Abstract
Nucleotide sequencing of the entire genomes was completed in the 1980s for most members of the Paramyxoviridae. It then became a new common task with challenge for researchers in the field to establish a system to recover the virus entirely from cDNA, thereby allowing reverse genetics (free manipulation of the viral genome). Using Sendai virus, we established a system of incomparable virus recovery efficiency early on. This technology was then fully exploited in answering a series of long-held questions. In particular, two accessory genes whose functions had remained enigmatic were demonstrated to encode special functions critical in viral in vivo pathogenesis producing fatal pneumonia in mice, although dispensable in virus replication at the in vitro cellular level. Their in vivo functions were found to counteract the two respective facets of the antiviral state induced by interferons and an interferon regulatory factor 3-dependent but yet unknown effector. These achievements appear to have facilitated a scientific trend where the accessory genes are a focus of active investigation in studies on other paramyxoviruses and opened up a new common ground shared between virology and immunology.
Keywords: paramyxovirus, Sendai virus, accessory genes, reverse-genetics, interferon, innate immunity
Introduction
SeV, also known as murine parainfluenza virus type 1 or hemagglutinating virus of Japan, is an enveloped virus with a negative-strand RNA genome. SeV is a member of the family Paramyxoviridae in the order Mononegavirales, which is comprised of a wide range of “classic” viruses of medical or veterinary significance that include MeV, MuV and NDV, and “new” deadly emergent agents such as NiV and HeV. Three other families (Rhabdo-, Filo- and Bornaviridae) in the Mononegavirales also include both classic and new deadly agents such as rabies, Ebola and Marburg viruses.1)
Reverse genetics has been established for SeV and most of its relatives (mononegaviruses), thus allowing the viral genome to be engineered at will and the outcomes assessed not only in terms of virus proliferation at the cellular level but also in the context of the viral pathogenesis in the entire host organism. This technological innovation has played a prominent role in settling outstanding issues and resolving long-held enigmas that had been impenetrable using conventional virology or forward genetics. Viral accessory genes are generally dispensable (non-essential) at cellular level proliferation and can be knocked out. Their functions can be best understood by comparing the phenotypes of the knockout viruses and the parental virus at the cellular level and in the susceptible host organism. Thus, accessory genes have become one of the most valuable research-subjects in reverse genetics.
The following two conditions need to be fulfilled for the successful application of reverse genetics in studying accessory genes. First, the efficiency of virus recovery from engineered cDNA needs to be sufficiently high, and that is because even if non-essential at the cellular level, accessory gene products often play great auxiliary roles and if they are deleted, it can cause, while not complete, profound loss of infectivity. Second, an authentic host animal system must be available, in which the outcomes of accessory gene deletion and manipulation can be readily assessed.
The efficiency of SeV recovery in our initial system already proved to be significantly higher (50∼100-fold) than those for other paramyxoviruses established by other parties,2) with the efficiency of our current system being even higher (10∼100-fold) than the initial one.3) The susceptible host for SeV is the mouse, in which the virus causes severe acute pneumonia. The outcomes of SeV engineering can be assessed with relative ease using laboratory mice with a large variety of genetic background. In contrast, in vivo studies of most other paramyxoviruses are considerably more difficult because their susceptible hosts are often humans, non-human primates or other large animals and also because the mouse- or other small animal models do not always satisfactorily mimic natural infections and diseases. Hence the SeV-mouse system represents a good experimental paradigm for use of reverse genetics in understanding the roles of accessory genes. The success of reverse genetics being applied in understanding the roles of accessory genes in the SeV-mouse experimental paradigm will now be described.
Fundamentals of SeV structure and biology
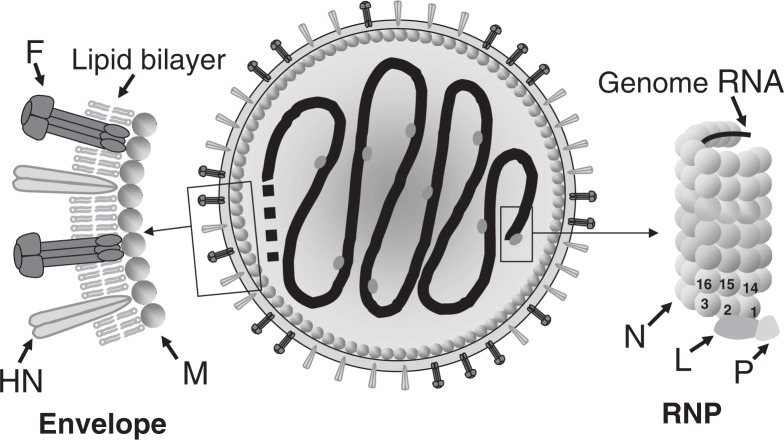
The SeV particle (virion) is spherical, with a diameter of approximately 200 nm. It consists of an inner RNP complex and outer envelope (Fig. 1). The RNP has a left-handed helical structure 1.1 μm in length and 18 nm in diameter, with a pitch of 5 nm, in which the 15,384-base RNA genome is encapsidated with the nucleocapsid (N) proteins. It has 13 N proteins per turn. Because one N protein binds to every 6 RNA bases, the entire RNP complex can be calculated as containing 2,564 (= 15,384/6) of N protein units. The large (L) and phospho- (P) proteins, the subunits of the RNA polymerase complex involved in genome transcription and replication, attach to the 3′-terminal site as well as occasionally to other sites in the RNP complex.
Fig. 1.
Schematic diagram of an SeV particle. (Adapted from Ref. 7 with permission.)
The envelope contains two types of glycoproteins protruding in the form of spikes that are anchored within a lipid bilayer derived from the plasma membrane of host cells (Fig. 1). One of the spike glycoproteins has hemagglutinin (HA) and neuraminidase (NA) activities and is named HN. The HN protein recognizes terminal sialic acids in the sugar chains as the receptor on the target cell surface. Its NA activity is essential for the release of progeny virus from the cell surface. The other spike glycoprotein is named F because it induces fusion of the virus envelope with the host cell plasma membrane, thereby introducing viral RNP into the cytoplasm. The F protein is produced as an inactive precursor molecule F0, which is processed by host proteases into the biologically active (fusion-competent) form, F1 and F2, which remain disulfide bridged. The HN protein is a tetramer, and the F protein a trimer.
The matrix (M) protein is located between the envelope and the RNP (Fig. 1). The M protein serves as an anchor to stabilize the spike glycoprotein molecules floating in the lipid bilayer by binding to their cytoplasmic tails and also binds to RNP, thus cross-linking the inner and outer structures. The M protein is indispensable for structural proteins to assemble into virus particles as well as for viral budding from the plasma membrane. The genes coding for the above six structural proteins lie in the order of 3′-N-P-M-F-HN-L-5′ (Fig. 2). For reviews see Refs. 4, 5, 6 and 7.
