast milk responses (8/54, 15%) versus those born to women with undetectable responses (1/19, 5.3%, P =0.4) at 1 month of age. We also did not find a difference in set-point HIV viral load between HIV-infected infants born to mothers with detectable and undetectable breast milk responses (data not shown).
Discussion
To our knowledge, this is the first demonstration of an association between breast milk cellular immune responses and protection from vertical transmission. Breast milk responses were independently associated with an approximately 70% lower odds of infant HIV infection at 1 month of life. Importantly, the breadth of breast milk response most significantly influenced transmission: infant infection was reduced by a third for each additional HIV-gag peptide pool targeted, which suggests that a broad response against multiple epitopes may be more protective against peripartum transmission than a narrow response. The level of breast milk MIP-1β, an effector molecule secreted by IFN-γ-producing CD8+ T cells [28,29], was also independently associated with protection from infection. Lastly, detection of these responses in women was not associated with detection of responses in HIV-exposed uninfected infants, or reduced viral replication in HIV-infected infants, suggesting their effect is concentrated in breast milk. Together, these data suggest a role for breast milk cell-mediated immunity in preventing vertical transmission of viruses and may explain why only a minority of infants exposed to HIV through breast milk become infected.
Breast milk IFN-γ responses could potentially decrease transmission by several mechanisms. Breast milk HIV-specific cytotoxic T cells (CTLs) may facilitate destruction of HIV-infected cells or lead to decreased production of infectious virus; this could occur either in the breast and/or at infant mucosal surfaces. This mechanism would be best validated by measuring the association between breast milk IFN-γ responses and cell-associated viral load. We observed correlations of breast milk IFN-γ with breast milk HIV-1 RNA, suggesting antigenic induction of IFN-γ responses rather than IFN-γ reduction of viral levels; however, we did not measure breast milk proviral DNA in this study. Our study supports a second potential mechanism by which breast milk HIV-specific CD8+ T cells could reduce transmission through secretion of HIV co-receptor binding chemokines that block infection. CD8+ T-cell secretion of MIP-1β has been associated with slower HIV disease progression [30,31]. We previously found breast milk MIP-1β levels to be associated with reduced risk of overall postpartum transmission during the first year of life [25]. We found MIP-1β detection in breast milk was associated with lower risk of infant HIV transmissions between delivery and 1 month postpartum, independent of breast milk viral load and breast milk IFN-γ responses. A large proportion of activated CD8+ T cells secrete both IFN-γ and MIP-1β [29,32], consistent with the correlation observed in our study between breast milk IFN-γ responses and MIP-1β levels. MIP-1β could protect against HIV entry by binding to CCR5 on target cells in either the breast milk and/or the infant gut. A third mechanism by which breast milk cellular immune responses may protect transmission is via passive transfer of immune cells to the infant. Animal studies have suggested that ingested breast milk cells can cross the neonatal gut and modify immune responses within the infant [33–35]. We addressed this hypothesis by evaluating HIV-specific immune responses in exposed uninfected infants and found no difference in the percentage of responses in uninfected children of mothers with breast milk responses versus those without. We also found no evidence to support the hypothesis that ingested breast milk cells in the infant circulation may affect HIV replication: HIV set-point was similar between HIV-infected infants of mothers with detectable and those with undetectable breast milk immune responses.
Three main factors were identified as correlates of breast milk IFN-γ responses: breast milk HIV viral load, chemokine levels, and subclinical mastitis (Na : K ratio). The association between breast milk viral load and breast milk HIV-specific IFN-γ responses was significant, although a low ρ value suggests viral load alone is not responsible for expansion of antigen-specific breast milk cells. Notably, breast milk HIV-specific responses were detected in some women with undetectable breast milk viral load in our study, which is consistent with the finding of a previous study [9]. The association between subclinical mastitis and breast milk IFN-γ responses could be explained by several mechanisms, including exudation of peripheral blood lymphocytes into the milk secondary to increased membrane permeability, proliferation of HIV-specific T cells in response to increased breast milk viral load, or increased homing of T cells to the milk in response to increased chemokine production during inflammation. As previously reported in this cohort, subclinical mastitis was associated with higher breast milk viral load and also with higher MIP-1α and MIP-1β chemokine levels [25].
