Abstract
Haemophilus ducreyi causes chancroid, a genital ulcer disease that facilitates the transmission of human immunodeficiency virus type 1. In humans, H. ducreyi is surrounded by phagocytes and must adapt to a hostile environment to survive. To sense and respond to environmental cues, bacteria frequently use two-component signal transduction (2CST) systems. The only obvious 2CST system in H. ducreyi is CpxRA; CpxR is a response regulator, and CpxA is a sensor kinase. Previous studies by Hansen and coworkers showed that CpxR directly represses the expression of dsrA, the lspB-lspA2 operon, and the flp operon, which are required for virulence in humans. They further showed that CpxA functions predominantly as a phosphatase in vitro to maintain the expression of virulence determinants. Since a cpxA mutant is avirulent while a cpxR mutant is fully virulent in humans, CpxA also likely functions predominantly as a phosphatase in vivo. To better understand the role of H. ducreyi CpxRA in controlling virulence determinants, here we defined genes potentially regulated by CpxRA by using RNA-Seq. Activation of CpxR by deletion of cpxA repressed nearly 70% of its targets, including seven established virulence determinants. Inactivation of CpxR by deletion of cpxR differentially regulated few genes and increased the expression of one virulence determinant. We identified a CpxR binding motif that was enriched in downregulated but not upregulated targets. These data reinforce the hypothesis that CpxA phosphatase activity plays a critical role in controlling H. ducreyi virulence in vivo. Characterization of the downregulated genes may offer new insights into pathogenesis.
INTRODUCTION
Haemophilus ducreyi is a Gram-negative facultative anaerobe that causes chancroid, a genital ulcer disease characterized by painful genital ulcers and regional lymphadenopathy. While uncommon in the United States, chancroid is endemic in the developing countries of Africa, Asia, and Latin America and is a risk factor for the acquisition and transmission of human immunodeficiency virus type 1 (HIV-1) (1). Because of syndromic management of genital ulcer disease, the global prevalence of chancroid is currently undefined (2). In the South Pacific, H. ducreyi is also reported to cause a chronic lower limb ulceration syndrome that is not sexually transmitted (3–5).
H. ducreyi is an obligate human pathogen with no known environmental reservoirs. The development of a human infection model has significantly contributed to our understanding of H. ducreyi pathogenesis (6). During experimental and natural infections, H. ducreyi resides in an abscess composed of neutrophils and macrophages (7, 8). Thus, H. ducreyi is likely exposed to multiple stresses in the human host, including antimicrobial peptides, the hypoxic environment of an abscess, and nutrient limitation. The fact that H. ducreyi is a successful pathogen suggests that this organism harbors mechanisms to sense and respond to stresses imposed by the host.
Gram-negative bacteria often use two-component signal transduction (2CST) systems to sense, respond to, and adapt to extracellular stresses. The Escherichia coli Cpx 2CST system allows bacteria to sense and respond to stresses affecting the cell envelope (9–12). The Cpx 2CST system consists of the histidine kinase CpxA and the response regulator CpxR. In response to envelope stress, CpxA autophosphorylates on a conserved histidine residue; phosphorylated CpxA donates its phosphoryl group to CpxR at a conserved aspartic acid residue (9). By binding to conserved recognition sequences, phosphorylated CpxR regulates the transcription of many genes involved in alleviating envelope stress in E. coli (13–15). In addition to kinase activity, CpxA also possesses CpxR-specific phosphatase activity (9). When grown in media containing glucose, CpxR accepts phosphoryl groups from acetyl phosphate (AcP) (16). When a cpxA deletion mutant is grown in the presence of glucose, CpxR is not readily dephosphorylated and the system is constitutively activated (16, 17).
The H. ducreyi genome (GenBank accession no. AE017143) contains homologues of CpxRA, which is the only obvious intact 2CST system in the genome. H. ducreyi is a fastidious organism and can only be grown in media containing glucose. Compared to its parent, a cpxR deletion mutant increases the transcription and expression of the lspB-lspA2 operon, which encodes proteins involved in the resistance of H. ducreyi to phagocytosis (18). The lspB-lspA2 promoter region contains putative CpxR recognition sequences, and recombinant CpxR binds to the promoter region of lspB-lspA2 in electrophoretic mobility shift assays. Taken together, these observations support the hypothesis that CpxR directly represses the expression of this operon (19). In contrast, a cpxA deletion mutant, which expresses the same level of CpxR as its parent, downregulates transcription from the lspB-lspA2 promoter (20, 21). Thus, CpxA regulates CpxR. If binding to the lspB-lspA2 promoter requires phospho-CpxR, then CpxA functions in H. ducreyi as if it were a net phosphatase (21), similar to what occurs when an E. coli cpxA deletion mutant is grown in media containing glucose (16, 17).
A previous microarray analysis showed that activation of the Cpx 2CST system by deletion of cpxA also downregulates the transcription of dsrA and the flp operon, which are both required for the virulence of H. ducreyi in humans (18); recombinant CpxR also binds to the dsrA and flp promoter regions in electrophoretic mobility shift assays, suggesting that phospho-CpxR also directly represses transcription from these promoters (18). Compared to its parent strain, a cpxA deletion mutant is avirulent (20), while a cpxR deletion mutant is fully virulent in human inoculation experiments (21). Taken together, these finding imply that CpxA likely functions primarily as a net phosphatase during infection in humans.
To better understand the role of the Cpx 2CST system in H. ducreyi pathogenesis, here we defined genes potentially regulated by CpxR in H. ducreyi by using RNA-Seq. To this end, we compared the RNA-Seq-defined transcriptomes of the parent, a cpxA mutant, and a cpxR mutant at different growth phases. We show that activation of the Cpx 2CST system by deletion of cpxA repressed the transcription of the majority of its targets, including seven known virulence determinants, and identified a CpxR binding motif that was enriched in the downregulated targets. In contrast, inactivation of the Cpx 2CST system by deletion of cpxR differentially regulated only a small number of genes and upregulated one gene encoding a known virulence determinant. These results are consistent with our previous observations that the cpxA mutant is fully attenuated while the cpxR mutant is fully virulent in humans and support the model in which CpxA acts primarily as a net phosphatase during H. ducreyi infection. Characterization of the downregulated genes may offer new insights into H. ducreyi pathogenesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The H. ducreyi strains were grown on chocolate agar plates supplemented with 1% IsoVitaleX at 33°C with 5% CO2 or in gonococcal (GC) broth supplemented with 5% fetal bovine serum (HyClone), 1% IsoVitaleX, and 50 μg/ml of hemin (Aldrich Chemical Co.) at 33°C. For RNA isolation, H. ducreyi strains were grown to the mid-log (optical density at 600 nm [OD660] of 0.2), transition (OD660 of 0.31), or early stationary phase (referred to here as stationary phase; OD660 of 0.35) in GC broth (see Fig. S1 in the supplemental material). E. coli strains were grown in Luria-Bertani medium at 37°C. When necessary, media were supplemented with streptomycin (50 μg/ml for H. ducreyi, 100 μg/ml for E. coli).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli DH5α | Strain used for general cloning procedures | Invitrogen |
| H. ducreyi strains | ||
| 35000HP | Human-passaged variant of strain 35000; parental strain | 56 |
| 35000HPΔcpxR | 35000HPcpxR in-frame deletion mutant containing a chloramphenicol resistance cartridge | 19 |
| 35000HPΔcpxA | 35000HPcpxA unmarked, in-frame deletion mutant | 20 |
| Plasmids | ||
| pRB157 | pLS88 derivative containing an ΩAmp cartridge, followed by a BglII site for insertion of promoter sequences and a promoterless GFP cassette derived from pGreenTIR | 20 |
| pKF1 | pRB157 derivative containing the lspB promoter region | 20 |
| pKF2 | pRB157 derivative containing the dsrA promoter region | 20 |
| pDG2 | pRB157 derivative containing the HD0182 promoter region | This study |
| pDG3 | pRB157 derivative containing the fimA promoter region | This study |
| pDG4 | pRB157 derivative containing the HD0427 promoter region | This study |
| pDG5 | pRB157 derivative containing the HD1123 promoter region | This study |
| pDG6 | pRB157 derivative containing the HD1278 promoter region | This study |
| pDG7 | pRB157 derivative containing the fis promoter region | This study |
RNA isolation and quality assessment.
Total RNA was extracted from 35000HP, 35000HPΔcpxA (20), and 35000HPΔcpxR (19) in the mid-log, transition, and stationary growth phases using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. RNA isolation was performed on four independent bacterial cultures for each strain in each growth phase. RNA was treated twice with the TURBO DNA-free DNase (Ambion). The integrity and the concentration of RNA were determined using the Agilent 2100 Bioanalyzer (Agilent Technologies) and the NanoDrop ND-1000 spectrophotometer (Thermo Scientific), respectively. The efficacy of DNase treatment was confirmed by reverse transcriptase PCR (RT-PCR) analysis of dnaE with the primer pair P1/P2 (see Table S1 in the supplemental material) and the QuantiTect SYBR green RT-PCR kit (Qiagen).
mRNA enrichment.
The removal of 23S, 16S, and 5S rRNA from total RNA was performed with the Ribo-Zero Magnetic kit (Gram-negative bacteria) (Epicenter Biotechnologies) by following the manufacturer's instructions. The removal of rRNA from total RNA was confirmed by the Agilent 2100 Bioanalyzer.
Preparation of RNA-Seq libraries and sequencing.
The TruSeq RNA sample preparation kit (Illumina, Inc.) was used to prepare RNA-Seq libraries by following the manufacturer's instructions. Briefly, approximately 10 to 100 ng of the enriched mRNA was fragmented and randomly primed for first-strand cDNA synthesis, followed by second-strand cDNA synthesis. The double-stranded cDNA was end repaired, adenylated, and ligated to adapters. The adapter-ligated cDNA library was then PCR enriched. Finally, the enriched RNA-Seq library was validated with the Agilent 2100 Bioanalyzer and quantitative RT-PCR (qRT-PCR). Clusters were generated on the cBOT automated cluster-generating system with the TruSeq PE Cluster kit (Illumina). Libraries were sequenced with the Illumina HiSeq 2500 sequencer with the TruSeq SBS kit (Illumina) for paired-end sequencing with read lengths of 100 bp in the Biomedical Genomics Core facility at Nationwide Children's Hospital (Columbus, OH). Image analysis and base calling were performed with the HiSeq Control software and the Real Time Analysis software. Demultiplexing was performed with the Illumina CASAVA software.