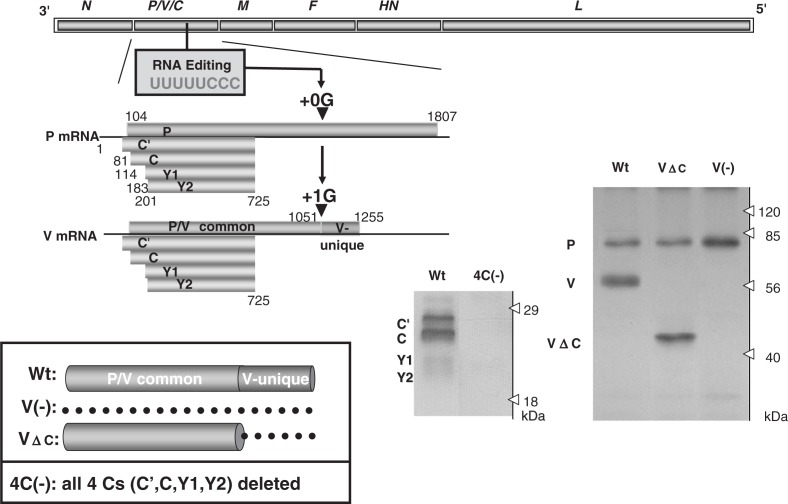
Fig. 2.
Expression of C and V proteins from SeV P gene and recovery of knockout viruses; V(-) with the entire V protein deleted, VΔC with the V-unique region deleted and 4C(-) with all four C proteins (C′, C, Y1 and Y2) deleted. For details see the text. Deletion of the respective proteins was verified by Western blotting of cells infected with the recovered knockout viruses. Wt, cells infected with wild-type SeV. The dotted line in the insert indicates deletion. (Adapted from Ref. 7 with permission.)
Coding strategies of the putative accessory genes of SeV
It has been suggested that the SeV P gene produces two putative accessory proteins, V and C, in addition to the basal gene product P (Fig. 2). The V and C proteins are widespread among the members of the subfamily Paramyxovirinae of the family Paramyxoviridae but are not as ubiquitous as the six major structural proteins, and therefore have been regarded as non-essential “accessory” gene products. However, the central question of how they contribute to actual viral replication and pathogenesis has remained unanswered since they were first described three decades ago (reviewed in Ref. 8). Even the non-essentiality of these proteins was yet to be established.
An ORF, which is +1 shifted from that of the P protein, gives rise to four proteins: C′, C, Y1, and Y2, collectively referred to as the C proteins. This diversity is accomplished by the use of translation initiation codons starting at different points (nt 81, 114, 183 and 201) (Fig. 2). The frames for the C proteins therefore are accessed thorough “ribosomal choice”. The ORFs terminate in the same position (nt 725). C is the major species of the four C proteins as it is expressed in infected cells at a molar ratio several-fold higher than those of the other three. The number of expressed C proteins differs among the paramyxoviruses, ranging from 1 to 4, and their primary structures are poorly conserved. However, they are, being common to all, basic and relatively short with approximately 200 amino acid residues (reviewed in Refs. 6 and 8).
The V protein, on the other hand, is produced by a transcriptional frameshift (“co-transcriptional RNA editing”) (Fig. 2). The P mRNA is a faithful copy of the P gene, accounting for about three-fourths of the total transcripts and gives rise to the P protein, the smaller subunit of the RNA polymerase complex. The remaining transcripts (one-fourth) are V mRNA, and carry a single guanine residue (+1G) (at nt 1052) inserted by the polymerase stuttering at a specific editing site (UUUUUCCC) located midway down the P gene, which then causes the frameshift (Fig. 2). The V protein therefore consists of a P/V common region (the N-terminal approximately three-quarters before the frameshift) and the V unique region (the C-terminal approximately one-quarter after the frameshift) (Fig. 2). The V unique region is highly conserved in the amino acid sequence among paramyxoviruses with a perfect preservation of seven cysteine residues that form a zinc finger-like motif, and indeed binding two atoms of Zn2+ (reviewed in Refs. 6 and 8).9)
Establishing the non-essentiality of the putative accessory genes of SeV
Without affecting the P ORF, mutations were either introduced into the editing site (UUUUUCCC → UUCUUUCC) or a stop codon was introduced just downstream of the editing site. Mutants with the total V protein deleted [V(-)] and only the V unique C terminal region deleted (VΔC) were respectively recovered without any difficulty (Fig. 2).10),11) These mutants replicated in cells as efficiently as the parental SeV, thus establishing that the V gene is nonessential.
Through a rather difficult process, a viable clone, 4C(-) SeV expressing none of the four C proteins (C′, C, Y1, and Y2) was recovered by disrupting the respective ORFs, thus indicating that SeV C proteins also fall into the category of nonessential accessory gene products (Fig. 2).12) The difficulty of recovery was the result of reducing the 4C(-) virus infectivity by approximately 100-fold, while the success of the virus rescue was attributable to the high efficiency of the SeV rescue system.2) The characterization of the 4C(-) virus and a series of other C mutants with one or two C proteins deleted along with the plasmid-based expression studies of the C proteins demonstrated the extreme versatility of the C protein.
They were found to block the induction of the anti-viral state with type I IFN, IFN-α/β13) and type II IFN, IFN-γ.14) The inhibitory magnitude of virus growth was much greater with IFN-α/β than with IFN-γ. Subsequent studies therefore focused on the blocking of the action of IFN-α/β. The blocking was caused by inhibition of STAT2 phosphorylation at the cellular level.15),16) STAT2 is a key component and its phosphorylation a key event in the IFN-α/β signaling pathway. The Y2 (175 residues), the smallest of the 4 C proteins, was sufficient to exhibit the capacity to counteract the antiviral action of IFN-α/β.14) The region responsible for this IFN antagonism was narrowed down to the 106-residue-long C terminal half.17) The SeV C proteins further inhibited IFN-β production.18)
Although abundantly expressed in cells, the C proteins are nonstructural proteins that are not incorporated into virus particles. They nevertheless were found to make a large contribution to the viral morphogenesis by budding from the plasma membrane,19) possibly by escorting ESCRT to the site of that budding.20) ESCRT is thought to be required in pinching off of budding plasma membrane patches, the immediate precursors of a viral envelope.
