A decrease in Cdc42 activation during mitotic exit is necessary to allow localization of key cytokinesis regulators and proper septum formation.
Abstract
The role of Cdc42 and its regulation during cytokinesis is not well understood. Using biochemical and imaging approaches in budding yeast, we demonstrate that Cdc42 activation peaks during the G1/S transition and during anaphase but drops during mitotic exit and cytokinesis. Cdc5/Polo kinase is an important upstream cell cycle regulator that suppresses Cdc42 activity. Failure to down-regulate Cdc42 during mitotic exit impairs the normal localization of key cytokinesis regulators—Iqg1 and Inn1—at the division site, and results in an abnormal septum. The effects of Cdc42 hyperactivation are largely mediated by the Cdc42 effector p21-activated kinase Ste20. Inhibition of Cdc42 and related Rho guanosine triphosphatases may be a general feature of cytokinesis in eukaryotes.
Introduction
During cytokinesis, Rho GTPases orchestrate a dramatic rearrangement of the cortical cytoskeleton. In fungi and animals, Polo kinase triggers activation of Rho1/RhoA at the cell equator during late anaphase, which in turn promotes contractile actin ring (CAR) assembly and contraction (Yoshida et al., 2006; Petronczki et al., 2008; Green et al., 2012). In contrast to the central positive role of Rho1/RhoA in cytokinesis, the role of other Rho GTPases, including Rac1 and Cdc42, is less well understood. Recent work suggests that Rac1 inhibition is important for normal cytokinesis, at least in part by preventing PAK1-dependent adhesion at the division site (Canman et al., 2008; Bastos et al., 2012). Cdc42’s function is especially unclear. In many cell types, including budding yeast, Cdc42 is dispensable for cytokinesis; however, in other experiments, constitutively active Cdc42 mutants compromise cytokinesis (Jantsch-Plunger et al., 2000; Tolliday et al., 2002; Iwase et al., 2006; Rincon et al., 2007; Canman et al., 2008; Jordan and Canman, 2012). Whether inhibition of Cdc42 contributes to cytokinesis is unknown.
In budding yeast, Cdc42 is essential in late G1 for actin polarization and for the assembly of a septin collar between the mother and newly forming daughter bud (Adams et al., 1990; Gladfelter et al., 2002; Caviston et al., 2003; Bi and Park, 2012). Although current models and indirect evidence suggest that Cdc42 is activated during bud emergence, this has never been measured experimentally (Moffat and Andrews, 2004; Knaus et al., 2007; McCusker et al., 2007; Sopko et al., 2007; Bi and Park, 2012). Cdc42 also has an incompletely understood role in activating the mitotic exit network signaling pathway that promotes mitotic exit (Höfken and Schiebel, 2002, 2004; Seshan et al., 2002; Monje-Casas and Amon, 2009). After cytokinesis, local inhibition of Cdc42 at the bud neck is required to position the site of polarized bud growth in the subsequent cell cycle (Tong et al., 2007; Meitinger et al., 2013).
Here, we demonstrate that in budding yeast Cdc42 activity is inhibited during cytokinesis, and this inhibition is at least partly mediated by Cdc5/Polo kinase. Forced activation of Cdc42, at a level comparable to Cdc5-dependent inhibition, impairs the normal accumulation of important cytokinesis proteins at the division site. The resulting cytokinesis defect is mediated by the Cdc42 effector p21-activated kinase (PAK) Ste20. Together, our findings define a new Polo kinase–dependent regulatory circuit that inactivates Cdc42 in parallel with the activation of Rho1 during cytokinesis.
Results and discussion
GTP-Cdc42 levels peak at G1/S and anaphase but fall during cytokinesis
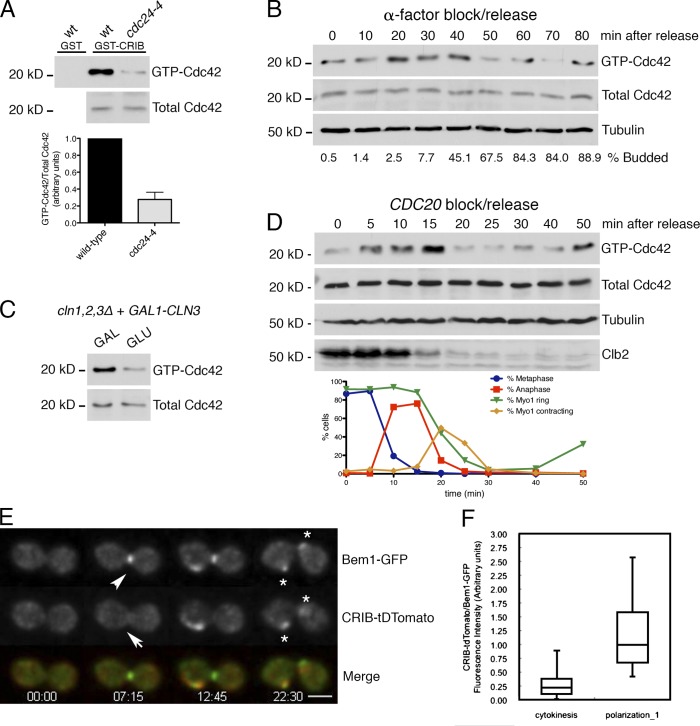
We monitored GTP loading of Cdc42 from synchronized cells using a Cdc42/Rac interactive binding (CRIB) domain pull-down assay. Control experiments established that the assay faithfully reports Cdc42 activation: for example, GTP-Cdc42 levels were significantly reduced after inactivation of the sole Cdc42–guanine nucleotide exchange factor Cdc24 using a temperature-sensitive (ts) allele compared with controls (Figs. 1 A and S1, A–D).
Figure 1.
Cdc42 activity peaks at G1/S and anaphase and is suppressed during cytokinesis. (A) Wild-type (wt) and cdc24-4 ts cells were shifted to 37°C for 2.5 h, and lysates were assayed for GTP-Cdc42 levels by GST-CRIB binding. Graph shows means ± SEM from three experiments, P < 0.01 by unpaired two-tailed t test. (B) bar1Δ cells were released from an α-factor block, and GTP-Cdc42 was measured at the indicated time points. (C) cln1,2,3Δ GAL1-CLN3 cells were split into media containing either galactose (GAL) or glucose (GLU), in which CLN3 expression is repressed. The example is representative of three experiments. (D) GALL-CDC20 GFP-TUB1 MYO1-GFP cells were arrested at metaphase and released. Samples were processed to measure GTP-Cdc42 activation and for microscopy to determine cell cycle stage. The example is representative of three experiments. (E) Time-lapse imaging of a CRIB-tdTomato BEM1-GFP cell undergoing mitotic exit. Image 00:00 (minutes and seconds) is 1 h after release from hydroxyurea. Bar, 5 µm. During cytokinesis, little CRIB-tdTomato (arrow) localizes to the bud neck relative to Bem1-GFP (arrowhead); both localize to the subsequent bud sites (asterisks). (F) Quantification of CRIB-tdTomato/Bem1-GFP intensity ratio during cytokinesis and polarization. Lines are medians, top and bottom edges indicate 25th and 75th quartiles, and whiskers are the highest and lowest measurements.
