Abstract
In recent years, the dramatic increase in community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections has become a significant health care challenge. Early detection of CA-MRSA is important because of its increased virulence associated with the arginine catabolic mobile element (ACME), Panton-Valentine leukocidin (PVL), and other toxins that may contribute to disease severity. In particular, the USA300 epidemic clone has emerged and now represents the cause of as much as 98% of CA-MRSA skin and soft tissue infections in the United States. Current diagnostic assays used to identify CA-MRSA strains are based on complex multiplex PCRs targeting the staphylococcal cassette chromosome mec (SCCmec) DNA junction, a multitude of genes, and noncoding DNA fragments or on a number of lengthy sequence-typing methods. Here, two nucleotide polymorphisms, G88A and G2047A, that were found to be in strict linkage disequilibrium in the S. aureus penicillin-binding protein 3 (pbp3) gene were also found to be highly associated with the USA300 clone of CA-MRSA. Clinical isolates that contained this pbp3 allele were also positive for the presence of SCCmec type IV, the ACME, and the PVL toxin gene and matched the t008 or t121 molecular spa types, which are associated specifically with the USA300 CA-MRSA clone. A single allele-specific PCR targeting the G88A polymorphism was developed and was found to be 100% sensitive and specific for the detection of USA300 CA-MRSA and 91.5% sensitive and 100% specific for the detection of all CA-MRSA isolates in this study.
INTRODUCTION
Staphylococcus aureus is a leading cause of skin and soft tissue infections (SSTIs) (1), surgical site infections (2), and invasive disease in the health care industry (3–6). The risk of S. aureus infections is high in hospitals, nursing homes, and other health care facilities, where patients' primary line of immunity, the skin, is breached; open wounds (including surgical wounds), catheter ports, and weakened immune systems (4, 5) allow S. aureus to establish infections. Methicillin-resistant S. aureus (MRSA) strains were first detected shortly after the introduction of methicillin in the early 1960s. The mechanism of resistance to β-lactam antibiotics involves integration of the staphylococcal cassette chromosome mec (SCCmec) transposon into the S. aureus genome. Specifically, SCCmec DNA integrates into the S. aureus orfX gene, described as the recombination hot spot (7). The antibiotic resistance associated with SCCmec is due to alternative penicillin-binding protein 2a (PBP2a), which is encoded by the mecA gene. This PBP2a has significantly lower affinity for β-lactam antibiotics (8, 9).
There are four predominant SCCmec types of MRSA in the United States, namely, types I to IV. Types I to III typically are considered hospital-associated MRSA (HA-MRSA), and type IV commonly is linked to community-associated MRSA (CA-MRSA) (10–12). CA-MRSA strains frequently are associated with the Panton-Valentine leukocidin (PVL) toxin, which may confer an increase in virulence; however, the exact role of PVL is of debate (13–16). In the United States between 1997 and 1999, strains of MRSA from community sources emerged in otherwise healthy people with no known health care-associated risks or exposures (17). Subsequently, outbreaks of CA-MRSA disease (primarily SSTIs) have been observed in locations with high densities of people, such as prisons and athletic team locker rooms (18, 19). Unlike SCCmec types I to III, SCCmec type IV confers little to no fitness cost and allows for more-robust growth, which likely is a factor in its success in community-associated isolates (20).
Once an individual is infected with MRSA, the choice of effective antibiotic therapies is limited. HA-MRSA and CA-MRSA strains generally possess different antibiotic susceptibility and virulence profiles. HA-MRSA strains often are resistant to multiple classes of antibiotics, in comparison with CA-MRSA strains. However, CA-MRSA infections tend to be more virulent (16, 21). Therefore, rapid accurate MRSA identification and type characterization may aid physicians in the development of successful treatment strategies for patients and might facilitate infection control measures such as isolation of infected or colonized patients. Additionally, the distinction between hospital- and community-associated isolates might help health care facilities monitor the efficacy of infection control measures and understand the epidemiology of the strains responsible for disease. To address this need, researchers are continuously developing novel methods to detect and to classify MRSA. However, there are significant challenges for the development of successful diagnostic approaches for MRSA characterization, due to the sequence complexity of SCCmec and the genetic variability of the organism. Assay costs, assay complexity, and turnaround times are confounding factors for rapid MRSA-typing diagnostic assays.
Culture-based methods for the detection of MRSA, such as the use of mannitol salt agar-cefoxitin (22) and MRSA CHROMagar media, are inexpensive and selective but can be time-consuming, and positive results are considered only presumptive (23). Results may be complicated by low MRSA concentrations, mixed infections, and small colony variants, leading to delays in the diagnosis of MRSA infections and the prescription of proper antibiotic therapy for patients. Culture-based methods also fail to help distinguish between community-associated and hospital-associated strains. Furthermore, as more is understood about the genetics of these strains, these assays do not provide useful epidemiological information for use in tracking outbreaks, determining antibiotic susceptibility, and understanding the virulence of MRSA strains.
Multiple kits for the detection of MRSA are available commercially. The BinaxNOW PBP2a assay (Alere) is an immunochromatographic assay that uses an antibody to the alternative PBP2a protein responsible for methicillin resistance (24). The BacLite rapid MRSA assay (3M) uses adenylate kinase activity to detect viable cells after growth in selective enrichment broths containing antibiotics, capture with antibodies linked to magnetic beads, and cell lysis with lysostaphin (25). This assay provides results within 5 h; however, it fails to detect ciprofloxacin-sensitive MRSA strains, and it generates a number of false-positive and false-negative results (sensitivity, 90.4%; specificity, 95.7% [25]). Both assays are incapable of differentiating between HA-MRSA and CA-MRSA, which may limit infection control measures and the tracking of outbreaks (26).
Several laboratory-based methods exist to provide detailed genetic information about MRSA strains. A common method used by many laboratories is spa typing, in which segments of the spa gene are sequenced and analyzed for repeats, which can be linked to epidemiologically distinct MRSA clones characterized in public databases (http://spaserver.ridom.de) (27). Pulsed-field gel electrophoresis (PFGE) separates large pieces of the S. aureus genome and compares the banding pattern with those of characterized laboratory-studied strains for similarities (28). Multilocus sequence typing requires sequencing of multiple housekeeping genes and comparison of the alleles with established clones in a database (29). All of these methods provide very useful information but are highly time and labor intensive and require a high level of skill to perform and to interpret. Thus, none of these laboratory-based methods is ideal for high-throughput clinical testing.