Sequence mapping and quantification of transcript levels.
The sequenced reads were mapped to the H. ducreyi 35000HP genome (GenBank accession no. AE017143) with the Burrows-Wheeler Alignment tool BWA (22), allowing up to two base mismatches. Reads that failed to map to any gene in the chromosome and reads that mapped to multiple locations in the genome were removed before quantifying the transcript levels. The total number of reads corresponding to the coding region of each gene was determined with the NGSUtils suite (23).
Identification of differentially expressed genes.
Differential expression of genes across different strains and growth phases was determined with edgeR software, a Bioconductor package for the differential expression analysis of digital gene expression data based on a negative binomial distribution (24). Because many genes were tested, we used a prespecified false-discovery rate (FDR) of ≤0.1 as the criterion for differential transcript expression; the FDR is less stringent than determining a familywise type 1 error with adjustment for multiple comparisons (25). We also chose a 2-fold change as a threshold for differential transcript expression, reasoning that this may favor the identification of genes that are biologically more significant than those with smaller changes. The differentially expressed genes were organized into known or putative operons with the computationally predicted operon structures from DOOR (Database for prOkaryotic OpeRons) (26). The differentially expressed genes or operons were functionally classified by using the annotations and pathway information from the sequenced H. ducreyi genome (R. S. Munson, Jr., et al., 2004, unpublished data), EcoCyc (27), and KEGG (28). We also determined if any biological pathways were enriched among the genes or operons differentially expressed in the cpxA and cpxR mutants compared to 35000HP by using the pathway annotations from BioCyc (29). A pathway was defined as enriched if the percentage of its differentially expressed genes was higher than that of all of the other pathways and if the P value for the pathway was <0.05 by Fisher's exact test. We also used the functional annotation-clustering algorithm of the DAVID (Database for Annotation, Visualization, and Integrated Discovery) bioinformatics resources (http://david.abcc.ncifcrf.gov/) to identify the biological pathways enriched in the cpxA and cpxR mutants relative to 35000HP (30). A pathway was considered enriched if the Fisher's exact P value for the cluster was < 0.05, the enrichment score for the cluster was greater than two, and the cluster involved greater than 5% of the genes on the submitted list (12).
qRT-PCR.
qRT-PCR was performed with the QuantiTect SYBR green RT-PCR kit (Qiagen) in an ABI Prism 7000 sequence detection system (Applied Biosystems). The primer pairs P1/P2 to P35/P36 were used to amplify internal gene-specific fragments ranging from 70 to 200 bp (see Table S1 in the supplemental material). For all qRT-PCR experiments, the amplification efficiency was determined for each primer pair; all primer pairs had greater than 95% efficiency. The expression levels of target genes were normalized to that of dnaE with the primer pair P1/P2. The fold change in expression was calculated as follows: [ratio = (Etarget)ΔCTtarget (35000HPΔcpxA-35000HPΔcpxR)/(Ereference)ΔCTreference (35000HPΔcpxA-35000HPΔcpxR)], where E is the amplification efficiency (equal to 10−1/slope) and ΔCT is the change in cycle threshold (20).
Reporter assays.
Reporter assays were performed as previously described (20). Approximately 200 bp of the HD0182, fimA, HD0427, HD1123, HD1278, and fis promoter regions were amplified by PCR with primer pairs P37/P38, P39/P40, P41/P42, P43/P44, P45/P46, and P47/P48, respectively (see Table S1 in the supplemental material). The amplified PCR products were ligated to pRB157 by using the BglII restriction site preceding a promoterless green fluorescent protein (GFP) cassette. The orientation of the insert with respect to the gfp cassette was confirmed by PCR with a promoter specific forward primer and the reverse primer P49 that hybridized to a region of the gfp cassette downstream of the BglII site. The final construct containing the promoter region preceding the gfp cassette was confirmed by sequencing. The reporter constructs were then electroporated into 35000HP, 35000HPΔcpxA, and 35000HPΔcpxR. Reporter strains generated previously were used for the analysis of the lspB and dsrA promoter activity (20). Whole-cell lysates were prepared from each transformant harvested at the stationary growth phase and analyzed by Western blot assays with monoclonal antibodies specific to GFP (Clontech) and the peptidoglycan-associated protein (PAL) (20). For each strain, the level of expression of GFP normalized to PAL was determined by densitometry with ImageJ software (31). Densitometry data were analyzed by two-tailed Student t test; a P value of <0.05 was considered statistically significant.
Identification of a putative CpxR binding motif.
The Multiple EM for Motif Elicitation (MEME) algorithm was used for de novo identification of the CpxR binding consensus sequence in H. ducreyi (32). Because of the lack of experimentally validated CpxR-dependent promoter motifs for H. ducreyi, the 450-bp upstream promoter regions of the 66 downregulated genes or operons were used for de novo motif identification by the MEME algorithm restricting the motif length to 15 to 50 bp. A similar analysis was also performed on the upregulated genes or operons. The identified motif was then encoded as a position-specific scoring matrix (PSSM). To test the significance of the discovered motif, matching scores were calculated on the basis of the promoter sequence and the motif feature characterized by the PSSM by using a previously published strategy (33). The score cutoff was determined by maximizing the enrichment of identified motif in the promoter regions of downregulated genes/operons compared to the promoter regions of unaffected genes.
Site-directed mutagenesis of the lspB promoter.
Mutations were introduced into the lspB promoter region in pKF1 with the QuikChange II XL (Agilent Technologies) site-directed mutagenesis kit by following the manufacturer's instructions. The lspB promoter in pKF1 contains four putative CpxR binding sites. The first two nucleotides in the first conserved region of each motif were mutated from G to T and from T to G. Mutagenic primers were designed with the QuikChange Primer Design Program (Agilent Technologies). The primer pairs P50/P51, P52/P53, P54/P55, and P56/P57 were used to mutagenize binding motifs 1 to 4, respectively (see Table S1 in the supplemental material). All of the mutations were confirmed by sequencing. The plasmids containing the mutated lspB promoter sequences were electroporated into 35000HPΔcpxA and 35000HPΔcpxR; the transformants were grown to mid-log phase, and the GFP/PAL expression ratio was measured as described above. Densitometry data were analyzed by two-tailed Student t test. Following Bonferroni adjustment, a P value of <0.0125 was considered statistically significant.
RESULTS
Transcriptome analysis.
In H. ducreyi, activation of the CpxRA system represses the transcription of several virulence determinants and cripples the ability of the organism to infect humans (18, 20). To more precisely define the role of the Cpx-regulated genes in virulence, we compared the transcriptomes of 35000HP, which has physiological levels of Cpx activation, 35000HPΔcpxA, which has a constitutively activated Cpx system due to loss of CpxA phosphatase activity, and 35000HPΔcpxR, which has no Cpx activity (18). We grew the strains to the mid-log, transition, and stationary growth phases (see Fig. S1 in the supplemental material), isolated RNA, and determined their transcriptomes with RNA-Seq.
Four biological replicates were included for each strain in each growth phase, summing to a total of 36 samples. The percentage of total reads aligned with the reference genome from all strains, growth phases, and replicates ranged from 98.5 to 99.3% (see Table S2 in the supplemental material). The percentage of reads aligned with the coding regions ranged from 63.9 to 83.6% (see Table S2). The average coverage per nucleotide ranged from 1.5 to 22.9 (see Table S2). As expected, approximately equal percentages of reads were found in the sense and antisense strands (see Table S2). The coefficients of determination (R2) between the samples from all strains, growth phases, and replicates ranged from 0.98 to 0.99, showing that there was high reproducibility of the RNA-Seq data.
Identification of genes regulated by CpxRA.
To identify genes potentially regulated by CpxRA in H. ducreyi, we calculated the fold change in the expression of genes in the cpxA mutant relative to the cpxR mutant and the fold change in the expression of genes in the cpxA and cpxR mutants relative to the parent. As discussed in Materials and Methods, we used an FDR of ≤0.1 and a 2-fold change as criteria for differential transcript expression. All genes considered differentially regulated by these criteria had P values of <0.01. Where appropriate, we grouped the differentially regulated genes into operons so that individual genes within a transcriptional unit were reported as one target.
As expected, comparison of 35000HPΔcpxA to 35000HPΔcpxR identified the most differences, with 86, 68, and 94 targets differentially regulated by the Cpx 2CST system in the mid-log, transition, and stationary growth phases, respectively; 63, 59, and 70% of these targets were downregulated (see Fig. S2 and Table S3 in the supplemental material). Comparison of 35000HPΔcpxA to 35000HP identified 52, 43, and 78 targets differentially regulated in the mid-log, transition, and stationary growth phases, respectively; 76, 82, and 69% of these targets were downregulated (see Fig. S2 and Table S4 in the supplemental material). Comparison of 35000HP to 35000HPΔcpxR identified only 12, 7, and 23 targets differentially regulated by the Cpx 2CST system in the mid-log, transition, and stationary growth phases, respectively; 33, 29, and 61% of these targets were downregulated (see Fig. S2 and Table S5 in the supplemental material). For the majority of the differentially regulated targets in all comparisons, the fold change was maximal in the stationary phase, intermediate in the mid-log phase, and lowest in the transition phase (see Tables S3, S4, and S5 in the supplemental material). These results suggest that the Cpx 2CST system is activated in a growth phase-dependent manner; most of the differential regulation occurs in the stationary phase.
To identify the overlap between the genes or operons that were differentially regulated by activation and inactivation of the Cpx 2CST system, we calculated the fold changes in the expression of genes in 35000HPΔcpxA and 35000HPΔcpxR relative to 35000HP and plotted the log10-transformed fold changes in 35000HPΔcpxA/35000HP against 35000HPΔcpxR/35000HP. This analysis clearly shows that activation of the system by deletion of cpxA serves to repress many targets, while repression of the system by deletion of cpxR increases the expression of a much smaller number of targets (Fig. 1). Few genes or operons were differentially regulated in both the cpxA and cpxR mutants relative to parent, suggesting that activation and inactivation of the system control unique sets of genes (Fig. 1).
Fig 1.
