Abstract
Hippocampal organotypic cultures are a highly reliable in vitro model for studying neuroplasticity: in this paper, we analyze the early phase of the transcriptional response induced by a 20 µM gabazine treatment (GabT), a GABA-Ar antagonist, by using Affymetrix oligonucleotide microarray, RT-PCR based time-course and chromatin-immuno-precipitation. The transcriptome profiling revealed that the pool of genes up-regulated by GabT, besides being strongly related to the regulation of growth and synaptic transmission, is also endowed with neuro-protective and pro-survival properties. By using RT-PCR, we quantified a time-course of the transient expression for 33 of the highest up-regulated genes, with an average sampling rate of 10 minutes and covering the time interval [10∶90] minutes. The cluster analysis of the time-course disclosed the existence of three different dynamical patterns, one of which proved, in a statistical analysis based on results from previous works, to be significantly related with SRF-dependent regulation (p-value<0.05). The chromatin immunoprecipitation (chip) assay confirmed the rich presence of working CArG boxes in the genes belonging to the latter dynamical pattern and therefore validated the statistical analysis. Furthermore, an in silico analysis of the promoters revealed the presence of additional conserved CArG boxes upstream of the genes Nr4a1 and Rgs2. The chip assay confirmed a significant SRF signal in the Nr4a1 CArG box but not in the Rgs2 CArG box.
Introduction
Cognitive processes such as learning and memory originate from plastic modifications in the central nervous system CNS: these plastic changes affect the structure and the functions of neurons and of synapses and lead to experience-dependent alterations in neural network wiring and behavior. The introduction of high-throughput assays and large-scale approaches in neuroplasticity has contributed to encompass the broad extent of this phenomenon, which involves the cooperative interplay of numerous cellular processes that not only regulate the synaptic transmission itself but also cell survival [1], neuronal growth [2] and neurogenesis [3].
The modulation of gene transcription has proven to be playing a key role in neuroplasticity: increased synaptic activity leads to calcium influx into the post-synaptic spines, dendrites and soma, which activates calcium dependent signaling pathways that in turn regulate transcription factors within the nucleus [4] [5] [6]. In our previous work with dissociated rat neuronal cultures [5] we combined transcriptome profiling with electrophysiological recordings in order to describe the role of different calcium sources in the regulation of gene expression changes. The variations of calcium dynamics driven by synaptic activity, as well as the resulting activation/deactivation changes in the relative signaling pathways, have shown to be tightly regulated both in time [7] [8] and space [9] [10] [11]. For instance, the modulation of the neurotrophin Bdnf (brain derived neurotropic factor) gene expression, following synaptic activity, requires a series of phosphorylation/dephosphorylation steps of the transcription factors CREB, MEF2 and MEcp2 in order to keep the Bdnf expression bound to the desired dynamics [12]. The expression level of many other plasticity-related genes is governed by sophisticated controls of dynamics [13]: this result is often achieved thanks to the interplay of a large number of transcription factors and is often related to signaling changes which are triggered within a time-scale of minutes [14] [15] [16].
Alterations in the dynamical pattern of activity-induced programs may result in pathological states: for example, the removal of the phosphatase MKP-1/DUSP1 negative feedback loop on the kinase JNK alters the proper JNK-activation dynamics and leads to the inability of forming new axonal branching during mice cortex development [17]. Despite the importance of the dynamical aspects of transcriptional changes, the information currently available is limited to time-courses with low temporal resolution, i.e. a few time points, and/or concerning a reduced number of genes, such as [15] [18] [19]. The purpose of the present study is to trace with high temporal resolution the early transcriptional dynamics associated with plasticity, using the gabazine treatment of rat organotypic cultures as hippocampal plasticity model: organotypic culture preparation has the advantage of retaining the general morphological and functional properties of the intact hippocampus [20] [21]. Besides, if compared to acute slices, organotypic cultures are able, within one week, to remodel the synaptic connections altered by the slicing procedure, which is not possible for acute slices [22]. In this work we will begin with a preliminary microarray-based assessment of the transcriptional response of hippocampal cultures to a 20 µM gabazine (also known as SR95531, a GABA-A receptor antagonist) treatment: the aim of this step is to obtain a general outline of the cellular activities involved in the response to GABA-A blocking. GABA-A channels are ionotropic channels that, upon binding of Gaba molecules, exert an inhibitory effect on neuronal excitability by specifically increasing the chloride conductance. Drugs such as gabazine, bicuculline or picrotoxin (PTX) act as GABA-A antagonists and therefore induce an increase of the overall neuronal excitability: these drugs have been extensively used as models for various types of plasticity (epilepsy, long term potentiation, homeostatic plasticity etc.), according to the tissue, dosage, duration of the treatment and possible concomitant stimuli. The 20 µM dosage was adopted in accordance to the evidences provided in [5], where we have previously studied the electrophysiological effects of a 20 µM GabT in dissociated hippocampal cultures.
Following the microarray assay, we will then quantify and analyze a high temporal resolution time course comprising a large set (33) of plasticity-related genes and we will relate the main features of the dynamical profiles with the putative biological functions of the relative genes/proteins. Following that, we will link one cluster of genes to a SRF-dependent regulation, by means of statistical and in silico analysis, and we will finally develop a chip (chromatin immunoprecipitation) assay in order to gain novel information about the role of SRF in the early phase of activity-dependent regulation of gene expression.
Results
Microarray analysis
A transcriptome profiling of a GABA-A receptor antagonist treatment is still lacking in the case of organotypic hippocampal cultures. Therefore, we decided to start the analysis with a preliminary, microarray-based, assessment of the response of rat organotypic hippocampal cultures to a 20 µM gabazine treatment (GabT): the purpose of this step was to obtain a complete profile of the tissue reaction to a prolonged GABA-A receptor blockade, which is strictly associated with a sudden and powerful increase in the tissue synaptic activity and in the intensity of calcium dynamics [6] [23] [24].
Three independent biological replicas were collected and analyzed on the Affymetrix rat 230.2 chip; for each replicas the expression of the gabazine-treated sample was then compared to the control-untreated sample and the probes/genes of the chip were arranged in ascending up-regulation/p-value score. The results of a GO enrichment analysis, performed considering the genes with an up-regulation value higher than 2, approximately corresponding to a p-value≤0.005, are presented in table 1. The complete list of probes/genes data used in the present and in the subsequent analysis is provided in the table A in file S1.
Table 1. Table presenting the principal families of GO terms found to be enriched in the microarray-based analysis of gabazine treatment.
| GO term | num. genes | p-value |
| regulation of synaptic transmission | 7 | 1.90·10−4 |
| nucleus | 14 | 1.20·10−2 |
| regulation of transcription | 19 | 8.60·10−4 |
| positive regulation of transcription | 15 | 3.70·10−7 |
| learning or memory | 9 | 3.40·10−7 |
| feeding behavior | 5 | 6.40·10−7 |
| regulation of calcium ion transport | 5 | 3.60·10−4 |
| transmembrane protein | 6 | 1.20·10−1 |
| regulation of apoptosis | 11 | 2.20·10−3 |
| negative regulation of apoptosis | 6 | 2.80·10−2 |
| positive regulation of cell death | 6 | 2.80·10−2 |
The first column specifies the GO term, the second column contains the number of genes associated with the GO term and the last column presents the p-value score of the enrichment.
The sudden increase of synaptic activity induces the up-regulation of a variety of genes involved in several cellular processes and localized into different cellular compartments. A significant component (p-value≤1.90·10−4, modified Fisher Exact P-value) of the up-regulated genes, including for instance the effectors Arc and Rgs2, is involved in the regulation of synaptic transmission itself, by acting directly in axon terminals and dendritic spines. Another group of genes (p-value≤1.90·10−5) consists in a large pool of transcription factors, like for example Cfos and Klf4, that is responsible for driving the second wave of cellular responses, possibly related to longer lasting changes in neuron metabolism, morphology and functions [7]. Interestingly, the same group of transcription factors results to be highly enriched in the positive regulation of transcription term (p-value≤3.70·10−7): this indicates that, despite the presence of transcriptional repressors, such as Icer and Nfil3, the longer lasting changes are mainly based on the activation of not-expressed genes rather than on the suppression of already expresses ones. A consistent (p-value≤2.20·10−3) component of genes is involved in the regulation of cell survival: interestingly, according to the GO, they appear to influence the survival in both a positive and a negative manner. However, it appears that GabT treatment induces a strong push (p-value≤1.7·10−2) towards growth, neurogenesis and neuritogenesis. Finally, it is worth mentioning that the MAPK signaling pathway as well as the small-gtpase family are confirmed as the most important mediators of the aforementioned processes (p-value≤2.9·10−2).
To verify the up-regulation values observed in the microarray assay, we selected a group of 33 genes among the highest up-regulated ones and we measured their expression level in gabazine vs. untreated samples by RT-PCR. These 33 transcripts correspond to the top-fifty up-regulated probes deprived of those pointing to “predicted” transcripts and deprived of those characterize by low values of mRNA abundance (i.e. intensity of microarray signal). The latter ones were excluded mainly because their low amounts of mRNA were causing the RT-PCR data to be excessively noisy. The final list of transcripts whose up-regulation was verified by RT-PCR is presented in Table 1, while the RT-PCR data is presented in table C in file S1.
As a next step, we wanted to validate the previous Gene Ontology analysis. The functions associated to the genes in the Gene Ontology database (www.geneontology.org) are often derived from bioinformatics predictions, such as inference from sequence ortology or from common expression patterns: these kind of predictions, although likely reliable, have not been verified experimentally. In order to assess the consistency of our GO analysis, we proceeded by creating a manually compiled “vocabulary” of gene functions for each of the genes belonging to the set confirmed by RT-PCT; this vocabulary was based on an extensive search in the literature and built by considering only the most reliable results. More precisely, we preferentially considered only functional evidences derived from hippocampal tissues such as organotypic slices, acute slices, dissociated cultures or in vivo conditions. When hippocampus-based studies were lacking, we collected proofs from other types of nervous tissues, such as cortical neurons, dorsal root ganglion cells or glioma tissue. The complete list of gene/protein roles extracted from the literature is available in file S2, while a brief summary of them is available in Table 2.