Molecular methods are a recent development for the identification and characterization of MRSA. PCR is a valuable tool; however, detection of the mecA gene alone is not sufficient for reliable MRSA screening, because the gene is found on a mobile genetic element that can be found in other less-virulent staphylococcal species. Staphylococcus epidermidis, for example, is a common commensal bacterial species on skin and often possesses SCCmec and mecA (30). To be MRSA-specific, PCR diagnostic assays may require selective culturing from a patient sample and confirmation of S. aureus by a conventional microbiological identification method, adding considerable time to the diagnosis. Other molecular methods use complex multilocus PCR assays to detect and to confirm the mecA integration within the S. aureus strain (31, 32). A MRSA PCR assay that utilizes the SCCmec-orfX junctions and improves on earlier molecular methods was developed. The PCR assay is designed with one primer that anneals to the S. aureus-specific orfX gene and a second primer that anneals to the right arm of the SCCmec transposon, resulting in an amplification product only when the SCCmec DNA is integrated into the S. aureus genome (33). This removes the possibility of false-positive results from non-S. aureus species containing mecA. Complex multiplex PCR assays utilizing this technology have been designed to differentiate between SCCmec types and to help differentiate CA-MRSA and HA-MRSA strains (34). Adding further complexity, SCCmec transposons have been shown to lose the mecA gene, thus providing false-positive results for MRSA (35–37). The Xpert MRSA assay (Cepheid) provides an integrated automated system for detection of MRSA by identification of SCCmec transposon types, using integrated chips to perform SCCmec-orfX junction PCRs, but requires expensive equipment and software as well as repeated purchases of new chips (38). A recent study explored the use of multiple molecular markers for detection of USA300 and obtained high sensitivity and specificity (98% and 97%, respectively) by simultaneously detecting the arcA arginine deiminase gene and the PVL-encoding genes within a single isolate (39). However, since arcA and PVL are not genetically linked, there is the potential for these genes to be present in mixed-infection samples containing additional S. aureus or coagulase-negative staphylococcal strains. Therefore, this method requires separation of individual S. aureus isolates to avoid the generation of false-positive results. Furthermore, culture and isolation methods add significant time to the diagnosis.
USA300 is the predominant clone of CA-MRSA in the United States, accounting for up to 98% of MRSA isolates from SSTIs (1) and 67% of invasive CA-MRSA isolates (3). Due to its high virulence and transmissibility (40), this clone has been speculated to be responsible for the surge in community-associated SSTIs observed between 2004 and 2008 (1). USA300 also has been shown to cause bacteremia, endocarditis, severe necrotizing pneumonia, and osteomyelitis (41). USA300 isolates were initially designated based on SmaI-digested PFGE profiles. Now, using more-modern molecular methods, they commonly are described as SCCmecIV (1, 6, 13, 21, 39, 42, 43), PVL-positive (6, 13, 21, 39, 43), multilocus sequence type 8 (ST-8) (39, 43), and spa type t008 (39, 43) or t121 (44) and possess the arginine catabolic mobile element (ACME) (13, 20, 39). The ACME is thought to have been acquired horizontally from Staphylococcus epidermidis and allows for greater tolerance to acidic environments, such as human skin (42).
Recently, single-nucleotide polymorphisms (SNPs) in the penicillin-binding protein 4 (pbp4) gene have been discovered to play a role in β-lactam and vancomycin resistance as well as disease progression in S. aureus (45–47). Gygax et al. (48) observed similar effects on pbp4 in relation to penicillin tolerance in group B streptococci. These findings suggest that penicillin-binding proteins other than PBP2 can play significant roles in disease and antimicrobial resistance, while also serving as biomarkers. A search for novel diagnostic biomarkers of S. aureus antibiotic resistance was conducted, and two previously uncharacterized nucleotide polymorphisms (G88A and G2047A) in the S. aureus genomic pbp3 gene were found in many CA-MRSA strains. Upon further investigation, these polymorphisms in pbp3 were found to be highly associated with the USA300 clone, the most prevalent CA-MRSA strain in North America (1, 6). A simple single allele-specific PCR was developed for detection of the G88A pbp3 polymorphism and presents a practical, cost-effective, high-throughput, rapid method for the detection of USA300 CA-MRSA. The optimal design for detection of USA300 directly from patient samples would be a single PCR assay that utilizes a stable S. aureus genomic marker rather than mobile genetic elements, which would allow more predictable and rapid detection of MRSA (specifically USA300 CA-MRSA). Such an assay would be rapid and eliminate the high costs associated with complicated multiplex PCRs and other technically difficult typing assays.
(This work was presented in part at the 50th Interscience Conference on Antimicrobial Agents and Chemotherapy, Boston, MA, 2010.)
MATERIALS AND METHODS
Bacterial strains and growth.
Clinical S. aureus strains in this study were obtained from deidentified patient samples sent to our clinical laboratory by physicians for patients suspected of having skin and soft tissue infections due to MRSA. Swabs were inoculated on mannitol salt agar plates (Becton, Dickinson, Sparks, MD), and the plates were incubated at 37°C for 24 to 48 h. Liquid cultures were grown in tryptic soy broth (Affymetrix, Cleveland, OH) at 37°C for 24 h. American Type Culture Collection (ATCC) (Manassas, VA) control strains BAA-38, BAA-39, BAA-1556, BAA-1747, BAA-1764, BAA-1766, 43300, 25923, and 29213 and Microbiologics (St. Cloud, MN) strain 0158p also were used.
Characterization of S. aureus isolates.
To obtain genomic DNA, a 1-μl loopful of S. aureus colonies was resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) and boiled for 5 min. Colonies were tested for the nuc gene (S. aureus-specific nuclease), the mecA gene (methicillin resistance), the SCCmec type (MRSA transposon type), and the specific ccrAB allele (SCCmecIV-specific allele) with an in-house multiplex PCR assay (S. E. Gygax, C. L. Overmyer, L. A. Desalvia, M. E. Adelson, and E. Mordechai, U.S. provisional patent application 13/068,331). PCR products were visualized using agarose gel electrophoresis with a 2% agarose gel. The mecA amplicon produced was 169 bp, nuc was 280 bp, SCCmecIV-specific ccrAB was 380 bp, SCCmecI produced a 566-bp typing amplicon, SCCmecII and SCCmecIV produced a 668-bp amplicon, and SCCmecIII produced a 622-bp amplicon. Methicillin-susceptible S. aureus (MSSA) strains amplified only the nuc gene. The presence of the PVL gene was detected with an in-house real-time PCR assay (Gygax et al., U.S. provisional patent application 13/068,331). HA-MRSA strains were defined as SCCmecI, -II, -III, or -IV and PVL-negative, and CA-MRSA was defined as SCCmecIV and PVL-positive (12, 50). MSSA possessed the nuc gene but lacked mecA.
spa typing of S. aureus isolates and arcA ACME detection.
