Abstract
Escherichia coli, a commensal bacterium from the intestinal tracts of humans and vertebrate animals, has been used as one of two bacterial indicators of fecal contamination, along with intestinal enterococci, to monitor the microbiological quality of water. However, water environments are now recognized as a secondary habitat where some strains can survive. We investigated the survival of E. coli isolates collected from bodies of water in France exhibiting distinct profiles of contamination, defined according to the following criteria: vicinity of the point sources of contamination, land use, hydrology, and physicochemical characteristics of the receiving water. We selected 88 E. coli strains among a collection of 352 strains to carry out a microcosm experiment in filtered estuarine water for 14 days at 10°C. The relationship between the survival of E. coli strains and genotypic and phenotypic characteristics was analyzed. This work showed that distinct E. coli survival types, able to survive from between 7 and 14 days to less than 2 days, coexisted in the water. E. coli isolates that rapidly lost their culturability were more frequently isolated in water recently contaminated by fecal bacteria of human origin, and most were multiresistant to antibiotics and harbored several virulence factors. In contrast, persistent strains able to survive from 4 to 14 days were more often found in water with low levels of fecal bacteria, belonged mainly to the B1 phylogroup, often harbored only one virulence factor, kspE or ompT, and were able to grow at 7°C.
INTRODUCTION
In order to assess the public health risk associated with surface water, Escherichia coli isolates in combination with intestinal enterococci were selected as relevant indicators of the presence of waterborne human pathogens of fecal origin. Thus, although E. coli strains can be both pathogenic and nonpathogenic, epidemiological studies led to international regulations of the World Health Organization and the European Union (2006/7/EC) defining reliable threshold values for concentrations of E. coli, above which there is a risk of gastrointestinal disease.
E. coli is a commensal bacterium found in the gastrointestinal tracts of humans and vertebrate animals, but it can also possess virulence factors involved in intestinal (InPEC) or extraintestinal (ExPEC) infection, causing serious illness in humans and domestic animals (1–3). The E. coli species have been divided into at least four main phylogenetic groups, A, B1, B2, and D (3–5), and recently, strains phenotypically indistinguishable but genetically distinct from E. coli have been reported and named Escherichia clades I to V (6–9). Although the distribution of the commensal E. coli phylogroups depends on the diet or the climate, strains belonging to phylogroups A and B1 are highly adapted to humans and vertebrate animals, the A phylogroup strains being predominant in humans and the B1 strains in animals (10–12). Interestingly, some strains from the B1 phylogroup persist in water environments (13, 14). Furthermore, Escherichia clade strains seem to be rarely isolated in humans (7) and to be environmentally adapted (9).
The substantial genetic and phenotypic diversity observed in E. coli populations explains the versatile behavior of these bacteria, which exhibit a biphasic lifestyle (11, 14). Much has been written recently about their primary habitat, corresponding to an animal/human host, as opposed to water, sediment, or soil, described as a nonhost or secondary habitat (15–18). However, survival of E. coli in a water environment requires the ability to overcome environmental stresses, such as nutrient deprivation, low temperature, salinity, exposure to solar radiation, competition with autochthonous microbial communities, and protozoan grazing (18–20). Beyond the persistence of a strain, “naturalized” E. coli strains have been described as being able to grow efficiently in oligotrophic conditions, which should be enhanced by an iron acquisition system (18). An association with biotic (algae, plants) or abiotic features promotes the survival of E. coli in water, due mainly to more efficient uptake of nutrients from the water column (21, 22).
The survival of E. coli has been found to range from 1 day in saline water in the presence of grazers to up to several months for pathogenic E. coli O157:H7 in sediment or soil (see reviews in references 18 and 19). This substantial variation can be explained partly by the various experimental approaches used to obtain these findings: (i) the type of experiments (in situ chamber or microcosm experiment with various designs); (ii) the exposure to stresses such as temperature, salinity, and light; and (iii) the method by which survival was monitored, by plating on specific medium to assess the ability to grow or by determination of enzymatic activity to estimate cell viability. Moreover, it has been reported that the survival of E. coli is also greatly influenced by previous exposure to stress (19). Thus, beyond the variance related to the design of experiments, the obtained results also illustrate the extensive genetic and phenotypic diversity exhibited within E. coli populations, which could explain the different survival abilities in aquatic environments (3, 8, 23, 24).
The transfer of E. coli of animal/human origin into surface water is related mainly to the effluents of wastewater treatment plants (WWTPs) or to the direct discharge of wastewater; both soil leaching and runoff release E. coli from humans and livestock and, to a lesser extent, from wildlife. The anthropogenic pressure exerted on the watershed, as well as the stream order of the river, influences not only the level of contamination of surface water but also the diversity of the E. coli population, as previously described by the rep-PCR DNA fingerprinting of E. coli isolated from water from tropical or Canadian watersheds (17, 25). The most probable hypothesis is that, once released into water, the environmental conditions exert selective pressure on the E. coli populations originating from human and animal microbiota (primary habitat), involving the loss of part of the population in the secondary habitat and the persistence of other E. coli strains with an adaptive advantage in the environment (8, 13, 14, 26). Thus, the structure of an E. coli population in water reflected both the origin and pathway of fecal bacteria, combined with the survival characteristics of each strain.
In order to test the latter hypothesis, the aim of this study was to investigate (i) the survival of E. coli from four water environments exhibiting distinct profiles of contamination by fecal bacteria, according to the vicinity of the point sources of contamination, land use, hydrology, and physicochemical characteristics of the receiving water, and (ii) the relationship between the survival of E. coli strains and genotypic and phenotypic characteristics. For this purpose, 58 E. coli strains were selected among 164 strains previously isolated from these four water contamination profiles for a microcosm experiment in filtered estuarine water for 14 days at 10°C. A similar comparative study was carried out with 30 strains selected among 188 strains from human and bovine origin. Each E. coli strain was further characterized by (i) the ability to metabolize different carbon sources, (ii) the capacity to overcome environmental stresses, such as low temperature and osmolarity, (iii) phylogroup affiliation, virulence genotypes, and antibiotic resistance phenotypes, (iv) biofilm formation on a plastic surface, and (v) predation by an amoeba, Dictyostelium discoideum.
MATERIALS AND METHODS
Sampling strategy.
A collection of 164 E. coli strains (see Table S1 in the supplemental material) were isolated from water (i.e., displaying four distinct contamination profiles of E. coli) in a highly anthropized estuary (Seine estuary, France) and one of its tributaries (La Risle) extensively studied within the framework of the Seine-Aval multidisciplinary scientific program (http://seine-aval.crihan.fr/web/) or in a well-studied experimental creek river (13). The watershed of the Seine River (Northwest European continental shelf) has a drainage surface area of 79,000 km2. Its inhabitants, including those in Paris, represent one-third of the French population. Agricultural and industrial activities in the basin amount to 40% of all French activities and are concentrated mainly downstream from Paris (27). Along the estuary, the Seine River receives waters from seven major tributaries and is also the receptacle of the effluents from 16 WWTPs with a treatment capacity of >104 inhabitant equivalents (28, 29). The flow rate of the Seine River was 519 m3 · s−1 when water was sampled, and the E. coli density was 1.5 × 102 ± 0.2 × 102 CFU · 100 ml−1. The contamination profile of water was defined according to the following proxy determinants of water vulnerability to microbial contamination: (i) the nature and the vicinity of the source of contamination, that is, a point source (outlet of a WWTP) or a nonpoint source after a rainfall event (soil leaching or runoff) or in a dry period; (ii) the human or bovine population densities within the watershed; and (iii) physicochemical characteristics of the receiving water, such as salinity and suspended particulate matter. To allow comparison of data, 50 strains from gray water from a hospital (87 beds) and a retirement home (180 beds) were sampled, at two discharge points before the main sewer, as human fecal contamination controls as previously described in reference 30. In addition, in order to simulate the input of E. coli of bovine origin resulting from a runoff event in the watershed of the creek river, 138 E. coli strains were isolated from a mixture of fresh and dry cowpats.