Table 2. Table containing the list of 33 genes whose up-regulation was confirmed by RT-PCR.
| GENE NAME | ROLE | FUNCTION(S) | REFERENCES |
| Arc | EF | NEUROGENESIS/SURVIVAL/anti-GROWTH/Reg.Syn.trasmission | [25] [26] [27] [28] [29] [30] [31] [32] [33] [34] |
| Atf3 | TF | SURVIVAL | [1] [35] [36] |
| Bdnf | EF | NEUROGENESIS/SURVIVAL/GROWTH/Pos.Reg.Syn.transmission | [37] [38] [39] [40] [41] [42] [43] [44] [45] [46] |
| Btg2 | TF | NEUROGENESIS/SURVIVAL | [1] [3] [47] [48] [49] |
| Cfos | TF | GROWTH/Pos.Reg.Syn.transmission | [18] [50] [51] [52] [53] [54] [55] [56] [57] |
| Cited2 | TF | [58] [59] [60] | |
| Crem/Icer | TF | anti-SURVIVAL/Neg.Reg.Syn.transmission | [61] [62] [63] [64] [65] [66] [67] |
| Cyr61 | EF | GROWTH | [68] [69] [70] |
| Dusp1 | EF | erk inactivation/anti-GROWTH | [17] [71] [72] |
| Dusp5 | EF | erk inactivation | [73] [74] |
| Dusp6 | EF | erk inactivation | [75] [76] |
| Egr1 | TF | Pos.Reg.Syn.transmission | [77] [78] [79] [80] [81] [82] |
| Egr2 | TF | [83] [84] | |
| Egr3 | TF | Pos.Reg.Syn.transmission | [82] [85] [86] [87] |
| Egr4 | TF | [88] [89] [90] | |
| Gadd45b | EF | NEUROGENESIS/SURVIVAL/GROWTH | [1] [2] |
| Homer1a | EF | Reg.Syn.trasmission/GROWTH | [91] [92] [93] [94] [95] |
| Irs2 | EF | GROWTH/Reg.Syn.trasmission | [96] [97] [98] |
| Klf4 | TF | anti-GROWTH | [99] [100] [101] [102] [103] |
| Mapk10 | EF | SURVIVAL/GROWTH/Reg. Syn. Transmission | [104] [105] [106] [107] [108] [109] [110] [111] [112] [113] [114] |
| Nfil3 | TF | SURVIVAL | [115] [116] [117] |
| Nptx2 | EF | Neg.Reg.Syn.Transmission | [118] [119] [120] [121] |
| Npy1r | EF | NEUROGENESIS/Pos.Reg.Syn.trasmission | [122] [123] [124] [125] [126] |
| Nr4a1 | TF | SURVIVAL/anti-GROWTH | [1] [127] [128] |
| Nr4a2 | TF | SURVIVAL/Reg.Syn.transmission | [81] [128] [129] [130] [131] [132] |
| Nr4a3 | TF | SURVIVAL/GROWTH | [19] [128] [133] [134] |
| NTF3 | EF | GROWTH/SURVIVAL | [135] [136] [137] [138] [139] [140] [141] [142] |
| Pcdh8 | EF | Neg.Reg.Syn.Transmission | [143] [144] |
| Plk2 | EF | Neg.Reg.Syn.Transmission | [24] [145] |
| Ptgs2 | EF | SURVIVAL/Pos.Reg.Syn.Transmission | [146] [147] [148] [149] [150] [151] |
| Rasl11b | EF | [152] | |
| Rgs2 | EF | Neg.Reg.Syn.Transmission | [153] [154] [155] [156] |
| Srf | TF | Reg.Syn. Transmission | [12] [69] [72] [157] [158] [159] [160] [161] |
The first column contains the official gene symbol, the second column assigns the role of EF, effector, or TF, transcription factor, while the last column summarizes the validated gene functions, which always refer to tissues or experimental conditions coherent with the present work. For the complete details please refer to the survey presented in the supplementary file S2.
Since it is well established that certain genes/proteins listed in Table 2 can exert different roles according to the cellular context [1] [110] [162] (see file S2 for more details), we also tried to avoid considering functional results obtained from excessive pathological stimuli, which could alter the physiological native role of a gene/protein. For instance in [60] the neurons were treated with Camptothecin to cause DNA damage and the Cbp/p300-interacting transactivator 2, also known as Cited2, was related to the activation of apoptosis: we found these circumstances too dissimilar from the gabazine-treatment of the present work and therefore we decided not to consider this as a functional evidence. The Fig. 1 represents the distribution of the literature-extrapolated functions with respect to the cellular compartments. The similarity between the functions/processes highlighted by GO and those derived from selected literature appears to be good, nonetheless we can make at least two considerations:
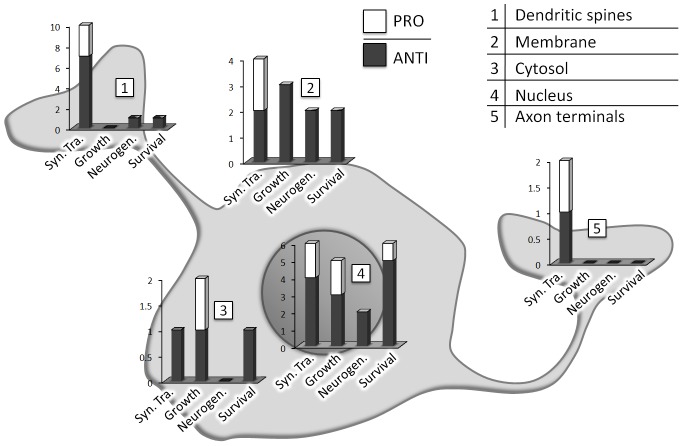
Figure 1. Cellular localization of the functions attributable to the genes up-regulated by GABAr blockage.
Gene functions, extracted from findings reported in the literature, are categorized in four main groups, indicated by the terms: Syn.Tra, regulation of synaptic transmission, growth, regulation of growth, neurogen., regulation of hippocampal neurogenesis, survival, regulation of survival. Each gene is counted as PRO when it positively regulates these processes, while it is counted as ANTI when it negatively affects these processes. The cellular compartment of action for each protein(gene) is chosen according to the indications reported in the literature. For example, it has been shown that the pentraxin Nptx2 (neuronal activity–regulated pentraxin) is localized in the excitatory synapses, where it exerts a homeostatic effect by recruiting AMPAr, AMPA receptors, at excitatory projections onto gabaergic interneurons [118]–[121].
In regard to the equilibrium of pro and anti-survival genes emerged from the GO, we must point out that the resulting situation from the literature analysis is quite different: instead of an equilibrium, we can actually notice a substantial shift towards pro-survival genes in response to gabazine. This difference arises from a different attribution of functions to the genes Nr4a1, Ptgs2, Arc, Atf3, Gadd45β and Nfil3. More precisely, all of these genes have proven, in the past years, to consistently promote neuron survival by protecting them from various oxidative, genotoxic and exitotoxic stresses; see file S2 for a complete review. (In short, we can confirm that a strong neuroprotective shield is induced by the synaptic activity associated with GABA-Ar blockage.)
Fig. 1 depicts more clearly how the effector early genes induced by the GABA-A blockade are mainly involved in the regulation of synaptic transmission and are localized in the synaptic terminals. Vice versa, those genes with growth, survival and neurogenesis promoting effects are mainly acting in the nucleus as transcription factors, thus their effects will realize only in conjunction with the subsequent wave of up-regulated genes.
Gene expression time course
To gain better insights into the mechanisms of the transcriptional response to GabT we decided to investigate whether the up-regulation value found after 1.5 hours (for the genes induced by gabazine) is reached following different temporal dynamics or, on the contrary, all genes share the same induction pattern.
Previous studies [50] [163] [164] have already suggested that, following episodes of synaptic activity or during synaptic plasticity processes, the induced immediate-early-genes (IEGs) are characterized by different up-regulation dynamics: nonetheless, the time-course data collected so far in the literature is mainly obtained by microarray analysis, such as [7] [165], and not by a reliable and accurate RT-PCR analysis: more precisely, the information currently available is limited to time-courses with low temporal resolution, i.e. a few time points, and/or concerning a reduced number of genes, such as [15] [18] [19]. In all of these cases the time-course measurement was not the main aim of the paper, but it was rather an instrument to verify the effects of certain blockers/conditions, therefore a particularly high temporal resolution was simply not needed.
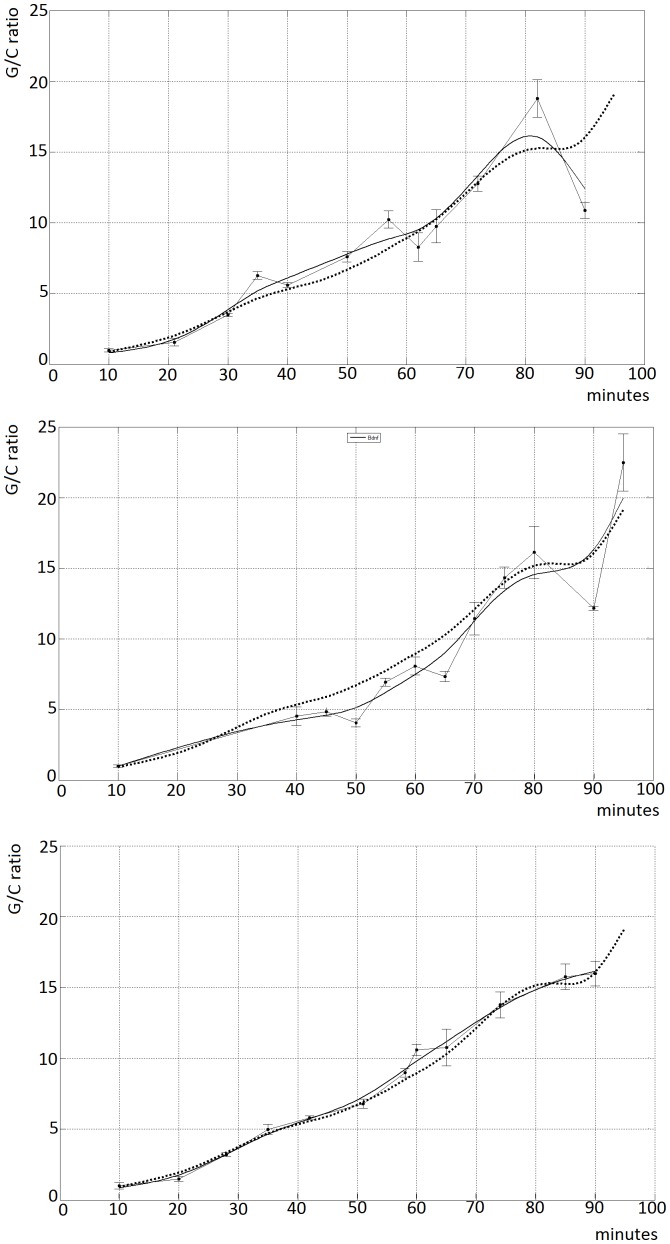
The rat organotypic hippocampal cultures were subjected to a 20 µM gabazine treatment and the total Rna was gathered at 12 different time points spanning from 10 minutes to 95 minutes, with an average inter-sample time (sampling period) of 10 minutes. The procedure was eventually repeated three times, each time with a different twin rats couple, in order to obtain three independent replicas of the time-course, and RT-PCR was then performed for every gene in order to measure the up-regulation values at the different time points. The genes included in the time course analysis are those presented in Table 2. The time points of each replicas were then interpolated with a smoothing spline in order to emphasize the major trend underlying the up-regulation process; afterwards, the three interpolations derived from the replicas were combined into an average one, which was considered as the reference trend in all of the subsequent analysis. As an example, the resulting time course for the Bdnf gene (exon IV ) is shown in Fig. 2, together with the original and interpolated results for each replicas.