To determine the clonality of the strains in this study, spa typing was performed as described by Shopsin et al. (27). Briefly, the reaction mixture contained 2.5 μl USB 10× PCR buffer (with 2.5 mM MgCl2), 1.5 μl of 10 μM primers spa-F and spa-R (Table 1), 15.37 μl nuclease-free water, 2.0 μl deoxynucleoside triphosphates (dNTPs) (2.5 mM), 0.13 μl FideliTaq (Affymetrix, Santa Clara, CA), and 2.0 μl S. aureus DNA template, and the thermal profile consisted of 94°C for 3 min, 30 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min, followed by 10 min at 72°C; the reaction mixture was then held at 4°C until sequencing. Amplicons were purified using the Promega Wizard SV gel and PCR Clean-Up system (Promega, Madison, WI), using the manufacturer's guidelines. A cycle sequencing reaction was set up using 8 μl Terminator Ready Reaction Mix v1.1 (Applied Biosystems, Foster City, CA), 3 μl of 1 μM spa-F primer, 4 μl purified spa PCR product, and 5 μl nuclease-free H2O. DNA sequences were obtained using an ABI 3130 genetic analyzer (Applied Biosystems). The spa type was determined by examining specific repeats and aligning them using the Ridom SpaServer online tool (http://spaserver.ridom.de). PCRs for the ACME arcA gene were performed as described previously (20), to characterize HA-MRSA and CA-MRSA strains further.
Table 1.
Oligonucleotide primers used in this study
| Primers and application | Oligonucleotide sequence, 5′ to 3′ | Position in gene (bp)a | Nucleotide position of gene | Reference or source |
|---|---|---|---|---|
| spa typing of MRSA isolates | ||||
| spa-F | GCCAAAGCGCTAACCTTTTA | 785–804 | 128109–129623 | 27 |
| spa-R | TCCAGCTAATAACGCTGCAC | 1478–1497 | 128109–129623 | 27 |
| ACME-specific arcA testing for detection of USA300 | ||||
| arcA-F | CTAACACTGAACCCCAATG | 561–579 | 71552–74116 | 20 |
| arcA-R | GAGCCAGAAGTACGCGAG | 2489–2506 | 71552–74116 | 20 |
| Amplification and sequencing of pbp3 regions | ||||
| pbp3-G88A-F | TCAAATGATGAAATCGTTCAAAA | 25–43 | 1662330–1664405 | This work |
| pbp3-G88A-R | TCCGATTGTGTTGTTTTTCG | 283–302 | 1662330–1664405 | This work |
| pbp3-G2047A-F | AAAACGGAGAGCCAAGAGTT | 1877–1896 | 1662330–1664405 | This work |
| pbp3-G2047A-R | TTTTACAACCATGCGCTACA | +19–+38 | 1662330–1664405 | This work |
| Allele-specific PCR to detect G88 and G88A sequences | ||||
| CaMRSA-For-AA | CTTTATATTTGGTGTGATAA | 69–88 | 1662330–1664405 | This work |
| CaMRSA-For-AG | CTTTATATTTGGTGTGATAG | 69–88 | 1662330–1664405 | This work |
| CaMRSA-Rev-4 | GATTTTCCGATTTTCGATAAC | 486–506 | 1662330–1664405 | This work |
Sequencing was based on the sequences for GenBank accession no. CP000730.1.
Sequencing of S. aureus pbp3.
S. aureus pbp3 was amplified from our clinically obtained isolates via conventional PCR methods for sequence analysis. The reaction mixture contained 2.5 μl USB 10× PCR buffer (with 2.5 mM MgCl2 added; Affymetrix, Santa Clara, CA), 1.5 μl of 10 μM primers pbp3-G88A-F and pbp3-G88A-R (Integrated DNA Technologies, Coralville, IA) (Table 1), 15.37 μl nuclease-free water, 2.0 μl dNTPs (2.5 mM), 0.13 μl FideliTaq (Affymetrix), and 2.0 μl S. aureus DNA template. The thermal profile consisted of 94°C for 2 min, 30 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min, and then 72°C for 10 min, with a hold step at 4°C. A second reaction was carried out similarly, using the pbp3-G2047A-F and pbp3-G2047A-R primers (Table 1). The 278-bp (flanking position 88) and 238-bp (flanking position 2047) amplicons were gel purified as described above.
Sequencing of pbp3 amplicons was carried out as described above, using 1 μM stock solutions of the pbp3-G88A-F or pbp3-G2047A-F primer. Sequences were aligned, and the nucleotides present at positions 88 and 2047 within the pbp3 gene were determined.
Sequences for publicly available laboratory strains were found at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov). The penicillin-binding protein 3 sequences for strains COL, N315, Mu50, MRSA252, JH1, JH9, Mu3, MSSA476, NCTC8325, RF122, Newman, USA300_FPR3757, USA300_TCH1516, 11819-97, M013, M10/0061, T-0131, JKD6159, JKD6008, ED133, S0385, TW20, and MW2 were analyzed with the clinical data set (see Table 3).
Table 3.
Analysis of public and laboratory S. aureus pbp3 sequences
| Strain | SNP 1 (position 88) | SNP 2 (position 2047) | SCCmec typea | Description | Source location |
|---|---|---|---|---|---|
| NCBI-deposited sequence strains | |||||
| HA-MRSA | |||||
| COL | G | G | I | ST-250 | England |
| N315 | G | G | II | ST-5 | Japan |
| Mu3 | G | G | II | ST-5 | Japan |
| Mu50 | G | G | II | ST-5 | Japan |
| MRSA252 | G | G | II | ST-36 | UK |
| JH1 | G | G | II | ST-105 | Baltimore, MD |
| JH9 | G | G | II | ST-105 | Baltimore, MD |
| T0131 | G | G | III | ST-239 | China |
| JKD6008 | G | G | III | ST-239 | New Zealand |
| TW20 | G | G | III | ST-239 | London |
| M10/0061 | G | G | XI | ST-130 | Ireland |
| ED133 | G | G | NA | ST-133 | France |
| MSSA | |||||
| NCTC8325 | G | G | NA | ST-8 | USA |
| MSSA476 | G | G | NA | ST-1 | UK |
| RF122 | G | G | NA | ST-151 | Ireland |
| Newman | G | G | NA | ST-8 | England, 1952 |
| CA-MRSA | |||||
| 11819-97 | G | G | IV | ST-80 | Europe |
| JKD6159 | G | G | IV | ST-93 | Australia |
| S0385 | G | G | V | ST-398 | Netherlands |
| M013 | G | G | V | ST-59 | Taiwan |
| MW2 | G | G | IV | ST-1 | North Dakota |
| USA300_FPR3757 | A | A | IV | ST-8 | San Francisco, CA |
| USA300_TCH1516 | A | A | IV | ST-8 | Texas |
| Laboratory strains | |||||
| HA-MRSA | |||||
| BAA-38 | G | G | I | Archaic | Denmark |
| 0158p | G | G | II | ||
| 43300 | G | G | II | Kansas | |
| BAA-39 | G | G | III | Hungarian | Hungary |
| MSSA | |||||
| 25923 | G | G | NA | Seattle, WA | |
| 29213 | G | G | NA | Wichita, KS | |
| CA-MRSA | |||||
| BAA-1556 | A | A | IV | USA300 | San Francisco, CA |
| BAA-1747 | G | G | IV | USA1000 | Vermont |
| BAA-1764 | G | G | IV | USA1100 | Alaska |
| BAA-1766 | G | G | V | USA700 | Wisconsin |
NA, not applicable.