To examine Cdc42 activity in cells progressing from early G1 through START, the time of actin polarization, we measured Cdc42 activity and budding index in synchronized cells at intervals after release from a G1 arrest. Cdc42 activity peaked around 30 min after release, slightly before bud emergence (Fig. 1 B). Furthermore, cells arrested by depletion of G1 cyclins had lower Cdc42 activity than an asynchronous culture (Fig. 1 C), supporting the idea that cells in early G1 have low Cdc42 activity (Sopko et al., 2007).
To determine whether Cdc42 activity changes later in the cell cycle, we measured Cdc42 activity and mitotic progression in synchronized cells released from a metaphase (CDC20) block. We observed a transient increase in Cdc42 activity coinciding with anaphase (10–15 min after release) followed by a sharp drop during cytokinesis (20–25 min), as judged by spindle breakdown, contraction of the type II myosin Myo1 (VerPlank and Li, 2005), and cyclin B Clb2 degradation (Fig. 1 D). Cdc42 activity remained low through early G1 until cells entered the next cell cycle (50 min).
Cdc42, Cdc24, and the polarity scaffold Bem1 localize to the bud neck during cytokinesis (Gulli et al., 2000; Richman et al., 2002), but the extent to which this pool of Cdc42 is active is unclear. We compared localization of a marker for active Cdc42, CRIB-tdTomato (Tong et al., 2007), to Bem1-GFP. Strikingly, in every cell, relatively little CRIB-tdTomato localized to the bud neck during cytokinesis compared with Bem1-GFP. In contrast, both fluorescent proteins robustly localized at the new budding site (n = 17; Fig. 1, E and F). Comparing CRIB-tdTomato to GFP-Cdc42 gave a similar result, but we focused on Bem1-GFP because GFP-Cdc42 is only partially functional. These data indicate that although Cdc42 localizes to the bud neck after mitotic exit, Cdc42 is largely inactive at this site and only becomes activated at the adjacent bud site when cells polarize in the next cell cycle (Tong et al., 2007; Meitinger et al., 2013).
Overall, we identified two peaks of Cdc42 activation at G1/S and anaphase, with troughs during G2 and cytokinesis/early G1. Although current models and indirect evidence suggest that Cdc42 is activated during polarization, this has never been directly measured. Our data provide experimental support for this idea. The anaphase peak may reflect Cdc42’s upstream regulation of the mitotic exit network (Höfken and Schiebel, 2002, 2004; Seshan et al., 2002; Monje-Casas and Amon, 2009). Consistent with our results, the activity of the Cdc42 effector PAK Cla4 is periodic and is highest in late mitosis (Benton et al., 1997; Tjandra et al., 1998).
Cdc5/Polo inhibits Cdc42 activity
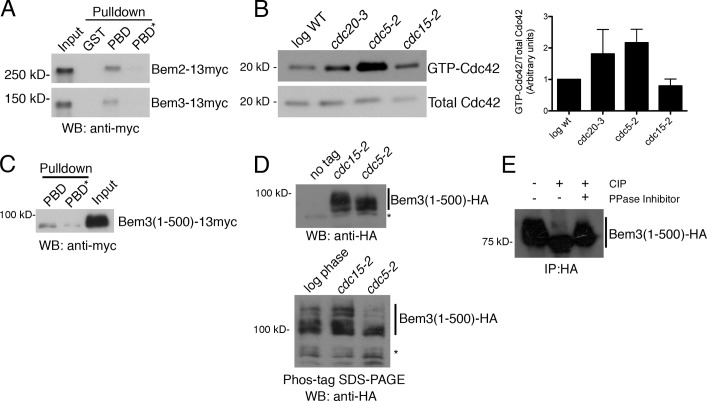
Because Cdc5/Polo controls Rho1 activation during late anaphase and because we found that the Cdc42 GTPase-activating proteins (GAPs) Bem2 and Bem3 interact with the Polo-box domain of Cdc5 in a canonical manner for Cdc5 substrates (Fig. 2 A; this study; Elia et al., 2003; Yoshida et al., 2006), we hypothesized that Cdc5/Polo may regulate Cdc42 reciprocally with its regulation of Rho1. To determine whether loss of Cdc5 affects the activation of Cdc42, cells were arrested at the end of mitosis by conditional inactivation of Cdc15 (cdc15-2 cells, which arrest with high Cdc5 kinase activity; Cheng et al., 1998) or Cdc5 (cdc5-2), and GTP loading of Cdc42 was measured. For comparison, GTP-Cdc42 was also measured in control asynchronous cells or cells arrested in mitosis by conditional inactivation of Cdc20 (cdc20-3). At the mitotic exit block, cells that contain active Cdc5 (cdc15-2) had significantly lower levels of GTP-Cdc42 relative to cells lacking functional Cdc5 (cdc5-2; Fig. 2 B).
Figure 2.
Cdc5/Polo kinase suppresses Cdc42 activation. (A) Lysates from the indicated strains were incubated with GST, GST-PBD, or a control “pincer” mutant PBD* (Elia et al., 2003). Bound GAPs were detected by Western blotting. (B) Log phase wild-type (WT), cdc20-3, cdc5-2, and cdc15-2 cells were shifted to 37°C for 3 h and processed for Cdc42 activation. The example is representative of three experiments. Graph shows means ± SEM. The difference between cdc5-2 and cdc15-2 was statistically significant (P < 0.05 by unpaired two-tailed t test). (C) Lysate from a Bem3(1–500)-13myc cdc15-2 strain (arrested 2.5 h at 35°C) was incubated with purified PBD or PBD* as in A. (D) The indicated strains were arrested as in C; shown is a Western blot to detect altered mobility of Bem3(1–500)-3HA. (bottom) Phos-tag SDS-PAGE was used for increased resolution of phosphorylated bands. Asterisks mark nonspecific bands. (E) Bem3(1–500)-3HA was immunoprecipitated from cdc15-2 cell lysates arrested in telophase as in C and incubated with calf intestinal phosphatase (CIP) and/or phosphatase (PPase) inhibitors. IP, immunoprecipitation; WB, Western blot.