E. coli isolation.
β-d-Galactosidase- and β-d-glucuronidase-positive E. coli cells were enumerated and characterized on the basis of activities using membrane filtration methods (0.45 μm HA047; Millipore, Bedford, MA) with a selective medium specific for E. coli (RAPID'E. coli 2; Bio-Rad) complemented with a selective supplement for water samples (RAPID'E. coli 2 supplement; Bio-Rad) and incubated for 24 h at 37°C. The threshold value for the enumeration of E. coli CFU in water was 5 CFU · 100 ml−1. Isolates were streaked a second time on this chromogenic medium to confirm the purity of the strain and finally for a third time on nonselective medium (LB) before storage on the Cryobead system (AES Chemunex).
Determination of the E. coli phylogroups, Escherichia cryptic lineages, O types, and the presence of virulence factors.
The phylogenetic groups to which the E. coli isolates belong were determined by the triplex PCR-based method targeting the chuA, yjA, and TspE4.C2 genes (4) for the entire collection of 352 strains. The detection of the Escherichia clades was also performed on the entire collection as described in reference 7. Among the collection of 352 strains, a subsample of 88 E. coli and Escherichia clade V strains were randomly selected for microcosm experiments. These strains were selected on the basis of the water profile contamination and their phylogroup distribution in order to reflect the diversity of the whole collection (see Table S1 in the supplemental material). In the 88 selected E. coli and Escherichia clade strains, classification into the C phylogroup, which appears as A by the Clermont method but which is closely related to the B1 group (31), was also determined by an allele-specific assay as described previously (32). Each of the 88 strains was tested for extraintestinal virulence factors (sfa foc, iutA, iroN, fyuA, hlyC, cnf1, kspE, ompT, papC) and intraintestinal virulence factors (afaD, eae, bfpA, ipaH, stx1, stx2, eltB, estA, aaiC, aatA) by PCR detection (33). All 88 strains were tested for O1, O2a, O4, O6a, O7, O8a, O12, O15, O16, O18, O25a, O25b, O26, O40, O45b, O55, O75, O78, O81, O86, O88, O102, O103, O104, O111, O128, O150, and O157 types by using the PCR-based method (34) with the primers shown in Table S2 in the supplemental material.
Characterization of alleles of the uidA gene of E. coli.
Partial uidA sequences (600 bp) of the 88 strains were sequenced after PCR amplification (uidAR, 5′-CCA-TCA-GCA-CGT-TAT-CGA-ATC-CTT-3′; uidAF, 5′-CAT-TAC-GGC-AAA-GTG-TGG-GTC-AAT-3′). Thirty-five microliters of PCR product, containing an estimated 100 ng · μl−1 DNA, was sequenced in both forward and reverse directions by Cogenics (Meylan, France). The sequence was determined by aligning the forward sequence with the reverse complement of the reverse sequence. Alleles of uidA were determined by comparison of the uidA sequences found in the MLST database Pasteur (www.pasteur.fr/cgi-bin/genopole/PF8/mlstdbnet.pl?file=Eco_profiles.xml).
Water microcosm.
The survival ability of the 88 strains was studied in duplicate in nondestructive water microcosms at 10°C for 14 days. For each experiment, water from the Seine estuary at kilometric point (kp) 243 (pH 8.2, zero salinity) was filtered (filter pore size, 0.22 μm) to remove suspended particles and microorganisms, and an aliquot of 100 ml was added in sterile plastic flasks (250 ml). Each microcosm was inoculated with one E. coli strain in the exponential growth phase at a final density of 107 CFU · ml−1 and immediately incubated in the dark at 10°C ± 1°C without shaking for 14 days. Throughout the microcosm experiment, all flasks were daily inverted by hand in order to prevent biofilm formation on the bottom of the flask. Four sets of microcosms were prepared independently. The change in culturable cell density over time was determined at regular intervals by plating a 100-μl aliquot of water in the microcosm onto R2A agar plates (AES Laboratory, France). The plates were incubated overnight at 37°C, and the number of colonies on each plate was determined. The life span of each strain was determined, as described in reference 9, by fitting an exponential decay function to the number of culturable cells at time t (in days), that is, Nt = N0e−μt, where N0 is the initial number of cells and μ is the mortality rate (1/μ = life span of a cell in days).
Growth at low temperature.
A sample of 500 μl of an overnight culture of each of the 88 strains kept at 37°C in Luria broth (LB; AES Laboratories, France) medium was used to inoculate 5 ml of R2A broth (AES Laboratories, France), corresponding to a final density of 5 × 105 · ml−1 in triplicate, and was then incubated at 7°C, 10°C, or 20°C for 48 h. The result was considered positive when visible growth was accompanied by an increase of turbidity (D0580 nm) to greater than 0.3.
Phenotype characterization by the Biolog system.
Biolog GEN III microplates (AES Chemunex, France) containing 71 carbon sources and assays of sensitivity to salinity were used to study the pattern of substrate utilization and osmotic tolerance of the 88 strains in duplicate. Bacterial colonies obtained after 24 h at 37°C on LB agar (AES, France) were resuspended in NaCl (0.9%, wt/vol) to inoculate wells of the Biolog microplates with a cell suspension of 5 × 105 CFU · ml−1 obtained from a serial dilution according to the manufacturer's procedure and then incubated for 24 h at 37°C. For the assays of carbon source utilization, the color intensities in wells were normalized against that of a negative-control well. All wells visually resembling the control well were scored as negative. Wells with a purple color (reduction of tetrazolium dye) more intense than that of the negative-control well, combined with a visual increase in turbidity, were considered positive, whereas purple wells without an increase in turbidity were considered negative. For assays related to the sensitivity to NaCl, only the wells showing a purple color without an increase in turbidity, similar the positive control, were considered positive.
Biofilm formation.
The ability of the 88 strains to form a biofilm on a plastic surface was assessed as previously described (35) with the following modifications. A volume of 150 μl of filtered estuarine water (0.2 μm cellulose acetate filter; Sartorius) was inoculated with 10 μl of an overnight culture (LB at 37°C) of each strain and then randomly deposited on a 96-well microtiter plate and incubated for 5 days at 10°C. Eight independent assays (repeats) were carried out, and the remaining wells on each plate acted as negative controls. After incubation, estuarine medium was removed and the wells were rinsed twice with 200 μl of sterile saline solution (0.9%, wt/vol) in order to remove all nonadhering bacterial cells. The remaining adherent cells were stained by adding 160 μl of crystal violet (0.1%, wt/vol). After 15 min of staining, excess crystal violet was removed from the wells, and the wells were then rinsed twice using 200 μl of a saline solution (0.9%, wt/vol). Ethanol (95%) was added to every well in order to destain the adherent bacterial cells. After 5 min of destaining, the amount of crystal violet in the ethanol was quantified by determining the absorbance at 590 nm using a PR 2100 scanning spectrophotometer (Sanofi Diagnostics Pasteur). When absorbance (optical density at 590 nm [OD590]) measured in the test well, corresponding to the mean of eight values obtained by well assay, was four times the mean value of absorbance observed in the eight negative controls, cells were considered highly adherent and scored as “positive.” When absorbance (OD590) was below four times the absorbance measured in the negative controls, the assay was scored as “negative,” corresponding to electrostatic interactions or to an absence of metabolic activity at 10°C.