Allele-specific PCR to detect the G88A polymorphism in CA-MRSA isolates.
Allele-specific PCRs were used to detect either the G88 sequence or the G88A polymorphism of the S. aureus pbp3 gene. The allele-specific reaction mixture contained 12.5 μl Quanta Perfecta SuperMix for iQ (Quanta Biosciences), 9.0 μl double-distilled water, 0.5 μl 50 μM forward (CaMRSA-For-AA or CaMRSA-For-AG) and reverse (CaMRSA-Rev-4) primers (Table 1), and 2.5 μl template DNA. Forward primers were designed with a penultimate T-A nucleotide mismatch to create primer 3′ instability and increased allele specificity. The G and A 3′-terminal nucleotides of the allele-specific forward primers annealed to the G88 and G88A nucleotides of the pbp3 gene to detect HA-MRSA/MSSA and CA-MRSA, respectively. The reverse primer was designed to anneal to a highly conserved S. aureus-specific region of the pbp3 gene. The thermal profile consisted of 95°C for 3 min and 35 cycles of 95°C for 30 s, 54°C for 30 s, and 72°C for 30 s. The reaction mixture was held at 72°C for 5 min and then kept at 4°C until needed. Both G88 and A88 reactions were performed for CA-MRSA (strain BAA-1556), HA-MRSA (strains BAA-38, BAA-39, and 0158p), MSSA (strain 29213), Streptococcus agalactiae strains O90R, NEM316, and A909, Streptococcus pyogenes ATCC 19615, Escherichia coli ATCC 11303, and Staphylococcus epidermidis ATCC 12228 to determine the specificity of the reaction.
Assay validation.
The analytical sensitivity, analytical specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated with GraphPad InStat3 software, using 2-way contingency table analysis and Fisher's exact test with 95% confidence intervals.
RESULTS
Characterization of clinical S. aureus strains.
A total of 125 S. aureus strains were isolated from deidentified patient swabs submitted to our clinical laboratory from patients with suspected MRSA infections. Colonies from the mannitol salt agar plates were SCCmec-typed using an in-house multiplex PCR assay (Gygax et al., U.S. provisional patent application 13/068,331), and a separate real-time PCR assay was performed for detection of the Panton-Valentine leukocidin (PVL) lukS-PV and lukF-PV genes (51). Nineteen of the clinical strains were determined to be HA-MRSA (2 SCCmecI, 8 SCCmecII, 3 SCCmecIII, and 6 SCCmecIV and PVL−), 96 strains were determined to be common CA-MRSA (SCCmecIV and PVL+), and 10 strains were MSSA (mecA-negative and nuc-positive) (12).
Characterization of S. aureus strains by spa typing and ACME detection.
spa typing was performed for 40 representative strains (10 MSSA, 10 HA-MRSA, and 20 CA-MRSA strains), for better understanding of the epidemiology or clonality of our isolates and for testing of the prevalence of these polymorphisms using a database of standard molecularly typed S. aureus strains. As detailed in Table 2, the 10 MSSA strains were spa types t922, t701 (two strains), t9602, t3169, t177, t008, t002, and t127 (two strains). The 10 HA-MRSA strains were spa types t012, t127, t002 (four strains), t242, t064, t1473, and t008. These results suggest that the MSSA and HA-MRSA isolates were highly diverse. Conversely, 18 of the 20 CA-MRSA strains were spa type t008. This spa type, which was supported by the presence of SCCmecIV and the PVL toxin, is indicative of the most common CA-MRSA strain, USA300. One CA-MRSA isolate was spa type t121, which has also been linked to the USA300 genotype (44). The remaining CA-MRSA strain was spa type t692, which has not been linked to the USA300 genotype. Significantly less diversity was observed for the CA-MRSA isolates, which has been previously reported (52).
Table 2.
Characterization of clinical S. aureus isolates
| Strain | SCCmec typea | PVL | ACME | SNP 1 (position 88) | SNP 2 (position 2047) | spa type |
|---|---|---|---|---|---|---|
| MSSA-1 | NA | − | − | G | G | t922 |
| MSSA-2 | NA | − | − | G | G | t701 |
| MSSA-3 | NA | − | − | G | G | t9602 |
| MSSA-4 | NA | − | − | G | G | t3169 |
| MSSA-5 | NA | − | + | G | G | t177 |
| MSSA-6 | NA | − | − | G | G | t008 |
| MSSA-7 | NA | − | − | G | G | t002 |
| MSSA-8 | NA | − | − | G | G | t701 |
| MSSA-9 | NA | + | − | G | G | t127 |
| MSSA-10 | I, mecA− | − | − | G | G | t127 |
| HA-MRSA-1 | I | − | − | G | G | t012 |
| HA-MRSA-2 | I | − | − | G | G | t127 |
| HA-MRSA-3 | II | − | − | G | G | t002 |
| HA-MRSA-4 | II | − | − | G | G | t242 |
| HA-MRSA-5 | II | − | − | G | G | t002 |
| HA-MRSA-6 | III | − | − | G | G | t002 |
| HA-MRSA-7 | III | − | − | G | G | t002 |
| HA-MRSA-8 | IV | − | − | G | G | t064 |
| HA-MRSA-9 | IV | − | − | G | G | t1473 |
| HA-MRSA-10 | IV | − | − | G | G | t008 |
| CA-MRSA-1 | IV | + | + | A | A | t008 |
| CA-MRSA-2 | IV | + | + | A | A | t008 |
| CA-MRSA-3 | IV | + | + | A | A | t008 |
| CA-MRSA-4 | IV | + | + | A | A | t008 |
| CA-MRSA-5 | IV | + | + | A | A | t008 |
| CA-MRSA-6 | IV | + | + | A | A | t008 |
| CA-MRSA-7 | IV | + | + | A | A | t121 |
| CA-MRSA-8 | IV | + | + | A | A | t008 |
| CA-MRSA-9 | IV | + | + | A | A | t008 |
| CA-MRSA-10 | IV | + | + | A | A | t008 |
| CA-MRSA-11 | IV | + | + | A | A | t008 |
| CA-MRSA-12 | IV | + | + | A | A | t008 |
| CA-MRSA-13 | IV | + | + | A | A | t008 |
| CA-MRSA-14 | IV | + | + | A | A | t008 |
| CA-MRSA-15 | IV | + | + | A | A | t008 |
| CA-MRSA-16 | IV | + | + | A | A | t008 |
| CA-MRSA-17 | IV | + | + | A | A | t008 |
| CA-MRSA-18 | IV | + | + | A | A | t008 |
| CA-MRSA-19 | IV | + | − | A | A | t008 |
| CA-MRSA-20 | IV | + | − | G | G | t692 |
NA, not applicable.