The SeV C protein was initially recognized as a down-modulator of viral RNA synthesis.21),22) This was supported through use of 4C(-) SeV.12),19) It was also previously shown that the inhibition was stronger on an internally deleted DI genome than on a copy-back DI genome, suggesting some promoter specificity.22),23) Using 4C(-) and other C mutants it has recently been established that the C proteins suppress the leader (antigenomic) promoter but not the trailer (genomic) promoter late in the life cycle, thereby facilitating accumulation of the genomic RNA of negative polarity so that the released viruses mainly contain negative-sense RNA genomes.24) It was surprising that an accessory gene plays such a fundamental role as guaranteeing that SeV is a “negative-strand” RNA virus. It would be intriguing to study how genome polarity is determined in other negative strand RNA viruses. Furthermore, C proteins appear to encode anti-apoptosis function.25)
Pivotal roles of SeV accessory proteins in viral growth and pathogenesis in mice
(i). The C protein as an antagonist of IFN signaling
As described above, SeV causes fatal pneumonia in mice. When intranasally inoculated under our experimental conditions, the virus titers in the lung increase exponentially during the initial one or two days. This is followed by a period of high viral load of up to 7–9 days, during which the lung lesion (consolidation) score increases (see Fig. 4). All mice eventually die of severe hemorrhagic pneumonia.
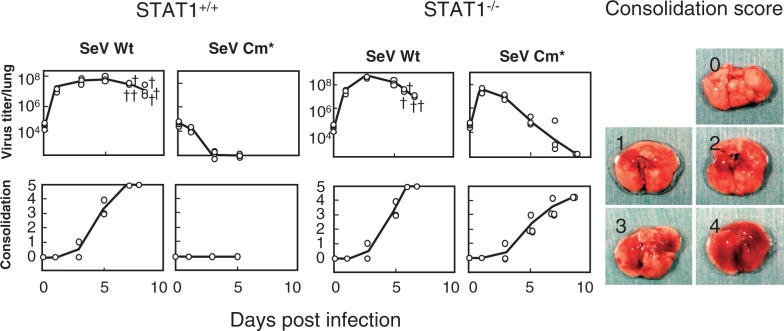
Fig. 4.
Growth and pathogenicity of SeV Cm* in wild-type and STAT1−/− mice. Five-week-old wild-type STAT1+/+ 129S6 and STAT1 deficient (STAT1−/−) 129S6 mice were infected intranasally with 107 CIU of SeV Wt or SeV Cm*. Two or three mice from each group were sacrificed at the intervals indicated and examined for virus infectivity in the lung (top) and lung consolidation (bottom). The symbol “†” indicates a dead mouse. Actual macroscopic views of the lung corresponding to the consolidation scores of 0 (no lesion) through 4 (maximum) are presented as references (right). When a mouse died, one point was added to the consolidation score (score 5) (Modified from Ref. 26).
As noted above, SeV C proteins are multifunctional. We identified some amino acids in the C protein crucial to the different functions and were able to generate a mutant Cm* that remained incapable of blocking IFN-α/β signaling but that did restore the other two functions (promoter-dependent RNA synthesis regulation and contribution to morphogenesis) to almost normal levels.26)
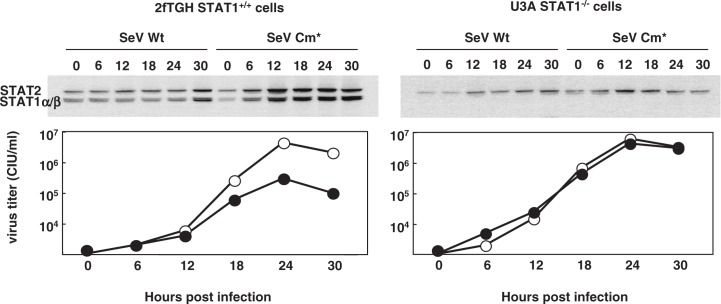
After binding to the receptor, IFN-α/β signaling is transduced through the Jak/STAT pathway. STAT1 is one of the key molecules in this signal transduction. The properties of the wild-type and Cm* SeV were studied using STAT1+/+ normal human 2fTGH cells and 2fTGH derived STAT1 deficient (STAT−/−) cells (Fig. 3).26) The lack of STAT1 expression in the latter cells was confirmed using immunoblotting. STAT1 and STAT2 are products induced by IFN-α/β. Both wild-type and Cm* SeV stimulate IFN-α/β production. In STAT1+/+ normal cells infected with SeV Cm*, the induction of STAT1 and STAT2 becomes detectable at 6 hr post infection (p.i.) and continues up to 30 hr, whereas little induction was observed in the same cells infected with wild-type SeV (Fig. 3). These results confirmed that the intact C expressed from wild-type SeV had the capacity to block IFN-α/β signaling, whereas the Cm* mutant had lost the IFN antagonism. Accordingly, the growth of SeV Cm* was self limiting in normal cells and comparable to that of wild-type SeV in STAT1−/− cells (Fig. 3). It remains to be elucidated whether or not SeV Cm* retains the anti-apoptosis and anti-IFN synthesis functions.
Fig. 3.
Replication of SeV Cm* in wild-type cells and in STAT1 deficient cells. Parental SeV Wt (wild type) and SeV Cm* were inoculated to the parental STAT1+/+2fTGH cells (left) and to its STAT1 deficient (STAT1−/−) derivative, U3A cells (right) at an m.o.i. of 5. Cells were collected at the hours indicated. The cell lysates were immunoblotted with a mixture of anti STAT1 and anti STAT2 antibodies (top). Titers of SeV Wt (open circle) and SeV Cm* (closed circle) in the culture fluid are shown (bottom) (Modified from Ref. 26).
Using the Cm* virus the attempt was made to verify whether anti IFN-α/β capacity of SeV C protein would indeed play a role in in vivo pathogenesis.26) In the lungs of STAT1+/+ normal mice, SeV Cm* did not grow at all and produced no lung lesions, in striking contrast to the wild-type SeV that grew vigorously with increasing consolidation scores and killed all the mice (Fig. 4). In STAT1−/− mice, however, SeV Cm* grew far better than in STAT1+/+ mice. And although the virus did not kill the STAT1−/− mice, the consolidation scores reached as high as 4 at day 9 (Fig. 4). These results clearly indicate that the IFN-α/β inducible antiviral state is indeed a key host defense mechanism at the forefront when coping with infecting SeV and that the SeV C protein plays a key role in the viral evasion of the antiviral state, thereby facilitating virus growth in the lung and causing severe pneumonia.
Recently SeV C proteins were found to inhibit PKR.27) This may play an additional role in counteracting the antiviral state induced by IFN-α/β.