An N-terminal fragment of Bem3 was sufficient to bind the Polo-box domain (Fig. 2 C). Interestingly, this fragment of Bem3 was phosphorylated in a Cdc5-dependent manner (detected by SDS-PAGE and Phos-tag gels; Fig. 2, D and E), suggesting that Bem3 may be a Cdc5 substrate. We were unable to detect a mobility shift for Bem2, possibly because of its large (∼250 kD) size. Although the details of the mechanism by which Cdc5 controls Cdc42 activity remain to be elucidated, these findings raise the possibility that Cdc5 may regulate Cdc42, at least in part, through its GAPs. Most importantly, our findings show that negative regulation of Cdc42 during mitotic exit is integrated into the cell cycle circuitry that controls cytokinesis.
Genetic evidence that Cdc42 inhibits cytokinesis
In budding yeast, CAR contraction guides Chs2 (chitin synthase II)-dependent primary septum (PS) formation (Shaw et al., 1991; Schmidt et al., 2002b; VerPlank and Li, 2005; Nishihama et al., 2009; Fang et al., 2010). Subsequently, a secondary septum is deposited surrounding the PS. Both CAR assembly and septum formation depend upon the septin cytoskeletal scaffold, which splits into two rings that delimit the region of cytokinesis (Hartwell, 1971; Dobbelaere et al., 2003). In many yeast strains, the CAR is not essential for cytokinesis, but the cells survive because septum formation, although abnormal, is sufficient to divide mother and daughter cells (Bi et al., 1998; Vallen et al., 2000). However, cells lacking both genes required for CAR assembly and genes required for septum formation cannot survive.
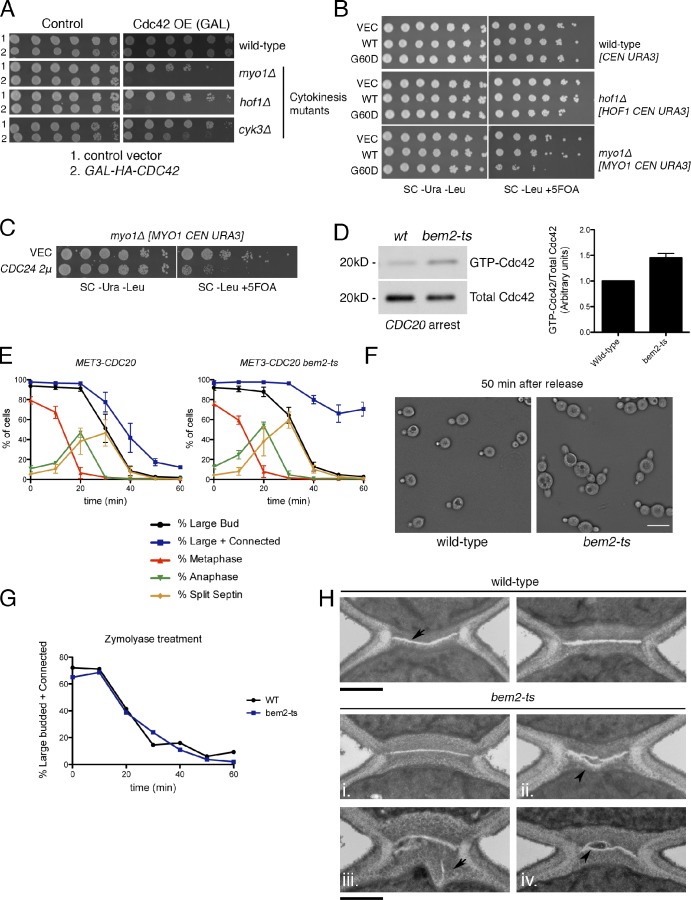
Because Cdc42 is suppressed during cytokinesis, we hypothesized that Cdc42 inhibition may be required for normal cytokinesis. Consistently, Cdc42 overexpression killed cytokinesis mutants that affect either CAR assembly or septation (Fig. 3 A). Interestingly, expression of a recessive but biochemically hyperactive allele, cdc42G60D, from a centromeric (CEN) plasmid was extremely toxic to cells defective for CAR assembly (myo1Δ) but much less so to cells lacking Hof1, a gene that does not affect CAR assembly but is required for normal septation (Figs. 3 B and S2 F; Vallen et al., 2000; Caviston et. al., 2002). Further consistent with the conclusion that Cdc42 activation interferes with cytokinesis, overexpression of the Cdc42–guanine nucleotide exchange factor Cdc24 was extremely toxic to myo1Δ cells (Fig. 3 C), cells lacking both Myo1 and the Cdc42-GAP Bem2 are inviable, and overexpression of Cdc42-GAPs suppresses mutants defective in primary septum formation (Wang and Bretscher, 1995; see Onishi et al. in this issue).
Figure 3.
Active Cdc42 interferes with cytokinesis and cell separation. (A) Cells transformed with vector (VEC) or GAL1-HA-CDC42 plasmids were spotted in fivefold dilutions on the indicated media (3 d at 25°C). OE, overexpression; GAL, galactose. (B) Expression of cdc42G60D from the CDC42 promoter on a CEN plasmid (one to three copies per cell) is toxic to myo1Δ cells. Cells were grown on the indicated media (3 d at 25°C). Synthetic lethality is detected on 5-fluoroorotic acid (5FOA) media as a counterselection for the indicated URA3-containing plasmids. (C) myo1Δ cells transformed with control or CDC24 2μ plasmids were grown on plates as in B. SC, synthetic complete. (D) Wild-type (wt) and bem2-ts cells were arrested in metaphase by CDC20 depletion, shifted to the restrictive temperature (37°C at 1.5 h), and processed for Cdc42 activation. Graph shows means ± SEM from three experiments. (E) Cells were arrested in metaphase, shifted to 37°C for 1 h, and released at 37°C. The percentage of cells with the indicated morphology is plotted as means ± SEM from three experiments. Connected indicates two adjacent cell bodies showing evidence of rebudding or repolarization. (F) Bright-field image of cells 50 min after release. Bar, 15 µm. (G) Cells from E were treated with zymolyase to digest the cell wall, and the percentage of large-budded or connected cells was scored. (H) Wild-type cells have a PS (arrow) sandwiched by secondary septa; bem2-ts cells have misaligned, multiple, or otherwise abnormal PSs (arrowheads). Bars, 500 nm.
Activation of Cdc42 during mitosis causes a defect in cell separation
Septins are essential for cytokinesis in budding yeast, and Cdc42 hyperactivation might indirectly affect cytokinesis by impairing normal septin assembly (Gladfelter et al., 2002). To exclude the possibility that the aforementioned genetic interactions could be an indirect consequence of septin assembly defects early in the cell cycle, we activated Cdc42 in cells released from a metaphase block. Cdc42 activation was achieved using a novel ts allele of the gene encoding the Cdc42-GAP BEM2. Although budding yeast has several Cdc42 GAPs, Bem2 appears to be the most important (Wang and Bretscher, 1995; Marquitz et al., 2002; Knaus et al., 2007). Importantly, metaphase-arrested bem2-ts cells had a modest (∼1.5-fold) increase in GTP-Cdc42 that is comparable in magnitude to down-regulation of Cdc42 that occurs during cytokinesis and is also comparable to the degree of inhibition of Cdc42 by Cdc5 (Fig. 3 D).