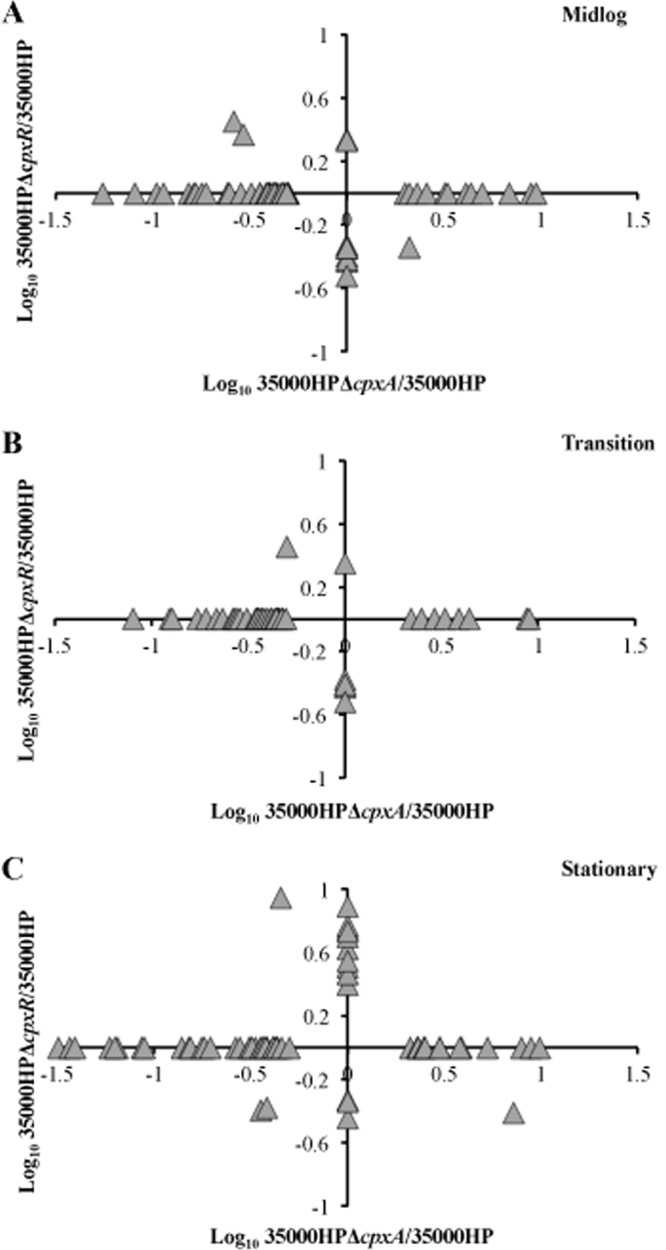
Scatter plots showing fold changes in the expression of genes or operons differentially expressed in 35000HPΔcpxA and 35000HPΔcpxR compared to 35000HP. The scatter plots were generated by plotting the log10-transformed fold changes in 35000HPΔcpxA versus 35000HP against 35000HPΔcpxR versus 35000HP at different growth phases. Each triangle in the graph indicates a single gene or operon.
qRT-PCR confirms the fold changes estimated by RNA-Seq.
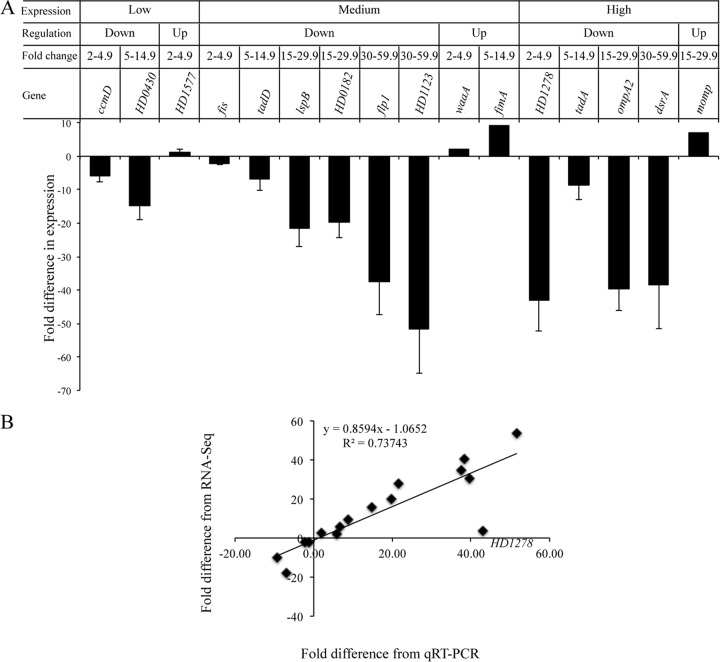
Since the most profound differential regulation was noted in the 35000HPΔcpxA-versus-35000HPΔcpxR comparison in the stationary phase, we concentrated on this data set for the remainder of this study. We validated the differentially regulated targets by qRT-PCR. The targets were divided into three groups on the basis of their expression levels (high, medium, and low); the expression levels were determined by the read abundance, which is the average log-transformed concentration for a gene across 35000HPΔcpxA and 35000HPΔcpxR (Fig. 2A). Genes from each expression level were then subgrouped into up- and downregulated genes (Fig. 2A). The up- and downregulated genes were stratified on the basis of their fold changes (2 to 4.9, 5 to 14.9, 15 to 29.9, and 30 to 60) (Fig. 2A). Representative genes were selected arbitrarily from each category; a total of 16 genes were evaluated by qRT-PCR. Our findings showed that the qRT-PCR analysis confirmed the expression of 15/16 targets identified by RNA-Seq (Fig. 2A). In general, the fold changes derived from RNA-Seq were in good agreement with the fold changes derived from qRT-PCR, except for HD1278, which was 3.7-fold downregulated by RNA-Seq and 43-fold downregulated by qRT-PCR (Fig. 2B); the reason for this discrepancy is unclear.
Fig 2.
qRT-PCR validation of the RNA-Seq data. (A) Fold changes in the expression of target genes in 35000HPΔcpxA relative to 35000HPΔcpxR in the stationary phase. The criteria used for selecting the targets for qRT-PCR validation are outlined at the top. The expression levels of target genes were normalized to that of dnaE. The data represent the mean ± SD from four independent experiments. (B) Correlation between the fold changes derived from qRT-PCR and RNA-Seq. The gene HD1278, for which the fold changes derived from the qRT-PCR data were not in agreement with the fold changes derived from the RNA-Seq data, is labeled on the graph.
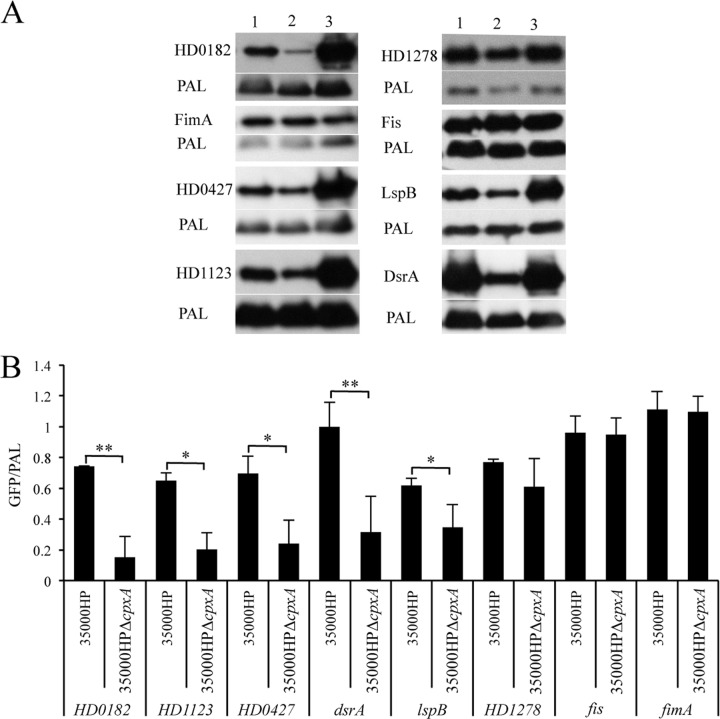
Reporter assays confirm the CpxR regulation of HD0182, HD0427, HD1123, lspB, and dsrA.
We determined the Cpx dependence of selected target gene promoters in 35000HP, 35000HPΔcpxA, and 35000HPΔcpxR grown to the mid-log, transition, and stationary phases. As the most profound differences were found in the stationary phase, these data are presented in Fig. 3. The expression levels of HD0182, HD0427, HD1123, lspB, and dsrA were downregulated in the cpxA mutant compared to its parent and the cpxR mutant, suggesting CpxR regulation of these targets (Fig. 3A and B). However, the expression of fimA, fis, and HD1278 was unaltered in 35000HP, 35000HPΔcpxA, and 35000HPΔcpxR (Fig. 3A and B). A previous study showed that recombinant CpxR does not bind to the fimA promoter region (18); thus, the regulation of fimA is likely indirect. RNA-Seq and qRT-PCR showed only 2.0- and 2.4-fold upregulation, respectively, of fis transcripts in the cpxA mutant compared to the cpxR mutant; the GFP reporter assay may not be sensitive enough to detect this level of upregulation. Alternatively, the Cpx 2CST system may regulate the expression of fimA, fis, and HD1278 at the posttranscriptional level. Overall, the results of the reporter assays correlated well with the RNA-Seq and qRT-PCR data.
Fig 3.
Promoter-reporter analysis of the Cpx targets identified by RNA-Seq. (A) The in vivo transcriptional activity of the Cpx-regulated promoters in 35000HP (lane 1), 35000HPΔcpxA (lane 2), and 35000HPΔcpxR (lane 3) grown to the stationary phase. For assessment of promoter activity, whole-cell lysates of H. ducreyi strains containing the reporter constructs were probed with an anti-GFP monoclonal antibody and anti-PAL monoclonal antibody 3B9, which served as a loading control. (B) Densitometry analysis of the Western blot assays in panel A. The Cpx dependence of target promoters was assessed by normalizing the GFP/PAL ratio in 35000HP and 35000HPΔcpxA to that in 35000HPΔcpxR, which was set to a value of 1. The data are means ± standard deviations from three independent experiments. *, P < 0.05; **, P < 0.01.
Activation of the Cpx 2CST system downregulates several H. ducreyi virulence determinants required for infection of humans.
We assessed if activation of the Cpx 2CST system affects the expression of genes tested for virulence in humans (Table 2). Activation of the Cpx 2CST system by deletion of cpxA downregulated the expression of genes in the flp-tad and lspB-lspA2 operons, dsrA, ncaA, and hgbA, which are all absolutely required for abscess formation in human volunteers (Table 2) (6, 34). Activation of the Cpx 2CST system downregulated dltA, which is partially required for abscess formation (Table 2) (6). These genes all encode secreted or outer membrane proteins (OMPs) that play direct roles in virulence such as nutrient acquisition, adherence, and resistance to phagocytic and serum killing. In contrast, deletion of cpxR had no effect on the transcription of virulence determinants, except for upregulation of lspB-lspA2. Surprisingly, deletion of cpxR downregulated dltA in all phases of growth (Table 2; see Table S5 in the supplemental material). These data are consistent with the attenuation of the cpxA mutant and the virulence of the cpxR mutant in humans and support the hypothesis that cpxA acts as a net phosphatase in vivo (18, 20).