(ii). The V protein as an antagonist of innate virus clearance through IRF3 activation, but not via IFNs
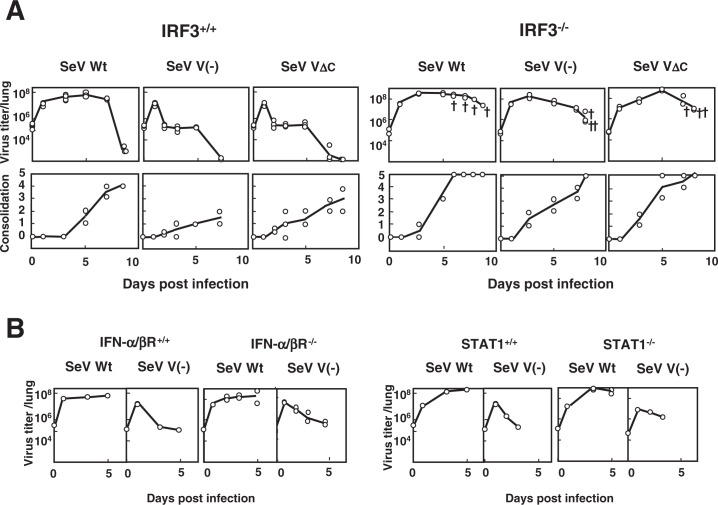
Both V(-) and VΔC proliferated in the lungs of normal mice that included IRF3+/+ mice as efficiently as the parental wild-type SeV until day 1 post infection. However, they were then rapidly cleared and produced only moderate consolidation scores, in contrast to the wild-type SeV, which maintained both a high viral load and high consolidation scores up to 7–9 days (Fig. 5A). The wild-type SeV was usually lethal for most mouse strains so far tested, whereas V(-) and VΔC were never lethal.10),11) Similar attenuation was observed with several mutants with the zinc finger-like motif disrupted and no capacity to bind zinc.28),29) The V protein, which is fully dispensable in the viral life cycle, was therefore found to encode a special function required for pathogenesis. The V unique region, particularly its zinc binding capacity, appeared to be crucial for this function.
Fig. 5.
Replication and pathogenicity of SeV V(-) and SeV VΔC in IRF3−/− mice. (A) Five-week-old IRF3+/+ C57BL/6J and IRF3 deficient (IRF3−/−) C57BL/6J mice were infected intranasally with 107 CIU of SeV Wt, SeV V(-) or SeV VΔC. Two or three mice from each group were sacrificed at the indicated intervals and examined for virus infectivity in the lung and lung consolidation. The symbol “†” indicates a dead mouse. (B) Six- to seven-week-old IFN-α/β R+/+ A129/Sv and IFN-α/β receptor deficient (IFN-α/β R−/−) A129/Sv mice were infected intranasally with 107 CIU of SeV Wt or SeV V(-) (left). Five-week-old STAT1+/+ 129S6 and STAT1 deficient (STAT1−/−) 129S6 mice were infected intranasally with 107 CIU of SeV Wt or SeV V(-) (right). Viral infectivity in the lung was measured at the indicated time intervals after infection (Modified from Ref. 30).
Of the various mice of high genetic diversity so far tested, IRF3−/− mice were found to be highly and almost equally susceptible to V(-), VΔC and wild-type viruses (Fig. 5A).30) The unique, rapid clearance of V(-) and VΔC viruses could no longer be observed. Instead, they grew as efficiently as the wild type and caused severe pneumonia and death almost as rapidly as the wild type. IFN-β is one of the well-known IRF3 inducible gene products, and IFN-α/β is subsequently amplified via IRF7 activation.31) It is therefore a reasonable assumption that the SeV V protein is also a type I IFN (IFN-α/β) antagonist.
As shown in Fig. 5B, however, no such active growth beyond 1 day after infection as seen in IRF3−/− mice was found for the V(-) virus in IFN-α/β-receptor −/− mice and STAT1−/− mice, where IFN-α/β mediated antiviral-state is not inducible. V(-) appeared to be cleared in these mice as efficiently as in normal mice.30) The V(-) virus was also efficiently cleared in IFN-γ−/− mice, indicating that type II IFN is not involved, either. It was also rapidly cleared in ISG15 (another IRF3 dependent product) deficient mice and various other mouse strains deficient in different facets of innate and adaptive immunity. The latter included NK cell deficient beige mice and T cell deficient nu/nu mice.30) More recently grouped in the category that is characterized by early clearance of V(-) virus were mouse strains deficient in NKT, TNFα, IL6, iNOs, TRIF and MyD88 (Kiyotani et al., manuscript in preparation).
These results predict the presence of a hitherto unknown (novel), antiviral innate-immunity mechanism induced following IRF3 activation.
The SeV V protein appears to interact with MDA5, thereby inhibiting IRF3 activation and IFN production.32) However, this IFN synthesis inhibition did not appear to be profound; quantitative differences in IFN in the mouse lung were about only 2.5-fold between infections with V(-) and the wild-type SeV during the early period when their viral loads were similar.30) The wild-type SeV displayed no appreciable difference in growth capacity in normal and MDA5−/− mice. The growth of V(-) virus showed no appreciable difference, either, in normal and MDA5−/− mice. The V protein interfering with the virus-sensing MDA5 may therefore not have an appreciable influence upon in vivo virus growth (Kiyotani et al., manuscript in preparation).