Figure 2. Bdnf time course: Graphs representing the three independent replicas of the Bdnf mRna time course.
Time (minutes) on the x-axis, ratio Gabazine/Control on the y-axis. The error-bar plot refers directly to the RT-PCR data. In each of the sub-pictures, it is presented the trend resulting from the single biological replica of the RT-PCR time-course, which is superimposed to the averaged one, calculated as the mean of the three replicates. Thanks to this representation, it is possible to directly compare the single measures (biological replicates) to the averaged one, which eventually was the one used for all of the subsequent clustering analysis. The neurotrophin Bdnf, one of the master regulators of learning and memory, will prove in the end to be up-regulated according to a pattern which is representative for almost 50% of the genes of the set under study.
The first step of analysis that we carried out was a clustering of the temporal data, aimed to unveil the existence of distinct temporal patterns. Given that the measured time series are highly non stationary, we decided to discard correlation-based methods in favor of a k-means clustering algorithm based on Euclidean-distance; after a preliminary normalization, which reduced all the expression values of each gene to the interval [0∶1], the Euclidean-distance method proved to be able to correctly group together genes sharing a similar temporal pattern, regardless the absolute values of up-regulation. This methodology is the same applied in [164].
The main drawback of the k-means algorithm is the necessity to manually set k, i.e. the number of desired clusters [166]. The ability of the algorithm to distinguish among potential different temporal dynamics increases as k increases, but, on the contrary, the Z-score of the grouping outcome becomes less significant at higher k values, which means that a random grouping would have produced similar results, as illustrated in Fig. 3A.
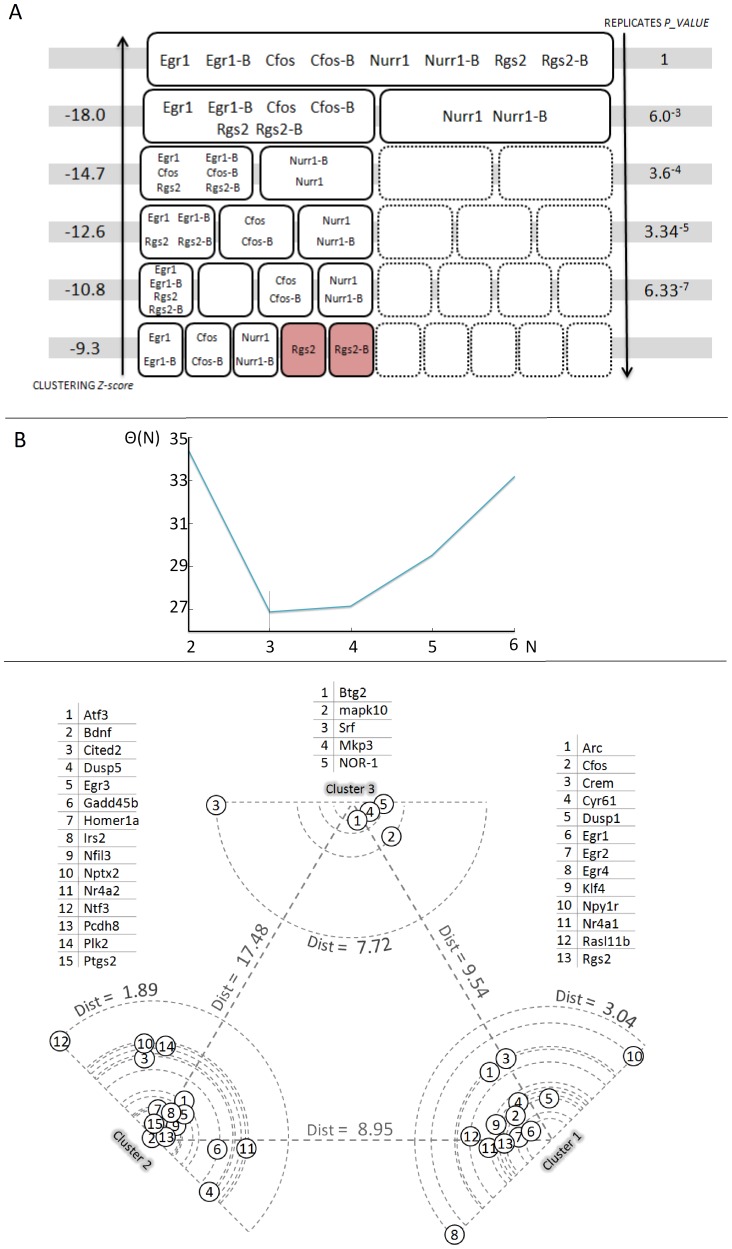
Figure 3. Analysis of the clustering quality for the time course data.
A) Outcome of the clustering algorithm, with progressive increase in the number of clusters k: the picture represents, at each different k, the grouping of the 4 couples of alternative primers pointing to the same gene. For k = 2,4,6,8 the alternative primers were correctly grouped together. The “replicas p-value”, on the right, indicates the statistical consistency of the alternative primer grouping, which reaches it maximum value when the algorithm is forced to split the 33 genes into 8 different clusters. On the left, the Z-value of the global clustering, indicating the consistency of the temporal dynamics discrimination. B) Outcome of the algorithm aimed at determining the optimal value for k. The number of clusters N is plotted against a function Θ(N): the minimum of Θ(N), i.e. N = 3, coincides with the optimal value for k. See Materials and Methods section for further details. C) Visual representation, with k = 3, of the distances between trajectories and cluster centroids for all the 33 genes. For each cluster, the genes are disposed at increasing distances from the centroid, proportionally to their normalized Euclidean distances. The distance of the farthest gene is indicated in the proximity of the outer circle. The orientation of the genes reflects the proximity to the remaining two clusters. The distances between the cluster centroids are also indicated.
To further test the consistency of the clustering procedure, we designed four new control primers for the genes Egr1, Cfos, Rgs2 and Nurr1: these alternative primers point to different exons and different exon-exon junctions with respect to the original ones. With k = 2 the control primers were correctly grouped together with their counterparts, as highlighted in figure 3. Most importantly, even at higher fragmentation levels, with k = 4, k = 6 and k = 8, the control primers remain associated to the proper original ones: the probability that this correct grouping might be due to chance is p = 6.33·10−7 when k = 8.
We decided to use the approach described in [167] to determine the optimal value for k in a unsupervised manner; the method is based on the minimization of a function Θ(N), where N is the number of clusters. Intuitively, the minimum of Θ(N) coincides with the number of clusters where the addition of a further one does not reduce significantly the average intra-cluster distance. More details about this approach are presented in the Materials and Methods section. The final result, presented in Fig. 3B, indicates that k = 3 is the optimal value for the cluster number. In Fig. 3C the outcome of the clusterization process with k = 3 is represented in a two-dimensional plane.
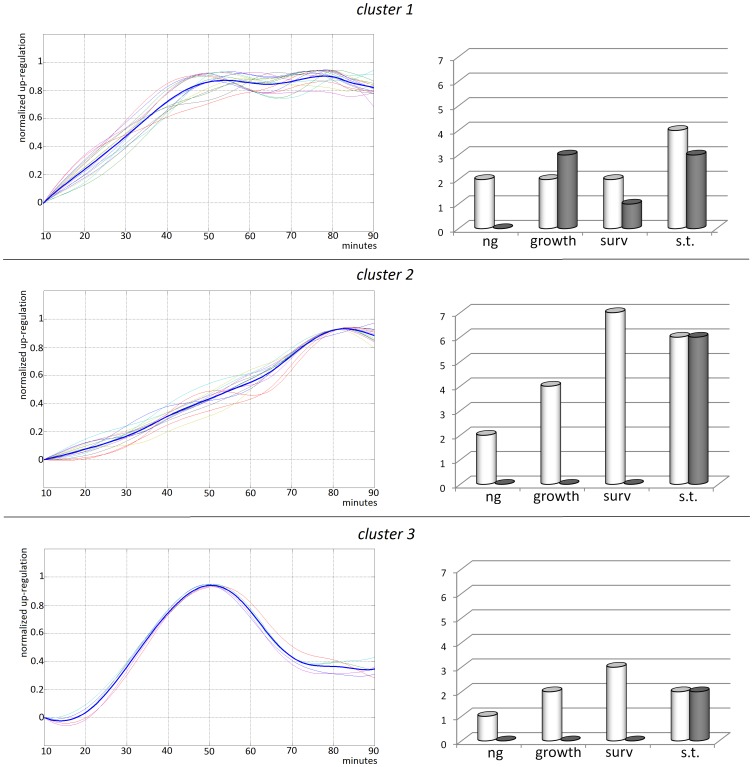
Cluster 1, which comprises genes such as Arc, cFos and Klf4, is characterized by a fast rise in the expression values, which peak at about 50 minutes and subsequently remains steady till the end of the measurement. The Arc gene was reported in several works to be rapidly induced by episodes of synaptic activity, with a peak within the first 60 minutes. Thus, for the Arc gene, our result is coherent with [168], [169] and [170]; furthermore, it extends the results to the other 12 IEGs characterized with the same dynamic of Arc, thus suggesting the existence of a common regulation system responsible for the induction of these faster-rising IEGs. Cluster 2, which comprises genes such as Bdnf, Irs2 and Homer1a, is characterized instead by a slower but constant increase, almost linear up to 90 minutes. The differential dynamics characterizing the Bdnf gene (cluster 2) with respect to the Cfos and Egr1 genes (cluster 1) are coherent with a previous study [15] of Schaffer-collateral HFS-induced LTP: again, here we extend the results to other 25 IEGs which result to be similar to Cfos/Egr1 or to Bdnf dynamics. Besides, the longer lasting duration of Cited2 (cluster 2) mRna up-regulation with respect to the faster and shorter up-regulation timings of Cfos (cluster 1) and NOR-1 (cluster 3) also recalls the results obtained in [19] with an ECS stimulation of the Dentate gyrus. The last cluster, which is smaller than the previous ones and comprises genes such as NOR-1 and Btg2, presents a marked peak which is concurrent to cluster 1 peak, but that is successively followed by a pronounced decrease of the expression value.