Table 2.
Regulation of genes tested for virulence in humans by the Cpx 2CST system
| Human challenge modela | Function | 35000HPΔcpxA/35000HPb | 35000HPΔcpxR/35000HPc |
|---|---|---|---|
| Attenuated | |||
| flp1-flp2-flp3 | Adherence and microcolony formation | −31.3 | —d |
| dsrA | Major role in serum resistance | −27.4 | — |
| lspA1, lspA2 | Escape from phagocytosis | −15.4 | 2.8 |
| ncaA | Collagen binding | −7.2 | — |
| tadA | Adherence and microcolony formation | −6.9 | — |
| hgbA | Heme and/or iron uptake | −2.6 | — |
| pal | Outer membrane stability | — | — |
| sapBC | Resistance to antimicrobial peptides | — | — |
| hfq | RNA-binding chaperone | — | — |
| Partially attenuated | |||
| dltA | Partial role in serum resistance | −2.8 | −2.7 |
| wecA | Initiates synthesis of putative glycoconjugate | — | — |
| luxS | Quorum sensing | — | — |
| fgbA | Fibrinogen binding | — | — |
| sapA | Resistance to antimicrobial peptides | — | — |
| csrA | Posttranscriptional regulation | — | — |
| Virulent | |||
| hhdBe | Lysis of fibroblasts | −16.9 | — |
| ompP2A, ompP2B | Encode classical trimeric porins | −11.2 (ompP2A), 15.9 (ompP2B) | — |
| sodC | Detoxifies ROSf | −2.4 | — |
| momp | OmpA homolog; minor role in fibronectin binding | 7.2 | — |
| losB | Extends LOSg beyond KDOh-triheptose-glucose | — | — |
| lst | Adds sialic acid to LOS | — | — |
| cdtCe | Toxic for T cells, epithelial cells, and fibroblasts | — | — |
| ftpA | Pilus | — | — |
| tdX/tdhA | Heme uptake | — | — |
| glu | Adds glucose to KDO-triheptose LOS core | — | — |
| ompP4 | Outer membrane lipoprotein | — | — |
| neuA | Enables sialic acid addition to LOS | — | — |
Mutants of genes that have been tested in human volunteers and classified as attenuated, partially attenuated, or virulent.
Fold change in 35000HPΔcpxA relative to 35000HP in stationary phase.
Fold change in 35000HPΔcpxR relative to 35000HP in stationary phase.
—, no change in expression.
hhdB cdtC double mutant is also virulent in humans.
ROS, reactive oxygen species.
LOS, lipooligosaccharide.
KDO, 2-keto-3-deoxyoctulosonic acid.
The transcription of pal, sapBC, and hfq, which are also absolutely required for virulence, and fgbA, wecA, sapA, luxS, and csrA, which are partially required for virulence, was not affected by activation of the Cpx 2CST system (6, 35–37) (unpublished data; Table 2). pal encodes a structural component of the cell envelope and may have an indirect role in virulence (36, 38). sapA and sapBC encode periplasmic and inner membrane proteins that facilitate the uptake and degradation of antimicrobial peptides (36, 39). hfq and csrA encode cytoplasmic proteins that are involved primarily in posttranscriptional regulation. fgbA encodes an OMP that binds fibrinogen in ligand blot assays (40). wecA encodes the first enzyme involved in the synthesis of the enterobacterial common antigen; however, the glycoconjugate synthesized by this pathway in H. ducreyi has not been identified (41). luxS encodes an enzyme that produces an autoinducer-2-like molecule (35) Thus, virulence determinants not regulated by the Cpx 2CST system were generally those that were not secreted extracellularly or incorporated into the outer envelope.
Activation of the Cpx 2CST system downregulated the expression of hhdB, ompP2A, and sodC, which are not required for virulence in humans (Table 2) (6). sodC encodes a periplasmic copper-zinc superoxide dismutase, hhdB is involved in the secretion of hemolysin, and ompP2A encodes a porin (42–44). Thus, downregulation of the expression of OMPs or secreted proteins by CpxRA does not necessarily imply a role in virulence. Activation of the Cpx 2CST system upregulated the expression of ompP2B, momp, neuA, and lst, which are also not required for virulence (Table 2) (6, 45). neuA and lst encode enzymes involved with sialylation of lipooligosaccharide (45), while the remaining genes encode structural OMPs.
Functional classification of genes or operons differentially expressed in the cpxA mutant compared to the cpxR mutant.
With the annotations and pathway information from the sequenced H. ducreyi genome (Munson et al., 2004, unpublished), EcoCyc (27), and KEGG (28), the identified Cpx targets were classified into several functional categories, including amino acid biosynthesis, cell surface structures and associated proteins, cofactor biosynthesis, generation of precursor metabolites and energy, membrane transport and uptake, regulation and cell signaling, stress survival, translation, and hypothetical proteins (Table 3). The most notable biologically significant transcriptome changes in the cpxA mutant compared to cpxR mutant in the stationary phase are described below.
Table 3.
Functional classification of genes or operons differentially regulated by activation or inactivation of the Cpx 2CST system in H. ducreyi
| Parameter | Mid-log |
Transition |
Stationary |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 35000HP-ΔcpxA/35000HP-ΔcpxRa | 35000HP-ΔcpxA/35000HPb | 35000HP-ΔcpxR/35000HPc | 35000HP-ΔcpxA/35000HP-ΔcpxRa | 35000HP-ΔcpxA/35000HPb | 35000HP-ΔcpxR/35000HPc | 35000HP-ΔcpxA/35000HP-ΔcpxRa | 35000HP-ΔcpxA/35000HPb | 35000HP-ΔcpxR/35000HPc | |
| % Downregulated targets | 63 | 76 | 67 | 59 | 82 | 71 | 70 | 69 | 39 |
| % Upregulated targets | 37 | 24 | 33 | 41 | 18 | 29 | 30 | 31 | 61 |
| Functional categories: | |||||||||
| Amino acid biosynthesis | 2 | 1 | —d | 3 | 1 | — | 3 | 2 | — |
| Cell surface structures and associated proteins | 21 | 18 | 5 | 19 | 13 | 2 | 30 | 23 | 11 |
| Cell wall biosynthesis and remodeling | 2 | — | — | 3 | — | — | 1 | — | — |
| Cofactor biosynthesis | 2 | 2 | — | 1 | 1 | — | 4 | 2 | 1 |
| Degradation/assimilation/utilization | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| DNA replication, recombination, and repair | 1 | 1 | — | 1 | — | — | — | — | — |
| Fatty acid biosynthesis | — | — | — | — | — | — | 1 | 1 | — |
| Generation of precursor metabolites and energy | 10 | 7 | 1 | 8 | 6 | 1 | 7 | 7 | — |
| Hypothetical proteins | 22 | 9 | 3 | 16 | 11 | 3 | 16 | 20 | 5 |
| Lipid biosynthesis | 1 | 1 | — | — | — | — | — | 1 | — |
| Membrane transport/uptake | 11 | 8 | 3 | 9 | 3 | 1 | 10 | 7 | 2 |
| Nucleoside and nucleotide metabolism | 2 | — | — | 1 | — | — | 1 | 1 | — |
| Phage-associated proteins | — | — | — | 1 | 5 | — | 1 | 1 | 2 |
| Protein folding and degradation | 1 | 1 | — | 1 | 1 | — | 1 | 1 | — |
| Regulation and cell signaling | 8 | 5 | 0 | 4 | 3 | 0 | 9 | 8 | 0 |
| Stress survival | 3 | — | — | 1 | — | — | 3 | 2 | — |
| Translation | — | — | — | — | — | — | 6 | 1 | 2 |
Genes or operons differentially expressed in 35000HPΔcpxA versus 35000HPΔcpxR.
Genes or operons differentially expressed in 35000HPΔcpxA versus 35000HP.
Genes or operons differentially expressed in 35000HPΔcpxR versus 35000HP.
—, no genes or operons were found to be differentially expressed.
Amino acid biosynthesis.
Activation of the Cpx 2CST system downregulated the expression of homologues of genes involved in the biosynthesis of cysteine (cysK) and selenocysteine (selD) and upregulated the expression of homologues of genes involved in arginine biosynthesis (argC-argB) and regulation (argR) (Table 4; see Table S3 in the supplemental material).
Table 4.