Accessory genes of other paramyxoviruses
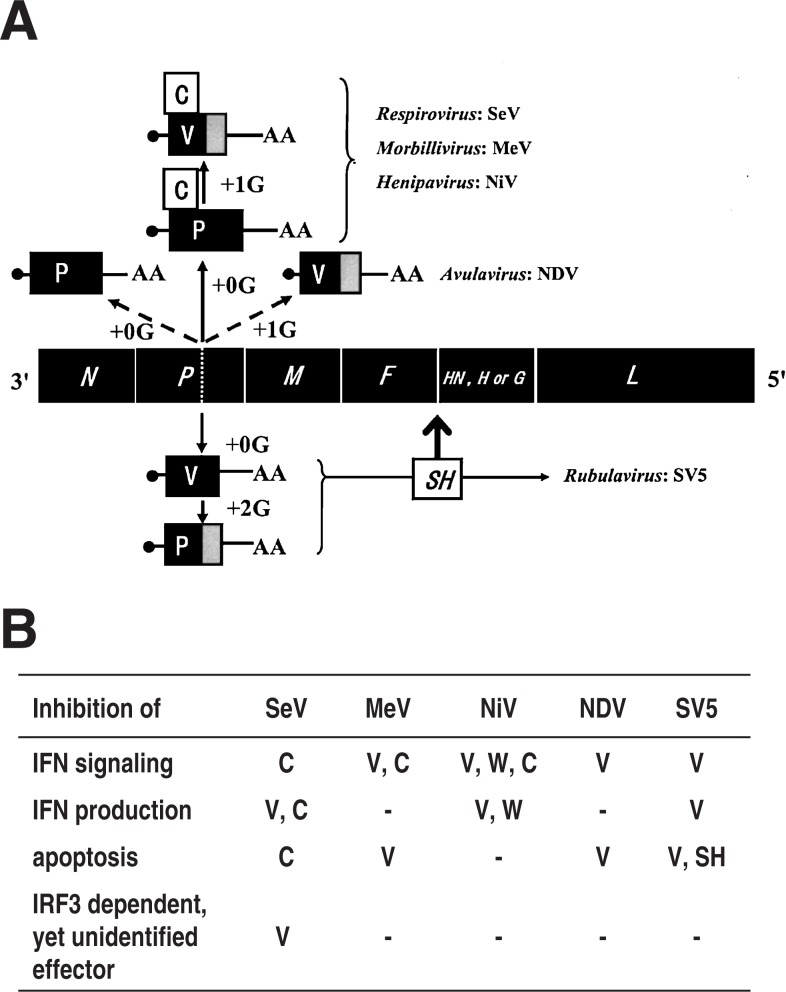
The subfamily Paramyxovirinae consists of five genera (Respiro-, Morbilli-, Henipa-, Avula- and Rubulavirus). All the members of these five genera encode the V protein, whereas the C protein is only encoded by the members of three genera, Respiro-, Morbilli- and Henipavirus. The way they express the respective accessory genes are schematically presented using a virus species representative of each genus in Fig. 6A. Mainly using the data obtained with these representative species, the nature of the V and C proteins in terms of dispensability, functions assigned in vitro cell culture and their verification in vivo will now be discussed.
Fig. 6.
(A) Expression of accessory genes from members of the subfamily Paramyxovirinae. The fifth gene encoding the receptor binding protein is named HN for Respiro-, Avula- and Rubulavirus, H for Morbillivirus and G for Henipavirus. Dotted line in the P gene, editing site;•, cap of mRNA; AA, 3′ polyadenylic acid of mRNA. SH, inserted between the F and HN genes (Modified from Ref. 8). (B) The coding assignment of the individual inhibitory functions. –, data not available; W, encoded by the mRNA generated by +2G insertion. For details refer to the text and Refs. 6, 8 and 39.
(i). Dispensability
The dispensability of both the V and C proteins appears to have been established for the Respirovirus and Morbillivirus including for SeV and MeV, respectively (reviewed in Ref. 8). Reverse genetics was relatively recently established for the Henipavirus,33) and the data are not yet available for the dispensability or indispensability of the V and C proteins of this genus.
For the Rubulavirus, the V mRNA is a faithful copy of the P gene, whereas the indispensable P mRNA is generated by +2G insertion editing (reviewed in Refs. 6 and 8) (Fig. 6A). Hence the rescue of recombinant SV5 that would express only P mRNA but not V mRNA may not be as easy as for other genera that express the V protein from edited mRNA. And in fact it has been unsuccessful.34) Thus deletion of the C-terminal V-unique domain was attempted to generate VΔC by changing the sixth and eighth codons downstream of the editing site to stop codons. This mutant grew poorly in cells and interestingly, the two stop codons introduced could not be stably maintained and were instead converted to sense codons, although not identical to the original ones, thus generating a pseudo-wild type.34) This indicates the presence of strong pressure to repair the truncated V to a full-length V. An analogous VΔC virus created from another rubulavirus hPIV2 was also shown to have critically impaired infectivity in cell cultures with highly anomalous virion morphology.35) The constraint due to V deletion did not appear to be fully attributable to the capacity of the Rubulavirus V protein to inhibit IFN signaling because strong constraint and the repair were still observed in Vero cells, which do not produce IFN.34) And hence dispensability or indispensability is yet to be established for the Rubulavirus V protein.
Avulavirus NDV V mRNA is generated by +1G editing (Fig. 6A). Severely impaired infectivity was also observed for its editing-minus mutant as well as VΔC version.36)–38) However, the constraint was much smaller in the Vero cell line, an IFN non-producer, indicating that it was largely due to increased IFN sensitivity caused by V deletion.38) And hence the Avulavirus V protein falls into the category of nonessential gene product.
(ii). IFN antagonism as a common role shared with V and C genes of the Paramyxovirinae
Inhibition of IFN-α/β mediated signaling to induce the antiviral state was demonstrated to be a major role in common with the paramyxovirus accessory gene products (reviewed in Refs. 6, 8 and 39) (Fig. 6B). This capacity to evade the antiviral state in host animals is encoded by either the V or C protein or by both, depending on the virus species (Fig. 6B). The inhibition mechanisms are highly diverse. The inhibition is caused through proteasome-dependent degradation of STAT1 or STAT2 by the Rubulavirus V protein,40),41) an irrelevant complex formation of the Morbillivirus C protein or Henipavirus V protein with STAT1 and STAT2,42),43) or inhibition of STAT2 phosphorylation by the Respirovirus SeV C protein.15),16)
Furthermore, inhibition of IFN-α/β production appeared to be encoded by the Respirovirus, Henipavirus and Rubulavirus44) (Fig. 6B). The mechanisms responsible for the inhibition of IFN production by the accessory proteins are poorly understood. With SV5 and SeV, the interaction of the V protein with virus-sensing MDA5 may be involved.32),45)
(iii). Other functions of accessory genes to evade host defense
Virus infection often causes apoptosis, which limits further virus spread in the body. Probably to evade this host defense, not all but many genera of the Paramyxovirinae appear to encode the anti-apoptosis function (Fig. 6B). SV5 encodes anti-apoptosis in not only the V gene but also another accessory gene, namely the SH gene.46),47) The latter is a small gene that evolved through insertion between the F and HN genes (Fig. 6A) in two members (SV5 and MuV) of the Rubulavirus and encodes 44- and 57-residue-long SH proteins, respectively (reviewed in Ref. 8).
Mice appeared to be capable of resisting SeV through not only exerting the IFN system but also inducing the IRF3 dependent, but as-yet-unidentified effector(s). Identification of the effector in this puzzling IRF3 dependent innate immunity is of critical importance. It would also be intriguing to decide whether a similar innate immunity is present in other virus-host systems.