E. coli antibiotic resistance testing.
Antibiotic resistance was determined on the 88 strains by the agar diffusion method by using 17 antibiotic discs (bioMérieux, France) as previously described (30), including amoxicillin (25 μg), amoxicillin-clavulanic acid (20 μg-10 μg), ticarcillin (75 μg), ticarcillin-clavulanic acid (75 μg-10 μg), cefalotin (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), imipenem (10 μg), tetracycline (30 μg), trimethoprim-sulfamethoxazole (1.25 μg-23.75 μg), nalidixic acid (30 μg), and ciprofloxacin (5 μg). The bacteria were classified as sensitive, intermediate, or resistant, according to the French National Guidelines (36). The E. coli CIP 7624 (ATCC 25922) strain was used as a control. Two classes of antibiotic-resistant bacteria have been defined: the first one corresponds to E. coli strains resistant to one or two antibiotics, and the second one corresponds to strains resistant to at least three antibiotics. Strains resistant to at least three antibiotics were defined as multiresistant strains.
Grazing by Dictyostelium discoideum.
Grazing resistance was assessed in triplicate by the coculture of D. discoideum (AX3) and each of the 88 strains in HL5 medium (5 g · liter−1 proteose peptone, 5 g · liter−1 thiotone E peptone, 10 g · liter−1 glucose, 5 g · liter−1 yeast extract, 0.35 g · liter−1 Na2HPO4 7H2O, 0.35 g · liter−1 KH2PO4, pH 6.5) incubated for 6 days at 24°C as described previously (37). A volume (300 μl) of an E. coli isolate culture grown overnight was plated on HL5 agar petri dishes and allowed to dry for 20 min under a sterile airflow. The same volume of amoeba culture (D. discoideum AX3) was added on these plates and allowed to dry for 20 min under a sterile airflow. Plates were covered with Parafilm and incubated for 6 days at 24°C. Three replicates were performed for each experiment. Plates were screened at day three and day six to assess the occurrence of bacterial lysis plaques formed by D. discoideum.
Statistical analyses.
Chi-square tests were used to determine the statistical significance of differences in frequencies of various phenotypic and genotypic traits and strain origins. To investigate the relationships among survival abilities and genotypic and phenotypic traits, correspondence factorial analysis (CFA) was performed using a contingency table resulting from a binary table with discriminating bacterial traits of the 88 strains. A chi-square test was computed to test if the rows and columns were independent. As the P value was lower than the significance level (0.05), we concluded that the rows and columns were not independent, which means there was a relationship between the rows and the columns. The bacterial traits were those corresponding to the four E. coli phylogenetic groups (A, B1, B2, D) and Escherichia clade V, four classes of virulence factors (VF: 0, absence of virulence factor; VF: 1, presence of only one virulence factor; VF: 2 to 3, presence of 2 or 3 virulence factors; VF: 4 to 8, presence of 4 to 8 virulence factors), macromolecule degradation (three classes as follows: M: 1, degradation of one macromolecule; M: 2, degradation of two macromolecules; M: 3, degradation of three macromolecules), antibiotic susceptibility (three classes), and growth at 20°C and 7°C. Tests were carried out using XLSTATS version 6.0 (Addinsoft).
RESULTS
Survival abilities of E. coli strains isolated from water with distinct contamination profiles.
In order to carry out a microcosm experiment to study whether E. coli strains vary in terms of survival according to their environmental origins, strains were collected from water displaying distinct contamination profiles (164 strains), in comparison with control samples of human (medical center wastewater) and bovine origin (188 strains). Four contrasting profiles of water contaminated by E. coli were defined on the basis of the main proxies influencing microbiological quality, related to (i) the human or animal pressure exerted on the watershed, (ii) the treatment of waste effluent, and (iii) the hydrology and physicochemical characteristics of the receiving water (Table 1; see also Table S3 in the supplemental material). Contamination profiles 1 and 2 corresponded to the same stream water from a small, well-studied watershed (10 km2) exhibiting a low level of contamination in a dry period (profile 1) and for which the microbial quality decreased due to an input of fecal bacteria through surface runoff and septic tank overflow (profile 2). Water contamination profile 3 corresponded to the confluence of a river (order 4) with the Seine estuary sampled at low tide, that is, corresponding to river water not mixed with estuarine water, and sampled 6 km upstream of a WWTP discharge. Water contamination profile 4 corresponded to the highly anthropized estuary (Seine; order 8), sampled in a high-turbidity zone, combining different sources of upstream contamination with a nearby WWTP discharge (1 km).
Table 1.
Contamination profiles of water by E. coli from vulnerability proxiesh
| Surface water (xa) | Vulnerability proxy |
E. coli density [±SD] (CFU · 100 ml−1) (min- max)c | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Receiving water |
Human or bovine source |
Amt of rainfall (mm) | ||||||||||
| Water contamination profile | Conductivity (μS · cm−1) | Salinity (min-max) | SPM (g · liter−1) | DOM (mg · liter−1) | No. of inhabitants | Head of cattle | Wastewater input |
|||||
| No. of septic tanks | WWTPb treatment capacity | Distance from the closest discharge | ||||||||||
| Creek water (1) | 1 | 345 | 0 | 0.01 | 0.1 | 147 | 172 | 49d | 400 m | 0 | 6.2 [±0.6] × 102 (NR) | |
| 2 | 557 | 0 | 0.07 | ND | 50e | 4.0 [±0.7] × 104 (NR) | ||||||
| Riverf (4) | 3 | 531 | 0.3 (0–15) | 1.1 | <0.5 | 1.8 × 105 | >3.0 × 103 | 1.5 × 104 | 6 km upstream | 10g | 2.0 [±0.3] × 102 (2.0 × 102-4.0 × 103) | |
| Anthropized estuary (8) | 4 | 7,500 | 0.5 (0–25) | 0.74 | 3.16 | 6.0 × 105 | >105 | 7 × 102 | 1 km | 10g | 1.5 [±0.2] × 102 (3.0 × 101-5.0 × 104) | |
x, Strahler stream order.
Treated effluent from wastewater treatment plant.
Min-max (minimum-maximum) data were collected during 4 years of bimonthly sampling.
There was one malfunctioning septic tank located 400 m from the sampling point.
Rainfall event on day of sampling.
Confluence river-estuary sampling during low tide.
Three days before sampling river-estuary confluence.
SPM, suspended particulate matter; DOM, dissolved organic matter; NR, not relevant; ND, not determined.
Among a total of 352 glucuronidase-positive E. coli strains, that is, excluding glucuronidase-negative strains such as E. coli O157:H7, 13 to 15 strains were selected from each water profile and control samples, resulting in 88 strains (see Table S1 in the supplemental material). The proportion of strains belonging to the B1 phylogroup was significantly higher in water contamination profile 1 than in the other three profiles (chi-square test, P < 0.0001, α = 0.05). The proportion of strains belonging to the A phylogroup was significantly lower in the water contamination profiles 1 and 2 than in water profile 4 (chi-square test, P < 0.0001, α = 0.05). In order to take into account the initial structure of E. coli populations, which differs depending on the sample source, these 88 strains were randomly selected within each original sample on the basis of the distribution into the four main phylogenetic groups (A, B1, B2, and D) and the Escherichia clades (see Table S1 in the supplemental material). Two E. coli strains of human origin which were previously characterized as belonging to the A phylogroup were further identified as E. coli phylogroup C (31, 32). When considering both the sample origin and the genetic diversity based on phylogroup belonging, uidA allele, O type, and virulence factor content (see below) of the strains, the 88 strains can be clustered in 68 distinct epidemiological types (see Table S4 in the supplemental material).