We observed an obvious cell separation defect in synchronized bem2-ts cultures released from a CDC20 block at 37°C (Figs. 3, E and F; and S2 C). Anaphase spindle elongation, spindle breakdown and septin ring splitting all occurred on schedule, demonstrating that this level of Cdc42 activation did not impair mitotic exit (Cid et al., 2001; Lippincott et al., 2001). As expected from previous work, bem2-ts cells had a defect in axial bud site selection (Kim et al., 1994). The connected bem2-ts cells were resolved by zymolyase, indicating a cell separation defect rather than a complete cytokinesis block (Fig. 3 G). Similarly, short-term conditional expression of GTP-locked Cdc42 (CDC42Q61L) during mitosis also caused cells to remain connected after mitotic exit (Fig. S2, A and B). Thus, Cdc42 activation causes a defect in cell separation.
The Bem2-GAP domain promotes GTP hydrolysis by both Cdc42 and Rho1 in vitro (Peterson et al., 1994; Marquitz et al., 2002). Several lines of evidence strongly suggest that Bem2’s effect on cell separation and cytokinesis (see following section) is caused by activation of Cdc42, not Rho1. Rho1- and Pkc1-dependent phosphorylation of Mpk1 was unaffected in synchronized bem2-ts cells at 37°C (Fig. S2 D), consistent with earlier work (Schmidt et al., 2002a), and the percentage of cells with bud neck localization of a marker for active Rho1 was not elevated in bem2-ts cells undergoing cytokinesis (Fig. S2 E); indeed, it was modestly reduced. Finally, Rho1 activation promotes rather than compromises cytokinesis, particularly in mutants with cytokinesis defects (Yoshida et al., 2009; Meitinger et al., 2013; Onishi et al., 2013).
The cell separation defect of bem2-ts cells is caused by a defect in cytokinesis/septum formation
Multiple mechanisms could explain the cell separation defect of bem2-ts cells: a defect in cytokinesis that indirectly compromises cell separation, abnormal signaling through the regulation of Ace2 and morphogenesis network, or abnormal secretory vesicle delivery to the bud neck (Weiss, 2012). The latter two explanations are strongly disfavored because the regulation of Ace2 and morphogenesis network target Ace2 and the secreted chitin synthases Chs2 and Chs3 localized normally in bem2-ts cells (Fig. S3, A–C). Because we observed aberrant chitin staining at the bud necks of many connected bem2-ts cells (Fig. S3 D), consistent with a defect in septum formation, we focused on the possibility that bem2-ts cells undergo defective cytokinesis.
EM was performed to determine whether bem2-ts cells have a defect in septum formation. Wild-type cells had well-organized trilaminar septa, with an electron-lucent PS sandwiched by secondary septum (n = 34/36; Fig. 3 H; Cabib et al., 1974). In contrast, ∼31% of the bem2-ts cells had misorganized PSs (n = 11/35) that were misaligned, bifurcated, and/or contained extra PS (Fig. 3 H). In some cases, the adjacent secondary septum appeared to be thickened or enlarged (e.g., 3Hiii). Thus, bem2-ts cells have a defect in septum formation (Cid et al., 1998), and this defect is not explained by early cell cycle defects in the assembly of the septin scaffold (Fig. 3 E).
Inefficient localization of Iqg1 and Inn1 to the bud neck in bem2-ts cells
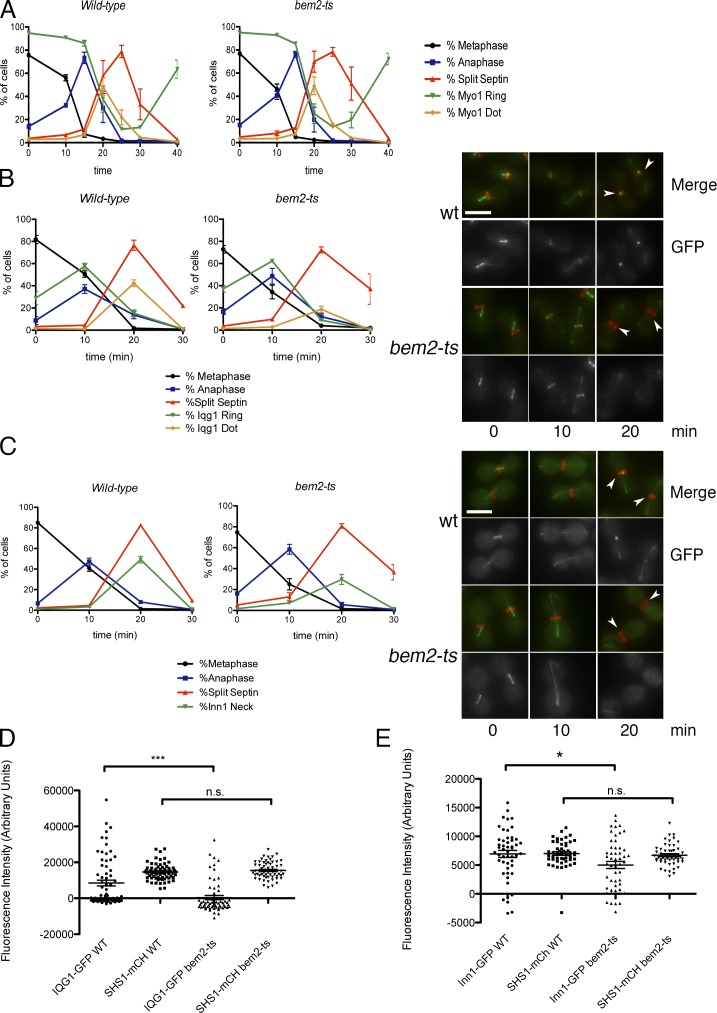
Myo1 localizes as a ring at the bud neck from late G1 until cytokinesis, when it undergoes contraction and disassembly (Bi et al., 1998; Lippincott and Li, 1998). Myo1 underwent apparent contraction with normal timing in synchronized bem2-ts cells, arguing that the effects of Cdc42 activation are not likely to be mediated by compromised Myo1 function (Fig. 4 A). The F-BAR protein Hof1 also localized normally in bem2-ts cells (Fig. S3 E).
Figure 4.