(iv). In vivo verification of the in vitro assigned functions
Many of the inhibitory mechanisms described above have been suggested by plasmid-based expression of the accessory genes and their variants or by the phenotypes of respective gene knockout viruses at the cellular level, and await in vivo verification to decide whether in vitro assigned functions indeed have grave consequences on in vivo virus growth and pathogenicity.
In this context, at least three different situations were encountered with the SeV-mouse system. First, the in vitro assessed property (IFN antagonism by the C protein) indeed brought about a far-reaching consequence in vivo as strongly supported by the results obtained with the SeV Cm* in STAT1−/− and normal mice (Fig. 4). Second, an in vitro assigned function (of the V protein to interfere with virus sensing MDA5) did not appear to have an appreciable consequence on in vivo virus growth as suggested by the results obtained with SeV V(-) in MDA5−/− mice (see above). Third, even if no phenotype was found in vitro following deletion of an accessory gene, the consequence of deletion was quite grave as shown by the case of SeV V(-) in normal and IRF3−/− mice (Fig. 5A). These results indicate that the gap between in vitro and in vivo experimental outcomes can sometimes be significant. Careful consideration and further experimentation are needed to bridge that gap.
In other virus-host systems, in vivo verification would be even more difficult. However, several mutants with the accessory genes deleted grew poorly and were less pathogenic in host animals including rhesus macaques48)–53) or grew better in STAT1−/− mice than in normal mice,34),46) agreeing with the concept that the in vitro assigned functions are of in vivo significance.
Concluding remarks
Genome nucleotide sequencing of the Paramyxoviridae was completed in the 1980s for most of the members and then for the newly emergent members. One of the important issues revealed by that sequencing was the identification of ORFs encoding putative accessory proteins. These accessory genes were then focused upon in active investigation using reverse genetics established in the mid 1990s.
The SeV-mouse system probably assumed the lead role in that line of research because of the extremely high efficiency of the reverse genetics system and the availability of mice of diverse genetic backgrounds as susceptible hosts. Research conducted using this model revealed that in the realm of in vivo pathogenesis, accessory genes were not mere accessories or minor players, but rather central players. Mice appear to resist SeV by exerting at least two facets of innate immunity. One is the IFN system and the other is through IRF3 dependent, but as-yet-unidentified effector(s). To counteract these two antiviral mechanisms and persist in nature, SeV appears to have acquired the capacity to express both the C and the V proteins in the evolutionary process. Hence accessory genes should be highly relevant to virus biology in natural settings or virus ecology.
This view appears to be well shared by the members of the subfamily Paramyxovirinae. The accessory genes of another subfamily Pneumovirinae appear to have evolved in a rather different way (insertion before or between the basal six genes), but play essentially the same roles; i.e. evasion of innate host defense, in particular, the IFN system (reviewed in Refs. 8 and 39).
The P gene, except for its V unique region, is the least conserved of the six basal genes in the members of the subfamily Paramyxovirinae, and thus might be most subject to change during the evolutionary process. Apparently the same important occupation, inhibiting the IFN signaling, is encoded by the C protein in Respirovirus SeV and by the V protein in Avulavirus NDV and Rubulavirus SV5, whereas Morbillivirus MeV and Henipavirus NiV encode the same function in both the V and C proteins (Fig. 6B). These differences in coding strategies of the accessory function may be related to the concept that Morbillivirus and Henipavirus are phylogenetically close and map between the Respirovirus and Rubulavirus and that Avulavirus maps between Rubulavirus and the Morbilli-Henipa group.8),54)
The P/V common region of NiV was as active as the full-length V in inhibiting IFN signaling.55) In the inhibition by MeV, not only the P/V common region but also the P protein was as active as the V protein.56) On the other hand, not the P/V common region but the V unique region is generally essential, although not sufficient, to carry out the IFN antagonism for the Rubulavirus (reviewed in Ref. 8). The different coding patterns of IFN antagonism depending on the genus may be related to the different underlying mechanisms as noted above. The different patterns further suggest that there could be considerable options in the way to encode IFN antagonism in the paramyxovirus P gene. Although highly speculative, the ancestral P gene might encode multiple functions including a role in RNA synthesis, IFN antagonism and others. V ORF might then open and become widespread in taking over the IFN antagonism as its principal occupation and some additional other jobs. In some other members, the C gene further evolved to take over some of the numerous functions of V gene. Respirovirus SeV may have evolved so as to encode the capacities to evade the two different facets of host defense (IFN system and IRF3 dependent, yet unknown function) in two separate ORFs, V and C.
Abbreviations used:
[Virus species]
- HeV
Hendra virus
- hPIV2
human parainfluenza virus type 2
- MeV
measles virus
- MuV
mumps virus
- NiV
Nipah virus
- NDV
Newcastle disease virus
- SeV
Sendai virus
- SV5
simian virus 5
[Miscellanea]
- CIU
cell infectious units
- DI
defective interfering
- ESCRT
endosome-associated complexes required for transport
- IFN
interferon
- IL
interleukin
- iNOs
inducible nitric-oxide synthase
- IRF
interferon regulatory factor
- ISG
interferon-stimulated gene
- Jak
Janus kinase
- MDA5
melanoma differentiation-associated gene 5
- m.o.i
multiplicity of infection
- MyD88
myeloid differentiation factor 88
- NK
natural killer
- NKT
natural killer T
- nt
nucleotide positions
- nu
nude
- ORF
open reading frame
- PKR
protein kinase R
- RNP
ribonucleoprotein
- SH
small hydrophobic
- STAT
signal transducer and activator of transcription
- TRIF
Toll/interleukin-1 receptor domain-containing adaptor inducing interferon-β
- TNFα
tumor necrosis factor alpha
Profile
Yoshiyuki Nagai was born in Gifu Prefecture, Japan, in 1939. He graduated from the Faculty of Medicine at Nagoya University in 1965, started his research career in the field of virology at the graduate school of the same university and earned his Ph. D. in 1973. He made a research stay at the Institut fuer Virologie, Giessen, Germany from 1974 to 1976 as a Scholarship Investigator of Alexander von Humboldt Foundation. His academic career involves assignment to Professor of the Research Institute for Disease Mechanism and Control, Nagoya University in 1984, Professor of the Institute of Medical Science, the University of Tokyo in 1993 and Director of AIDS Research Center, National Institute of Infectious Diseases in 1998. He further served in the field of public health as Director of Toyama Institute of Health from 2001 to 2005 and returning to Tokyo in 2005, he is taking an active part in constructing Japan’s Asian-African research network for infectious diseases as Director of the Center of Research Network for Infectious Diseases (CRNID), RIKEN. For his great deal of scientific contribution, especially to the understanding of molecular basis of paramyxovirus replication and pathogenicity, he was awarded Humboldt Research Award in 1990, Hideyo Noguchi Memorial Award for Medical Sciences in 1994, Chunichi Cultural Award in 1995, Professor Emeritus of Nagoya University and Takeda Prize for Medical Sciences in 2000, the Medal with Purple Ribbon in 2001 and the Japan Academy Prize in 2008. His administrative roles have also been greatly appreciated as exemplified by the assumption to President of the Japanese Society for Virology (2004–2005) and inauguration to Vice President of the International Union of Microbiological Societies this year (2008).