The loss of culturability of each of the 88 strains was monitored for 14 days at 10°C in a filtered estuarine water microcosm under dark conditions (Fig. 1). In these experimental conditions, survival of E. coli ranged from less than 2 days up to 14 days. E. coli strains could be clustered into four survival phenotypes based on their life span: 10 strains were able to survive between 7 and 14 days (group S1), 34 strains between 4 and 6 days (group S2), 21 strains between 2 and 3 days (group S3), and 23 strains less than 2 days (group S4).
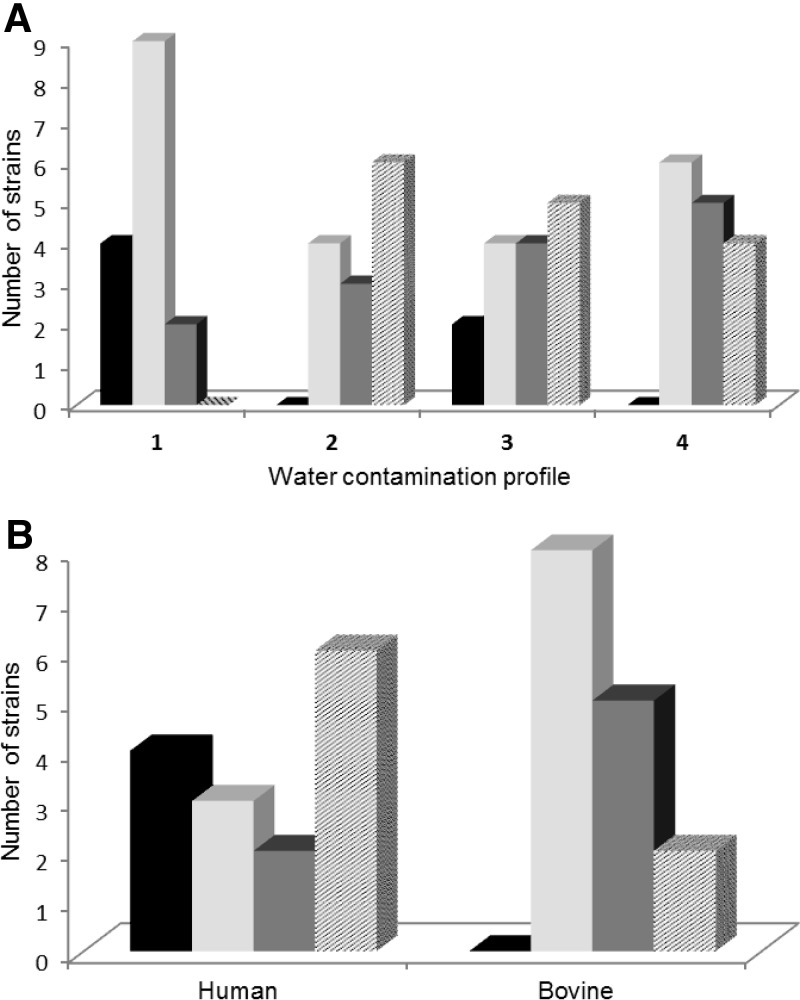
Fig 1.
Survival of 88 E. coli and Escherichia clade strains in a water microcosm related to water contamination profile origin. Strains were isolated from bodies of water (A) and human and bovine feces (B). S1 to S4 correspond to the four survival groups described in Results: black bars, S1 survival group (between 7 to 14 days); light-gray bars, S2 survival group (between 4 to 6 days); dark-gray bars, S3 survival group (between 2 to 4 days); striped bars, S4 survival group (less than 2 days). The water contamination profiles (from 1 to 4) correspond to those described in Materials and Methods.
The number of E. coli strains belonging to both the S1 (4/15) and S2 (9/15) survival groups was significantly higher than that for the S4 survival group (0/15) in water contamination profile 1 than in the three other water contamination profiles (chi-square test, P = 0.003, α = 0.05). Thus, in water exhibiting a low level of contamination of E. coli (water contamination profile 1), namely, creek water during a dry period, the circulating E. coli strains were the most persistent. In contrast, in the same creek water but after a rainfall event, in which there was a high level of fecal contamination of human origin (water contamination profile 2), E. coli strains (6/13) belonging to the S4 survival group were predominant, while persistent E. coli strains (group S1) were absent (Fig. 1A). A similar pattern was observed in water impacted by very close input of treated wastewater effluent (water contamination profile 4). In water continuously impacted by past and distant input of fecal bacteria from soil runoff or point sources (water contamination profile 3), the four E. coli survival phenotypes were found.
All survival phenotypes were represented in E. coli from a human origin isolated in gray water. In cowpat samples (a mix of fresh and dry), the proportion of E. coli strains losing their culturability after 4 to 6 days (group S2) was significantly higher than in the S1 survival group (chi-square test, P = 0.023) (Fig. 1B). These results suggest that, once released into water, E. coli strains of bovine and human origin were selected on the basis of their survival ability: the strains belonging to groups S1 and S2 persisted in water, while others rapidly lost their culturability (groups S3 and S4). Thus, in water contaminated with a low level of E. coli, survival groups S1 and S2 should be selected on the basis of their phenotypic traits (water contamination profile 1).
Phenotypic traits of E. coli strains according to sample origin and survival abilities.
Thus, in order to study a possible link between the survival abilities of E. coli strains in water and their phenotypes, we first investigated the characteristics that could favor their survival in water. The phenotypic patterns of each strain were characterized on the basis of their abilities as follows: (i) to metabolize various carbon substrates at 37°C, (ii) to overcome osmotic and temperature stresses, and (iii) to adhere to a surface, that is, to form a biofilm on a plastic surface after 5 days at 10°C in estuarine water. Among 71 carbon substrates of various types, the following 12 compounds allowed growth of all the E. coli strains: dextrin, mannose, inosine, glycerol, l-serine, d-galacturonic acid, d-gluconic acid, d-glucuronic acid, d-glucose-6-phosphate, l-alanine, l-lactic acid, and glucuronamide (see Fig. S1 in the supplemental material). In contrast, 59 carbon substrates allowed growth in some cases, but no significant difference between the four survival groups was observed within the entire E. coli collection. However, pectin, gelatin, and Tween 40 are of particular interest, because the hydrolysis of these macromolecules requires the production of exoenzymes, which could provide E. coli with a selective advantage in surviving in a water environment. The percentage of strains unable to metabolize one of these compounds was lower in the control collection than in the water E. coli collection, even if the level was higher in the bovine collection than in the human one (Table 2). Nevertheless, no relationship was found between the ability to degrade macromolecules as a carbon source and the survival group.
Table 2.