Inefficient localization of Iqg1 and Inn1 during cytokinesis in bem2-ts cells. (A) MET3-CDC20 GFP-TUB1 SHS1-mCherry MYO1-GFP strains with or without bem2-ts were synchronized as in Fig. 3 E. Graph shows means ± SEM for three experiments. (B) Similar strains as in A but with IQG1-GFP were synchronized as in Fig. 3 E. IQG1-GFP, if present at the bud neck, was scored as either being a ring or contracted dot. The percentage of cells with the indicated localization pattern is plotted as means ± SEM from three experiments. (right) Representative images of cells. Arrowheads show Iqg1 bud neck localization. (C) Similar strains as in A but with INN1-GFP. Percentage of Inn1 neck represents percentage of cells with Inn1-GFP at the bud neck. (right) Representative images of cells from the indicated time points. Arrowheads show Inn1 bud neck localization. (D) Scatter plot of Iqg1-GFP and Shs1-mCherry background-corrected fluorescence intensity at the bud neck in bem2-ts cells 20 min after release from CDC20 arrest in SHS1-mCherry IQG1-GFP cells. Note that negative values after background subtraction indicate either increased cytoplasmic background or completion of cytokinesis with a corresponding loss of background signal in the bud neck area. Lines indicate means, and error bars represent SEMs; n > 60 cells. The difference for Iqg1-GFP intensity between wild type (WT) and bem2-ts was statistically significant (P < 0.0001 by unpaired two-tailed t test), whereas the difference for Shs1-mCherry (mCh) was not significant. (E) Inn1-GFP fluorescence intensity at the bud neck was measured as in D; n > 50 cells. The difference for Inn1-GFP was significant (P < 0.05 by unpaired two-tailed t test). Bars, 5 µm.
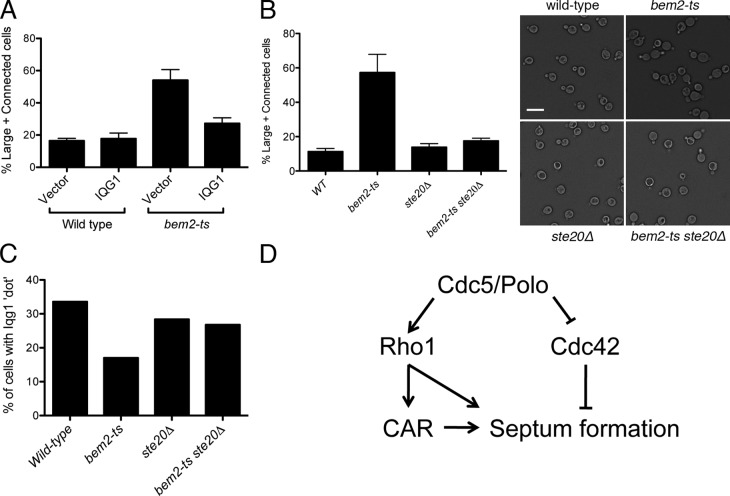
Interestingly, the IQGAP homologue Iqg1, a protein involved in both CAR assembly and septum formation (Osman and Cerione, 1998; Shannon and Li, 1999; Ko et al., 2007; Fang et al., 2010), and Inn1, a protein involved in PS formation (Sanchez-Diaz et al., 2008; Jendretzki et al., 2009; Nishihama et al., 2009; Meitinger et al., 2010), were inefficiently localized to the bud neck in bem2-ts cells undergoing mitotic exit. The initial anaphase ring of Iqg1-GFP formed normally in bem2-ts cells, but Iqg1-GFP was localized to the bud neck of approximately twofold fewer cells during the cytokinesis peak of 20 min after release, when Iqg1 localization appears as a contracted “dot” (Fig. 4 B). The defect in Iqg1 localization was more obvious after measurement of the fluorescence intensity of Iqg1 at the bud neck (Fig. 4 D). A similar but more modest defect was observed for Inn1-GFP in bem2-ts cells (Fig. 4, C and E). These data suggest that persistent Cdc42 activation during mitotic exit leads to a failure to maintain normal amounts of two key proteins required for septum formation. This localization defect appears to be functionally significant because Iqg1 overexpression suppressed the cell separation defect of bem2-ts cells to nearly wild-type levels (Fig. 5 A).
Figure 5.
Ste20 and Iqg1 mediate the inhibitory effect of active Cdc42 on cytokinesis and cell separation. (A) Cells transformed with 2μ IQG1 or a control vector were synchronized as in Fig. 3 E, and the percentage of large-budded or connected cells was determined 50 min after release. Graph shows means ± SEM from three experiments using independent 2μ-containing isolates (B) Cells were synchronized and scored as in A. Graph shows means ± SEM for three experiments. Bar, 10 µm. (C) Percentage of cells with contracting Iqg1-GFP dot 20 min after release from CDC20 arrest. Data are representative of two experiments; n > 200 cells. WT, wild type.
Iqg1 has been suggested to coordinate CAR constriction and PS formation (Fang et al., 2010). Therefore, defective Iqg1 localization could explain the observed septation defect of bem2-ts cells. Although Inn1 localization is under complex control (Jendretzki et al., 2009; Nishihama et al., 2009; Meitinger et al., 2010), in some strain backgrounds, Inn1 localization depends on Iqg1, suggesting that Cdc42 could affect Inn1 indirectly (Sanchez-Diaz et al., 2008). Overall, these data suggest that the mechanism by which Cdc42 activation impairs septum formation involves mislocalization of Iqg1 and Inn1.
Cdc42 effector Ste20 mediates Cdc42’s inhibitory effect on cytokinesis
As a first step toward identifying the signaling mechanism linking Cdc42 to Iqg1 and Inn1, we determined whether known Cdc42 effectors are required to mediate the effect of persistent Cdc42 activation on cytokinesis. We focused on the Cdc42 effector PAKs because of their known roles in mitotic exit. Interestingly, deletion of the gene for the PAK STE20 significantly reversed the cell separation defect of bem2-ts cells (Fig. 5 B), suggesting that Ste20 is a key mediator of the adverse effects of hyperactive Cdc42. Moreover, loss of Ste20 in bem2-ts cells largely restored Iqg1 localization to the bud neck as a contracted dot during cytokinesis (Fig. 5 C).
The regulatory effect of Ste20 could be mediated by direct phosphorylation of Iqg1 or by indirect mechanisms. Because of the previously described, but functionally unclear, interaction between Cdc42 and Iqg1, we do not exclude the possibility of direct effects of Cdc42 on Iqg1 (Osman and Cerione, 1998; Shannon and Li, 1999). Our data, combined with previous work in Schizosaccharomyces pombe and mammalian cells, suggest that negative regulation of cytokinesis by PAKs is highly conserved (Loo and Balasubramanian, 2008; Bastos et al., 2012).
