Abstract
We examined the environmental dissemination of Acinetobacter nosocomialis multilocus sequence typing clonal complex 260/71 in Rio de Janeiro, Brazil, including water from a dam and food samples. The increasing use of sequence based methods has demonstrated a large, previously unpredicted, dissemination of bacteria that may serve as opportunistic pathogens.
TEXT
Acinetobacter nosocomialis has emerged in the health care setting and may serve as a reservoir of resistance determinants. Clonal spread of A. nosocomialis was suspected since the 1980s, but the dissemination of specific types remained debatable until the use of DNA gel band pattern techniques (1–5).
The development of multilocus sequence typing (MLST) schemes evidenced the population structure of A. baumannii (6–8). However, only three studies applied MLST to define lineages of A. nosocomialis (6, 8, 9). In the first study (8), 20 clinical isolates obtained in Europe and the United States from 1985 to 1996 were included in 18 different sequence types (ST) of the MLST scheme hosted at University of Oxford (described from here on as the UO scheme). The second study typed seven A. nosocomialis isolates used as outgroups in phylogenetic analysis of A. baumannii by the MLST scheme hosted at Institut Pasteur (described from here on as the IP scheme) (9). Two clusters were observed among the seven A. nosocomialis isolates: (i) ST68/IP, including one isolate obtained in Nijmegen, Netherlands, in 1984 and one obtained in Odense, Denmark, during the 1980s, and (ii) ST71/IP, including three isolates from different cities in the Netherlands (Utrecht, Leiden, and Rotterdam) during 1987-2000 (6). The third study included 234 patients admitted to an intensive care unit (ICU) of Rio de Janeiro, Brazil, screened for Acinetobacter sp. colonization in 2007-2008 (9). A single A. nosocomialis RAPD type was found in surveillance cultures of 24 (10%) patients and caused one episode of bloodstream infection. This type belonged to ST260/UO (9). The aim of the present study was the assess the extent of environmental dissemination of A. nosocomialis ST260/UO.
Screening for Acinetobacter sp. was performed in samples collected from three sources: inside the ICU (n = 28), including the hands of health care staff, sinks, faucets, benches, bed rails, mattresses, and top of tables; lettuce (Lactuca sativa) (n = 65) bought at different markets in the city of Rio de Janeiro; and water samples (n = 4) and swab specimens (n = 61) from the surroundings of the main water reservoir of Rio de Janeiro. This reservoir belongs to a complex of dams providing water to almost 90% of the people of Rio de Janeiro city. Imprints from the dominant hand were made on blood agar plates from health care workers. Specimens from environmental surfaces were collected with swabs, inoculated onto MacConkey broth, and incubated at 35°C overnight. Lettuce specimens (10 μl each) were obtained from serial dilutions of 10 g of the leaves (dry weight) homogenized in saline solution. Water specimens (100 ml) were membrane filtered. Samples were subcultured onto MacConkey agar at 35°C for 48 h. Gram-negative coccobacilli, catalase-positive, oxidase-negative, nonmotile and glucose nonfermenters were classified as Acinetobacter sp. and speciated by partial rpoB gene sequencing analysis as previously described (10). Antimicrobial susceptibility was determined by disk diffusion for amikacin, cefepime, ceftazidime, ciprofloxacin, gentamicin, imipenem, meropenem, piperacillin-tazobactam, tobramycin, and trimethoprim-sulfamethoxazole and by microdilution method for colistin (11). Isolates were defined as multidrug-resistant (MDR) when resistant to three or more antimicrobial classes (12).
MLST was performed with the UO scheme (http://pubmlst.org/abaumannii/), and the IP scheme (www.pasteur.fr/mlst) as described previously (6, 7). goeBURST (http://www.phyloviz.net/wiki/) was used to determine relationships between STs in the Acinetobacter databases (http://pubmlst.org/abaumannii/ and www.pasteur.fr/mlst) (13). A. nosocomialis isolates were typed by pulsed-field gel electrophoresis (PFGE) as described based on DNA ApaI digestion (14). Band patterns were analyzed for similarity with GelCompar version 4.01 (Applied Maths, Kortrijk, Belgium) based on Dice coefficient and the unweighted pair group method with arithmetic averages. The study was approved by the Institutional Review Board of the hospital.
A total of 11 Acinetobacter isolates, all non-A. baumannii species, were obtained from environmental specimens. Environmental isolates included four from lettuce (two Acinetobacter berezinae and one each A. nosocomialis and Acinetobacter rhizosphaerae); four from ICU nonclinical specimens (two A. nosocomialis from furniture surfaces and one from the hand of a staff member, and one Acinetobacter schindleri from ICU bench); and three from water (two A. nosocomialis and one Acinetobacter radioresistens).