Phenotypic traits, phylogroups, and virulence factors according to origin and survival abilities of 88 E. coli and Escherichia clade strainsb
| Phenotypic trait | Bodies of water (58 strains) |
Controls (30 strains) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. (%) | No. of strains in survival group: |
Total no. (%) | No. of strains in human survival group: |
No. of strains in bovine survival group: |
||||||||||
| S1 (n = 6) | S2 (n = 23) | S3 (n = 14) | S4 (n = 15) | S1 (n = 4) | S2 (n = 3) | S3 (n = 2) | S4 (n = 6) | S1 (n = 0) | S2 (n = 8) | S3 (n = 5) | S4 (n = 2) | |||
| Metabolism of macromolecules: pectin, gelatin, Tween 40 | ||||||||||||||
| None | 20 (34.5) | 1 | 9 | 6 | 4 | 15 (50) | 1 | 0 | 1 | 1 | 0 | 5 | 5 | 2 |
| 1 | 15 (25.8) | 2 | 5 | 2 | 6 | 6 (20) | 0 | 1 | 1 | 3 | 0 | 1 | 0 | 0 |
| 2 | 14 (24.2) | 0 | 6 | 5 | 3 | 8 (26.7) | 2 | 2 | 0 | 2 | 0 | 2 | 0 | 0 |
| 3 | 9 (15.5) | 2 | 3 | 2 | 2 | 1 (3.3) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. coli phylogroup | ||||||||||||||
| A | 3 | 5 | 3 | 6 | 2 | 1 | 1 | 3 | 0 | 0 | 1 | 0 | ||
| B1 | 3 | 12 | 5 | 4 | 1 | 0 | 0 | 0 | 0 | 7 | 4 | 2 | ||
| B2 | 0 | 0 | 2 | 1 | 1 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | ||
| C | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| D | 0 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ||
| Clade V | 0 | 3 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Virulence gene(s)a | ||||||||||||||
| No virulence factor | 10 (17.2) | 0 | 4 | 1 | 5 | 7 (23.3) | 0 | 1 | 0 | 1 | 0 | 1 | 2 | 2 |
| 1 virulence factor | 18 (31) | 3 | 6 | 7 | 2 | 3 (10) | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 |
| 2 or 3 virulence factors | 23 (39.7) | 3 | 13 | 1 | 6 | 15 (50) | 3 | 1 | 1 | 2 | 0 | 5 | 3 | 0 |
| 4 to 8 virulence factors | 7 (12.1) | 0 | 0 | 5 | 2 | 5 (26.7) | 1 | 1 | 0 | 3 | 0 | 0 | 0 | 0 |
Virulence factors involved in intraintestinal infection were absent in all strains.
n = total number of strains in each collection.
In order to assess the ability of E. coli strains to overcome osmotic and temperature stresses, NaCl tolerance in the range of 1% to 8% was tested according to the Biolog GEN III system procedure, and a growth experiment was carried out at a temperature below the optimum temperature of 37°C. Electron transfer activity (reduction of tetrazolium dye) was detected for the entire E. coli collection in the presence of 1% to 4% (wt/vol) NaCl and for 85% (75/88) of the strains at 8% (wt/vol) NaCl. These results suggest that E. coli maintained viability for at least 24 h at 37°C in a high NaCl concentration, namely, up to 4%, but no significant difference was observed between the four survival groups at the selective concentration of 8% NaCl.
All E. coli strains collected from bodies of water were able to grow at 20°C, and among them 37 strains still grew at 10°C, with only 4 isolates from water growing at 7°C, corresponding to the minimal reported growth temperature of E. coli (38).
Regardless of the origin of E. coli isolates, a small number of strains (9/58 from the bodies of water and 7/30 from the controls) formed a biofilm on a plastic surface at 10°C in estuarine water. These results suggest that these conditions did not favor the formation of biofilm regulated by quorum sensing, which requires active metabolic cells.
Phylogroup, virulence factors, and antibiotic resistance of E. coli strains according to sample origin and survival abilities.
For further analysis, the relationship between the survival abilities of E. coli strains in water and their genotypic characteristics was investigated. The four main phylogenetic groups, the additionally identified C phylogroup (31), and Escherichia clades I to V were determined for each survival group (Table 2). In water collection, E. coli strains belonging to phylogroups A and B1 were present in the four E. coli survival groups. However, the E. coli strains from the B1 phylogroup were more abundant in more persistent survival groups (S2 and S1) than in isolates from the human control. Interestingly, in the bovine control, E. coli strains from the B1 phylogroup were also predominant in the S2 survival group, probably due to the contribution of the persistent strain from dry cowpats. Escherichia clade V strains were observed only in the S2, S3, and S4 survival groups.
E. coli isolates were further screened for the virulence factors associated with intraintestinal or extraintestinal diseases (Table 2; see also Table S5 in the supplemental material). This involved carrying out the molecular detection of 9 extraintestinal virulence factors (sfa foc, iutA, iroN, fyuA, hlyC, cnf1, kspE, ompT, papC) and 10 intraintestinal virulence factors (afaD, eae, bfpA, ipaH, stx1, stx2, eltB heat-labile enterotoxin, estA heat-stable enterotoxin, aaiC, aatA). Overall, in the selected 88 strains, including the controls, the 10 intraintestinal virulence factors were never detected, with the exception of one E. coli strain of water origin possessing the afaD gene. Among the E. coli strains isolated from the bodies of water, no significant correlation could be observed between the occurrence of the detected virulence genes and their survival capacities (see Table S5 in the supplemental material). However, in the strains collected from bodies of water, the abundance of E. coli harboring a single virulence gene was significantly higher than in control samples (chi-square test, P = 0.028, α = 0.05), due mainly to the presence of the kspE or ompT gene (Table 2). Within strains collected from the bodies of water, the occurrence of E. coli strains harboring 4 to 8 virulence genes was significantly lower in persistent strains (groups S1 and S2) than in those that rapidly lost their culturability (group S3; Table 2) (chi-square test, P = 0.01, α = 0.05). These results suggest that strains harboring several virulence genes would be less suited to survive in a water environment.
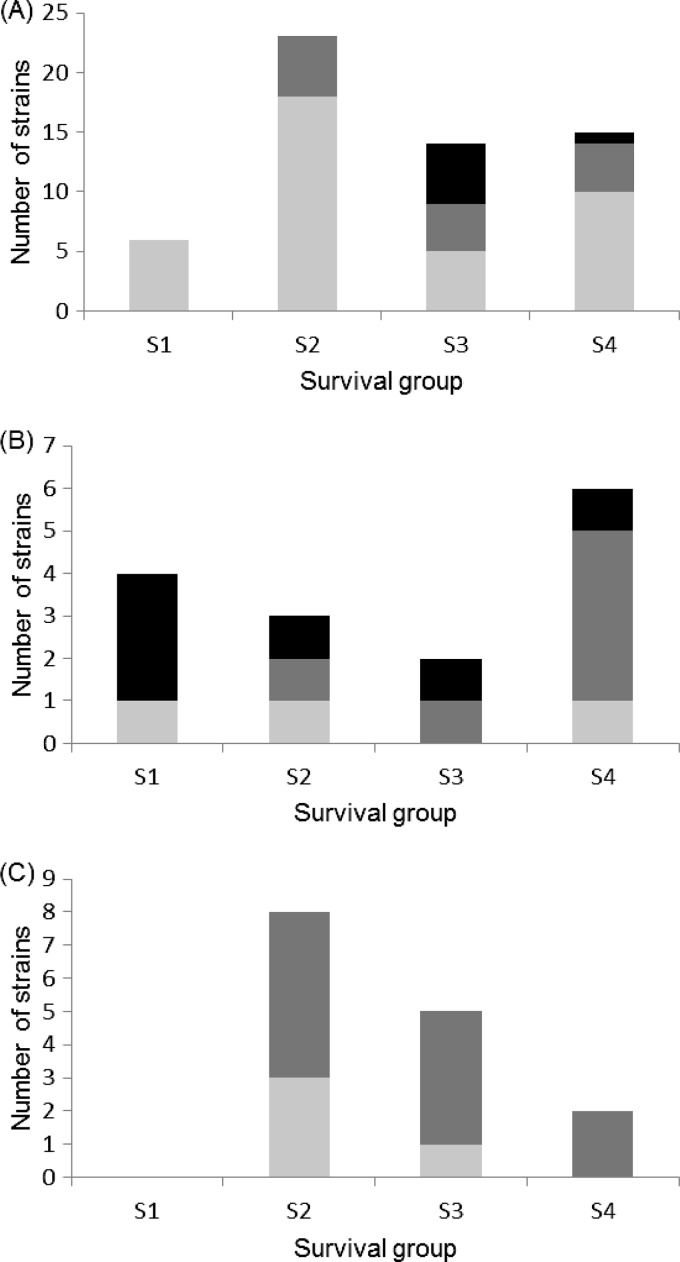
The antibiotic resistance profiles of E. coli strains were further determined using 17 antibiotics used in human and veterinary medicine in France (Fig. 2). Among the strains collected from bodies of water, the proportion of E. coli strains susceptible to the 17 antibiotics (67.2%) was higher than in control samples (23.3%), while the proportions of E. coli resistant (22.4% compared to 56.7% for the control samples) and multiresistant strains were the lowest (10.4% compared to 20% for the control samples). Strains resistant to multiple antibiotics were significantly more abundant in survival groups that lost their culturability before 4 days (survival group S3) (Fig. 2A; chi-square test, P = 0.001, α = 0.05). These results are consistent with the number of sensitive strains observed in water related to the contamination profile. Indeed, in cases with a low level of contamination (water profile 1), all the E. coli strains (15/15) were sensitive to the 17 antibiotics tested, whereas the number of E. coli strains resistant to at least one antibiotic increased in water that was recently or continuously contaminated (water profile 2, 5/13 strains; water profile 3, 3/15 strains) or that was located very close to a site of wastewater discharge (water profile 4, 9/15 strains).