In this study, we have characterized the cell cycle dynamics of Cdc42 activation in budding yeast. Cdc42 activity drops during cytokinesis, and Cdc5/Polo kinase is one upstream negative regulator of Cdc42 activity (Fig. 5 D). Forced activation of Cdc42, to levels comparable to that during normal mitosis, suggests that negative regulation of Cdc42 is required for normal cytokinesis. Because Cdc42, Polo kinase, IQGAP, and PAKs are highly conserved, we speculate that a similar negative regulatory loop may be important for cytokinesis in other organisms.
Materials and methods
Strains, media, and molecular biology
Standard approaches were used for molecular biology and genetic manipulation of yeast (Sherman, 1991). All yeast strains are isogenic or congenic with the S288c-derived BY4741 (MATa his3Δ leu2Δ met15Δ ura3Δ). Gene deletion or modification was confirmed by PCR. For a complete list of yeast strains and plasmids, see Tables S1 and S2.
Cell synchronization
Synchronization experiments were performed essentially as previously described (Amon, 2002). For α-factor block/release, 0.5 µg/ml α-factor (Zymo Research) was added to log phase cultures until >90% of cells were arrested. To release the cells from the arrest, the cells were washed and resuspended in media containing 50 µg/ml protease type XIV (Sigma-Aldrich). For MET3-CDC20 block/release, cultures grown to log phase in SD (synthetic complete media containing dextrose)-Met media were transferred to YEPD (yeast extract/peptone/dextrose) supplemented with 2 mM methionine until >90% of cells were large budded. For temperature shift experiments, cells were transferred to fresh prewarmed media. To release the cells into the cell cycle, the cells were washed with three culture volumes of SD-Met and resuspended in SD-Met.
Biochemical methods
Protein isolation and Western blotting were performed using standard approaches. Rat anti–α-tubulin antibody (Accurate Chemical & Scientific Corporation), rabbit anti-Cdc42 antibody (y-191; Santa Cruz Biotechnology, Inc.), rabbit anti-Clb2 antibody (y-180; Santa Cruz Biotechnology, Inc.), rabbit anti-Mpk1 antibody (y-244; Santa Cruz Biotechnology, Inc.), and rabbit P-p44/42 MAPK antibody (Cell Signaling Technology) were obtained from commercial sources. Densitometry was performed using ImageJ software (National Institutes of Health).
For phosphatase treatment, cell lysates were prepared in ice-cold PBS supplemented with phosphatase inhibitor cocktail (Roche). Bem3-3HA was immunoprecipitated from the lysates by addition of the anti-HA antibody (12CA5) followed by protein A–Sepharose beads (GE Healthcare). The beads were split into three aliquots; one was mock treated, the second was treated with calf intestinal alkaline phosphatase (New England Biolabs, Inc.), and the third was treated with calf intestinal alkaline phosphatase and phosphatase inhibitor cocktail. After 30-min incubation at 37°C, samples were boiled and analyzed by Western blotting using the anti-HA antibody (12CA5).
Cdc42 activation assay (CRIB pull-down)
The assay was adapted for use in yeast based on previous methods (Benard and Bokoch, 2002). Yeast lysates were prepared by bead beating in lysis buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 12 mM MgCl2, and 1 mM DTT) supplemented with protease inhibitors (Complete Mini EDTA-free; Roche) and 0.2 mM PMSF using a cell disruptor (Mini-BeadBeater-16; BioSpec Products) followed by addition of 1% Nonidet P-40. Clarified lysates were incubated with purified GST-CRIB for 30 min at 4°C. The beads were washed three times, boiled in SDS sample buffer, and subjected to Western blotting. Wild-type and cdc24-4 control samples were always processed in parallel as a quality control for the beads.
Isolation of bem2-ts mutants
bem2-ts mutants were isolated by a mutagenic PCR and gap repair strategy similar to that used for cdc42 mutants (Moskow et al., 2000). In brief, mutants were generated using error-prone PCR of the Bem2 GAP domain and recombined into a low-copy plasmid by gap repair (homologous recombination in yeast). Mutants were screened for those that could complement the synthetic lethality of cla4Δ bem2Δ and rga1Δ bem2Δ strains at 24°C but not 37°C. The tight bem2-84 mutant was sequenced and found to carry two amino acid substitutions (K1986R and I2041N) in the GAP domain.
Microscopy and image analysis
Cells were imaged using an automated microscope (AX10; Carl Zeiss) equipped with a 63×, 1.4 NA Plan Apochromat objective at room temperature. Images were acquired with a camera (CoolSNAP HQ; Roper Scientific). The microscope and camera were controlled by SlideBook (Intelligent Imaging Innovations) software, which was also used for image analysis. Photoshop (Adobe) was used for assembling figures. Cells were fixed by the addition of 3.7% formaldehyde directly to the culture for 5–10 min followed by washing three times with PBS. Fixed cells were vortexed for 90 s before imaging. Cells were imaged at 0.3-µm intervals in the z plane to characterize localization of proteins. For scoring cell morphology, cells were considered connected if two equivalent-sized cell bodies were directly adjacent and showed evidence of repolarization or rebudding. More than 200 cells were scored for each time point for each synchronization experiment. Zymolyase treatment and calcofluor white staining were performed as previously described (Pringle, 1991; Tolliday et al., 2002). In brief, for zymolyase treatment, cells were fixed for 1 h with 3.7% formaldehyde, washed three times with PBS and twice with KS buffer (1.2 M sorbitol and 50 mM KHPO4), and then resuspended in KS containing 0.2 mg/ml zymolyase (Zymo Research) and 2 mM DTT until >90% of cells lost their refractive appearance. To visualize chitin, cells were fixed with 3.7% formaldehyde, washed three times with PBS, incubated with 1 mg/ml calcofluor white (Fluorescent Brightener 28; Sigma-Aldrich) for 5 min, and then washed three times with PBS before imaging. For fluorescence microscopy images, unless otherwise noted, a maximum-intensity projection of a z stack is shown.
For quantification of CRIB-tdTomato and Bem1-GFP fluorescence intensity, cells were released from a hydroxyurea arrest for ∼1 h and imaged using 30 z planes per time point. Images were deconvolved, and for each cell in a video, we picked two time points: (1) when the neck Bem1-GFP signal was brightest (cytokinesis) and (2) when the subsequent polarized Bem1-GFP was brightest (polarization). Whichever daughter polarized first was used for the analysis. A region of interest (neck or polarity site) was selected, and the mean intensity of the Gic2 CRIB-tdTomato probe was divided by the mean intensity of the Bem1-GFP probe (after subtracting background in each case). For transmission EM, cells were released from a metaphase arrest, and samples from 20 and 30 min after release were pooled and fixed with 2.5% glutaraldehyde for 1 h, washed twice with water, resuspended in 4% potassium permanganate, washed twice with water, and incubated with 7% uranylacetate for 30 min followed by dehydration and embedding, as described previously (Meitinger et al., 2010).