Relationship between clustering and function
Since previous studies have already supported the notion that temporally clustered genes are likely involved in the same biological functions [165] [171], we next wanted to determine whether it was possible to relate the different temporal profiles previously extracted with particular inherent functions. Therefore, for each temporal cluster of gene expression we performed an enrichment analysis of functional evidence collected in the manually compiled vocabulary, introduced in the “microarray analysis” section.
The recent developments in the study of hippocampal plasticity have consolidated the idea that episodes of intense physiological synaptic activity strongly promote neurogenesis [3] [34] [37], growth [2] [97] [136] and survival [1] [36] [150]. Our data confirm the up-regulation of numerous genes endowed with these properties already in the early phase (10–90 minutes) of transcriptional regulation, see Fig. 4, indicating that a strong neuroprotective shield is quickly activated by synaptic activity in organotypic cultures, together with an increase in the Dentate Granule Cells neurogenesis and an increase in the growth rate of neurons and synaptic connections. However, also genes with negative effects on growth (namely Icer, Klf4, Nr4a1 and Mkp-1) are induced in association with the mentioned majority of positive regulators, see Fig. 4. Interestingly, these four genes were all grouped in the cluster 1 temporal pattern, thus making the cluster 1 significantly enriched with anti-growth properties (p-value≤0.017, Fisher's exact test). Vice versa, the cluster 2 comprises only genes providing a positive effect on growth.
Figure 4. Graphs of time-course data and their associated functions.
The bold line, representing the average temporal pattern of each cluster, is superimposed to the patterns of the single genes. The histograms depicts the amounts of positive (white bars) and negative (black bars) regulators of the indicated processes for each cluster: n.g., regulation of hippocampal neurogenesis, growth, regulation of growth, surv., regulation of survival, S.T., regulation of synaptic transmission.
In the past decade the mechanisms involved in the homeostatic regulation of synaptic strength have emerged as a fundamental complement to Hebbian plasticity [172] [173] [174]. In the present work we report that in rat organotypic cultures, following chronic blockade of GABA-Ar, many genes involved in homeostatic-scaling (weakening) processes, namely Narp/Nptx2, Arc, rgs2, arcadlin, plk2, Homer1a, Icer, Dusp1, Dusp3, Dusp6, of which the single contributes to plasticity have been partially unveiled [24] [33] [67] [91] [121] [144] [154], are induced in concert already in the first minutes of synaptic activity, thus suggesting the existence of a sensitive and fast feedback mechanism that is activated almost contextually to the perturbation. As illustrated in Fig. 4, the homeostatic genes are equally spread among the three clusters (p-value≤0.43, Fisher's exact test) indicating that there is no particular relationship between the homeostatic function and the up-regulation timings in the early phase (0–90 min.) of the hippocampal response to perturbation. Interestingly, we noticed that the homeostatic genes are tightly associated, in every cluster, with genes exerting the opposite function, i.e. the potentiation of synaptic transmission, as depicted in Fig. 4. Therefore, differently from the survival and growth functions, for the regulation of synaptic transmission we observe a functional equilibrium between homeostatic-plasticity (weakening) genes and Hebbian-plasticity (potentiation) genes.
Another crucial step of the homeostatic response is the re-establishment of the basal level of active MAPKs [15] [111]; this process is carried out mainly by means of a negative feedback loop involving the MAPKs themselves, together with the Dusp family of phosphatases [72] [76] [134]. Here we report that Dusp1, Dusp5 and Dusp6 are induced together by GabT, but with different temporal patterns, since they are grouped into different clusters, see Fig. 4. This result, which is coherent with previous studies [175] [176], indicates that each of the DUSPs is dynamically tied to a different group of genes: in this way, each cluster of the induced genes is synchronized with a relative homeostatic feedback to the MAPKs.
The peculiar distribution of the Dusp family members, as well as the in-cluster balance between homeostatic and Hebbian plasticity genes, led us to notice that, concerning the regulation of synaptic transmission, genes endowed with different, but at the same time complementary/counterbalancing, functions seem to be bound together into the same temporal dynamics in order to favor global robustness of the system: indeed in this case the dysregulation of a pathway caused by a pathological state would not create excessive imbalances since the inner genes compensate each other. It is interesting to point out that the present observation about global stability recalls the conclusions of previous work [177], in which a bioinformatic analysis of the CA1 hippocampal intracellular pathways [178] revealed the existence of robustness, stability and adaptability properties.
Relationship between clustering and regulation
In order to investigate the possible relationships between the different temporal patterns of gene induction and the regulators of gene transcription, we performed an accurate and extensive literature research aimed to recreate the complete network of pathways involved in the regulation of hippocampal gene transcription: the complete survey is available in file S2. By a cross comparison between pathways and transcription factors on one side and time-course patterns on the other side, it emerged that cluster 1, which was characterized by a fast increase in the expression values followed by a flat/stationary state, is particularly enriched in SRF, serum response factor, dependent regulations (p-value≤0.02, Fisher's exact test). On the contrary, the cluster 2 does not present any SRF dependent regulation (p - value≤0.05 ). This data indicates that the SRF dependent regulation is consistently biased towards the cluster 1, which is the cluster of genes such as Arc, Cfos, Cyr61, Egr1 and Egr2, all of which have shown to be regulated by Serum Response Factor in various plasticity models [69] [179] [180] [181] [182] [183].
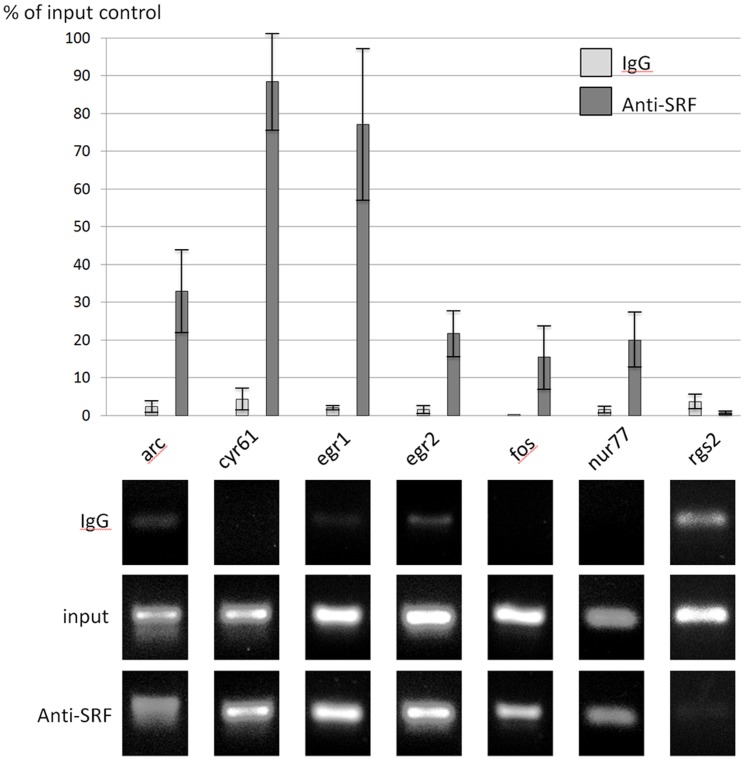
To assess the validity of the above mentioned SRF regulatory evidence for the genes Arc, Cfos, Cyr61, Egr1 and Egr2 in our model of hippocampal plasticity, i.e. GabT of organotypic cultures, we performed chip (chromatin immunoprecipitation) experiments to detect SRF binding levels in their promoters during GabT. Besides, we carried out an in silico analysis of the promoters of the remaining 8 genes belonging to the same cluster in order to detect other possible active CArG boxes, the DNA sequence motif CC[A/T]6GG that has a high affinity for SRF. As a result, CArG boxes conserved among humans, rats and mice were found in the upstream region of RGS2 and NR4A1 genes, respectively at −5 kb and −123-111; therefore, those genes were included in the chip experiment together with the previous ones. The results of chip, presented in Fig. 5, show that a strong SRF signal was detected in Arc, Cfos, Cyr61, Egr1 and Egr2 and Nr4a1 while no significant signal was found for RGS2, indicating that the latter gene is likely not to be regulated by SRF in our plasticity model.
Figure 5. Analysis of SRF binding sites by chromatin immunoprecipitation.
Chromatin fragments of hippocampal organotypic cultures were immunoprecipitated with anti-SRF antibody. A) Immunoprecipitation levels normalized to input control: the s.e.m. is calculated over three different replicas. B) Immunoprecipitation of each promoter region, together with input control and IgG antibody, was amplified by PCR. Each sample is derived from three independent replicas.
Discussion
The present article identifies three different dynamical patterns in the early-phase (10–90 min) of the transcriptional response induced by GabT of organotypic hippocampal cultures and provides novel information about the role of Serum Response Factor. The blockage of GABA-A ionotropic channels by means of gabazine/bicuculline/PTX is a widespread [1] [145] [184] [185] model of plasticity where the increased synaptic activity triggered by GabT leads to the up-regulation of a plethora of activity-dependent genes. While the electrophysiology of GABA-A antagonists in organotypic hippocampal cultures has been extensively studied [6] [24], the relative variations in the transcriptome have been so far conducted in dissociated cultures [1] [5] [7] [186]. This last aspect prompted us to develop a preliminary assessment with a microarray-based transcriptome profiling.
The Gene Ontology analysis of microarray data revealed that the major functions of the 346 genes up-regulated by GabT (p-value≤0.005) are related to the regulation of synaptic transmission, calcium ions transport, transcription, apoptosis, feeding behavior, learning and memory. With respect to apoptosis regulation, the GO analysis further indicates that both positive and negative regulators of survival are up-regulated in organotypic cultures and therefore the general effects of GabT on cell fate seems not to be predictable. Nevertheless, a manual annotation of the gene functions actually revealed that GabT promotes a push towards survival, neurogenesis and neuroprotection, confirming the results obtained in dissociated cultures [1] [185] and extending them to the case of hippocampal organotypic cultures.