Fig 2.
Antibiotic resistance according to origin and survival abilities of 88 E. coli and Escherichia clade strains. Strains were isolated from bodies of water (A) and human (B) and bovine (C) feces. S1 to S4 correspond to the four survival groups described in Results. Antibiotic susceptibility: strains susceptible to the 17 antibiotics tested (light-gray bars), resistant to one or two antibiotics (dark-gray bars), and resistant to at least 3 antibiotics (multiresistant strains) (black bars).
Protozoan predation on E. coli strains according to sample origin and survival abilities.
Although the microcosm experiment was carried out with filtered estuarine water to prevent rapid decay due to grazing by protozoans, we investigated the susceptibility of each E. coli strain to predation by D. discoideum, which could also be an explanatory factor behind better survival in a water environment. The results showed that, in the samples collected from bodies of water, the number of strains resistant to predation by this amoeba was higher than that in the control strains (67.2% and 46.6%, respectively) (see Table S6 in the supplemental material). Nevertheless, no significant difference was observed among the survival groups, probably because these groups were defined in our microcosm experiment, in which predation was absent.
Survival abilities of E. coli strains related to the phylogroups, virulence gene content, and phenotypic traits.
To carry out a correspondence factorial analysis (CFA) based on the 58 E. coli and Escherichia clade water isolates and to understand which parameters are associated with the survival time of E. coli in water, we retained from the previous analyses the following variables as putative significant descriptors: phylogroups, virulence factors, minimal growth temperature, macromolecule hydrolysis, and antibiotic resistance (Fig. 3). These variables were retained because they increased the discrimination of E. coli and Escherichia strains according to the CFA analysis.
Fig 3.
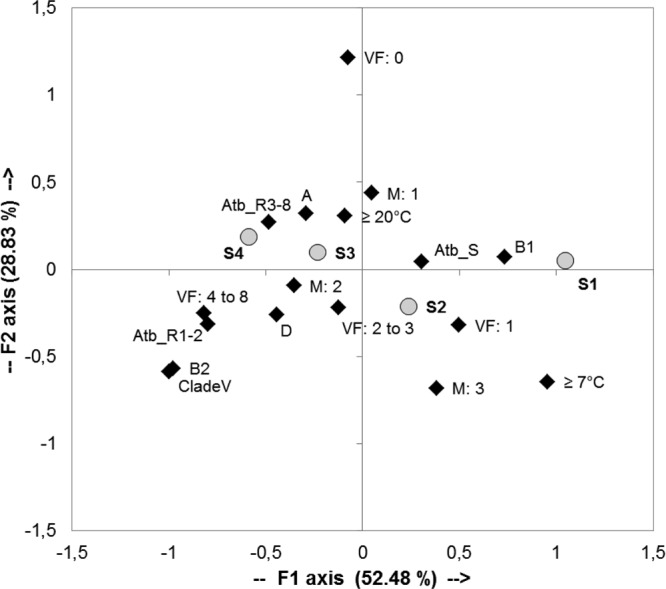
Correspondence factorial analysis of 58 E. coli and Escherichia clade water sample strains. Projection on the F1/F2 plane of the 17 bacterial traits (black diamonds) and 4 survival groups (gray circles): S1, 7 to 14 days; S2, 4 to 6 days; S3, 2 to 3 days; S4, <2 days. Phylogroups A, B1, B2, and D and clade V (Escherichia clade V); minimal growth temperatures, ≥ 20°C and ≥ 7°C. VF: 0, absence of virulence factor; VF: 1, presence of only one virulence factor; VF: 2 to 3, presence of 2 and 3 virulence factors; VF: 4 to 8, presence of 4 to 8 virulence factors; Atb_R3-8, strains resistant to at least 3 antibiotics (multiresistant strains); Atb_R1-2, resistant to one or two antibiotics; Atb_S, susceptible to the 17 antibiotics tested; M: 1, degradation of one macromolecule; M: 2, degradation of two macromolecules; M: 3, degradation of three macromolecules.
The projection of the variables on the F1/F2 plane accounted for 81.31% of the total variance and distinguished clearly the four survival groups on the first axis, which accounted for 52.48% of the total variance. For survival group S1, encompassing strains surviving 7 to 14 days, and to a lesser extent survival group S2, the ability to grow at 7°C, belonging to the B1 phylogroup, harboring only one virulence factor (VF: 1; kspE or ompT), the degradation of three macromolecules (pectin, gelatin, Tween 40), and the sensitivity to antibiotics were distinguished by positive values of F1. In contrast, E. coli isolates with the loss of culturability in less than 2 days (survival group S4) and, to a lesser extent, survival group S3 were distinguished by negative values of the F1 axis, with resistance to antibiotics, the presence of four to eight virulence factors, and belonging to the B2 and D phylogroups and Escherichia clade V. The positive values of the F2 axis distinguished the lack of virulence factors. This analysis confirmed that these specific parameters were associated with the survival categories. It suggested that glucuronidase-positive E. coli strains of human origin, which are antibiotic resistant and belong to the B2 phylogroup, lose their culturability more rapidly once released into water.
DISCUSSION
In this work, a comparative microcosm experiment was carried out with E. coli strains collected from various types of water. It showed that the distribution of distinct survival phenotypes within E. coli populations was related to the water profile of contamination by fecal bacteria. Indeed, once released into water close to a site of discharge, bovine/human host-associated E. coli strains displaying distinct survival phenotypes could be found. In recently contaminated water, the number of E. coli strains that rapidly lost their culturability in less than 2 days was higher. In contrast, in water with a low level of contamination in a dry period, the proportion of strains able to survive at least 14 days increased. These results are consistent with the existence of both E. coli strains that persist in water and sediment and strains from untreated wastewater effluent that lose their culturability within 2 days (18, 20, 22, 39). Similar selection of stress-tolerant E. coli strains by the soil environment has been observed, highlighting the importance of the natural environment in the selection of culturable E. coli strains from human or animal microbiota and the emergence of E. coli strains better adapted to the nonhost environment (18, 40, 41).