This work complements our previous study examining breast milk HIV-specific responses in 53 women from the same cohort using recombinant vaccinia virus (rVV) expressing HIV-gag and env [12]. We note several important distinctions from our earlier work. First, overall prevalence of both breast milk and blood responses was greater in the current study. We hypothesize that the use of peptide pools resulted in higher sensitivity to detect low-level responses as opposed to rVV vectors, which require infection, antigen processing, and presentation. Also, the current study was designed to have adequate power to detect an effect of immune responses on transmission and focused on infections occurring during the first month of life, when the rate of transmission, the cellularity of breast milk, and, therefore, the relevance of protective breast milk responses were expected to be greatest.
Limitations of this study include inability to definitively distinguish intrapartum transmission from early breast milk transmission, low cell numbers recovered in breast milk, and the inability to evaluate late transmission events due to the combined effect of decreased breast milk cellularity and rarity of late infections. Colostrum and early milk are highly cellular, whereas the number of cells in mature breast milk are approximately 1 log10 lower than those in early milk [14,36–38]; we, thus, speculated that if breast milk T-cell responses were associated with protection from HIV transmission, their effects would be most discernable in the early postpartum period. Although HIV DNA and RNA were undetectable in all infants at birth, some transmissions could have occurred either late in utero or intrapartum; since breast milk T cells would not be expected to affect these routes of transmission, our data may underestimate the protective effect of HIV-specific T-cell responses. Although the measurement of IFN-γ secretion in response to one HIV protein does not fully describe the anti-HIV cellular immune response present in milk, HIV-gag peptide pools were used exclusively due to limited cellularity; previous studies have correlated breadth of IFN-γ gag responses with viral control [39–44]. Finally, because PBMCs include CD4 T and natural killer cells, we can determine neither the source of IFN-γ nor the proportion of CD8+ T cells capable of MIP-1β and IFN-γ secretion. Regardless of the cellular source, HIV-specific IFN-γ-secretion in the breast milk was associated with reduced HIV transmission.
In conclusion, in this large study, HIV-infected mothers frequently had HIV-specific IFN-γ responses detected in breast milk that were associated with decreased infant HIV infection. Cellular immune responses in breast milk may play a role in protecting infants from mucosal exposures to viral pathogens. More generally, these data support the relevance of the HIV-gag-specific responses, and the breadth of responses, in the development of an HIV vaccine.
Supplementary Material
Acknowledgments
We thank the women and children who participated in the CTLs and Prevention of Breast milk HIV study in Nairobi and the clinical and research staff involved in the study. We thank Sandy Emery for technical assistance with obtaining viral load data and the Kizazi Working Group for helpful discussions and feedback during manuscript preparation.
This work was supported by the National Institutes of Health HD-23412; HD-054314; HD-42949 to GJ-S. B.L.P., J.A.S., and S.M. were scholars in the International AIDS research and Training Program supported by Fogarty International Center D43-TW00007. The University of Washington Center for AIDS Research grant P30 AI027757 provided infrastructure and administrative logistics for this study in Seattle and Nairobi.
Footnotes
Conflicts of interest
There are no conflicts of interest.
B.L.P., G.J.-S., and D.M.-N. conceived and obtained funding for the study; J.A.S. and B.A.R. analyzed the data; S.M., B.L.-P., J.O., and S.R.-J. designed and executed the laboratory portions of the study; C.F., E.M.-O., D.C.W., and D.M.-N. developed the clinic site and cohort, managed recruitment, follow-up, tracing, and care of the study participants. C.F. designed and executed the chemokine studies and provided data analysis and interpretation. The manuscript was co-written by B.L.-P. and J.A.S. All authors have contributed edits to this paper and have approved this final submission.