To further investigate the dynamics underlying the early-phase of the regulation of activity-dependent genes, we developed the quantification of a high temporal resolution time course, ranging from 10 to 90 minutes, with an average inter-sample time of 10 minutes. The trajectories of the 33 genes included in the time-course were subjected to a unsupervised k-means clustering: the unsupervised clustering identified three different dynamical patterns, as depicted in Fig. 3 and Fig. 4. By crossing the cluster grouping with the gene functions listed in the manually compiled vocabulary (see file S2) we found that the group of genes characterized by a fast rise to a plateau value (cluster 1) seems to be significantly (p-value<0.05) provided with anti-growth and anti-survival properties. Since this cluster is characterized by the fastest response, peaking already at 50 minutes, this data suggests that a rapid activation of negative regulators of growth, possibly involved in the initial disassembly of existent structures, is subsequently followed by an induction of growth promoting genes (cluster 2, slow up-regulation).
Besides, we also found that cluster 1 is also enriched in SRF, serum response factor, dependent regulations (p-value≤0.02, Fisher's exact test). Interestingly, in a previous work [187] with dissociated cultures we showed that the genes Nr4a1, Arc, Egr1, Egr2 and Egr3, which belong, in the present paper, to cluster 1 (with the exception of Egr3), are characterized by a marked dependence on MAPK-dependent regulation when compared to Bdnf and Homer1a, which instead here belong to cluster 2. Moreover, a strong dependence Dusp1 and Fos, which again belong to cluster 1, on MAPK regulation was previously emphasized in rat neuroendocrine cells [188] [189]. These data suggest that the cluster is particularly dependent on SRF/MAPK and motivated us to investigate whether the aforementioned SRF dependent regulations, which were extrapolated from the literature and derived from different experimental conditions, are still valid in the case of GabT of organotypic cultures.
To this end, we performed chip, chromatin immunoprecipitation, for detecting SRF binding levels during GabT and we found that Cyr61, Egr1, Egr2, Fos and Arc present a significant SRF binding signal. While genes Fos and Egr1 have already been reported to be regulated by SRF in hippocampal organotypic cultures [179], ours is the first report for genes Cyr61, Arc and Egr2.
To complete the survey of working CArG boxes in cluster 1, we analyzed the sequences upstream of TSS for the remaining genes and we found conserved CArG boxes also upstream of Rgs2 and Nr4a1. Eventually, the chip assay revealed that the Nr4a1 CArG box presents a significant SRF signal while no signal was found for Rgs2. This result is interesting in particular for Nr4a1 gene, for which the functionality of the aforementioned CArG box has so far provided motley evidences. In fact, in serum stimulation of NIH-3T3 fibroblasts [190] and platelet-derived growth factor (PDGF) stimulation of T98G-glioblastoma [181] the CArG box has proven to be functional but in hippocampal neuronal cultures [191] [192], cerebellar cortex [127] and in vivo [193] conditions general findings are in favor of a CREB and MEF2 determinant role. Therefore, our latter result suggests that, in organotypic cultures, SRF may play a role in the regulation of Nr4a1 gene during the intense synaptic activity triggered by GabT.
In conclusion, this study provides novel insights into the early dynamics of transcriptional regulation in a plasticity model, showing how a large group of co-expressed activity-dependent genes is characterized by consistently different patterns of induction in the first 90 minutes of tissue response and linking these patterns to different inherent functions and regulatory mechanisms. We believe that unveiling the finest tuning in the regulatory dynamics of plasticity is the key step to gain a more quantitative awareness of the phenomenon.
Materials and Methods
Ethics Statement
Rat hippocampi were dissected from Wistar rats (P4–P5), in accordance with the regulations of the Italian Animal Welfare Act, and the procedure was approved by the local authority veterinary service (Dr. R Zucca). Every possible effort was taken in order to minimize both the number and the suffering of used animals. The experiments were carried out in accordance with the European Communities Council Directive of 24 November 1986 (86/609/EEC) and formal approval for experimental procedures was provided by the Ministry of Health(protocol 13/97–A).
Tissue, pharmacology and Rna extraction
Rat hippocampi were dissected from Wistar rats (P4–P5). Organotypic cultures were prepared following the roller tube method [194]. Gabazine was purchased from Tocris (Bristol, UK). Gabazine treatment (GabT) for microarray samples consisted in treating the cultures for 90 min with 20 µM of gabazine, a specific GABA-A receptor antagonist [195]. Gabazine treatment (GabT) for time course samples consisted in treating the cultures with 20 µM of gabazine for a variable time with time samples ranging from 10 minutes to 90 minutes. The total RNA for the microarray samples and the time-course samples was extracted using the TRIzol reagent (Sigma, Milano, Italy) according to the manufacturer's instructions followed by a DNase I (Invitrogen, Carlsbad, California, USA) treatment to remove any genomic DNA contamination. The total RNA was further purified using RNeasy Mini Kit Column (Qiagen, Valencia, CA) and subsequently quantified by ND-1000 Nanodrop spectrophotometer (Agilent Technologies, Palo Alto, CA).
Analysis of Microarray data and P-value calculation
For the microarray data, three biological replicas were collected at 90 min of GabT and Standard Affymetrix protocols were applied for amplification and hybridization. Gene profiling was carried out with the Affymetrix RAT2302 GeneChip containing 31099 probes, corresponding to 14181 probes with a gene symbol. Low level analysis was performed using an Robust Multi-array Average (RMA) algorithm [196] directly on the scanned images.
Data were organized in matrices “m×n” (m, number of genes; n, number of replicas). Two samples were considered: an untreated culture (Cij: i = 1,..,n j = 1,..,m), a culture treated with gabazine (Gij). Data were analyzed by considering log2 changes of gene expression in each replicas against its own untreated control, that is, log2 (Gij/Cij). Thus, from the microarray data we obtained an “m×n” ratio-matrix for each treatment. Considering the three replicas as independent variables, this matrix was treated as a multivariate variable in three dimensions. We derived the empirical cumulative distribution function with upper and lower bounds of the multivariate variable, using the Kaplan–Meier estimator (Kaplan and Meier, 1958) so to assign a p-value to all the genes and select the most significant ones. The microarray data can be found in the GEO database, accession number: GSE46864.
GO enrichment analysis
GO enrichment analysis for microarray data was performed with Gene David [197] (http://david.abcc.ncifcrf.gov/). GO analysis for the manually annotated vocabulary was performed according to the following formulas:
The probability to have exactly 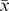 genes characterized with a certain “GO term” (for example, “SRF regulation” or “positive regulation of synaptic transmission”), in a cluster of dimension n, is
genes characterized with a certain “GO term” (for example, “SRF regulation” or “positive regulation of synaptic transmission”), in a cluster of dimension n, is
Where N is the total number of genes(elements), n is the dimension of the cluster, k is the total number of genes(elements) which present the “GO term” under consideration. The cumulative probability to have an amount of terms equal or higher than 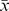 , in a cluster of dimension n, is
, in a cluster of dimension n, is
Quantitative RT-PCR and time-course analysis
For the time course experiment, the expression level of the target mRNA was quantified bt RT-PCR. RNA (250 ng) was reverse-transcribed using SuperScript II reverse transcriptase and random hexamer (Invitrogen). qRT-PCR was performed using iQ SYBR Green supermix (Bio-Rad, Munich, Germany) and the iQ5 LightCycler (Bio-Rad). Gene specific primers were designed using Primer3 [198](http://frodo.wi.mit.edu/). The thermal cycling conditions comprised 3 min at 95C, and 45 cycles of 10 sec for denaturation at 95C and 45 sec for annealing and extension at 58C. The expression level of the target mRNA was normalized to the relative ratio of the expression of Gapdh mRNA. Fold change calculations were made between treated and untreated samples at each time point using the DDCT method. Three organotypic cultures were used for each sample. The 36 primers used for the time course analysis are provided in table B in file S1.
The resulting time-course data-set consists of three biological replicas, each one containing 12 time points ranging from 10 m to 90 m. Each raw data time-course replicas obtained from RT-PCR data was independently fitted with a smoothing spline (Matlab environment) and normalized to the [0∶1] interval. Subsequently, the three replicas were jointed together and analyzed via a k-means clustering, based on Euclidean distance (same method as [[164]]). To identify the optimal number of clusters we adopted the approach proposed in [167]. Briefly, a function
is computed at every k, i.e. cluster number. N is the number of clusters, dist(ci) is the intra-cluster distance, i.e. the scaled average squared distance between shapes in the cluster ci and α is a parameter controlling the grain of the clustering. The minimum of the function Θ(N) corresponds to the optimal number of clusters.
The enrichment score for the transcription factors regulatory evidences was computed using the same approach described in one of the previous section, “Analysis of Microarray data”.
Identification of upstream sequences and transcription factor binding sites
The 10 k-bp upstream regions for mouse, rat and human of the cluster 1 genes were extracted from mapviewer (http://www.ncbi.nlm.nih.gov/mapview/). To identify the putative transcription factor binding sites within each upstream sequence, a preliminary verification of the conserved regions among mouse, rat and human was performed by aligning the sequences with blast-bl2seq (http://blast.ncbi.nlm.nih.gov/Blast.cgi), using a word letter size 16. To refine the blast results a further analysis was carried out with Evoprinter (http://evoprinter.ninds.nih.gov/) [199]. Finally, conserved domains were analyzed with Jaspar [200] (http://jaspar.cgb.ki.se/), using the MA0083.1 SRF binding matrix with a threshold score of 0.8.
Chromatin immunoprecipitation
The chromatin immunoprecipitation assay was performed using the MAGnify Chromatin Immunoprecipitation System (Invitrogen, Catalog Number49-2024) according to the manufacturer's instructions with slight modifications. Briefly, organotypic cultures (ten cultures per condition) were cross-linked at room temperature, immediately after the GabT, using a PBS solution with formaldehyde 1%. Shearing was performed with a MSE Soniprep 150 (7 pulses of 5 seconds) to yield an average length of 300 bp. Samples were immunoprecipitated with 10 ug of anti-SRF antibody (Santa Cruz Biotechnology, Heidelberg, Germany, cat.no sc-335x) and with 1 ug of anti-rabbit IgG negative control antibody. Promoter specific primers were used for amplification:
Nr4a1 Forward: 5′-TTAAGAGGTGGGTCGGGTTC-3′
Reverse: 5′-GCAATCCTTCTCGCACACTA-3′
C-fos: Forward: 5′-CTCGCCTTCTCTGCCTTTC-3′
Reverse: 5′-GTAGGATTTCGGGGATGGTT-3′
Egr1: Forward: 5′-TGGGAGGTCTTCACGTCACT-3′
Reverse: 5′-GAATCGGCCTCTATTTCAAGG-3′
Egr2: Forward: 5′-ATGTGACCGGCAAAAGCTAC-3′
Reverse: 5′-AATGAATCGCTGCTCTCTCAG-3′
Cyr61: Forward: 5′-TCAAGAATGCCTTGTGGTTG-3′
Reverse: 5′-ACGGGGTAGAAGGAGGTGAT-3′
Rgs2: Forward: 5′-TGCCACCCCAGTAGTTACG-3′
Reverse: 5′-TTTGCCGAGAGATGAACAGA-3′
Arc: Forward: 5′-GTGGGGAAGCTCCTTGCT-3′
Reverse: 5′-CCAGTTAGAGGGCGGTGTT-3′
Supporting Information
Supplementary tables. - Supplementary Table A: The complete list of microarray probes/genes. - Supplementary Table B: List of primers used for the time course analysis. - Supplementary Table C: RT-PCR validation data.