The densities and diversities of E. coli strains have already been shown to be linked to stream order and land use (17). In this work, it was shown that in watersheds where humans and bovine cattle are the major sources of input of fecal bacteria, a typology of the water can be defined and is indirectly associated with the residence time of circulating E. coli. This typology was based on (i) the human or animal pressure exerted on the watershed, (ii) the kinds of treatment and the proximity to wastewater discharge, and (iii) the hydrology and physicochemical characteristics and stream order of the receiving water.
Because E. coli populations have extensive genetic and phenotypic diversities (3, 8, 23, 24), we tried to explain these different survival abilities by studying the genotypic and phenotypic traits involved in strain persistence in aquatic environments. The presence of an E. coli genotype related to persistent and naturalized strains was already described in temperate and tropical freshwater conditions (15, 17, 26, 42). Here, we showed that E. coli B1 strains seemed to be the most persistent strains in water. These results were consistent with recent studies showing that E. coli B1 strains can persist longer in water than strains of the other phylogroups (13, 14). The prevalence of B1 isolates has also been observed in other environmental samples, such as manure and drinking water, or associated with plant colonization (see the reference list in reference 43).
In this study, persisting E. coli strains harboring only one extraintestinal virulence factor were predominant in water in the absence of recent contamination, suggesting that the bacteria with fewer virulence factors would have the ability of utilizing more energy in order to adapt to the environmental factor. Molecular detection of virulence genes of E. coli has also been reported after an enrichment culture procedure from estuarine water (44). However, no correlation has been observed between the presence of virulence genes and the number of E. coli strains in estuarine water, brackish water, and freshwater (44).
A strong association between phenotypes and E. coli phylogenetic groups was observed for E. coli isolated from plants, with B1 isolates more likely harboring traits indicative of a higher ability to colonize plants, such as higher biofilm formation and higher frequency of utilization of sucrose and an aromatic compound (43). Thus, the ability to utilize sucrose could provide a substantial advantage in the environment (43, 45). However, in this study, no relationship between carbon degradation and the survival ability of isolates was observed. This work showed that the ability to grow at a low temperature could be a physiological advantage by which E. coli strains that can persist in water with a low level of contamination are selected. The gene regulation of these strains may occur across a broad temperature range compared with that of the host-associated strains (18). Thus, the growth capacity of E. coli strains at temperatures below their optimum was a factor that contributed to better adaptation to the environment, as previously described in similar studies in water and soil (16, 40). As observed in freshwater recently contaminated by fecal bacteria from hospital effluent, E. coli strains resistant to multiple antibiotics more rapidly lost their culturability, leading to a selective decrease of these populations during their transfer into a river (30). In contrast to the water column, sediment environments, as proven for the case of coastal sediment from the Adriatic Sea, may be a suitable environment for the survival of antibiotic-resistant E. coli strains of phylogenetic group A (46).
Interaction with particles is a major factor influencing the fate of E. coli in water and sediment (20, 47, 48). Nevertheless, in this study, the ability to form a biofilm on a plastic surface did not explain the extended survival abilities of E. coli strains in water, which was probably because the test did not recreate the environmental conditions. The formation of biofilm depends on curli synthesis and/or quorum sensing regulation, which requires metabolically active cells, while association with organic mineral particles is mainly a passive phenomenon related to physical and chemical interactions (20, 22, 49).
These results highlight the role of the environment in structuring populations of E. coli by exerting a selection on the strains exhibiting a better survival, and thus the corresponding well-adapted phenotype, during their transfer from their primary habitat to a water environment, which is considered a secondary habitat. Indeed, once released into water, E. coli strains of bovine and human origin could be selected on the basis of their survival ability, and the resulting population differed from the original one in terms of phenotypic traits, including virulence factors and antibiotic resistance (17, 18, 41, 42). In response to a recent fecal contamination event or at the vicinity of an input of fecal bacteria related to point sources, threshold values of fecal indicators defined according to the World Health Organization and the European Union (2006/7/EC) are pertinent to evaluating the risk of gastrointestinal disease. The results of this study, however, show the presence of persistent circulating E. coli strains in environmental water in the absence of recent fecal contamination or vicinity of point sources of contamination.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the scientific program FLASH-Seine Aval (http://seine-aval.crihan.fr/web/), the CNRS EC2CO, and the SFR SCALE. M.R. held a research grant from the Haute-Normandie Regional Council (France).
We are grateful to Matthieu Fournier for his assistance in statistical analysis.
The work presented here was carried out through a collaboration among all the authors. T.B. and F.P. defined the research theme. M.R., T.B., and F.P. defined the sampling strategy, and all authors designed the methods and the experiments. M.R. carried out the microcosm experiments. Phenotypic and molecular characterizations of E. coli strains collected from the environment were carried out by M.R. and O.C. All authors analyzed the data and interpreted the results. All authors contributed to, saw, and approved the final manuscript.
Footnotes
Published ahead of print 31 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00698-13.
REFERENCES
- 1. Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8:26–38 [DOI] [PubMed] [Google Scholar]
- 2. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 3. Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217 [DOI] [PubMed] [Google Scholar]
- 4. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gordon DM, Clermont O, Tolley H, Denamur E. 2008. Assigning Escherichia coli strains to phylogenetic groups: multilocus sequence typing versus the PCR triplex method. Environ. Microbiol. 10:2484–2496 [DOI] [PubMed] [Google Scholar]
- 6. Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, Whittam TS. 2009. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75:6534–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clermont O, Gordon DM, Brisse S, Walk ST, Denamur E. 2011. Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ. Microbiol. 13:2468–2477 [DOI] [PubMed] [Google Scholar]
- 8. Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. 2011. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc. Natl. Acad. Sci. U. S. A. 108:7200–7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ingle DJ, Clermont O, Skurnik D, Denamur E, Walk ST, Gordon DM. 2011. Biofilm formation by and thermal niche and virulence characteristics of Escherichia spp. Appl. Environ. Microbiol. 77:2695–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventre A, Elion J, Picard B, Denamur E. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671–1676 [DOI] [PubMed] [Google Scholar]
- 11. Gordon DM, Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586 [DOI] [PubMed] [Google Scholar]
- 12. Skurnik D, Bonnet D, Bernède-Bauduin C, Michel R, Guette C, Becker JM, Balaire C, Chau F, Mohler J, Jarlier V, Boutin JP, Moreau B, Guillemot D, Denamur E, Andremont A, Ruimy R. 2008. Characteristics of human intestinal Escherichia coli with changing environments. Environ. Microbiol. 10:2132–2137 [DOI] [PubMed] [Google Scholar]