References
- 1.John-Stewart G, Mbori-Ngacha D, Ekpini R, Janoff EN, Nkengasong JN, Read JS, et al. Breast-feeding and transmission of HIV-1. J Acquir Immune Defic Syndr. 2004;35:196–202. doi: 10.1097/00126334-200402010-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nduati R, John GC, Mbori-Ngacha D, Richardson BA, Overbaugh J, Mwatha A, et al. Effect of breastfeeding and formula feeding on transmission of HIV-1: a randomized clinical trial. JAMA. 2000;283:1167–1174. doi: 10.1001/jama.283.9.1167. [DOI] [PubMed] [Google Scholar]
- 3.Bosire R, Guthrie BL, Lohman-Payne BL, Mabuka JM, Majiwa M, Wariua G, et al. Longitudinal comparison of chemokines in breastmilk early postpartum among HIV-1 infected and uninfected Kenyan women. Breastfeeding Med. 2007;2:129–138. doi: 10.1089/bfm.2007.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawkes JS, Bryan D-L, Gibson RA. Cytokine production by human milk cells and peripheral blood mononuclear cells from the same mothers. J Clin Immunol. 2002;22:338–344. doi: 10.1023/a:1020652215048. [DOI] [PubMed] [Google Scholar]
- 5.Ichikawa M, Sugita M, Takahashi M, Satomi M, Takeshita T, Araki T, Takahashi H. Breast milk macrophages spontaneously produce granulocyte-macrophage colony-stimulating factor and differentiate into dendritic cells in the presence of exogenous interleukin-4 alone. Immunology. 2003;108:189–195. doi: 10.1046/j.1365-2567.2003.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becquart P, Petitjean G, Al Tabaa Y, Valea D, Huguet M-F, Tuaillon E, et al. Detection of a large T-cell reservoir able to replicate HIV-1 actively in breast milk. AIDS. 2006;20:1453–1462. doi: 10.1097/01.aids.0000233581.64467.55. [DOI] [PubMed] [Google Scholar]
- 7.Petitjean G, Becquart P, Tuaillon E, Al Tabaa Y, Valea D, Huguet M-F, et al. Isolation and characterization of HIV-1-infected resting CD4+ T lymphocytes in breast milk. J Clin Virol. 2007;39:1–8. doi: 10.1016/j.jcv.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Kourtis AP, Ibegbu C, Theiler R, Xu Y-X, Bansil P, Jamieson DJ, et al. Breast milk CD4+T cells express high levels of chemokine receptor 5 and CXC chemokine receptor 4 and are preserved in HIV-infected mothers receiving highly active antiretroviral therapy. J Infect Dis. 2007;195:965–972. doi: 10.1086/512082. [DOI] [PubMed] [Google Scholar]
- 9.Sabbaj S, Edwards BH, Ghosh MK, Semrau K, Cheelo S, Thea DM, et al. Human immunodeficiency virus-specific CD8+ T cells in human breast milk. J Virol. 2002;76:7365–7373. doi: 10.1128/JVI.76.15.7365-7373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabbaj S, Ghosh MK, Edwards BH, Leeth R, Decker WD, Goepfert PA, Aldrovandi GM. Breast milk-derived antigen-specific CD8+ T cells: an extralymphoid effector memory cell population in humans. J Immunol. 2005;174:2951–2956. doi: 10.4049/jimmunol.174.5.2951. [DOI] [PubMed] [Google Scholar]
- 11.Tuaillon E, Valea D, Becquart P, Al Tabaa Y, Meda N, Bollore K, et al. Human milk-derived B cells: a highly activated switched memory cell population primed to secrete antibodies. J Immunol. 2009;182:7155–7162. doi: 10.4049/jimmunol.0803107. [DOI] [PubMed] [Google Scholar]
- 12.Lohman BL, Slyker J, Mbori-Ngacha D, Bosire R, Farquhar C, Obimbo E, et al. Prevalence and magnitude of human immunodeficiency virus (HIV) type 1-specific lymphocyte responses in breast milk from HIV-1-seropositive women. J Infect Dis. 2003;188:1666–1674. doi: 10.1086/379374. [DOI] [PubMed] [Google Scholar]
- 13.Mahlokozera T, Kang HH, Goonetilleke N, Stacey AR, Lovingood RV, Denny TN, et al. The magnitude and kinetics of the mucosal HIV-specific CD8+ T lymphocyte response and viral RNA load in breast milk. PLoS One. 2011;6:e23735. doi: 10.1371/journal.pone.0023735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rousseau CM, Nduati RW, Richardson BA, John-Stewart GC, Mbori-Ngacha DA, Kreiss JK, Overbaugh J. Association of levels of HIV-1-infected breast milk cells and risk of mother-to-child transmission. J Infect Dis. 2004;190:1880–1888. doi: 10.1086/425076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otieno PA, Brown ER, Mbori-Ngacha DA, Nduati RW, Farquhar