Online supplemental material
Fig. S1 shows additional control experiments for the CRIB pull-down assay. Fig. S2 shows additional data for the cell separation defect of cells with hyperactive Cdc42. Fig. S3 shows localization of proteins involved in cytokinesis and cell separation in bem2-ts cells. Table S1 shows yeast strains used in this study. Table S2 shows plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201301090/DC1.
Supplementary Material
Acknowledgments
We thank E. Bi, C. Boone, F. Uhlmann, J. Heinisch, Y. Zheng, S. Valentini, M. Peter, and G. Fink for reagents; M. Onishi and J. Pringle for communicating unpublished results; F. Meitinger, G. Pereira, and the Harvard Medical School EM core for EM advice and assistance; and the Pellman laboratory, especially K. Kono, X. Su, K. Crasta, and S. Godinho for discussions and advice.
This work was supported by National Institutes of Health grants RO1 GM061345 and R01 GM62300.
Footnotes
Abbreviations used in this paper:
- CAR
- contractile actin ring
- CEN
- centromeric
- CRIB
- Cdc42/Rac interactive binding
- GAP
- GTPase-activating protein
- PAK
- p21-activated kinase
- PS
- primary septum
- ts
- temperature sensitive
References
- Adams A.E., Johnson D.I., Longnecker R.M., Sloat B.F., Pringle J.R. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111:131–142 10.1083/jcb.111.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A. 2002. Synchronization procedures. Methods Enzymol. 351:457–467 10.1016/S0076-6879(02)51864-4 [DOI] [PubMed] [Google Scholar]
- Bastos R.N., Penate X., Bates M., Hammond D., Barr F.A. 2012. CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis. J. Cell Biol. 198:865–880 10.1083/jcb.201204107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard V., Bokoch G.M. 2002. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 345:349–359 10.1016/S0076-6879(02)45028-8 [DOI] [PubMed] [Google Scholar]
- Benton B.K., Tinkelenberg A., Gonzalez I., Cross F.R. 1997. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol. 17:5067–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Park H.O. 2012. Cell polarization and cytokinesis in budding yeast. Genetics. 191:347–387 10.1534/genetics.111.132886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D.J., Salmon E.D., McMillan J.N., Yeh E., Pringle J.R. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142:1301–1312 10.1083/jcb.142.5.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E., Ulane R., Bowers B. 1974. A molecular model for morphogenesis: the primary septum of yeast. Curr. Top. Cell. Regul. 8:1–32 [DOI] [PubMed] [Google Scholar]
- Canman J.C., Lewellyn L., Laband K., Smerdon S.J., Desai A., Bowerman B., Oegema K. 2008. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science. 322:1543–1546 10.1126/science.1163086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J.P., Tcheperegine S.E., Bi E. 2002. Singularity in budding: a role for the evolutionarily conserved small GTPase Cdc42p. Proc. Natl. Acad. Sci. USA. 99:12185–12190 10.1073/pnas.182370299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J.P., Longtine M., Pringle J.R., Bi E. 2003. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 14:4051–4066 10.1091/mbc.E03-04-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Hunke L., Hardy C.F. 1998. Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p. Mol. Cell. Biol. 18:7360–7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid V.J., Cenamor R., Sánchez M., Nombela C. 1998. A mutation in the Rho1-GAP-encoding gene BEM2 of Saccharomyces cerevisiae affects morphogenesis and cell wall functionality. Microbiology. 144:25–36 10.1099/00221287-144-1-25 [DOI] [PubMed] [Google Scholar]
- Cid V.J., Adamiková L., Sánchez M., Molina M., Nombela C. 2001. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology. 147:1437–1450 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Gentry M.S., Hallberg R.L., Barral Y. 2003. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell. 4:345–357 10.1016/S1534-5807(03)00061-3 [DOI] [PubMed] [Google Scholar]
- Elia A.E., Rellos P., Haire L.F., Chao J.W., Ivins F.J., Hoepker K., Mohammad D., Cantley L.C., Smerdon S.J., Yaffe M.B. 2003. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 115:83–95 10.1016/S0092-8674(03)00725-6 [DOI] [PubMed] [Google Scholar]
- Fang X., Luo J., Nishihama R., Wloka C., Dravis C., Travaglia M., Iwase M., Vallen E.A., Bi E. 2010. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J. Cell Biol. 191:1333–1350 10.1083/jcb.201005134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A.S., Bose I., Zyla T.R., Bardes E.S., Lew D.J. 2002. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 156:315–326 10.1083/jcb.200109062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.A., Paluch E., Oegema K. 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28:29–58 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Gulli M.P., Jaquenoud M., Shimada Y., Niederhäuser G., Wiget P., Peter M. 2000. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 6:1155–1167 10.1016/S1097-2765(00)00113-1 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. 1971. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69:265–276 10.1016/0014-4827(71)90223-0 [DOI] [PubMed] [Google Scholar]
- Höfken T., Schiebel E. 2002. A role for cell polarity proteins in mitotic exit. EMBO J. 21:4851–4862 10.1093/emboj/cdf481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfken T., Schiebel E. 2004. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J. Cell Biol. 164:219–231 10.1083/jcb.200309080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M., Luo J., Nagaraj S., Longtine M., Kim H.B., Haarer B.K., Caruso C., Tong Z., Pringle J.R., Bi E. 2006. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Biol. Cell. 17:1110–1125 10.1091/mbc.E05-08-0793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V., Gönczy P., Romano A., Schnabel H., Hamill D., Schnabel R., Hyman A.A., Glotzer M. 2000. CYK-4: A Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149:1391–1404 10.1083/jcb.149.7.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendretzki A., Ciklic I., Rodicio R., Schmitz H.P., Heinisch J.J. 2009. Cyk3 acts in actomyosin ring independent cytokinesis by recruiting Inn1 to the yeast bud neck. Mol. Genet. Genomics. 282:437–451 10.1007/s00438-009-0476-0 [DOI] [PubMed] [Google Scholar]
- Jordan S.N., Canman J.C. 2012. Rho GTPases in animal cell cytokinesis: an occupation by the one percent. Cytoskeleton (Hoboken). 69:919–930 10.1002/cm.21071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Francisco L., Chen G.C., Marcotte E., Chan C.S. 1994. Control of cellular morphogenesis by the Ip12/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J. Cell Biol. 127:1381–1394 10.1083/jcb.127.5.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus M., Pelli-Gulli M.P., van Drogen F., Springer S., Jaquenoud M., Peter M. 2007. Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J. 26:4501–4513 10.1038/sj.emboj.7601873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko N., Nishihama R., Tully G.H., Ostapenko D., Solomon M.J., Morgan D.O., Pringle J.R. 2007. Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol. Biol. Cell. 18:5139–5153 10.1091/mbc.E07-05-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. 1998. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140:355–366 10.1083/jcb.140.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Shannon K.B., Shou W., Deshaies R.J., Li R. 2001. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J. Cell Sci. 114:1379–1386 [DOI] [PubMed] [Google Scholar]