References
- 1).Lamb, R.A., Collins, P.L., Kolakofsky, D., Maleno, J.A., Nagai, Y. and Oldstone, M.B. (2005) Paramyxoviridae. InVirus Taxonomy: Classification and Nomenclature of Viruses; Eighth Report of the International Committee on Taxonomy of Viruses (eds. Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U. and Ball, L.A.). Academic Press, London, pp. 655–668 [Google Scholar]
- 2).Kato, A., Sakai, Y., Shioda, T., Kondo, T., Nakanishi, M. and Nagai, Y. (1996) Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1, 569–579 [DOI] [PubMed] [Google Scholar]
- 3).Li, H.O., Zhu, Y.F., Asakawa, M., Kuma, H., Hirata, T., Ueda, Y., Lee, Y.S., Fukumura, M., Iida, A., Kato, A.et al. (2000) A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J. Virol. 74, 6564–6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Nagai, Y. (1993) Protease-dependent virus tropism and pathogenicity. Trends Microbiol. 1, 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Kolakofsky, D., Pelet, T., Garcin, D., Hausmann, S., Curran, J. and Roux, L. (1998) Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J. Virol. 72, 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Lamb, R.A. and Parks, G.D. (2006) Paramyxoviridae: the viruses and their replication. InFields Virology, 5th ed (eds. Knipe, D.M. and Howley, P.M.). Lippincott Williams & Wilkins, Philadelphia, pp. 1449–1496 [Google Scholar]
- 7).Nagai, Y., Inoue, M., Iida, A., Zhu, Y.-F., Hasegawa, M., Kato, A. and Matano, T. (2007) Sendai virus engineering: from reverse genetics to vector development. InVirus Expression Vectors (ed. Hefferon, K. L.). Transworld Research Network, Kerala, pp. 123–146 [Google Scholar]
- 8).Nagai, Y. and Kato, A. (2004) Accessory genes of the paramyxoviridae, a large family of nonsegmented negative-strand RNA viruses, as a focus of active investigation by reverse genetics. Curr. Top. Microbiol. Immunol. 283, 197–248 [DOI] [PubMed] [Google Scholar]
- 9).Li, T., Chen, X., Garbutt, K.C., Zhou, P. and Zheng, N. (2006) Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124, 105–117 [DOI] [PubMed] [Google Scholar]
- 10).Kato, A., Kiyotani, K., Sakai, Y., Yoshida, T. and Nagai, Y. (1997) The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16, 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Kato, A., Kiyotani, K., Sakai, Y., Yoshida, T., Shioda, T. and Nagai, Y. (1997) Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J. Virol. 71, 7266–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Kurotani, A., Kiyotani, K., Kato, A., Shioda, T., Sakai, Y., Mizumoto, K., Yoshida, T. and Nagai, Y. (1998) Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells 3, 111–124 [DOI] [PubMed] [Google Scholar]
- 13).Gotoh, B., Takeuchi, K., Komatsu, T., Yokoo, J., Kimura, Y., Kurotani, A., Kato, A. and Nagai, Y. (1999) Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-α/β-mediated responses. FEBS Lett. 459, 205–210 [DOI] [PubMed] [Google Scholar]
- 14).Kato, A., Ohnishi, Y., Kohase, M., Saito, S., Tashiro, M. and Nagai, Y. (2001) Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75, 3802–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Gotoh, B., Takeuchi, K., Komatsu, T. and Yokoo, J. (2003) The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J. Virol. 77, 3360–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Kato, A., Cortese-Grogan, C., Moyer, S.A., Sugahara, F., Sakaguchi, T., Kubota, T., Otsuki, N., Kohase, M., Tashiro, M. and Nagai, Y. (2004) Characterization of the amino acid residues of Sendai virus C protein that are critically involved in its interferon antagonism and RNA synthesis down-regulation. J. Virol. 78, 7443–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Kato, A., Ohnishi, Y., Hishiyama, M., Kohase, M., Saito, S., Tashiro, M. and Nagai, Y. (2002) The amino-terminal half of Sendai virus C protein is not responsible for either counteracting the antiviral action of interferons or down-regulating viral RNA synthesis. J. Virol. 76, 7114–7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Komatsu, T., Takeuchi, K., Yokoo, J. and Gotoh, B. (2004) C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325, 137–148 [DOI] [PubMed] [Google Scholar]
- 19).Hasan, M.K., Kato, A., Muranaka, M., Yamaguchi, R., Sakai, Y., Hatano, I., Tashiro, M. and Nagai, Y. (2000) Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J. Virol. 74, 5619–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Sakaguchi, T., Kato, A., Sugahara, F., Shimazu, Y., Inoue, M., Kiyotani, K., Nagai, Y. and Yoshida, T. (2005) AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J. Virol. 79, 8933–8941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Curran, J., Marq, J.B. and Kolakofsky, D. (1992) The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189, 647–656 [DOI] [PubMed] [Google Scholar]
- 22).Cadd, T., Garcin, D., Tapparel, C., Itoh, M., Homma, M., Roux, L., Curran, J. and Kolakofsky, D. (1996) The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J. Virol. 70, 5067–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Tapparel, C., Hausmann, S., Pelet, T., Curran, J., Kolakofsky, D. and Roux, L. (1997) Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J. Virol. 71, 9588–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Irie, T., Nagata, N., Yoshida, T. and Sakaguchi, T. (2008) Paramyxovirus Sendai virus C proteins are essential for maintenance of negative-sense RNA genome in virus particles. Virology 374, 495–505 [DOI] [PubMed] [Google Scholar]
- 25).Koyama, A.H., Irie, H., Kato, A., Nagai, Y. and Adachi, A. (2003) Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins. Microbes Infect. 5, 373–378 [DOI] [PubMed] [Google Scholar]
- 26).Kato, A., Kiyotani, K., Kubota, T., Yoshida, T., Tashiro, M. and Nagai, Y. (2007) Importance of the anti-interferon capacity of Sendai virus C protein for pathogenicity in mice. J. Virol. 81, 3264–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Takeuchi, K., Komatsu, T., Kitagawa, Y., Sada, K. and Gotoh, B. (2008) A role of Sendai virus C protein in restricting PKR activation: limitation of intracellular double stranded RNA generation. J. Virol. 82, 10102–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Huang, C., Kiyotani, K., Fujii, Y., Fukuhara, N., Kato, A., Nagai, Y., Yoshida, T. and Sakaguchi, T. (2000) Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J. Virol. 74, 7834–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Fukuhara, N., Huang, C., Kiyotani, K., Yoshida, T. and Sakaguchi, T. (2002) Mutational analysis of the Sendai virus V protein: importance of the conserved residues for Zn binding, virus pathogenesis, and efficient RNA editing. Virology 299, 172–181 [DOI] [PubMed] [Google Scholar]