Genes or operons differentially expressed by activation or inactivation of the H. ducreyi Cpx 2CST system in the stationary phase
| Function and gene identification codea | Gene(s)b | Description or homologc | Fold changed |
||
|---|---|---|---|---|---|
| 35000HP-ΔcpxA/35000HP-ΔcpxRe | 35000HP-ΔcpxA/35000HPf | 35000HP-ΔcpxR/35000HPg | |||
| Amino acid biosynthesis | |||||
| HD0577 | selD | Selenide, water dikinase | −2.2 | ||
| HD0890-2h | argC, argB | N-Acetyl-γ-glutamyl-phosphate reductase | 2.3 | 2.1 | |
| HD0896 | cysK | O-Acetylserine sulfhydrylase | −7.2 | −5.7 | |
| Cell surface structures and associated proteins | |||||
| Lipooligosaccharide biosynthesis | |||||
| HD0450-4h | waaA | 3-Deoxy-d-manno-octulosonic-acid transferase | 2.1 | −2.1 | |
| HD0653 | waaF | ADP-heptose-LPSj heptosyltransferase II | −2.3 | −2.3 | |
| HD1665 | d,d-Heptose 1,7-bisphosphate phosphatase | −4.3 | −3.6 | ||
| OMPs | |||||
| HD0045 | momp | Major OMP | 17.9 | 7.2 | −2.6 |
| HD0046-8h | ompA2, HD0047, HD0048 | Major OMP, OmpA2 | −30.5 | −25.6 | |
| HD0651-2h | oapA, oapB | Opacity-associated protein A | −2.2 | ||
| HD0746 | dltA | Endo-1,4-β-xylanase A | −2.8 | −2.5 | |
| HD0769 | dsrA | Serum resistance protein DsrA | −40.4 | −27.4 | |
| HD1017 | 24-kDa OMP; MltA-interacting MipA protein | −5.7 | −3.9 | ||
| HD1077-8h | ompP1 | OMP P1 | −2.3 | −2.7 | |
| HD1155-6h | lspB, lspA2 | Large supernatant protein exporter | −15.4 | ||
| HD1170 | ompP4 | OMP P4 | −2.1 | ||
| HD1278-80h | HD1278, HD1280 | Serine protease | −3.7 | −3.0 | |
| HD1326 | hhdB | Hemolysin activation/secretion protein | −20.1 | −16.9 | |
| HD1327 | hhdA | Hemolysin | −3.1 | −2.4 | |
| HD1433 | ompP2A | OMP P2-like protein | −9.8 | −11.2 | |
| HD1435 | ompP2B | OMP P2-like protein | 13.1 | 15.9 | |
| HD1856 | Possible OMP | −5.2 | −5.5 | ||
| HD1920 | ncaA | NcaA class II, necessary for collagen adhesion | −8.8 | −7.2 | |
| HD2025 | hgbA | Hemoglobin-binding protein HgbA | −3.3 | −2.6 | |
| Sialyltransferases, HD0685-6h | neuA, lst | Acylneuraminate cytidylyltransferase | 2.2 | ||
| Type I fimbriae, HD0281-5h | fimA, fimB, fimC, fimD, fimE | Fimbrial major pilin protein | 9.8 | 9.9 | |
| Type IVa pili/type II secretion/competence | |||||
| HD0182-5h | HD0182, HD0183, HD0184, HD0185 | Prepilin peptidase dependent protein A | −20.1 | −2.2 | 8.8 |
| HD0209 | comF | Competence protein F | −2.7 | 2.5 | |
| HD0427-34h | comA, HD0428, HD0429, HD0430, HD0431, HD0432, HD0433, comE | Competence protein A-like protein | −10.8 | 5.6 | |
| HD0650 | comEA | DNA uptake protein/type II secretion protein | −5.1 | 4.2 | |
| HD0732 | radC | DNA repair protein RadC | −5.2 | 5.0 | |
| HD1123-6h | pilA, pilB, pilC, pilD | Prepilin peptidase dependent protein D | −54.0 | −4.3 | 12.3 |
| HD1256 | rec2 | Recombination protein 2 | −10.2 | 5.9 | |
| HD1888 | smf | DNA processing chain A | −3.3 | 3.2 | |
| HD1921 | recG | ATP-dependent DNA helicase RecG | −2.5 | −2.3 | |
| HD1985 | DNA transformation protein, T(foX)/Sxy | −9.5 | −6.6 | ||
| Flp pili and associated proteins, HD1298-312h | flp1, flp2, flp3, orfBC, rcpAB, orfD, tadABCDEFG | flp operon protein Flp1 | −35.0 | −31.3 | |
| Cell wall biosynthesis and remodeling, HD1400 | Soluble lytic murein transglycosylase | 2.5 | |||
| Cofactor biosynthesis | |||||
| HD1389-92h | moaA, moaC, moaD, moaE | Molybdenum cofactor biosynthesis protein A | −2.8 | −3.2 | |
| HD1480 | bioD | Dithiobiotin synthetase | −2.5 | ||
| HD1495 | Putative dithiobiotin synthetase | −2.3 | −2.4 | ||
| HD1874 | ubiF | 2-Octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase | −3.5 | 2.9 | |
| Degradation/assimilation/utilization, HD1857 | ulaD | 3-Keto-l-gulonate-6-phosphate decarboxylase | −2.0 | ||
| Fatty acid biosynthesis, HD0181 | fabA | 3-Hydroxydecanoyl-ACP dehydratase | 3.6 | 2.3 | |
| Generation of precursor metabolites and energy | |||||
| ATP synthase, HD0003-11h | HD0003, atpB, atpE, atpF, atpH, atpA, atpG, atpD, atpC | ATP synthase F0F1 subunit A | −7.6 | −6.5 | |
| Anaerobic respiration | |||||
| HD0084 | lldD | l-Lactate dehydrogenase | −9.8 | −11.6 | |
| HD0344-47h | nrfA, nrfB | Cytochrome c552 | −2.2 | −2.7 | |
| HD1110-12h | fdhEi | Formate dehydrogenase accessory protein FdhE | 3.1 | 3.8 | |
| HD1393-4h | torY, torZ | Cytochrome c-type protein TorY | −2.7 | −2.4 | |
| HD1456-7h | ackA | Acetate kinase | −2.4 | ||
| HD1986 | fumC | Fumarate hydratase | −3.6 | −3.0 | |
| Lipid biosynthesis, HD1253 | dgkA | Diacylglycerol kinase | −2.0 | ||
| Membrane transport/uptake | |||||
| HD0025 | dcuB1 | Anaerobic C4-dicarboxylate transporter | 2.5 | ||
| HD0313-6h | dppB | Dipeptide transport system permease | 2.0 | 2.3 | |
| HD0614 | kefB | Glutathione-regulated K+ efflux system protein | 2.6 | 2.5 | |
| HD0766-8h | manX, manY, manZ | Phosphotransferase system mannose-specific transporter subunit IID | 2.0 | 2.3 | |
| HD0790 | ccmD | Heme exporter protein D | −2.2 | ||
| HD1074 | potD2 | Spermidine/putrescine-binding periplasmic protein | 2.2 | ||
| HD1109 | Oxalate/formate antiporter | 7.6 | 7.9 | ||
| HD1146 | glpF | Glycerol uptake facilitator protein | 3.3 | 3.0 | |
| HD1540-6h | HD1546i | Putative ABC transporter | −2.8 | ||
| HD1639 | Major facilitator superfamily transporter | 4.7 | 5.3 | ||
| HD1814-5h | HD1814 | Transporter/inner membrane protein YccA | −2.2 | ||
| HD1859-60h | HD1859i | Phosphotransferase system enzyme IIA permease | −3.3 | −2.8 | |
| Nucleoside and nucleotide metabolism, HD0888-9h | deoC, deoD | Deoxyribose-phosphate aldolase | 2.6 | 2.5 | |
| Phage-associated proteins | |||||
| HD0111 | Mu-like phage C protein | 2.0 | |||
| HD0531-8h | HD1535i | Tail assembly protein | 7.7 | ||
| HD1537-9h | HD1538i | Putative phage-related DNA-binding protein | −2.6 | −2.4 | |
| Protein folding and degradation, HD1900 | fkpA | FK506-binding protein-type peptidyl prolyl isomerase | −4.4 | −3.2 | |
| Regulation and cell signaling | |||||
| HD0261-2h | argR, HD0261 | Arginine repressor ArgR | 3.4 | 3.0 | |
| HD0280 | Putative GrlR family protein | 8.8 | 8.9 | ||
| HD0589-93h | HD0591i | LemA protein; GacS homolog | −2.5 | −3.8 | |
| HD0664-5h | HD0665 | LysM domain/BON superfamily protein | 2.5 | ||
| HD0738 | crp | Cyclic AMP regulatory protein | 3.2 | 2.5 | |
| HD0910-11h | hicA, hicB | HicA protein | −2.2 | −2.6 | |
| HD1028-31h | gcvA, HD1029 | DNA-binding transcriptional activator GcvA | −7.1 | −5.1 | |
| HD1096 | DNA-binding helix-turn-helix protein | −3.0 | −2.6 | ||
| HD1641-2h | purR | DNA-binding transcriptional repressor PurR | −2.8 | −2.8 | |
| Stress survival | |||||
| HD0848 | sodC | Superoxide dismutase [Cu-Zn] | −2.8 | −2.4 | |
| HD1665-8h | bcpi, queF | Thioredoxin-dependent thiol peroxidase | −3.0 | −2.3 | |
| HD1754-5h | ftnA | Ferritin | −2.1 | ||
| Translation | |||||
| HD0223-6h | miaB | (Dimethylallyl)adenosine tRNA methyl-thiotransferase | −2.3 | ||
| HD0318-9h | HD0318, rlmN | rRNA large subunit methyltransferase N | −7.5 | 5.4 | |
| HD0448-9h | dusB, fis | Dehydrogenase; tRNA-dihydrouridine synthase B | −2.9 | −2.3 | |
| HD1405-7h | efp | Elongation factor P | −2.3 | ||
| HD1257-8h | queAi | S-Adenosylmethionine-tRNA ribosyltransferase-isomerase | −4.6 | 3.5 | |
| HD1664 | tRNA/rRNA methyltransferase | −2.2 | |||
Does not include genes or operons that encode hypothetical proteins.
For genes that are members of a putative or known operon, only the differentially regulated genes are listed (ordered in the direction of transcription).
For genes that are members of a putative or known operon, the description corresponds to the first gene in the operon.
For genes that are members of a putative or known operon, the fold change corresponds to the first gene in the operon.
Genes or operons differentially regulated in 35000HPΔcpxA versus 35000HPΔcpxR.
Genes or operons differentially regulated in 35000HPΔcpxA versus 35000HP.
Genes or operons differentially regulated in 35000HPΔcpxR versus 35000HP.
Putative or known H. ducreyi operons.
Only genes located internal to the operon or toward the 3′ end of the operon were found to be differentially regulated; the description and the fold change correspond to the 5′ gene in the operon that was differentially regulated.
LPS, lipopolysaccharide.
Cell surface structures and associated proteins.
The Cpx 2CST system downregulated the expression of 14 genes or operons encoding or predicted to encode OMPs, secreted proteins, or enzymes involved in the assembly of cell surface structures (Table 4; see Table S3 in the supplemental material). Many of these genes and operons were discussed in the virulence determinant section (Table 2). The Cpx 2CST system also downregulated the expression of homologues of two genes that encode serine protease autotransporters (HD1278 and HD1280) (Table 4; see Table S3 in the supplemental material) (46). In addition to these genes, activation of the Cpx 2CST system downregulated the expression of waaF and HD1665, while it upregulated the expression of waaA; these genes are involved in the biosynthesis of lipooligosaccharide (Table 4; see Table S3 in the supplemental material). The Cpx 2CST system also downregulated the expression of homologues of genes involved in type IV pilus biogenesis/type II secretion/competence in other organisms (pilABCD, HD0427-comA-HD0428-HD0429-HD0430-HD0431-HD-432-HD0433-HD0434, HD0182-HD0183-HD0184-HD0185, comF, comE, radC, rec2, smf, and recG) and upregulated the expression of homologues of genes or operons involved in the biogenesis of type I fimbriae in other organisms (fimABCDE) and (Table 4; see Table S3 in the supplemental material). The majority of these genes were differentially regulated only in the stationary phase (Table 4; see Table S3). Whether type I fimbriae or type IV pili are expressed by H. ducreyi is unknown; the regulation of these genes by the Cpx system suggests that their expression and role in virulence should be explored.