(DOCX)
Supplementary notes.
(DOCX)
Funding Statement
The authors acknowledge the financial support of the SI-CODE project of the Future and Emerging Technologies (FET) programme within the Seventh Framework Programme for Research of the European Commission, under FET-Open grant number: FP7-284553.
References
- 1. Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, et al. (2009) Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, et al. (2009) Neuronal activity-induced gadd45b promotes epigenetic dna demethylation and adult neurogenesis. Science 323: 1074–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farioli-Vecchioli S, Saraulli D, Costanzi M, Leonardi L, Cinà I, et al. (2009) Impaired terminal differentiation of hippocampal granule neurons and defective contextual memory in pc3/tis21 knockout mice. PLoS One 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bading H, Ginty DD, Greenberg ME (1993) Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science 260: 181–186. [DOI] [PubMed] [Google Scholar]
- 5. Pinato G, Pegoraro S, Iacono G, Ruaro ME, Torre V (2009) Calcium control of gene regulation in rat hippocampal neuronal cultures. J Cell Physiol 220: 727–747. [DOI] [PubMed] [Google Scholar]
- 6. Mohajerani MH, Cherubini E (2005) Spontaneous recurrent network activity in organotypic rat hippocampal slices. Eur J Neurosci 22: 107–118. [DOI] [PubMed] [Google Scholar]
- 7. Pegoraro S, Broccard FD, Ruaro ME, Bianchini D, Avossa D, et al. (2010) Sequential steps underlying neuronal plasticity induced by a transient exposure to gabazine. J Cell Physiol 222: 713–728. [DOI] [PubMed] [Google Scholar]
- 8. Raymond CR (2007) Ltp forms 1, 2 and 3: different mechanisms for the “long” in long-term potentiation. Trends Neurosci 30: 167–175. [DOI] [PubMed] [Google Scholar]
- 9. Augustine GJ, Santamaria F, Tanaka K (2003) Local calcium signaling in neurons. Neuron 40: 331–346. [DOI] [PubMed] [Google Scholar]
- 10. Raymond CR, Redman SJ (2006) Spatial segregation of neuronal calcium signals encodes different forms of ltp in rat hippocampus. J Physiol 570: 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pinato G, Pegoraro S, Visentini M, Ruaro ME, Torre V (2009) Elevation of somatic ca2+ upregulates genes nr4a1 and egr2, but not bdnf and arc. Neuroreport 20: 869–874. [DOI] [PubMed] [Google Scholar]
- 12. Greer PL, Greenberg ME (2008) From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59: 846–860. [DOI] [PubMed] [Google Scholar]
- 13. Tabuchi A (2008) Synaptic plasticity-regulated gene expression: a key event in the long-lasting changes of neuronal function. Biol Pharm Bull 31: 327–335. [DOI] [PubMed] [Google Scholar]
- 14. Kotaleski JH, Blackwell KT (2010) Modelling the molecular mechanisms of synaptic plasticity using systems biology approaches. Nat Rev Neurosci 11: 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Racaniello M, Cardinale A, Mollinari C, D'Antuono M, De Chiara G, et al. (2010) Phosphorylation changes of camkii, erk1/2, pkb/akt kinases and creb activation during early long-term potentiation at schaffer collateral-ca1 mouse hippocampal synapses. Neurochem Res 35: 239–246. [DOI] [PubMed] [Google Scholar]
- 16. Ha S, Redmond L (2008) Erk mediates activity dependent neuronal complexity via sustained activity and creb-mediated signaling. Dev Neurobiol 68: 1565–1579. [DOI] [PubMed] [Google Scholar]
- 17. Jeanneteau F, Deinhardt K, Miyoshi G, Bennett AM, Chao MV (2010) The map kinase phosphatase mkp-1 regulates bdnf-induced axon branching. Nat Neurosci 13: 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sokolova OO, Shtark MB, Lisachev PD, Pustyl'nyak VO, Pan IV (2009) Time course of expression of “early” genes during long-term posttetanic potentiation in rat hippocampal ca1 field. Bull Exp Biol Med 148: 416–418. [DOI] [PubMed] [Google Scholar]
- 19. Sun W, Choi SH, Park SK, Kim SJ, Noh MR, et al. (2007) Identification and characterization of novel activity-dependent transcription factors in rat cortical neurons. J Neurochem 100: 269–278. [DOI] [PubMed] [Google Scholar]
- 20. Gähwiler BH, Capogna M, Debanne D, McKinney RA, Thompson SM (1997) Organotypic slice cultures: a technique has come of age. Trends Neurosci 20: 471–477. [DOI] [PubMed] [Google Scholar]
- 21. Holopainen IE (2005) Organotypic hippocampal slice cultures: a model system to study basic cellular and molecular mechanisms of neuronal cell death, neuroprotection, and synaptic plasticity. Neurochem Res 30: 1521–1528. [DOI] [PubMed] [Google Scholar]
- 22. De Simoni A, Griesinger CB, Edwards FA (2003) Development of rat ca1 neurones in acute versus organotypic slices: role of experience in synaptic morphology and activity. J Physiol 550: 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ivanov A, Pellegrino C, Rama S, Dumalska I, Salyha Y, et al. (2006) Opposing role of synaptic and extrasynaptic nmda receptors in regulation of the extracellular signal-regulated kinases (erk) activity in cultured rat hippocampal neurons. J Physiol 572: 789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seeburg DP, Sheng M (2008) Activity-induced polo-like kinase 2 is required for homeostatic plasticity of hippocampal neurons during epileptiform activity. J Neurosci 28: 6583–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guzowski JF, McNaughton BL, Barnes CA, Worley PF (1999) Environment-specific expression of the immediate-early gene arc in hippocampal neuronal ensembles. Nat Neurosci 2: 1120–1124. [DOI] [PubMed] [Google Scholar]
- 26. Kelly MP, Deadwyler SA (2002) Acquisition of a novel behavior induces higher levels of arc mrna than does overtrained performance. Neuroscience 110: 617–626. [DOI] [PubMed] [Google Scholar]
- 27. Wang KH, Majewska A, Schummers J, Farley B, Hu C, et al. (2006) In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell 126: 389–402. [DOI] [PubMed] [Google Scholar]
- 28. Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, et al. (2006) Arc/arg3.1 interacts with the endocytic machinery to regulate ampa receptor trafficking. Neuron 52: 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT (2006) Increased expression of the immediate-early gene arc/arg3.1 reduces ampa receptor-mediated synaptic transmission. Neuron 52: 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Messaoudi E, Kanhema T, Soulé J, Tiron A, Dagyte G, et al. (2007) Sustained arc/arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci 27: 10445–10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bramham CR, Alme MN, Bittins M, Kuipers SD, Nair RR, et al. (2010) The arc of synaptic memory. Exp Brain Res 200: 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, et al. (2006) Arc/arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52: 437–444. [DOI] [PubMed] [Google Scholar]
- 33. Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, et al. (2006) Arc/arg3.1 mediates homeostatic synaptic scaling of ampa receptors. Neuron 52: 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuipers SD, Tiron A, Soule J, Messaoudi E, Trentani A, et al. (2009) Selective survival and maturation of adult-born dentate granule cells expressing the immediate early gene arc/arg3.1. PLoS One 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang SJ, Buchthal B, Lau D, Hayer S, Dick O, et al. (2011) A signaling cascade of nuclear calcium-creb-atf3 activated by synaptic nmda receptors defines a gene repression module that protects against extrasynaptic nmda receptor-induced neuronal cell death and ischemic brain damage. J Neurosci 31: 4978–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Francis JS, Dragunow M, During MJ (2004) Over expression of atf-3 protects rat hippocampal neurons from in vivo injection of kainic acid. Brain Res Mol Brain Res 124: 199–203. [DOI] [PubMed] [Google Scholar]
- 37. Scharfman H, Goodman J, Macleod A, Phani S, Antonelli C, et al. (2005) Increased neurogenesis and the ectopic granule cells after intrahippocampal bdnf infusion in adult rats. Exp Neurol 192: 348–356. [DOI] [PubMed] [Google Scholar]
- 38. Pencea V, Bingaman KD, Wiegand SJ, Luskin MB (2001) Infusion of brain-derived neurotrophic factor into the lateral ventricle of the adult rat leads to new neurons in the parenchyma of the striatum, septum, thalamus, and hypothalamus. J Neurosci 21: 6706–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Danzer SC, Crooks KR, Lo DC, McNamara JO (2002) Increased expression of brain-derived neurotrophic factor induces formation of basal dendrites and axonal branching in dentate granule cells in hippocampal explant cultures. J Neurosci 22: 9754–9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu B, Gottschalk W, Chow A, Wilson RI, Schnell E, et al. (2000) The role of brain-derived neurotrophic factor receptors in the mature hippocampus: modulation of long-term potentiation through a presynaptic mechanism involving trkb. J Neurosci 20: 6888–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gärtner A, Polnau DG, Staiger V, Sciarretta C, Minichiello L, et al. (2006) Hippocampal long-term potentiation is supported by presynaptic and postsynaptic tyrosine receptor kinase b-mediated phospholipase cgamma signaling. J Neurosci 26: 3496–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vaillant AR, Mazzoni I, Tudan C, Boudreau M, Kaplan DR, et al. (1999) Depolarization and neurotrophins converge on the phosphatidylinositol 3-kinase-akt pathway to synergistically regulate neuronal survival. J Cell Biol 146: 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen MJ, Russo-Neustadt AA (2005) Exercise activates the phosphatidylinositol 3-kinase pathway. Brain Res Mol Brain Res 135: 181–193. [DOI] [PubMed] [Google Scholar]
- 44. Niu C, Yip HK (2011) Neuroprotective signaling mechanisms of telomerase are regulated by brain-derived neurotrophic factor in rat spinal cord motor neurons. J Neuropathol Exp Neurol 70: 634–652. [DOI] [PubMed] [Google Scholar]