A. nosocomialis typing by MLST with the UO scheme showed that one isolate obtained from lettuce belonged to ST260 (UO) and that another obtained from an ICU cabinet was a double-locus variant (DLV) of this ST. For one isolate obtained from ventilator equipment, it was not possible to amplify the gdhB gene fragment by PCR, and a complete ST profile was not determined. These results together with data from four A. nosocomialis clinical isolates from the previous study (9) are shown in Table 1. The four clinical isolates belong to RAPD [random(ly) amplified polymorphic DNA] genotype A and were obtained from three patients. Isolates An2a and An2b are from a single patient and had RAPD profiles with a slight difference. Two isolates were additionally typed by the IP scheme: one ST260 (UO) and one ST260 (UO) DLV. Both were classified as ST161/IP, a single-locus variant (SLV) of ST71/IP, forming CC71/IP. PFGE was performed on 9 of 10 A. nosocomialis study isolates; one was untypeable. Dendrogram of the PFGE bands is shown in Fig. 1. ST260/UO isolates and one of the DLVs of ST260/UO, corresponding to ST161/IP, formed a cluster with >95% similarity. Another DLV of ST260/UO had only 87% similarity with this cluster, and all other isolates had unique band patterns.
Table 1.
Characteristics of Acinetobacter nosocomialis isolatesa
| Type of sample | Isolate | Specimen source | Isolation date (day/mo/yr) | Allelic profile (UO scheme) | ST (UO/IP) | Resistance profile |
|---|---|---|---|---|---|---|
| Human clinical specimen | An2b | Tracheal secretion | 16/5/2007 | 11-65-14-21-37-105-15 | 260/161 | CAZ, CIP, GEN, SXT, TOB |
| An2a | Tracheal secretion | 6/7/2007 | 11-67-14-51-37-105-15 | NA(260DLV)/161 | CIP, GEN, SXT, TOB | |
| An8 | Blood | 13/7/2007 | 11-65-14-21-37-105-15 | 260/161 | AMI, CIP, FEP, GEN, TOB | |
| An7b | Tracheal secretion | 26/7/2007 | 11-65-14-21-37-105-15 | 260/161 | CIP, GEN, SXT, TOB | |
| ICU | CTI021 | ICU cabinet | 28/5/2008 | 11-70-14-21-37-4-15 | 264(260DLV) | None |
| CTI10 | Ventilator equipment | 28/5/2008 | 11-27-ND-21-37-113-15 | ND | SXT | |
| CTI15.3 | Hand of ICU staff | 28/5/2008 | 11-65-93- 61-25-112-15 | 263 | FEP, SXT, TZP | |
| Environment unrelated to hospital | PCA073 | Water reservoir | 13/12/2007 | 49- 68-119-53-41-115-58 | 262 | TOB |
| PCA111 | Water reservoir | 14/3/2008 | 42-36-92-30-40-114-57 | 261 | None | |
| A12.1 | Lettuce | 25/3/2008 | 11-65-14-21-37-105-15 | 260/161 | CIP |
ICU, intensive care unit; UO, University of Oxford MLST scheme; ND, not determined; ST, sequence type; IP, Institut Pasteur; NA, not assigned; DLV, double-locus variant; AMI, amikacin; CIP, ciprofloxacin; FEP, cefepime; GEN, gentamicin; TOB, tobramycin; CAZ, ceftazidime; SXT, sulfamethoxazole-trimethoprim; TZP, piperacillin-tazobactam.
Fig 1.
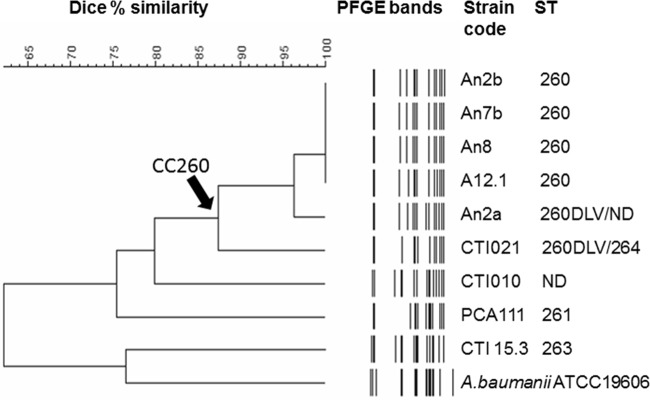
Dendrogram of PFGE band profiles of A. nosocomialis isolates and one outgroup A. baumannii ATCC 19606 reference strain. DLV, double locus variant; ND, not determined.
A. nosocomialis clinical isolates and the isolate from the hand of ICU staff member were MDR strains. Five isolates obtained from environment were susceptible to all or resistant to only one agent.