None of the HA-MRSA strains possessed arcA, an ACME-specific gene often found in USA300 CA-MRSA isolates. One MSSA strain, MSSA-5, possessed arcA, and 18/20 CA-MRSA strains possessed the arcA arginine deiminase (Table 2).
Identification of the pbp3 G88A/G2047 allele in clinical S. aureus isolates.
The pbp3 genes from the 10 MSSA, 10 HA-MRSA, and 20 CA-MRSA isolates were amplified by conventional PCR methods and sequenced. Sequencing revealed that 19 of 20 CA-MRSA isolates possessed G88A and G2047A polymorphisms, resulting in V30I and D683N amino acid substitutions in PBP3, respectively. The 10 MSSA and 10 HA-MRSA strains all possessed the sequences G88 and G2047 (Table 2). These data revealed that the G88A and G2047A polymorphisms were in strict linkage disequilibrium and were found only in CA-MRSA isolates. Sequencing of nucleotide 88 of the S. aureus pbp3 gene was performed on additional clinical isolates. Four additional HA-MRSA strains possessed the G88 sequence, and 47 additional CA-MRSA strains possessed the G88A polymorphism (data not shown).
Confirmation of pbp3 polymorphisms with sequences from the NCBI database and laboratory strains.
Using pbp3 sequences available in the NCBI database, Table 3 shows that the G88 and G2047 sequences were found in the following common laboratory strains: MSSA476 (MSSA), NCTC8325 (MSSA), RF122 (MSSA), Newman (MSSA), COL (HA-MRSA, SCCmecI), N315 (HA-MRSA, SCCmecII), Mu50 (HA-MRSA, SCCmecII), MRSA252 (HA-MRSA, SCCmecII), JH1 (HA-MRSA, SCCmecII), JH9 (HA-MRSA, SCCmecII), Mu3 (HA-MRSA), 11819-97 (ST-80, SCCmecIV), M013 (ST-59, SCCmecV), M10/0061 (ST-130, SCCmecXI), T-0131 (SCCmecIII), JKD6159 (Australian CA-MRSA clone, ST-39), JKD6008 (ST-239, SCCmecIII), ED133 (clonal complex 133, ovine), S0385 (SCCmecV, ST-398), TW20 (ST-239, SCCmecIII), and MW2 (CA-MRSA, SCCmecIV, USA400). The G88A and G2047A polymorphisms were found in both USA300_FPR3757 and USA300_TCH1516 (CA-MRSA, SCCmecIV, USA300). These two strains represent the most common USA300 subtype, USA300-0114, which is responsible for the vast majority of USA300 outbreaks (6, 42, 53).
Ten in-house laboratory strains were sequenced (Table 3). ATCC BAA-38 (HA-MRSA, SCCmecI), BAA-39 (HA-MRSA, SCCmecIII), BAA-1747 (CA-MRSA, SCCmecIV, USA1000), BAA-1764 (CA-MRSA, SCCmecIV, USA1100), BAA-1766 (CA-MRSA, SCCmecV, USA700), 43300 (HA-MRSA, SCCmecII), 25923 (MSSA, PVL+), 29213 (MSSA), and Microbiologics strain 0518p (HA-MRSA, SCCmecII) all possessed the G88 and G2047 sequences. BAA-1556, the USA300 CA-MRSA laboratory isolate, possessed the G88A and G2047A polymorphisms.
Creation of an allele-specific PCR to detect G88A and screening of clinical strains.
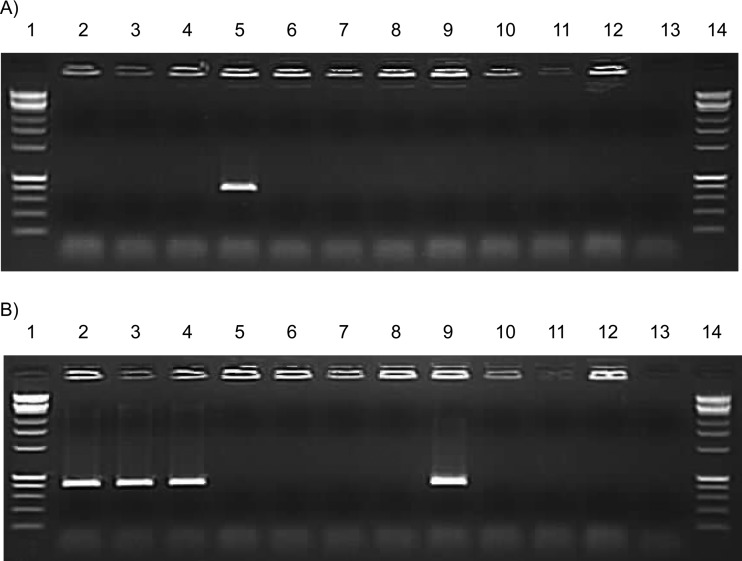
The G88A and G2047A polymorphisms were found to be highly associated with USA300 CA-MRSA and therefore make attractive candidates as diagnostic biomarkers. The G88A polymorphism was chosen as a target for an allele-specific PCR because the region surrounding this nucleotide was more favorable for primer design. Thus, a single allele-specific PCR was designed to amplify either the G88 sequence or the G88A polymorphism from samples that contained S. aureus. As shown in Fig. 1, CA-MRSA DNA from ATCC strain BAA-1556 (USA300) was the only DNA that generated a pbp3 amplicon using the G88A allele-specific set of primers, while HA-MRSA and MSSA DNA amplified the pbp3 amplicon using the G88 allele-specific set of primers. A panel of non-S. aureus bacteria demonstrated good specificity, since there was no cross-reactivity using these primer sets.
Fig 1.