C, Obimbo EM, et al. HIV-1 disease progression in breast-feeding and formula-feeding mothers: A prospective 2-year comparison of T cell subsets, HIV-1 RNA levels, and mortality. J Infect Dis. 2007;195:220–229. doi: 10.1086/510245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.John-Stewart GC, Mbori-Ngacha D, Lohman-Payne B, Farquhar C, Richardson BA, Emery S, et al. HIV-1-specific cytotoxic T lymphocytes and breast milk HIV-1 transmission. J Infect Dis. 2009;199:889–898. doi: 10.1086/597120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaffer N, Chuahchoowong R, Mock PA, Bhadrakom C, Siriwasin W, Young NL, et al. Short-course zidovudine for perinatal HIV-1 transmission in Bangkok, Thailand: a randomized clinical trial. Bangkok Collaborative Perinatal HIV Transmission Study Group. Lancet. 1999;353:773–780. doi: 10.1016/s0140-6736(98)10411-7. [DOI] [PubMed] [Google Scholar]
- 18.Emery S, Bodrug S, Richardson BA, Giachetti C, Bott MA, Panteleeff D, et al. Evaluation of performance of the Gen-Probe human immunodeficiency virus type 1 viral load assay using primary subtype A, C, and D isolates from Kenya. J Clin Microbiol. 2000;38:2688–2695. doi: 10.1128/jcm.38.7.2688-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeVange Panteleeff D, Emery S, Richardson BA, Rousseau C, Benki S, Bodrug S, et al. Validation of performance of the Gen-Probe human immunodeficiency virus type 1 viral load assay with genital swabs and breast milk samples. J Clin Microbiol. 2002;40:3929–3937. doi: 10.1128/JCM.40.11.3929-3937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mwau M, Cebere I, Sutton J, Chikoti P, Winstone N, Wee EG-T, et al. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccina virus Ankara (MVA) vaccines in humans. J Gen Virol. 2004;85:911–919. doi: 10.1099/vir.0.19701-0. [DOI] [PubMed] [Google Scholar]
- 21.Mwau M, McMichael AJ, Hanke T. Design and validation of an enzyme-linked immunospot assay for use in clinical trials of candidate HIV vaccines. AIDS Res Hum Retroviruses. 2002;18:611–618. doi: 10.1089/088922202760019301. [DOI] [PubMed] [Google Scholar]
- 22.Kaul R, Rowland-Jones SL. HIV Immunology Database. Los Alamos, NM: Theoretical Biology and Biophysics; 1999. Methods of detection of HIV-specific CTL and their role in protection against HIV infection; pp. 27–36. [Google Scholar]
- 23.DeVange Panteleeff D, John GC, Nduati R, Mbori-Ngacha D, Richardson BA, Kreiss JK, Overbaugh J. Rapid method for screening dried blood samples on filter paper for human immunodeficiency virus type 1 DNA. J Clin Microbiol. 1999;37:350–353. doi: 10.1128/jcm.37.2.350-353.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filteau S. The influence of mastitis on antibody transfer to infants through breast milk. Vaccine. 2003;21:3377–3381. doi: 10.1016/s0264-410x(03)00337-2. [DOI] [PubMed] [Google Scholar]
- 25.Farquhar C, Mbori-Ngacha DA, Redman MW, Bosire RK, Lohman BL, Piantadosi AL, et al. CC and CXC chemokines in breastmilk are associated with mother-to-child HIV-1 transmission. Current HIV Res. 2005;3:361–369. doi: 10.2174/157016205774370393. [DOI] [PubMed] [Google Scholar]
- 26.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 27.Rousseau CM, Nduati RW, Richardson BA, Steele MS, John-Stewart GC, Mbori-Ngacha DA, et al. Longitudinal analysis of human immunodeficiency virus type 1 RNA in breast milk and of its relationship to infant infection and maternal disease. J Infect Dis. 2003;187:741–747. doi: 10.1086/374273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattapallil JJ, Smit-McBride Z, McChesney MB, Dandekar S. Intestinal intraepithelial lymphocytes are primed for gamma interferon and MIP-1b expression and display antiviral cytotoxic activity despite severe CD4+ T-cell depletion in primary simian immunodeficiency virus infection. J Virol. 1998;72:6421–6429. doi: 10.1128/jvi.72.8.6421-6429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Appay V, Nixon DF, Donahoe SM, Gillespie GMA, Dong T, King A, et al. HIV-specific CD8+ T cells produce antiviral cytokines but are impaired in cytolytic function. J Exp Med. 2000;192:63–75. doi: 10.1084/jem.192.