Cofactor biosynthesis.
The Cpx 2CST system downregulated the expression of homologues of genes involved in the biosynthesis of molybdenum cofactor (moaACDE), biotin (bioD and HD1495), and ubiquinone (ubiF) (Table 4; see Table S3 in the supplemental material).
Generation of precursor metabolites and energy.
The Cpx 2CST system downregulated the expression of homologues of genes involved in ATP synthesis (HD0003-atpBEFHAGDC), anaerobic respiration (lldD, nrfAB, torYZ, and dcuB1), the tricarboxylic acid cycle (fumC), and degradation of AcP (ackA) (Table 4; see Table S3 in the supplemental material). Downregulation of ackA should lead to accumulation of AcP and thus foster the accumulation of phospho-CpxR, promoting a positive feedback loop for repression by CpxR. The Cpx 2CST system activated the expression of a homologue of a gene encoding FdhE, which serves as a proofreading chaperone for formate dehydrogenase, an enzyme involved in anaerobic respiration (Table 4; see Table S3).
Membrane transport and uptake.
The Cpx 2CST system downregulated the expression of homologues of genes encoding heme transporter (ccmD) and l-ascorbate transporter (HD1859-HD1860) (Table 4; see Table S3 in the supplemental material). Homologues of genes involved in dipeptide transport (dppB), glycerol transport (glpF), secondary sugar transport (manXYZ), polyamine transport (potD2), potassium/proton antiport (kefB), and oxalate-dependent generation of proton motive force (oxlT) were upregulated by the Cpx 2CST system (Table 4; see Table S3 in the supplemental material).
Protein folding and degradation.
The H. ducreyi Cpx 2CST system downregulated the expression of a homologue of a gene involved in protein folding (fkpA) (Table 4; see Table S3 in the supplemental material). However, the H. ducreyi Cpx 2CST system did not affect the expression of homologs of degP and dsbA, which are upregulated when the system is activated in E. coli (30). H. ducreyi lacks an obvious homolog of ppiA, which is also upregulated upon activation of the system in E. coli (30).
Regulation and cell signaling.
The Cpx 2CST system downregulated the expression of homologues of genes encoding or predicted to encode the transcriptional activator GcvA (gcvA), the transcriptional repressor PurR (purR), the toxin-antitoxin system HicAB (hicAB), a putative homolog of GacS (HD0591), and a putative homolog of DNA-binding helix-turn-helix protein (HD1096) (Table 4; see Table S3 in the supplemental material). The Cpx 2CST system upregulated the expression of homologues of genes encoding or predicted to encode the catabolite activator protein Crp (crp), a putative LysM domain/BON superfamily protein (HD0665), and a putative GrlR family protein (HD0280) (Table 4; see Table S3 in the supplemental material).
Stress survival.
In addition to sodC, activation of the Cpx 2CST system downregulated the expression of bcp and ftnA, which are involved in oxidative stress defense in other organisms (Table 4; see Table S3 in the supplemental material).
Translation.
The Cpx 2CST system downregulated the expression of homologues of many genes involved in protein synthesis (dusB, efp, miaB, queA, queF, rlmN, and HD1664), particularly those that are involved in tRNA and rRNA base modification (Table 4; see Table S3 in the supplemental material). Downregulation of the protein synthesis genes by the Cpx 2CST system was observed only in bacteria grown to the stationary phase (Table 4; see Table S3).
Hypothetical proteins.
The Cpx 2CST system downregulated the expression of 15 genes or operons predicted to encode hypothetical proteins, while it upregulated the expression of 14 genes or operons predicted to encode hypothetical proteins (see Table S3 in the supplemental material).
Pathway analysis.
Pathway enrichment analysis with annotations from both BioCyc and DAVID bioinformatics resources showed that genes encoding proteins involved in ATP synthesis and nucleoside and nucleotide metabolism were enriched among the genes differentially expressed in the cpxA mutant compared to 35000HP (data not shown).
De novo identification of the CpxR binding motif.
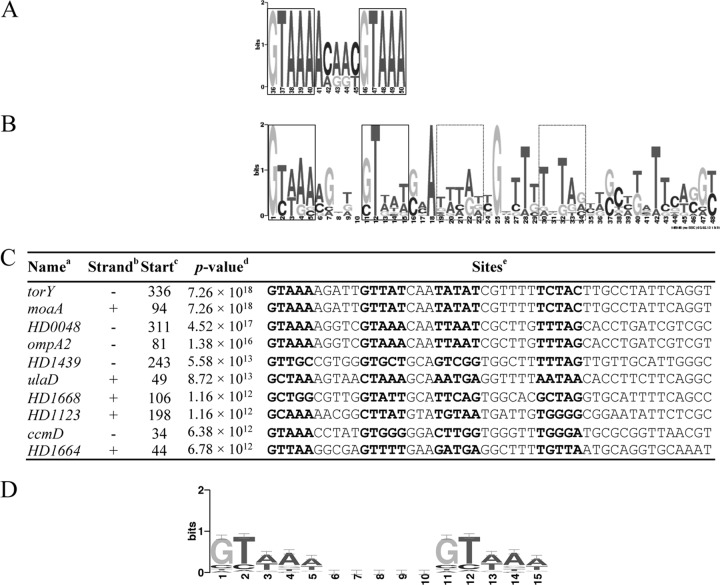
In E. coli, the consensus CpxR binding motif is the tandem repeat GTAAA(N5)GTAAA (Fig. 4A) (13). A de novo motif analysis of the 450-bp promoter regions of all upregulated and downregulated targets in the 35000HPΔcpxA-versus-35000HPΔcpxR comparison did not yield any significant motifs. However, analysis of the promoter regions from the downregulated targets identified an extended motif with an expect value of 6.7 × 10−9 (Fig. 4B). The extended motif consisted of two imperfect tandem repeats arranged in opposite orientations and separated by a 3-bp linker (Fig. 4B); each half of the extended motif bore some similarity to the E. coli tandem repeat. The first half of the extended motif contained imperfect 5-bp repeats separated by a 5-bp linker, while the second half of the extended motif contained imperfect 5-bp repeats separated by a 6-bp linker (Fig. 4B). However, the extended motif was found in only 10 out of 66 downregulated promoters (Fig. 4C), suggesting that it might not be required for CpxR binding.
Fig 4.
The H. ducreyi and E. coli CpxR binding motifs. (A) Sequence logo of the E. coli CpxR binding sites. The sequence logo was generated with the 450-bp promoter regions from 10 known Cpx target operons (12). The core regions in the motif are boxed. (B) The extended CpxR binding motif in H. ducreyi. The extended logo was generated by the MEME algorithm with the 450-bp upstream promoter sequences from the 66 downregulated targets identified in the 35000HPΔcpxA-versus-35000HPΔcpxR comparison in the stationary phase. The putative core regions in the extended motif are boxed. (C) Multiple-sequence alignment of the extended CpxR binding motif in the 10 promoter regions that contained the most significant matches. Superscript letters: a, genes in which the motif was identified; b, location of the extended motif on the template strand (+) or the complementary strand (−); c, start position of the binding site relative to the ATG start codon; d, P value for the extended motif; e, sequence of the predicted motif with the putative core regions in bold. (D) Sequence logo of H. ducreyi CpxR recognition weight matrix. The matrix frequency of a single GTAAA repeat is obtained by averaging the two parts of the first tandem repeat shown in Fig. 4B. The same nucleotide frequency was used for both GTAAA repeats. The 5-bp linker was assigned with a background frequency that was given by the MEME algorithm.
Next, we examined whether one tandem repeat was enriched in the downregulated promoters. A genome-wide search with a PSSM derived from the first tandem repeat in the extended motif with a 4- to 6-nucleotide gap (Fig. 4D) showed that this logo was present in the upstream promoter sequences of 51 (77%) of 66 downregulated targets (see Table S6 in the supplemental material), in 131 (64%) of 204 genes or operons whose expression levels were less than 2-fold downregulated (P = 0.03; odds ratio, 2), and in 395 (55%) of 718 genes or operons whose expression levels were not differentially regulated (P < 0.001; odds ratio, 3). Searches for enrichment of the single tandem repeat in the upregulated genes did not reveal any significant matches. The promoter regions of HD0182, HD0427, HD1123, lspB, and dsrA, which regulated the GFP reporter in response to Cpx activation, contained the CpxR logo (Fig. 4D), while the promoter regions of fis, fimA, and HD1278, which did not regulate the GFP reporter, did not (Fig. 3). These data suggest that the Cpx 2CST system probably directly regulates the majority of the downregulated targets by recognizing one tandem repeat and indirectly regulates the upregulated targets; the significance of the extended motif is unclear.
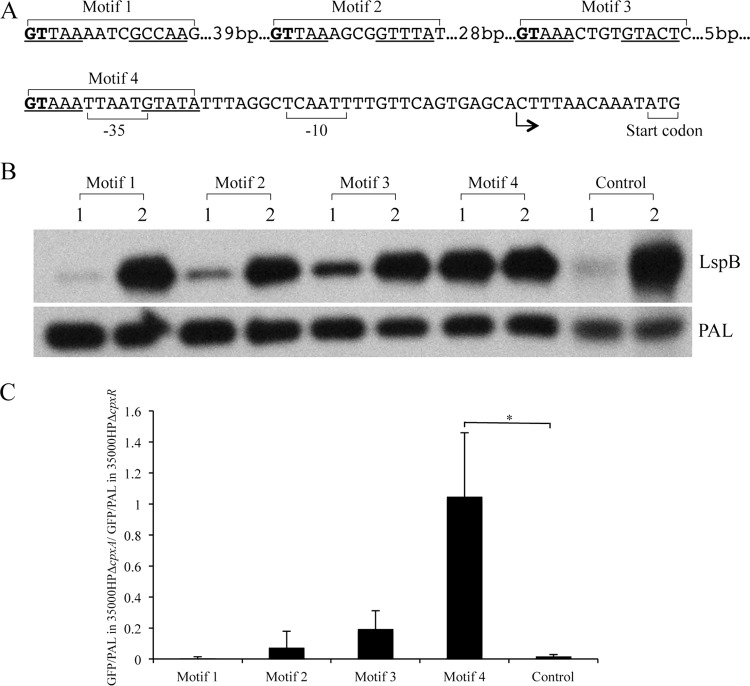
Site-directed mutagenesis confirms the predicted CpxR binding motif.