- 45. Lähteinen S, Pitkänen A, Saarelainen T, Nissinen J, Koponen E, et al. (2002) Decreased bdnf signalling in transgenic mice reduces epileptogenesis. Eur J Neurosci 15: 721–734. [DOI] [PubMed] [Google Scholar]
- 46. Ramsden M, Berchtold NC, Patrick Kesslak J, Cotman CW, Pike CJ (2003) Exercise increases the vulnerability of rat hippocampal neurons to kainate lesion. Brain Res 971: 239–244. [DOI] [PubMed] [Google Scholar]
- 47. Canzoniere D, Farioli-Vecchioli S, Conti F, Ciotti MT, Tata AM, et al. (2004) Dual control of neurogenesis by pc3 through cell cycle inhibition and induction of math1. J Neurosci 24: 3355–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Farioli-Vecchioli S, Tanori M, Micheli L, Mancuso M, Leonardi L, et al. (2007) Inhibition of medulloblastoma tumorigenesis by the antiproliferative and pro-differentiative gene pc3. FASEB J 21: 2215–2225. [DOI] [PubMed] [Google Scholar]
- 49. Passeri D, Marcucci A, Rizzo G, Billi M, Panigada M, et al. (2006) Btg2 enhances retinoic acid-induced differentiation by modulating histone h4 methylation and acetylation. Mol Cell Biol 26: 5023–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bisler S, Schleicher A, Gass P, Stehle JH, Zilles K, et al. (2002) Expression of c-fos, icer, krox-24 and junb in the whisker-to-barrel pathway of rats: time course of induction upon whisker stimulation by tactile exploration of an enriched environment. J Chem Neuroanat 23: 187–198. [DOI] [PubMed] [Google Scholar]
- 51. Dragunow M, Faull R (1989) The use of c-fos as a metabolic marker in neuronal pathway tracing. J Neurosci Methods 29: 261–265. [DOI] [PubMed] [Google Scholar]
- 52. Kadar E, Aldavert-Vera L, Huguet G, Costa-Miserachs D, Morgado-Bernal I, et al. (2011) Intracranial self-stimulation induces expression of learning and memory-related genes in rat amygdala. Genes Brain Behav 10: 69–77. [DOI] [PubMed] [Google Scholar]
- 53. VanElzakker M, Fevurly RD, Breindel T, Spencer RL (2008) Environmental novelty is associated with a selective increase in fos expression in the output elements of the hippocampal formation and the perirhinal cortex. Learn Mem 15: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, et al. (2003) Impaired long-term memory and nr2a-type nmda receptor-dependent synaptic plasticity in mice lacking c-fos in the cns. J Neurosci 23: 9116–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gass P, Fleischmann A, Hvalby O, Jensen V, Zacher C, et al. (2004) Mice with a fra-1 knock-in into the c-fos locus show impaired spatial but regular contextual learning and normal ltp. Brain Res Mol Brain Res 130: 16–22. [DOI] [PubMed] [Google Scholar]
- 56. Katche C, Bekinschtein P, Slipczuk L, Goldin A, Izquierdo IA, et al. (2010) Delayed wave of c-fos expression in the dorsal hippocampus involved specifically in persistence of long-term memory storage. Proc Natl Acad Sci U S A 107: 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Watanabe Y, Johnson RS, Butler LS, Binder DK, Spiegelman BM, et al. (1996) Null mutation of c-fos impairs structural and functional plasticities in the kindling model of epilepsy. J Neurosci 16: 3827–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bhattacharya S, Michels CL, Leung MK, Arany ZP, Kung AL, et al. (1999) Functional role of p35srj, a novel p300/cbp binding protein, during transactivation by hif-1. Genes Dev 13: 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bamforth SD, Bragana J, Eloranta JJ, Murdoch JN, Marques FI, et al. (2001) Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking cited2, a new tfap2 co-activator. Nat Genet 29: 469–474. [DOI] [PubMed] [Google Scholar]
- 60. Gonzalez YR, Zhang Y, Behzadpoor D, Cregan S, Bamforth S, et al. (2008) Cited2 signals through peroxisome proliferator-activated receptor-gamma to regulate death of cortical neurons after dna damage. J Neurosci 28: 5559–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Borlikova G, Endo S (2009) Inducible camp early repressor (icer) and brain functions. Mol Neurobiol 40: 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mouravlev A, Young D, During MJ (2007) Phosphorylation-dependent degradation of transgenic creb protein initiated by heterodimerization. Brain Res 1130: 31–37. [DOI] [PubMed] [Google Scholar]
- 63. Mioduszewska B, Jaworski J, Szklarczyk AW, Klejman A, Kaczmarek L (2008) Inducible camp early repressor (icer)-evoked delayed neuronal death in the organotypic hippocampal culture. J Neurosci Res 86: 61–70. [DOI] [PubMed] [Google Scholar]
- 64. Klejman A, Kaczmarek L (2006) Inducible camp early repressor (icer) isoforms and neuronal apoptosis in cortical in vitro culture. Acta Neurobiol Exp (Wars) 66: 267–272. [DOI] [PubMed] [Google Scholar]
- 65. Jaworski J, Mioduszewska B, Sánchez-Capelo A, Figiel I, Habas A, et al. (2003) Inducible camp early repressor, an endogenous antagonist of camp responsive element-binding protein, evokes neuronal apoptosis in vitro. J Neurosci 23: 4519–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kojima N, Borlikova G, Sakamoto T, Yamada K, Ikeda T, et al. (2008) Inducible camp early repressor acts as a negative regulator for kindling epileptogenesis and long-term fear memory. J Neurosci 28: 6459–6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Porter BE, Lund IV, Varodayan FP, Wallace RW, Blendy JA (2008) The role of transcription factors cyclic-amp responsive element modulator (crem) and inducible cyclic-amp early repressor (icer) in epileptogenesis. Neuroscience 152: 829–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chung KC, Ahn YS (1998) Expression of immediate early gene cyr61 during the differentiation of immortalized embryonic hippocampal neuronal cells. Neurosci Lett 255: 155–158. [DOI] [PubMed] [Google Scholar]
- 69. Kim KH, Min YK, Baik JH, Lau LF, Chaqour B, et al. (2003) Expression of angiogenic factor cyr61 during neuronal cell death via the activation of c-jun n-terminal kinase and serum response factor. J Biol Chem 278: 13847–13854. [DOI] [PubMed] [Google Scholar]
- 70. Sin WC, Bechberger JF, Rushlow WJ, Naus CC (2008) Dose-dependent differential upregulation of ccn1/cyr61 and ccn3/nov by the gap junction protein connexin43 in glioma cells. J Cell Biochem 103: 1772–1782. [DOI] [PubMed] [Google Scholar]
- 71. Owens DM, Keyse SM (2007) Differential regulation of map kinase signalling by dual-specificity protein phosphatases. Oncogene 26: 3203–3213. [DOI] [PubMed] [Google Scholar]
- 72. Davis S, Vanhoutte P, Pages C, Caboche J, Laroche S (2000) The mapk/erk cascade targets both elk-1 and camp response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. J Neurosci 20: 4563–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mandl M, Slack DN, Keyse SM (2005) Specific inactivation and nuclear anchoring of extracellular signal-regulated kinase 2 by the inducible dual-specificity protein phosphatase dusp5. Mol Cell Biol 25: 1830–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Volmat V, Camps M, Arkinstall S, Pouysségur J, Lenormand P (2001) The nucleus, a site for signal termination by sequestration and inactivation of p42/p44 map kinases. J Cell Sci 114: 3433–3443. [DOI] [PubMed] [Google Scholar]
- 75. Tárrega C, Nunes-Xavier C, Cejudo-Marín R, Martín-Pérez J, Pulido R (2010) Studying the regulation of map kinase by map kinase phosphatases in vitro and in cell systems. Methods Mol Biol 661: 305–321. [DOI] [PubMed] [Google Scholar]
- 76. Domercq M, Alberdi E, Sánchez-Gómez MV, Ariz U, Pérez-Samartín A, et al. (2011) Dual-specific phosphatase-6 (dusp6) and erk mediate ampa receptor-induced oligodendrocyte death. J Biol Chem 286: 11825–11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jones MW, Errington ML, French PJ, Fine A, Bliss TV, et al. (2001) A requirement for the immediate early gene zif268 in the expression of late ltp and long-term memories. Nat Neurosci 4: 289–296. [DOI] [PubMed] [Google Scholar]
- 78. Abraham WC, Dragunow M, Tate WP (1991) The role of immediate early genes in the stabilization of long-term potentiation. Mol Neurobiol 5: 297–314. [DOI] [PubMed] [Google Scholar]
- 79. Davis S, Bozon B, Laroche S (2003) How necessary is the activation of the immediate early gene zif268 in synaptic plasticity and learning? Behav Brain Res 142: 17–30. [DOI] [PubMed] [Google Scholar]
- 80. Ribeiro S, Mello CV, Velho T, Gardner TJ, Jarvis ED, et al. (2002) Induction of hippocampal long-term potentiation during waking leads to increased extrahippocampal zif-268 expression during ensuing rapid-eye-movement sleep. J Neurosci 22: 10914–10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zheng D, Butler LS, McNamara JO (1998) Kindling and associated mossy fibre sprouting are not affected in mice deficient of ngfi-a/ngfi-b genes. Neuroscience 83: 251–258. [DOI] [PubMed] [Google Scholar]
- 82. Cheval H, Chagneau C, Levasseur G, Veyrac A, Faucon-Biguet N, et al. (2011) Distinctive features of egr transcription factor regulation and dna binding activity in ca1 of the hippocampus in synaptic plasticity and consolidation and reconsolidation of fear memory. Hippocampus [DOI] [PubMed] [Google Scholar]
- 83. Inokuchi K, Murayama A, Ozawa F (1996) mrna differential display reveals krox-20 as a neural plasticity-regulated gene in the rat hippocampus. Biochem Biophys Res Commun 221: 430–436. [DOI] [PubMed] [Google Scholar]
- 84. Poirier R, Cheval H, Mailhes C, Charnay P, Davis S, et al. (2007) Paradoxical role of an egr transcription factor family member, egr2/krox20, in learning and memory. Front Behav Neurosci 1: 6–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li L, Yun SH, Keblesh J, Trommer BL, Xiong H, et al. (2007) Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Mol Cell Neurosci 35: 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Roberts DS, Hu Y, Lund IV, Brooks-Kayal AR, Russek SJ (2006) Brain-derived neurotrophic factor (bdnf)-induced synthesis of early growth response factor 3 (egr3) controls the levels of type a gaba receptor alpha 4 subunits in hippocampal neurons. J Biol Chem 281: 29431–29435. [DOI] [PubMed] [Google Scholar]