Allele-specific PCRs to detect G88A and G88 pbp3 alleles in S. aureus. Allele-specific PCR assays that detect the USA300 G88A (A) and non-USA300 G88 (B) pbp3 sequences were performed, and gel electrophoresis on a 1% agarose gel was used to detect amplification for the following bacterial strains: lanes 1 and 14, Affymetrix 1-kb Plus ladder; lane 2, HA-MRSA, SCCmecI, BAA-38; lane 3, HA-MRSA, SCCmecII, 0158p; lane 4, HA-MRSA, SCCmecIII, BAA-39; lane 5, CA-MRSA, SCCmecIV, BAA-1556; lane 6, Streptococcus agalactiae A909; lane 7, Streptococcus agalactiae NEM316; lane 8, Streptococcus agalactiae O90R; lane 9, methicillin-susceptible S. aureus ATCC 29213; lane 10, Streptococcus pyogenes ATCC 19615; lane 11, Escherichia coli ATCC 11303; lane 12, Staphylococcus epidermidis ATCC 12228; lane 13, no-template control.
The allele-specific PCR was applied to the rest of our clinical S. aureus strains. All five remaining HA-MRSA strains possessed the G88 sequence. The remaining 29 CA-MRSA strains all possessed the G88A polymorphism. Three sequenced MSSA strains were tested and reconfirmed for the G88 sequence using the allele-specific PCR. This allele-specific PCR assay also was able to detect G88A directly from a patient swab with confirmed CA-MRSA (data not shown). Further validation of this assay might prove useful for the detection of USA300 CA-MRSA strains directly from patient samples with high sensitivity and specificity.
Statistical analyses.
A total of 157 S. aureus pbp3 sequences were available for analysis, including the 125 clinical S. aureus strains from this study, 10 laboratory strains, and 23 NCBI-deposited sequences (one laboratory strain, BAA-1556, was also an NCBI-deposited sequence, USA300_FPR3757). The G88A polymorphism was not found in any of the clonally and geographically diverse HA-MRSA or MSSA strains in this study. G88A was found in 95/96 clinical CA-MRSA strains, 1/10 laboratory strains (BAA-1556, USA300_FPR3757), and 2/23 NCBI-deposited strains (USA300_FPR3757 and USA300_TCH1516).
Further analysis of CA-MRSA strains revealed that the G88A polymorphism was found in 97/106 (91.5%) of all available CA-MRSA pbp3 sequences. CA-MRSA strains lacking the G88A polymorphism included MW2 (USA400), BAA-1766 (USA700), BAA-1747 (USA1000), BAA-1764 (USA1100), 11819-97 (ST-80), M013 (ST-59), JKD6159 (ST-93), S0385 (ST-398), and one clinical strain (spa t692, ACME-negative). These 9 strains represent STs that are rare or unreported in the United States and are not related to the predominant U.S. clone, USA300 (ST-8). spa typing of a subset of our clinical isolates revealed that G88A was found in spa types t008 and t121, which are linked to the most prominent CA-MRSA strain, USA300 (1, 3, 6). These spa types were found in 19/20 CA-MRSA isolates, indicating minimal diversity among CA-MRSA isolates. Strain CA-MRSA-19 was SCCmecIV, PVL-positive, and spa type t008 but ACME-negative; thus, this strain was not confirmed to be a common USA300 strain according to our definitions, although ACME-negative USA300 strains have been reported (20, 39). USA300 strain sequences, including USA300_FPR3757 and USA300_TCH1516, demonstrated that G88A was found in 20/20 (100%) of all available confirmed epidemic USA300 strains. G88A was also seen in the CA-MRSA-19 strain, which is likely a minor variant clone of USA300. Thus, detection of G88A provides 91.5% sensitivity, 100% specificity, a positive predictive value (PPV) of 100%, and a negative predictive value (NPV) of 85% for the overall detection of CA-MRSA in this study. Furthermore, the sensitivity, specificity, PPV, and NPV are 100% when the criteria are narrowed to the detection of USA300 CA-MRSA.
DISCUSSION
Multiple methods have been developed for the detection and characterization of MRSA and for provision of additional epidemiological information (SCCmec typing, PFGE typing, and spa typing). Furthermore, the epidemiological distinction between HA-MRSA and CA-MRSA is difficult to determine with molecular tools. A major reason behind the complicated methodologies for the determination of CA-MRSA is that both the SCCmec transposon and the PVL toxin are encoded by mobile genetic elements that are also found in other staphylococcal species. An objective of this study was to identify a suitable genomic biomarker that would provide a simple tool for direct screening of patient samples for detection of CA-MRSA, specifically the predominant USA300 clone.
Examination of the sequences of multiple S. aureus strains revealed two strongly USA300 CA-MRSA-associated polymorphisms, G88A and G2047A, in the genomic pbp3 gene, leading to amino acid changes of V30I and D683N, respectively. Genomic biomarkers are preferable to those found on mobile genetic elements such as plasmids or transposons because they are more stable and can be traced to certain genetic lineages. The two identified polymorphisms were shown to be in strict linkage disequilibrium. A total of 157 pbp3 sequences were analyzed in this study, including 106 CA-MRSA, 35 HA-MRSA, and 16 MSSA strains. Of the 51 HA-MRSA and MSSA strains, all possessed the common G88 sequence. Of the 106 CA-MRSA isolates, 97 possessed the G88A polymorphism and 9, which were of non-USA300 CA-MRSA lineages, possessed the common G88 sequence. Twenty strains were shown to be representative common USA300 clones, and all possessed the G88A polymorphism. The remaining 86 clinical CA-MRSA isolates were deemed likely to be USA300 strains, because they possessed SCCmecIV, PVL, and the G88A polymorphism and were from patients within the United States, where USA300 is the predominant clone.
Additionally, a study by Kennedy et al. (52) examined 10 USA300 genomic sequences, using the NCBI-deposited USA300_FPR3757 sequence as a reference. The 10 genomes examined were from different locations across the United States (including California, Colorado, Georgia, and New York), from patients exhibiting different clinical infections (such as abscesses, endocarditis, bacteremia, or necrotizing pneumonia), and were collected between 2002 and 2005. As shown in our study, USA300_FPR3757 possesses G88A and G2047A SNPs (Table 3). Kennedy et al. (52) did not identify sequence differences at position 88 or 2047 in pbp3 in any of the 10 isolates, in comparison with USA300_FPR3757. Thus, it can be deduced that the 10 USA300 isolates possessed both G88A and G2047A SNPs in pbp3. Genomic conservation among the clonal USA300 isolates was demonstrated by the fact that these isolates differed from each other by only 11 to 477 bases.