Activation of the Cpx 2CST system by deletion of cpxA downregulates lspB expression, and recombinant CpxR binds to the lspB promoter region (18). Therefore, we used the reporter plasmid containing the lspB promoter sequences to validate the predicted CpxR binding site. With the PSSM derived from the first tandem repeat (Fig. 4D), the lspB promoter region in pKF1 was predicted to contain four CpxR binding motifs (see Table S6 in the supplemental material). To identify which of these motifs was required for CpxR binding, we mutagenized the first two nucleotides in the first 5-bp repeat of each binding motif separately (Fig. 5A), introduced the mutagenized plasmids into the cpxA and the cpxR mutants, and measured the GFP-to-PAL expression ratio. Repression of lspB promoter activity in the cpxA mutant was significantly relieved by mutagenesis of the first two nucleotides in motif 4 (Fig. 5B and C). However, mutations in motifs 1, 2, and 3 did not relieve the repression of the lspB promoter (Fig. 5B and C). Taken together, these data suggest that motif 4 constitutes the binding site for CpxR in the lspB promoter and that CpxR likely uses the predicted binding logo (Fig. 4D) to control its direct targets.
Fig 5.
The predicted CpxR binding logo is required for lspB promoter activity. (A) lspB promoter region showing the locations of the predicted CpxR binding motifs and the locations of the mutagenized nucleotides. Predicted CpxR binding motifs 1 to 4 are indicated, and the mutagenized nucleotides in the binding sites are in bold. The ATG start codon, the transcriptional start site (arrow) determined on the basis of the RNA-Seq data, and the predicted −10 and −35 regions are also indicated. (B) Effect of site-directed mutagenesis of the predicted CpxR binding sites on lspB promoter activity measured in the GFP reporter assay. Plasmids containing the wild-type and mutagenized lspB promoters were electroporated into the cpxA and cpxR mutants. Whole-cell lysates were probed with an anti-GFP monoclonal antibody and anti-PAL monoclonal antibody 3B9, which served as a loading control. Motif 1, motif 2, motif 3, and motif 4, reporters with nucleotide substitutions in each respective motif; control, reporter containing the wild-type lspB promoter region. Lane 1, cpxA mutant, lane 2, cpxR mutant. (C) Densitometry analysis of the Western blot assays in panel B. The GFP/PAL ratio in 35000HPΔcpxA is expressed relative to that in 35000HPΔcpxR, which was set to a value of 1. The data are means ± standard deviations from three independent experiments. *, P = 0.0123.
DISCUSSION
A previous microarray analysis showed that activation of CpxR by deletion of cpxA directly downregulates transcription and expression of several major virulence determinants (18). Consistent with these findings, an H. ducreyi cpxA deletion mutant is avirulent in humans (20). To better understand the role of the Cpx 2CST system in controlling virulence, we used RNA-Seq to further define genes potentially regulated by the Cpx 2CST system in H. ducreyi at different phases of growth. We showed that activation of the Cpx 2CST system downregulates the majority of its targets, including known and putative virulence factors. These data indicate that CpxA functions as a net phosphatase to maintain parental levels of expression of these virulence determinants during infection of humans.
Our RNA-Seq data showed a high correlation in the results obtained from biological replicates at different phases of growth. The fold changes derived from the RNA-Seq data were generally concordant with the fold changes derived from qRT-PCR. Reporter assays of selected Cpx target promoters further corroborated the transcriptome data.
By comparing the transcriptome of a cpxA mutant to that of its parent in the mid-log phase with a 2-fold change cutoff and P ≤ 0.05, a previous microarray analysis identified 165 genes that are differentially regulated by activation of CpxR (18). Of these 165 genes, 98 (68%) are upregulated, while 67 (32%) are downregulated. In the microarray study, the differentially regulated genes were not grouped by operon structure. Examination of our data obtained from the comparison of the cpxA mutant to its parent in the mid-log phase showed that 82 individual genes were differentially regulated. Of these 82 genes, only 17 (21%) were upregulated, while 65 (79%) were downregulated. In H. ducreyi, cpxA is contained in an operon whose gene order is mazG, cpxR, cpxA, and HD1471. The cpxA mutant used in the microarray study is an in-frame insertion/deletion mutant, while the cpxA mutant used in the RNA-Seq study is an in-frame, unmarked deletion mutant; since both mutants are in frame, they should have no effect on the transcription of HD1471 (20). The two studies used different media to grow the strains. We do not know if the differences in the results reflect the differences in the sensitivities of the techniques or differences in the strains and growth conditions.
In our study, activation of the H. ducreyi Cpx 2CST system by deletion of cpxA downregulated nearly 70% of its targets, including dsrA, hgbA, lspB-lspA2, ncaA, flp1-3, and tadA. We previously demonstrated that mutants in these genes were fully attenuated for virulence in humans (6, 34). Thus, attenuation of the cpxA mutant in humans is likely a consequence of downregulation of multiple virulence determinants. The H. ducreyi Cpx 2CST system also highly downregulated the expression of 15 genes or operons encoding hypothetical proteins and 24 genes in 11 operons encoding homologues of proteins involved in the biogenesis of type IV pili/type II secretion/competence in other organisms. The majority of the type IV pilus/type II secretion/competence genes were differentially regulated only in bacteria grown to the stationary phase and were not identified in the previous microarray analysis, which was performed with bacteria grown to the mid-log phase. Type IV pili are important for adherence, twitching motility, competence, and virulence in other organisms (47). H. ducreyi is not naturally competent (48); whether type IV pili are expressed by H. ducreyi is unclear. The regulation of these genes by the Cpx system suggests that their expression and role in virulence should be explored.
Unlike deletion of cpxA, deletion of cpxR differentially regulated only 23 genes or operons in stationary phase; 61% of these genes were upregulated. Deletion of cpxR did not affect the expression of any known virulence determinants, except for upregulation of lspB-lspA2 and downregulation of dltA; this is consistent with the observation that a cpxR mutant is fully virulent in humans (21). Given that deletions of cpxA and cpxR yield opposing behaviors with regard to virulence, the findings of this study indicate that activation of the Cpx 2CST system represses H. ducreyi virulence determinants and that phosphatase activity of CpxA is critical for H. ducreyi to establish and maintain infection in the human host.
Activation of the H. ducreyi Cpx 2CST system by deletion of cpxA downregulated the expression of homologues of many genes involved in protein synthesis, anaerobic respiration, and ATP synthesis. However, the H. ducreyi cpxA mutant showed growth kinetics similar to those of the parental strain. Consistent with this finding, activation of the Cpx 2CST system upregulated the expression of homologues of many genes involved in alternative metabolic, respiratory, and energy-generating pathways, including secondary sugar transport (manXYZ) (49), glycerol uptake (glpF) (50), dipeptide transport (dppBCDF) (51), anaerobic fumarate respiration (dcuB1, aspA) (52, 53), and oxalate/formate-dependent energy generation (oxlT) (54). Given that the activation of the Cpx 2CST system leads to repression of canonical respiratory and energy-generating pathways, we speculate that the upregulation of homologues of genes involved in alternative metabolic, respiratory, and energy-generating pathways probably reflects a compensatory response to maintain energy homeostasis and cell viability.
The H. ducreyi CpxA and CpxR proteins are 42 and 60% identical to their E. coli homologues, respectively. In E. coli, a variety of signals activate the Cpx 2CST system; the majority of these signals involve misfolded proteins in the periplasm as the common stimulus for activation (11). In H. ducreyi, the signals that activate the Cpx 2CST system are undefined. Deletion of H. ducreyi mtrC activates the Cpx 2CST system; the lack of MtrC may lead to accumulation of toxic products in the periplasm, which induces envelope stress and activates the system (55). In response to membrane stress, the E. coli Cpx 2CST system upregulates the expression of many genes involved in envelope protein folding and degradation, including degP and dsbA, downregulates inner membrane proteins, and coordinates with other regulators of the cell envelope (10, 12). However, consistent with a previous microarray analysis (18), activation of the H. ducreyi Cpx 2CST system did not affect the expression of degP and dsbA and had variable effects on the differential expression of inner membrane proteins. Thus, whether and how the H. ducreyi Cpx 2CST system controls envelope stress is unclear (18).
The E. coli Cpx 2CST system is activated upon entry into the stationary phase and by signals related to growth and metabolism (16). Comparison of the transcriptomes of the parent and cpxR mutant strains showed that the H. ducreyi Cpx 2CST system is partially activated in the parent at all growth phases, with maximal activation in the stationary phase. Specifically, the H. ducreyi Cpx 2CST system upregulated the expression of homologues of genes involved in relieving protein translocation stress (yccA), anaerobic respiration (dcuB1), polyamine metabolism (potD2), sulfur metabolism (HD1815), xylan utilization (HD0746), membrane integrity (momp), and lipooligosaccharide biosynthesis (waaA). The functional significance of the products of these genes in H. ducreyi is unknown. Given that the products of the majority of these genes are involved in basic cellular functions, the partial activation of the Cpx 2CST system in the parent probably reflects a physiological role for the system in H. ducreyi growth and metabolism.
Using the promoter regions from the downregulated genes identified in our study, we defined a putative H. ducreyi CpxR binding motif that is similar to the tandem repeat motif identified in E. coli (13). Genome-wide motif analysis showed that the H. ducreyi imperfect tandem repeat was enriched in the downregulated genes but not in upregulated genes, suggesting that the upregulated genes are regulated indirectly by CpxR in H. ducreyi. Site-directed mutagenesis showed that the tandem repeat motif is required for CpxR regulation of lspB promoter sequences. These findings suggest that the single tandem repeat motif is sufficient for CpxR binding.
We conclude that activation of the Cpx 2CST system serves primarily to repress its targets in H. ducreyi, including several major virulence determinants, and that CpxA phosphatase activity is critical for H. ducreyi to establish and maintain infection in the human host. Future studies will focus on characterizing the direct targets of CpxR and their contributions to H. ducreyi pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant AI27863 to S.M.S. from the National Institutes of Allergy and Infectious Diseases (NIAID). We have no relevant financial relationships to disclose.
We thank Margaret Bauer and David Nelson for their thoughtful criticism of the manuscript and the Hanson laboratory for sharing strains and plasmids.
Footnotes
Published ahead of print 31 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00372-13.