This is the first study to describe A. nosocomialis MLST typing by both UO and IP schemes. A. nosocomialis clone defined by MLST as CC260/UO and ST161/IP spread among several patients admitted to an ICU and its environment over a 9-month period. The clone was also present in the environment outside the hospital. Since the first description of the likely clonal spread of A. nosocomialis, its environmental dissemination was suspected (2). Later, dissemination of isolates belonging to a single ribotype and PFGE genotype between patients and the environment was clearly demonstrated during an outbreak in a neurosurgical ICU in the Netherlands (4). In the present study, we describe the environmental dissemination of isolates related to A. nosocomialis CC260/UO and ST71/IP. One isolate obtained from lettuce bought in a market unrelated to the hospital was indistinguishable from this clone by MLST and PFGE. The isolate from lettuce was susceptible to several antimicrobials, whereas clinical isolates were all MDR. These findings may indicate CC260/UO is widespread in the environment but able to colonize and cause human disease, acquiring resistant determinants after exposure to other resistant strains under the selection pressure in the hospital setting.
The observation that CC260/UO-ST161/IP isolates are a SLVs of ST71/IP, obtained in the Netherlands during 1987-2003, is remarkable and shows the intercontinental spread of an A. nosocomialis strain (6).
ACKNOWLEDGMENTS
This study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), the Comissão Fullbright-Brasil, the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) of Brazil, and the Fogarty International Program in Global Infectious Diseases (TW006563) of the National Institutes of Health.
We acknowledge the platform Genotyping of Pathogens and Public Health (Institut Pasteur) for coding MLST alleles and profiles. This publication also made use of the Acinetobacter baumannii MLST website (http://pubmlst.org/abaumannii/) developed by Keith Jolley at the University of Oxford.
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Gerner-Smidt P. 1987. Endemic occurrence of Acinetobacter calcoaceticus biovar anitratus in an intensive care unit. J. Hosp. Infect. 10:265–272 [DOI] [PubMed] [Google Scholar]
- 2. Dijkshoorn L, Aucken HM, Gerner-Smidt P, Kaufmann ME, Ursing J, Pitt TL. 1993. Correlation of typing methods for Acinetobacter isolates from hospital outbreaks. J. Clin. Microbiol. 31:702–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spence RP, Tower KJ, Henwood CJ, James D, Woodford N, Livermore DM. 2002. Population structure and antibiotic resistance of Acinetobacter DNA group 2 and 13TU isolates from hospitals in the UK. J. Med. Microbiol. 51:1107–1112 [DOI] [PubMed] [Google Scholar]
- 4. Van Dessel H, Kamp-Hopmans TEM, Fluit AC, Brisse S, de Smet AMGA, Dijkshoorn L, Troelstra A, Verhoef J, Mascini EM. 2002. Outbreak of a susceptible strain of Acinetobacter species 13 (sensu Tjernberg and Ursing) in an adult neurosurgical intensive care unit. J. Hosp. Infect. 51:89–95 [DOI] [PubMed] [Google Scholar]
- 5. McDonald A, Amyes SG, Paton R. 1999. The persistence and clonal spread of a single strain of Acinetobacter 13TU in a large Scottish teaching hospital. J. Chemother. 11:338–344 [DOI] [PubMed] [Google Scholar]
- 6. Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390 (Erratum, 45:2011, 2007.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wisplinghoff H, Hippler C, Bartual SG, Haefs C, Stefanik D, Higgins PG, Seifert H. 2008. Molecular epidemiology of clinical Acinetobacter baumannii and Acinetobacter genomic specie 13TU isolates using multilocus sequencing typing. Clin. Microbiol. Infect. 14:708–715 [DOI] [PubMed] [Google Scholar]
- 9. Martins N, Martins IS, Freitas WV, Matos JA, Girao VBC, Coelho-Souza T, Maralhaes ACG, Cacci LC, Figueiredo MP, Dias RCS, Costa-Lourenço APR, Ferreira ALP, Dalla-Costa L, Nouer SA, Santoro-Lopes G, Riley LW, Moreira BM. 2013. Imported and intensive care unit-born Acinetobacter baumannii clonal complexes: one-year prospective cohort study in intensive care patients. Microb. Drug Res. 19:216–223 [DOI] [PubMed] [Google Scholar]
- 10. La Scola B, Gundi VAKB, Khamis A, Raoult D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clinical and Laboratory Standards Institute 2012. Performance for antimicrobial susceptibility testing; 12th informational supplement. CLSI document M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Selling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18:268–281 [DOI] [PubMed] [Google Scholar]
- 13. Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 10:e1–e11. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seifert H, Dolzani L, Bressan R, Van Der Reijden T, Van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]