- Loo T.H., Balasubramanian M. 2008. Schizosaccharomyces pombe Pak-related protein, Pak1p/Orb2p, phosphorylates myosin regulatory light chain to inhibit cytokinesis. J. Cell Biol. 183:785–793 10.1083/jcb.200806127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquitz A.R., Harrison J.C., Bose I., Zyla T.R., McMillan J.N., Lew D.J. 2002. The Rho-GAP Bem2p plays a GAP-independent role in the morphogenesis checkpoint. EMBO J. 21:4012–4025 10.1093/emboj/cdf416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker D., Denison C., Anderson S., Egelhofer T.A., Yates J.R., III, Gygi S.P., Kellogg D.R. 2007. Cdk1 coordinates cell-surface growth with the cell cycle. Nat. Cell Biol. 9:506–515 10.1038/ncb1568 [DOI] [PubMed] [Google Scholar]
- Meitinger F., Petrova B., Lombardi I.M., Bertazzi D.T., Hub B., Zentgraf H., Pereira G. 2010. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J. Cell Sci. 123:1851–1861 10.1242/jcs.063891 [DOI] [PubMed] [Google Scholar]
- Meitinger F., Richter H., Heisel S., Hub B., Seufert W., Pereira G. 2013. A safeguard mechanism regulates Rho GTPases to coordinate cytokinesis with the establishment of cell polarity. PLoS Biol. 11:e1001495 10.1371/journal.pbio.1001495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J., Andrews B. 2004. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat. Cell Biol. 6:59–66 10.1038/ncb1078 [DOI] [PubMed] [Google Scholar]
- Monje-Casas F., Amon A. 2009. Cell polarity determinants establish asymmetry in MEN signaling. Dev. Cell. 16:132–145 10.1016/j.devcel.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskow J.J., Gladfelter A.S., Lamson R.E., Pryciak P.M., Lew D.J. 2000. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:7559–7571 10.1128/MCB.20.20.7559-7571.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R., Schreiter J.H., Onishi M., Vallen E.A., Hanna J., Moravcevic K., Lippincott M.F., Han H., Lemmon M.A., Pringle J.R., Bi E. 2009. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J. Cell Biol. 185:995–1012 10.1083/jcb.200903125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M., Ko N., Nishihama R., Pringle J.R. 2013. Roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J. Cell Biol. 202:311–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman M.A., Cerione R.A. 1998. Iqg1p, a yeast homologue of the mammalian IQGAPs, mediates cdc42p effects on the actin cytoskeleton. J. Cell Biol. 142:443–455 10.1083/jcb.142.2.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J., Zheng Y., Bender L., Myers A., Cerione R., Bender A. 1994. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J. Cell Biol. 127:1395–1406 10.1083/jcb.127.5.1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Lénárt P., Peters J.M. 2008. Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev. Cell. 14:646–659 10.1016/j.devcel.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Pringle J.R. 1991. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 194:732–735 10.1016/0076-6879(91)94055-H [DOI] [PubMed] [Google Scholar]
- Richman T.J., Sawyer M.M., Johnson D.I. 2002. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot. Cell. 1:458–468 10.1128/EC.1.3.458-468.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon S., Coll P.M., Perez P. 2007. Spatial regulation of Cdc42 during cytokinesis. Cell Cycle. 6:1687–1691 10.4161/cc.6.14.4481 [DOI] [PubMed] [Google Scholar]
- Sanchez-Diaz A., Marchesi V., Murray S., Jones R., Pereira G., Edmondson R., Allen T., Labib K. 2008. Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat. Cell Biol. 10:395–406 10.1038/ncb1701 [DOI] [PubMed] [Google Scholar]
- Schmidt A., Schmelzle T., Hall M.N. 2002a. The RHO1-GAPs SAC7, BEM2 and BAG7 control distinct RHO1 functions in Saccharomyces cerevisiae. Mol. Microbiol. 45:1433–1441 10.1046/j.1365-2958.2002.03110.x [DOI] [PubMed] [Google Scholar]
- Schmidt M., Bowers B., Varma A., Roh D.H., Cabib E. 2002b. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115:293–302 [DOI] [PubMed] [Google Scholar]
- Seshan A., Bardin A.J., Amon A. 2002. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 12:2098–2110 10.1016/S0960-9822(02)01388-X [DOI] [PubMed] [Google Scholar]
- Shannon K.B., Li R. 1999. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol. Biol. Cell. 10:283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J.A., Mol P.C., Bowers B., Silverman S.J., Valdivieso M.H., Durán A., Cabib E. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114:111–123 10.1083/jcb.114.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 10.1016/0076-6879(91)94004-V [DOI] [PubMed] [Google Scholar]
- Sopko R., Huang D., Smith J.C., Figeys D., Andrews B.J. 2007. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 26:4487–4500 10.1038/sj.emboj.7601847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra H., Compton J., Kellogg D. 1998. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr. Biol. 8:991–1000 10.1016/S0960-9822(07)00419-8 [DOI] [PubMed] [Google Scholar]
- Tolliday N., VerPlank L., Li R. 2002. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr. Biol. 12:1864–1870 10.1016/S0960-9822(02)01238-1 [DOI] [PubMed] [Google Scholar]
- Tong Z., Gao X.D., Howell A.S., Bose I., Lew D.J., Bi E. 2007. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein–mediated zone of inhibition. J. Cell Biol. 179:1375–1384 10.1083/jcb.200705160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E.A., Caviston J., Bi E. 2000. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 11:593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L., Li R. 2005. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol. Biol. Cell. 16:2529–2543 10.1091/mbc.E04-12-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Bretscher A. 1995. The rho-GAP encoded by BEM2 regulates cytoskeletal structure in budding yeast. Mol. Biol. Cell. 6:1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E.L. 2012. Mitotic exit and separation of mother and daughter cells. Genetics. 192:1165–1202 10.1534/genetics.112.145516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Kono K., Lowery D.M., Bartolini S., Yaffe M.B., Ohya Y., Pellman D. 2006. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 313:108–111 10.1126/science.1126747 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Bartolini S., Pellman D. 2009. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 23:810–823 10.1101/gad.1785209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.