- 13. Ratajczak M, Laroche E, Berthe T, Clermont O, Pawlak B, Denamur E, Petit F. 2010. Influence of hydrological conditions on the Escherichia coli population structure in the water of a creek on a rural watershed. BMC Microbiol. 10:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walk ST, Alm EW, Calhoun LM, Mladonicky JM, Whittam TS. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9:2274–2288 [DOI] [PubMed] [Google Scholar]
- 15. Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brennan FP, O'Flaherty V, Kramers G, Grant J, Richards KG. 2010. Long-term persistence and leaching of Escherichia coli in temperate maritime soils. Appl. Environ. Microbiol. 76:1449–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lyautey E, Lu Z, Lapen DR, Wilkes G, Scott A, Berkers T, Edge TA, Topp E. 2010. Distribution and diversity of Escherichia coli populations in the South Nation River drainage basin, eastern Ontario, Canada. Appl. Environ. Microbiol. 76:1486–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Elsas JD, Semenov AV, Costa R, Trevors JT. 2010. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 5:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rozen Y, Belkin S. 2001. Survival of enteric bacteria in seawater. FEMS Microbiol. Rev. 25:513–529 [DOI] [PubMed] [Google Scholar]
- 20. Pachepsky YA, Shelton DR. 2011. Escherichia coli and fecal coliforms in freshwater and estuarine sediments. Crit. Rev. Environ. Sci. Technol. 41:1067–1110 [Google Scholar]
- 21. Garcia-Armisen T, Servais P. 2009. Partitioning and fate of particle-associated E. coli in river waters. Water Environ. Res. 81:21–28 [PubMed] [Google Scholar]
- 22. Moreira S, Brown A, Ha R, Iserhoff K, Yim M, Yang J, Liao B, Pszczolko E, Qin W, Leung KT. 2012. Persistence of Escherichia coli in freshwater periphyton: biofilm-forming capacity as a selective advantage. FEMS Microbiol. Ecol. 79:608–618 [DOI] [PubMed] [Google Scholar]
- 23. Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Medigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sabarly V, Bouvet O, Glodt J, Clermont O, Skurnik D, Diancourt L, De Vienne D, Denamur E, Dillmann C. 2011. The decoupling between genetic structure and metabolic phenotypes in Escherichia coli leads to continuous phenotypic diversity. J. Evol. Biol. 24:1559–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goto DK, Yan T. 2011. Genotypic diversity of Escherichia coli in the water and soil of tropical watersheds in Hawaii. Appl. Environ. Microbiol. 77:3988–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Power ML, Littlefield-Wyer J, Gordon DM, Veal DA, Slade MB. 2005. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7:631–640 [DOI] [PubMed] [Google Scholar]
- 27. Guerrini M-C, Mouchel JM, Meybeck M, Penven MJ, Hubert G, Muxart T. 1998. Le bassin de la Seine: la confrontation du rural et de l'urbain, p 29–75 In Meybeck M, de Marsily G, Fustec É. (ed), La Seine en son basin: fonctionnement ècologique d'un système fluvial anthropisé, la fiche détaillée du livre. Elsevier, Paris, France [Google Scholar]
- 28. Servais P, Garcia-Armisen T, George I, Billen G. 2007. Fecal bacteria in the rivers of the Seine drainage network (France): sources, fate and modelling. Sci. Total Environ. 375:152–167 [DOI] [PubMed] [Google Scholar]
- 29. Touron A, Berthe T, Gargala G, Fournier M, Ratajczak M, Servais P, Petit F. 2007. Assessment of fecal contamination and the relationship between pathogens and fecal bacterial indicators in an estuarine environment (Seine, France). Mar. Pollut. Bull. 54:1441–1450 [DOI] [PubMed] [Google Scholar]
- 30. Oberlé K, Capdeville MJ, Berthe T, Budzinski H, Petit F. 2012. Evidence for a complex relationship between antibiotics and antibiotic-resistant Escherichia coli: from medical center patients to a receiving environment. Environ. Sci. Technol. 46:1859–1868 [DOI] [PubMed] [Google Scholar]
- 31. Moissenet D, Salauze B, Clermont O, Bingen E, Arlet G, Denamur E, Merens A, Mitanchez D, Vu-Thien H. 2010. Meningitis caused by Escherichia coli producing TEM-52 extended-spectrum beta-lactamase within an extensive outbreak in a neonatal ward: epidemiological investigation and characterization of the strain. J. Clin. Microbiol. 48:2459–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lescat M, Clermont O, Woerther PL, Glodt J, Dion S, Skurnik D, Djossou F, Dupont C, Perroz G, Picard B. 2013. Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host-dependent population structure. Environ. Microbiol. Rep. 5:49–57 [DOI] [PubMed] [Google Scholar]
- 33. Clermont O, Olier M, Hoede C, Diancourt L, Brisse S, Keroudean M, Glodt J, Picard B, Oswald E, Denamur E. 2011. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11:654–662 [DOI] [PubMed] [Google Scholar]
- 34. Clermont O, Johnson JR, Menard M, Denamur E. 2007. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: application to the O types involved in human septicemia. Diagn. Microbiol. Infect. Dis. 57:129–136 [DOI] [PubMed] [Google Scholar]
- 35. Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40:175–179 [DOI] [PubMed] [Google Scholar]
- 36. Comité de l'Antibiogramme de la Société Française de Microbiologie 2001. Communiqué du comité de l'antibiogramme de la société française de microbiologie, p 2–13 Société Française de Microbiologie, Paris, France [Google Scholar]
- 37. Adiba S, Nizak C, Van Baalen M, Denamur E, Depaulis F. 2010. From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS One 5:e11882. 10.1371/journal.pone.0011882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Membré JM, Leporq B, Vialette M, Mettler E, Perrier L, Thuault D, Zwietering M. 2005. Temperature effect on bacterial growth rate: quantitative microbiology approach including cardinal values and variability estimates to perform growth simulations on/in food. Int. J. Food Microbiol. 100:179–186 [DOI] [PubMed] [Google Scholar]
- 39. Dick LK, Stelzer EA, Bertke EE, Fong DL, Stoeckel DM. 2010. Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl. Environ. Microbiol. 76:3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gordon DM, Bauer S, Johnson JR. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513–1522 [DOI] [PubMed] [Google Scholar]
- 41. Bergholz PW, Noar JD, Buckley DH. 2011. Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Appl. Environ. Microbiol. 77:211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Byappanahalli MN, Whitman RL, Shively DA, Sadowsky MJ, Ishii S. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504–513 [DOI] [PubMed] [Google Scholar]
- 43. Méric G, Kemsley EK, Falush D, Saggers EJ, Lucchini S. 2013. Phylogenetic distribution of traits associated with plant colonization in Escherichia coli. Environ. Microbiol. 15:487–501 [DOI] [PubMed] [Google Scholar]
- 44. Masters N, Wiegand A, Ahmed W, Katouli M. 2011. Escherichia coli virulence genes profile of surface waters as an indicator of water quality. Water Res. 45:6321–6333 [DOI] [PubMed] [Google Scholar]
- 45. Janezic KJ, Ferry B, Hendricks EW, Janiga BA, Johnson T, Murphy S, Roberts ME, Scott SM, Theisen AN, Hung KF, Daniel SL. 2013. Phenotypic and genotypic characterization of Escherichia coli isolated from untreated surface waters. Open Microbiol. J. 7:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vignaroli C, Luna G, Rinaldi C, Di Cesare A, Danovaro R, Biavasco F. 2012. New sequence types and multidrug resistance among pathogenic Escherichia coli isolates from coastal marine sediments. Appl. Environ. Microbiol. 78:3916–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muirhead RW, Collins RP, Bremer PJ. 2006. Interaction of Escherichia coli and soil particles in runoff. Appl. Environ. Microbiol. 72:3406–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berthe T, Touron A, Leloup J, Deloffre J, Petit F. 2008. Fecal-indicator bacteria and sedimentary processes in estuarine mudflats (Seine, France). Mar. Pollut. Bull. 57:59–67 [DOI] [PubMed] [Google Scholar]
- 49. Droppo IG, Liss SN, Williams D, Nelson T, Jaskot C, Trapp B. 2009. Dynamic existence of waterborne pathogens within river sediment compartments. Implications for water quality regulatory affairs. Environ. Sci. Technol. 43:1737–1743 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.