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cocchi F, DeVico AL, Yarchoan R, Redfield R, Cleghorn F, Blattner WA, et al. Higher macrophage inflammatory protein (MIP)-1a and MIP-1b levels from CD8+ T cells are associated with asymptomatic HIV-1 infection. Proc Natl Acad Sci U S A. 2000;97:13812–13817. doi: 10.1073/pnas.240469997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferre AL, Hunt PW, Critchfield JW, Young DH, Morris MM, Garcia JC, et al. Mucosal immune responses to HIV-1 in elite controllers: a potential correlate of immune control. Blood. 2009;113:3978–3989. doi: 10.1182/blood-2008-10-182709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamin-Lewis R, Abdelwahab SF, Trang c, Baker A, DeVico AL, Gallo RC, Kewis GK. Perforin-low memory CD8+ cells are the predominant T cells in normal humans that synthesize the b-chemokine macrophage inflammatory protein-1b. Proc Natl Acad Sci U S A. 2001;98:9283–9288. doi: 10.1073/pnas.161298998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiler IJ, Hickler W, Sprenger R. Demonstration that milk cells invade suckling neonatal mice. Am J Repro Immunol. 1983;4:95–98. doi: 10.1111/j.1600-0897.1983.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 34.Jain L, Vidyasagar D, Xanthou M, Ghai V, Shimada S, Blend M. In vivo distribution of human milk leucocytes after ingestion by newborn baboons. Arch Dis Child. 1989;64:923–930. doi: 10.1136/adc.64.7_spec_no.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar SN, Stewart GL, Steven WM, Seelig LL. Maternal to neonatal transmission of T-cell mediated immunity to Trichinella sprialis during lactation. Immunol. 1989;68:87–92. [PMC free article] [PubMed] [Google Scholar]
- 36.Ogra SS, Ogra PL. Immunologic aspects of human colostrum and milk. II: Characteristics of lymphocyte reactivity and distribution of E-rossete forming cells at different times after the onset of lactation. J Pediat. 1978;92:550–555. doi: 10.1016/s0022-3476(78)80286-8. [DOI] [PubMed] [Google Scholar]
- 37.van de Perre P. Breast milk transmission of HIV-1. Laboratory and clinical studies. Ann N Y Acad Sci. 2000;918:122–127. doi: 10.1111/j.1749-6632.2000.tb05480.x. [DOI] [PubMed] [Google Scholar]
- 38.Southern S, Southern P. Cellular mechanism for milk-borne transmission of HIV and HTLV. In: David EA, editor. Integrating population outcomes, biological mechanisms and research methods in the study of human milk and lactation. New York: Kluwer Academic/Plenum Publishers; 2002. pp. 183–190. [Google Scholar]
- 39.Buseyne F, Le Chenadec J, Corre B, Porrot F, Burgard M, Rouzioux C, et al. Inverse correlation between memory gag-specific cytotoxic T lymphocytes and viral replication in human immunodeficiency virus-infected children. J Infect Dis. 2002;186:1589–1596. doi: 10.1086/345482. [DOI] [PubMed] [Google Scholar]
- 40.Chouquet C, Autran B, Gomard E, Bouley J-M, Calvez V, Katlama C, et al. Correlation between breadth of memory HIV-specific cytotoxic T cells, viral load and disease progression in HIV infection. AIDS. 2002;16:2399–2407. doi: 10.1097/00002030-200212060-00004. [DOI] [PubMed] [Google Scholar]
- 41.Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan MJ, Goepfert PA. Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J Virol. 2002;76:2298–2305. doi: 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mollet L, Li T-S, Samri A, Tournay C, Tubiana R, Calvez V, et al. Dynamics of HIV-specific CD8+ T lymphocytes with changes in viral load. J Immunol. 2000;165:1692–1704. doi: 10.4049/jimmunol.165.3.1692. [DOI] [PubMed] [Google Scholar]
- 43.Novitsky V, Gilbert P, Peter T, McLane MF, Gaolekwe S, Rybak N, et al. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J Virol. 2003;77:882–890. doi: 10.1128/JVI.77.2.882-890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patke DS, Langan SJ, Carruth LM, Keating SM, Sabundayo BP, Margolick JB, et al. Association of gag-specific T lymphocyte responses during the early phase of human immunodeficiency virus type 1 infection and lower virus set point. J Infect Dis. 2002;186:1177–1180. doi: 10.1086/343811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.