It is important to note that the G88A and G2047A polymorphisms were not found in the 6 strains that were SCCmecIV, ACME-negative, and PVL-negative (likely USA500 clones [54]) or in ATCC strain 25923 and clinical strain MSSA-9 (Table 2), which were PVL-positive MSSA strains. The polymorphisms also were not found in the MSSA-6 and HA-MRSA-10 strains (Table 2), which possessed spa type t008. The MSSA-5 strain possessed ACME but lacked PVL and mecA resistance and did not possess the G88A polymorphism. These data suggest that the two polymorphisms discussed are not linked specifically to the SCCmecIV transposon, ACME, PVL toxin, or spa type t008 or t121 alone but serve as markers of the USA300 lineage that encompass these aspects.
Standard sequencing is a low-throughput and labor-intensive process; therefore, detection of G88A or G2047A via sequencing would be too inefficient for screening of a large number of patient samples. Thus, allele-specific PCR primers were designed to amplify S. aureus strains that possessed the G88A polymorphism of pbp3, creating a simple single-reaction assay for detection of the USA300 clone. The primers were specific, allowing G88A primers to amplify only USA300 CA-MRSA. Moreover, primers targeting the wild-type G88 sequence amplified only HA-MRSA and MSSA isolates.
Early detection of the USA300 clone of CA-MRSA may be of particular importance because it is highly virulent and transmissible, it is increasing in prevalence across the United States, and it has begun spreading to other parts of the world (12). CA-MRSA strains should be contained early because they have been reported to cause outbreaks of disease after becoming established within health care facilities and replacing endemic strains. Recent screenings have shown that USA300 is the predominant CA-MRSA strain in circulation (1, 6, 21), accounting for up to 98% of all MRSA isolates from SSTIs (1). Of the common lineages of CA-MRSA, USA1000 displays a “sporadic” phenotype, USA1100 displays a “local outbreak” phenotype, and USA300 exhibits an “epidemic” phenotype, capable of wide spread (40).
MRSA colonization and infection bring large financial burdens to health care facilities and patients (2). As a measure to identify and to eradicate MRSA, hospitals have begun screening inpatients for the presence of MRSA and decontaminating them with prophylactic antibiotic treatments. With proper screening, high-risk patients can be isolated and decolonized and physicians can exercise extra caution to prevent further epidemic spread of MRSA infections, leading to significant savings (55, 56).
The S. aureus pbp3 polymorphisms identified in this study were found to be highly associated with USA300 CA-MRSA, especially the most common subtype, USA300-0114 (represented by NCBI sequences USA300_FPR3757 and USA300_TCH1516), which is responsible for the majority of USA300 outbreaks in the United States (6, 18, 21, 43, 57). The allele-specific PCR assay removes the variability of the PFGE method, which can provide similar banding patterns for vastly different isolates (58). The allele-specific PCR assay also proved to be highly specific for USA300 (notably USA300-0114) and may serve as a useful, cost-effective tool for rapid early detection and high-throughput screening for USA300 infection or colonization directly in patient samples, allowing better patient outcomes and fewer outbreaks caused by this epidemic strain.
ACKNOWLEDGMENTS
We are employed by Medical Diagnostic Laboratories, LLC. A provisional patent application has been filed for a method for detection of CA-MRSA and USA300 using the polymorphisms in S. aureus pbp3 (application 13/068,331).
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Talan DA, Krishnadasan A, Gorwitz RJ, Fosheim GE, Limbago B, Albrecht V, Moran GJ. 2011. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin. Infect. Dis. 53:144–149 [DOI] [PubMed] [Google Scholar]
- 2. Engemann JJ, Carmeli Y, Cosgrove SE, Fowler VG, Bronstein MZ, Trivette SL, Briggs JP, Sexton DJ, Kaye KS. 2003. Adverse clinical and economic outcomes attributable to methicillin resistance among patients with Staphylococcus aureus surgical site infection. Clin. Infect. Dis. 36:592–598 [DOI] [PubMed] [Google Scholar]
- 3. Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771 [DOI] [PubMed] [Google Scholar]
- 4. Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. 2006. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885 [DOI] [PubMed] [Google Scholar]
- 5. Klein E, Smith DL, Laxminarayan R. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 13:1840–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666–674 [DOI] [PubMed] [Google Scholar]
- 7. Hiramatsu K, Katayama Y, Yuzawa H, Ito T. 2002. Molecular genetics of methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 292:67–74 [DOI] [PubMed] [Google Scholar]
- 8. Hartman B, Tomasz A. 1981. Altered penicillin-binding proteins in methicillin-resistant strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 19:726–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pinho MG, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. U. S. A. 98:10886–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, Daum RS, Hiramatsu K. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chambers HF, DeLeo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McDougal LK, Fosheim GE, Nicholson A, Bulens SN, Limbago BM, Shearer JE, Summers AO, Patel JB. 2010. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob. Agents Chemother. 54:3804–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diep AD, Palazzolo-Balance AM, Tattevin P, Basuino L, Braughton KR, Whitney AR, Chen L, Kreiswirth BN, Otto M, DeLeo FR, Chambers HF. 2008. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS One 3:e3198. 10.1371/journal.pone.0003198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, Long RD, Dorward DW, Gardner DJ, Lina G, Kreiswirth BN, DeLeo FR. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761–1770 [DOI] [PubMed] [Google Scholar]
- 16. Diep AD, Otto M. 2008. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16:361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Centers for Disease Control and Prevention 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus–Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 282:707–710 [PubMed] [Google Scholar]
- 18. Kazakova SV, Hageman JC, Matava M, Srinivasan A, Phelan L, Garfinkel B, Boo T, McAllister S, Anderson J, Jensen B, Dodson D, Lonsway D, McDougal LK, Arduino M, Fraser VJ, Killgore G, Tenover FC, Cody S, Jernigan DB. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352:468–475 [DOI] [PubMed] [Google Scholar]
- 19. Okano JT, Bowler S. 2010. Are correctional facilities amplifying the epidemic of community-acquired methicillin-resistant Staphylococcus aureus? Nat. Rev. Microbiol. 8:83. [DOI] [PubMed] [Google Scholar]
- 20. Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages SA, Jones A, Palazzolo-Balance AM, Perdreau-Remington F, Sensabaugh GF, DeLeo FR, Chambers HF. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197:1523–1530 [DOI] [PubMed] [Google Scholar]