- 30).Kiyotani, K., Sakaguchi, T., Kato, A., Nagai, Y. and Yoshida, T. (2007) Paramyxovirus Sendai virus V protein counteracts innate virus clearance through IRF-3 activation, but not via interferon, in mice. Virology 359, 82–91 [DOI] [PubMed] [Google Scholar]
- 31).Honda, K. and Taniguchi, T. (2006) IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6, 644–658 [DOI] [PubMed] [Google Scholar]
- 32).Andrejeva, J., Childs, K.S., Young, D.F., Carlos, T.S., Stock, N., Goodbourn, S. and Randall, R.E. (2004) The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101, 17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Yoneda, M., Guillaume, V., Ikeda, F., Sakuma, Y., Sato, H., Wild, T.F. and Kai, C. (2006) Establishment of a Nipah virus rescue system. Proc. Natl. Acad. Sci. USA 103, 16508–16513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).He, B., Paterson, R.G., Stock, N., Durbin, J.E., Durbin, R.K., Goodbourn, S., Randall, R.E. and Lamb, R.A. (2002) Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303, 15–32 [DOI] [PubMed] [Google Scholar]
- 35).Kawano, M., Kaito, M., Kozuka, Y., Komada, H., Noda, N., Nanba, K., Tsurudome, M., Ito, M., Nishio, M. and Ito, Y. (2001) Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 284, 99–112 [DOI] [PubMed] [Google Scholar]
- 36).Mebatsion, T., Verstegen, S., De Vaan, L.T., Romer-Oberdorfer, A. and Schrier, C.C. (2001) A recombinant Newcastle disease virus with low-level V protein expression is immunogenic and lacks pathogenicity for chicken embryos. J. Virol. 75, 420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Huang, Z., Krishnamurthy, S., Panda, A. and Samal, S.K. (2003) Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J. Virol. 77, 8676–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Park, M.S., Garcia-Sastre, A., Cros, J.F., Basler, C.F. and Palese, P. (2003) Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 77, 9522–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Randall, R.E. and Goodbourn, S. (2008) Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89, 1–47 [DOI] [PubMed] [Google Scholar]
- 40).Didcock, L., Young, D.F., Goodbourn, S. and Randall, R.E. (1999) The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73, 9928–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Andrejeva, J., Young, D.F., Goodbourn, S. and Randall, R.E. (2002) Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76, 2159–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Rodriguez, J.J., Parisien, J.P. and Horvath, C.M. (2002) Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76, 11476–11483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Yokota, S., Saito, H., Kubota, T., Yokosawa, N., Amano, K. and Fujii, N. (2003) Measles virus suppresses interferon-α signaling pathway: suppression of Jak1 phosphorylation and association of viral accessory proteins, C and V, with interferon-α receptor complex. Virology 306, 135–146 [DOI] [PubMed] [Google Scholar]
- 44).Poole, E., He, B., Lamb, R.A., Randall, R.E. and Goodbourn, S. (2002) The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303, 33–46 [DOI] [PubMed] [Google Scholar]
- 45).Yoneyama, M., Kikuchi, M., Matsumoto, K., Imaizumi, T., Miyagishi, M., Taira, K., Foy, E., Loo, Y.M., Gale, M.Jr., Akira, S.et al. (2005) Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175, 2851–2858 [DOI] [PubMed] [Google Scholar]
- 46).He, B., Lin, G.Y., Durbin, J.E., Durbin, R.K. and Lamb, R.A. (2001) The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75, 4068–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Lin, Y., Bright, A.C., Rothermel, T.A. and He, B. (2003) Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J. Virol. 77, 3371–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Valsamakis, A., Schneider, H., Auwaerter, P.G., Kaneshima, H., Billeter, M.A. and Griffin, D.E. (1998) Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J. Virol. 72, 7754–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Tober, C., Seufert, M., Schneider, H., Billeter, M.A., Johnston, I.C., Niewiesk, S., ter Meulen, V. and Schneider-Schaulies, S. (1998) Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72, 8124–8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Durbin, A.P., McAuliffe, J.M., Collins, P.L. and Murphy, B.R. (1999) Mutations in the C, D, and V open reading frames of human parainfluenza virus type 3 attenuate replication in rodents and primates. Virology 261, 319–330 [DOI] [PubMed] [Google Scholar]
- 51).Takeuchi, K., Takeda, M., Miyajima, N., Ami, Y., Nagata, N., Suzaki, Y., Shahnewaz, J., Kadota, S. and Nagata, K. (2005) Stringent requirement for the C protein of wild-type measles virus for growth both in vitro and in macaques. J. Virol. 79, 7838–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).von Messling, V., Svitek, N. and Cattaneo, R. (2006) Receptor (SLAM [CD150]) recognition and the V protein sustain swift lymphocyte-based invasion of mucosal tissue and lymphatic organs by a morbillivirus. J. Virol. 80, 6084–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Devaux, P., Hodge, G., McChesney, M. B. and Cattaneo, R. (2008) Attenuation of V- or C-defective measles virus: Infection control by the inflammatory and interferon responses of rhesus monkeys. J. Virol. 82, 5359–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Lamb, R.A. and Kolakofsky, D. (2001) Paramyxoviridae: the viruses and their replication. InFields Virology, 4th ed (eds. Knipe, D.M. and Howley, P.M.). Lippincott Williams & Wilkins, Philadelphia, pp. 1305–1340 [Google Scholar]
- 55).Park, M.S., Shaw, M.L., Munoz-Jordan, J., Cros, J.F., Nakaya, T., Bouvier, N., Palese, P., Garcia-Sastre, A. and Basler, C.F. (2003) Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W and C proteins. J. Virol. 77, 1501–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Ohno, S., Ono, N., Takeda, M., Takeuchi, K. and Yanagi, Y. (2004) Dissection of measles virus V protein in relation to its ability to block alpha/beta interferon signal transduction. J. Gen. Virol. 85, 2991–2999 [DOI] [PubMed] [Google Scholar]