REFERENCES
- 1.Steen R. 2001. Eradicating chancroid. Bull. World Health Organ. 79:818–826 [PMC free article] [PubMed] [Google Scholar]
- 2.Spinola SM. 2008. Chancroid and Haemophilus ducreyi, p 689–699 In Holmes KK, Sparling PF, Stamm WE, Piot P, Wasserheit JN, Corey L, Cohen MS, Watts DH. (ed), Sexually transmitted diseases, 4th ed McGraw-Hill, New York, NY [Google Scholar]
- 3.McBride WJ, Hannah RC, Le Cornec GM, Bletchly C. 2008. Cutaneous chancroid in a visitor from Vanuatu. Australas. J. Dermatol. 49:98–99 [DOI] [PubMed] [Google Scholar]
- 4.Peel TN, Bhatti D, De Boer JC, Stratov I, Spelman DW. 2010. Chronic cutaneous ulcers secondary to Haemophilus ducreyi infection. Med. J. Aust. 192:348–350 [DOI] [PubMed] [Google Scholar]
- 5.Ussher JE, Wilson E, Campanella S, Taylor SL, Roberts SA. 2007. Haemophilus ducreyi causing chronic skin ulceration in children visiting Samoa. Clin. Infect. Dis. 44:e85–e87 [DOI] [PubMed] [Google Scholar]
- 6.Janowicz DM, Ofner S, Katz BP, Spinola SM. 2009. Experimental infection of human volunteers with Haemophilus ducreyi: fifteen years of clinical data and experience. J. Infect. Dis. 199:1671–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer ME, Goheen MP, Townsend CA, Spinola SM. 2001. Haemophilus ducreyi associates with phagocytes, collagen, and fibrin and remains extracellular throughout infection of human volunteers. Infect. Immun. 69:2549–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer ME, Townsend CA, Ronald AR, Spinola SM. 2006. Localization of Haemophilus ducreyi in naturally acquired chancroidal ulcers. Microbes Infect. 8:2465–2468 [DOI] [PubMed] [Google Scholar]
- 9.Raivio TL, Silhavy TJ. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179:7724–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogt SL, Raivio TL. 2012. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 326:2–11 [DOI] [PubMed] [Google Scholar]
- 11.Hunke S, Keller R, Muller VS. 2012. Signal integration by the Cpx-envelope stress system. FEMS Microbiol. Lett. 326:12–22 [DOI] [PubMed] [Google Scholar]
- 12.Raivio TL, Leblanc SK, Price NL. 2013. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J. Bacteriol. 195:2755–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Wulf P, McGuire AM, Liu X, Lin ECC. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652–26661 [DOI] [PubMed] [Google Scholar]
- 14.Price NL, Raivio TL. 2009. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 191:1798–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bury-Mone S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Gen. 5:e1000651. 10.1371/journal.pgen.1000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe AJ, Parikh N, Lima BP, Zemaitaitis B. 2008. Signal integration by the two-component signal transduction response regulator CpxR. J. Bacteriol. 190:2314–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lima BP, Thanh Huyen TT, Basell K, Becher D, Antelmann H, Wolfe AJ. 2012. Inhibition of acetyl phosphate-dependent transcription by an acetylatable lysine on RNA polymerase. J. Biol. Chem. 287:32147–32160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labandeira-Rey M, Brautigam CA, Hansen EJ. 2010. Characterization of the CpxRA regulon in Haemophilus ducreyi. Infect. Immun. 78:4779–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labandeira-Rey M, Mock JR, Hansen EJ. 2009. Regulation of expression of the Haemophilus ducreyi LspB and LspA2 proteins by CpxR. Infect. Immun. 77:3402–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spinola SM, Fortney KR, Baker B, Janowicz DM, Zwickl B, Katz BP, Blick RJ, Munson RS., Jr 2010. Activation of the CpxRA system by deletion of cpxA impairs the ability of Haemophilus ducreyi to infect humans. Infect. Immun. 78:3898–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labandeira-Rey M, Dodd D, Fortney KR, Zwickl B, Katz BP, Janowicz DM, Spinola SM, Hansen EJ. 2011. A Haemophilus ducreyi cpxR deletion mutant is virulent in human volunteers. J. Infect. Dis. 203:1859–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breese MR, Liu Y. 2013. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics 29:494–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57:289–300 [Google Scholar]
- 26.Mao F, Dam P, Chou J, Olman V, Xu Y. 2009. DOOR: a database for prokaryotic operons. Nucleic Acids Res. 37:D459–D463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39:D583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Pujar A, Shearer AG, Travers M, Weerasinghe D, Zhang P, Karp PD. 2012. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 40:D742–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 31.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- 33.Liu Y, Taylor MW, Edenberg HJ. 2006. Model-based identification of cis-acting elements from microarray data. Genomics 88:452–461 [DOI] [PubMed] [Google Scholar]
- 34.Janowicz DM, Cooney SA, Walsh J, Baker B, Katz BP, Fortney KR, Zwickl B, Ellinger S, Munson RS., Jr 2011. Expression of the Flp proteins by Haemophilus ducreyi is necessary for virulence in human volunteers. BMC Microbiol. 11:208. 10.1186/1471-2180-11-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labandeira-Rey M, Janowicz DM, Blick RJ, Fortney KR, Zwickl B, Katz BP, Spinola SM, Hansen EJ. 2009. Inactivation of the Haemophilus ducreyi luxS gene affects the virulence of this pathogen in human subjects. J. Infect. Dis. 200:409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinker SD, Gu X, Fortney KR, Zwickl BW, Katz BP, Janowicz DM, Spinola SM, Bauer ME. 2012. Permeases of the Sap transporter are required for cathelicidin resistance and virulence of Haemophilus ducreyi in humans. J. Infect. Dis. 206:1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gangaiah D, Li W, Fortney KR, Janowicz DM, Ellinger S, Zwickl B, Katz BP, Spinola SM. 2013. Carbon storage regulator A contributes to the virulence of Haemophilus ducreyi in humans by multiple mechanisms. Infect. Immun. 81:608–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortney KR, Young RS, Bauer ME, Katz BP, Hood AF, Munson RS, Jr, Spinola SM. 2000. Expression of peptidoglycan-associated lipoprotein is required for virulence in the human model of Haemophilus ducreyi infection. Infect. Immun. 68:6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mount KL, Townsend CA, Rinker SD, Gu X, Fortney KR, Zwickl BW, Janowicz DM, Spinola SM, Katz BP, Bauer ME. 2010. Haemophilus ducreyi SapA contributes to cathelicidin resistance and virulence in humans. Infect. Immun. 78:1176–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauer ME, Townsend CA, Doster RS, Fortney KR, Zwickl BW, Katz BP, Spinola SM, Janowicz DM. 2009. A fibrinogen-binding lipoprotein contributes to virulence of Haemophilus ducreyi in humans. J. Infect. Dis. 199:684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banks KE, Fortney KR, Baker B, Billings SD, Katz BP, Munson RS, Jr, Spinola SM. 2008. The enterobacterial common antigen-like gene cluster of Haemophilus ducreyi contributes to virulence in humans. J. Infect. Dis. 197:1531–1536 [DOI] [PubMed] [Google Scholar]
- 42.Bong CTH, Fortney KR, Katz BP, Hood AF, San Mateo LR, Kawula TH, Spinola SM. 2002. A superoxide dismutase C mutant of Haemophilus ducreyi is virulent in human volunteers. Infect. Immun. 70:1367–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer KL, Thornton AC, Fortney KR, Hood AF, Munson RS, Jr, Spinola SM. 1998. Evaluation of an isogenic hemolysin-deficient mutant in the human model of Haemophilus ducreyi infection. J. Infect. Dis. 178:191–199 [DOI] [PubMed] [Google Scholar]
- 44.Janowicz D, Luke NR, Fortney KR, Katz BP, Campagnari AA, Spinola SM. 2006. Expression of OmpP2A and OmpP2B is not required for pustule formation by Haemophilus ducreyi in human volunteers. Microb. Pathog. 40:110–115 [DOI] [PubMed] [Google Scholar]
- 45.Spinola SM, Li W, Fortney KR, Janowicz DM, Zwickl B, Katz BP, Munson RS., Jr 2012. Sialylation of lipooligosaccharides is dispensable for the virulence of Haemophilus ducreyi in humans. Infect. Immun. 80:679–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruiz-Perez F, Wahid R, Faherty CS, Kolappaswamy K, Rodriguez L, Santiago A, Murphy E, Cross A, Sztein MB, Nataro JP. 2011. Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 108:12881–12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66:493–520 [DOI] [PubMed] [Google Scholar]
- 48.Redfield RJ, Findlay WA, Bosse J, Kroll JS, Cameron AD, Nash JH. 2006. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol. Biol. 6:82. 10.1186/1471-2148-6-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plumbridge J. 1998. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 27:369–380 [DOI] [PubMed] [Google Scholar]
- 50.Beijer L, Nilsson RP, Holmberg C, Rutberg L. 1993. The glpP and glpF genes of the glycerol regulon in Bacillus subtilis. J. Gen. Microbiol. 139:349–359 [DOI] [PubMed] [Google Scholar]
- 51.Weinberg MV, Maier RJ. 2007. Peptide transport in Helicobacter pylori: roles of Dpp and Opp systems and evidence for additional peptide transporters. J. Bacteriol. 189:3392–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janausch IG, Garcia-Moreno I, Unden G. 2002. Function of DcuS from Escherichia coli as a fumarate-stimulated histidine protein kinase in vitro. J. Biol. Chem. 277:39809–39814 [DOI] [PubMed] [Google Scholar]
- 53.Kleefeld A, Ackermann B, Bauer J, Kramer J, Unden G. 2009. The fumarate/succinate antiporter DcuB of Escherichia coli is a bifunctional protein with sites for regulation of DcuS-dependent gene expression. J. Biol. Chem. 284:265–275 [DOI] [PubMed] [Google Scholar]
- 54.Anantharam V, Allison MJ, Maloney PC. 1989. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J. Biol. Chem. 264:7244–7250 [PubMed] [Google Scholar]
- 55.Rinker SD, Trombley MP, Gu X, Fortney KR, Bauer ME. 2011. Deletion of mtrC in Haemophilus ducreyi increases sensitivity to human antimicrobial peptides and activates the CpxRA regulon. Infect. Immun. 79:2324–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Tawfiq JA, Thornton AC, Katz BP, Fortney KR, Todd KD, Hood AF, Spinola SM. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 178:1684–1687 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.