- 21. Limbago B, Fosheim GE, Schoonover V, Crane CE, Nadle J, Petit S, Heltzel D, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Mu Y, Fridkin SK. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J. Clin. Microbiol. 47:1344–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smyth RW, Kahlmeter G. 2005. Mannitol salt agar-cefoxitin combination as a screening medium for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:3797–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diederen B, van Duijn I, van Belkum A, Willemse P, van Keulen P, Kluytmans J. 2005. Performance of CHROMagar MRSA medium for detection of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:1925–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Romero-Gómez MP, Quiles-Melero I, Navarro C, Paño-Pardo JR, Gómez-Gil R, Mingorance J. 2012. Evaluation of the BinaxNOW PBP2a assay for the direct detection of methicillin resistance in Staphylococcus aureus from positive blood culture bottles. Diagn. Microbiol. Infect. Dis. 72:282–284 [DOI] [PubMed] [Google Scholar]
- 25. Johnson G, Millar MR, Matthews S, Skyrme M, Marsh P, Barringer E, O'Hara S, Wilks M. 2006. Evaluation of BacLite Rapid MRSA, a rapid culture based screening test for the detection of ciprofloxacin and methicillin resistant S. aureus (MRSA) from screening swabs. BMC Microbiol. 6:83. 10.1186/1471-2180-6-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sturenburg E. 2009. Rapid detection of methicillin-resistant Staphylococcus aureus directly from clinical samples: methods, effectiveness and cost considerations. Ger. Med. Sci. 7:Doc06. 10.3205/000065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreisworth BN. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29:2240–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fang H, Hedin G. 2003. Rapid screening and identification of methicillin-resistant Staphylococcus aureus from clinical samples by selective-broth and real-time PCR assay. J. Clin. Microbiol. 41:2894–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reischl U, Linde HJ, Metz M, Leppmeier B, Lehn N. 2000. Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence PCR. J. Clin. Microbiol. 38:2429–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cuny C, Witte W. 2005. PCR for the identification of methicillin-resistant Staphylococcus aureus (MRSA) strains using a single primer pair specific for SCCmec elements and the neighbouring chromosome-borne orfX. Clin. Microbiol. Infect. 11:834–837 [DOI] [PubMed] [Google Scholar]
- 34. Huletsky A, Giroux R, Rossbach V, Gagnon M, Vaillancourt M, Bernier M, Gagnon F, Truchon K, Bastien M, Picard FJ, van Belkum A, Ouellette M, Roy PH, Bergeron MG. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blanc DS, Basset P, Nahimana-Tessemo I, Jaton K, Greub G, Zanetti G. 2011. High proportion of wrongly identified methicillin-resistant Staphylococcus aureus carriers by use of a rapid commercial PCR assay due to presence of staphylococcal cassette chromosome element lacking the mecA gene. J. Clin. Microbiol. 49:722–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Donnio PY, Oliveira DC, Faria NA, Wilhelm N, Le Coustumier A, de Lencastre H. 2005. Partial excision of the chromosomal cassette containing the methicillin resistance determinant results in methicillin-susceptible Staphylococcus aureus. J. Clin. Microbiol. 43:4191–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rupp J, Fenner I, Solbach W, Gieffers J. 2006. Be aware of the possibility of false-positive results in single-locus PCR assays for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wolk DM, Picton E, Johnson D, Davis T, Pancholi P, Ginocchio CC, Finegold S, Welch DF, de Boer M, Fuller D, Solomon MC, Rogers B, Mehta MS, Peterson LR. 2009. Multicenter evaluation of the Cepheid Xpert methicillin-resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J. Clin. Microbiol. 47:758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. David MZ, Taylor A, Lynfield R, Boxrud DJ, Short G, Zychowski D, Boyle-Vavra S, Daum RS. 2013. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton-Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. medical center. J. Clin. Microbiol. 51:814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pan ES, Diep BA, Charlebois ED, Auerswald C, Carleton HA, Sensabaugh GF, Perdreau-Remington F. 2005. Population dynamics of nasal strains of methicillin-resistant Staphylococcus aureus–and their relation to community-associated disease activity. J. Infect. Dis. 192:811–818 [DOI] [PubMed] [Google Scholar]
- 41. Tenover FC, Goering RV. 2009. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J. Antimicrob. Chemother. 64:441–446 [DOI] [PubMed] [Google Scholar]
- 42. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, and epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 43. Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, Dunman PM. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haran KP, Godden SM, Boxrud D, Jawahir S, Bender JB, Sreevatsan S. 2012. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. J. Clin. Microbiol. 50:688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. 2008. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 52:3955–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Navratna V, Nadig S, Sood V, Prasad K, Arakere G, Gopal B. 2010. Molecular basis for the role of Staphylococcus aureus penicillin binding protein 4 in antimicrobial resistance. J. Bacteriol. 192:134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. 2010. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level β-lactam resistance contains mutations in three genes. Antimicrob. Agents Chemother. 54:4900–4902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gygax SE, Prasad A, Adelson ME, Mordechai E. 2010. Detection of penicillin tolerance in group B Streptococcus: single nucleotide polymorphisms in penicillin binding protein 4. US Patent and Trademark Office publication 2010/0028884
- 49. Reference deleted.
- 50. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakagawa S, Taneike I, Mimura D, Iwakura N, Nakayama T, Emura T, Kitatsuji M, Fujimoto A, Yamamoto T. 2005. Gene sequences and specific detection for Panton-Valentine leukocidin. Biochem. Biophys. Res. Commun. 328:995–1002 [DOI] [PubMed] [Google Scholar]
- 52. Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Methema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. 2008. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc. Natl. Acad. Sci. U. S. A. 105:1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Highlander SK, Hulten KG, Qin X, Jiang H, Yerrapragada S, Mason EO, Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7:99. 10.1186/1471-2180-7-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 106:5883–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hansen D, Patzke PI, Werfel U, Benner D, Brauksiepe A, Popp W. 2007. Success of MRSA eradication in hospital routine: depends on compliance. Infection 35:260–264 [DOI] [PubMed] [Google Scholar]
- 56. van der Zee A, Hendriks WDH, Roorda L, Ossewaarde JM, Buitenwerf J. 2013. Review of a major epidemic of methicillin-resistant Staphylococcus aureus: the costs of screening and consequences of outbreak management. Am. J. Infect. Control 41:204–209 [DOI] [PubMed] [Google Scholar]
- 57. Carpaij N, Willems RJL, Rice TW, Weinstein RA, Hinds J, Witney AA, Lindsay JA, Bonten MJM, Fluit AC. 2011. Genetic variation in spatio-temporal confined USA300 community-associated MRSA isolates: a shift from clonal dispersion to genetic evolution? PLoS One 6:e16419. 10.1371/journal.pone.0016419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Larsen AR, Goering R, Stegger M, Lindsay JA, Gould KA, Hinds J, Sorum M, Westh H, Boye K, Skov R. 2009. Two distinct clones of methicillin-resistant Staphylococcus aureus (MRSA) with the same USA300 pulsed-field gel electrophoresis profile: a potential pitfall for identification of USA300 community-associated MRSA. J. Clin. Microbiol. 47:3765–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]