- 87. Kim JH, Roberts DS, Hu Y, Lau GC, Brooks-Kayal AR, et al. (2012) Brain-derived neurotrophic factor uses creb and egr3 to regulate nmda receptor levels in cortical neurons. J Neurochem 120: 210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ludwig A, Uvarov P, Soni S, Thomas-Crusells J, Airaksinen MS, et al. (2011) Early growth response 4 mediates bdnf induction of potassium chloride cotransporter 2 transcription. J Neurosci 31: 644–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Uvarov P, Ludwig A, Markkanen M, Rivera C, Airaksinen MS (2006) Upregulation of the neuron-specific k+/cl- cotransporter expression by transcription factor early growth response 4. J Neurosci 26: 13463–13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ludwig A, Uvarov P, Pellegrino C, Thomas-Crusells J, Schuchmann S, et al. (2011) Neurturin evokes mapk-dependent upregulation of egr4 and kcc2 in developing neurons. Neural Plast 2011: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sala C, Futai K, Yamamoto K, Worley PF, Hayashi Y, et al. (2003) Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein homer1a. J Neurosci 23: 6327–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sala C, Roussignol G, Meldolesi J, Fagni L (2005) Key role of the postsynaptic density scaffold proteins shank and homer in the functional architecture of ca2+ homeostasis at dendritic spines in hippocampal neurons. J Neurosci 25: 4587–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lynch G, Rex CS, Gall CM (2007) Ltp consolidation: substrates, explanatory power, and functional significance. Neuropharmacology 52: 12–23. [DOI] [PubMed] [Google Scholar]
- 94. Inoue Y, Udo H, Inokuchi K, Sugiyama H (2007) Homer1a regulates the activity-induced remodeling of synaptic structures in cultured hippocampal neurons. Neuroscience 150: 841–852. [DOI] [PubMed] [Google Scholar]
- 95. Celikel T, Marx V, Freudenberg F, Zivkovic A, Resnik E, et al. (2007) Select overexpression of homer1a in dorsal hippocampus impairs spatial working memory. Front Neurosci 1: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Martín ED, Sánchez-Perez A, Trejo JL, Martin-Aldana JA, Cano Jaimez M, et al. (2011) Irs-2 deficiency impairs nmda receptor-dependent long-term potentiation. Cereb Cortex [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Schubert M, Brazil DP, Burks DJ, Kushner JA, Ye J, et al. (2003) Insulin receptor substrate-2 deficiency impairs brain growth and promotes tau phosphorylation. J Neurosci 23: 7084–7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Irvine EE, Drinkwater L, Radwanska K, Al-Qassab H, Smith MA, et al. (2011) Insulin receptor substrate 2 is a negative regulator of memory formation. Learn Mem 18: 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B (1996) A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem 271: 31384–31390. [DOI] [PubMed] [Google Scholar]
- 100. Rowland BD, Peeper DS (2006) Klf4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer 6: 11–23. [DOI] [PubMed] [Google Scholar]
- 101. Rowland BD, Bernards R, Peeper DS (2005) The klf4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol 7: 1074–1082. [DOI] [PubMed] [Google Scholar]
- 102. Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, et al. (2009) Klf family members regulate intrinsic axon regeneration ability. Science 326: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhu S, Tai C, MacVicar BA, Jia W, Cynader MS (2009) Glutamatergic stimulation triggers rapid krüpple-like factor 4 expression in neurons and the overexpression of klf4 sensitizes neurons to nmda-induced caspase-3 activity. Brain Res 1250: 49–62. [DOI] [PubMed] [Google Scholar]
- 104. Weston CR, Davis RJ (2007) The jnk signal transduction pathway. Curr Opin Cell Biol 19: 142–149. [DOI] [PubMed] [Google Scholar]
- 105. Han D, Zhang QG, Yong-Liu, Li C, Zong YY, et al. (2008) Co-activation of gaba receptors inhibits the jnk3 apoptotic pathway via the disassembly of the glur6-psd95-mlk3 signaling module in cerebral ischemic-reperfusion. FEBS Lett 582: 1298–1306. [DOI] [PubMed] [Google Scholar]
- 106. Zhao J, Pei DS, Zhang QG, Zhang GY (2007) Down-regulation cdc42 attenuates neuronal apoptosis through inhibiting mlk3/jnk3 cascade during ischemic reperfusion in rat hippocampus. Cell Signal 19: 831–843. [DOI] [PubMed] [Google Scholar]
- 107. Bode AM, Dong Z (2007) The functional contrariety of jnk. Mol Carcinog 46: 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yu C, Minemoto Y, Zhang J, Liu J, Tang F, et al. (2004) Jnk suppresses apoptosis via phosphorylation of the proapoptotic bcl-2 family protein bad. Mol Cell 13: 329–340. [DOI] [PubMed] [Google Scholar]
- 109. Ventura JJ, Hübner A, Zhang C, Flavell RA, Shokat KM, et al. (2006) Chemical genetic analysis of the time course of signal transduction by jnk. Mol Cell 21: 701–710. [DOI] [PubMed] [Google Scholar]
- 110. Curran BP, Murray HJ, O'Connor JJ (2003) A role for c-jun n-terminal kinase in the inhibition of long-term potentiation by interleukin-1beta and long-term depression in the rat dentate gyrus in vitro. Neuroscience 118: 347–357. [DOI] [PubMed] [Google Scholar]
- 111. Liu MG, Wang RR, Chen XF, Zhang FK, Cui XY, et al. (2011) Differential roles of erk, jnk and p38 mapk in pain-related spatial and temporal enhancement of synaptic responses in the hippocampal formation of rats: multi-electrode array recordings. Brain Res 1382: 57–69. [DOI] [PubMed] [Google Scholar]
- 112. Zhu Y, Pak D, Qin Y, McCormack SG, Kim MJ, et al. (2005) Rap2-jnk removes synaptic ampa receptors during depotentiation. Neuron 46: 905–916. [DOI] [PubMed] [Google Scholar]
- 113. Barnat M, Enslen H, Propst F, Davis RJ, Soares S, et al. (2010) Distinct roles of c-jun n-terminal kinase isoforms in neurite initiation and elongation during axonal regeneration. J Neurosci 30: 7804–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Oliva AA, Atkins CM, Copenagle L, Banker GA (2006) Activated c-jun n-terminal kinase is required for axon formation. J Neurosci 26: 9462–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Junghans D, Chauvet S, Buhler E, Dudley K, Sykes T, et al. (2004) The ces-2-related transcription factor e4bp4 is an intrinsic regulator of motoneuron growth and survival. Development 131: 4425–4434. [DOI] [PubMed] [Google Scholar]
- 116. MacGillavry HD, Stam FJ, Sassen MM, Kegel L, Hendriks WT, et al. (2009) Nfil3 and camp response element-binding protein form a transcriptional feedforward loop that controls neuronal regeneration-associated gene expression. J Neurosci 29: 15542–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. MacGillavry HD, Cornelis J, van der Kallen LR, Sassen MM, Verhaagen J, et al. (2011) Genome-wide gene expression and promoter binding analysis identifies nfil3 as a repressor of c/ebp target genes in neuronal outgrowth. Mol Cell Neurosci 46: 460–468. [DOI] [PubMed] [Google Scholar]
- 118. O'Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, et al. (1999) Synaptic clustering of ampa receptors by the extracellular immediate-early gene product narp. Neuron 23: 309–323. [DOI] [PubMed] [Google Scholar]
- 119. O'Brien R, Xu D, Mi R, Tang X, Hopf C, et al. (2002) Synaptically targeted narp plays an essential role in the aggregation of ampa receptors at excitatory synapses in cultured spinal neurons. J Neurosci 22: 4487–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Reti IM, Baraban JM (2000) Sustained increase in narp protein expression following repeated electroconvulsive seizure. Neuropsychopharmacology 23: 439–443. [DOI] [PubMed] [Google Scholar]
- 121. Chang MC, Park JM, Pelkey KA, Grabenstatter HL, Xu D, et al. (2010) Narp regulates homeostatic scaling of excitatory synapses on parvalbumin-expressing interneurons. Nat Neurosci 13: 1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hökfelt T, Stanic D, Sanford SD, Gatlin JC, Nilsson I, et al. (2008) Npy and its involvement in axon guidance, neurogenesis, and feeding. Nutrition 24: 860–868. [DOI] [PubMed] [Google Scholar]
- 123. Olesen MV, Christiansen SH, Gotzsche CR, Nikitidou L, Kokaia M, et al. (2011) Neuropeptide y y1 receptor hippocampal overexpression via viral vectors is associated with modest anxiolytic-like and proconvulsant effects in mice. J Neurosci Res [DOI] [PubMed] [Google Scholar]
- 124. Brooks PA, Kelly JS, Allen JM, Smith DA, Stone TW (1987) Direct excitatory effects of neuropeptide y (npy) on rat hippocampal neurones in vitro. Brain Res 408: 295–298. [DOI] [PubMed] [Google Scholar]
- 125. Reibel S, Nadi S, Benmaamar R, Larmet Y, Carnahan J, et al. (2001) Neuropeptide y and epilepsy: varying effects according to seizure type and receptor activation. Peptides 22: 529–539. [DOI] [PubMed] [Google Scholar]
- 126. Decressac M, Wright B, David B, Tyers P, Jaber M, et al. (2011) Exogenous neuropeptide y promotes in vivo hippocampal neurogenesis. Hippocampus 21: 233–238. [DOI] [PubMed] [Google Scholar]
- 127. Shalizi A, Gaudillière B, Yuan Z, Stegmüller J, Shirogane T, et al. (2006) A calcium-regulated mef2 sumoylation switch controls postsynaptic differentiation. Science 311: 1012–1017. [DOI] [PubMed] [Google Scholar]
- 128. Volakakis N, Kadkhodaei B, Joodmardi E, Wallis K, Panman L, et al. (2010) Nr4a orphan nuclear receptors as mediators of creb-dependent neuroprotection. Proc Natl Acad Sci U S A 107: 12317–12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Peña de Ortiz S, Maldonado-Vlaar CS, Carrasquillo Y (2000) Hippocampal expression of the orphan nuclear receptor gene hzf-3/nurr1 during spatial discrimination learning. Neurobiol Learn Mem 74: 161–178. [DOI] [PubMed] [Google Scholar]
- 130. Colón-Cesario WI, Martínez-Montemayor MM, Morales S, Félix J, Cruz J, et al. (2006) Knockdown of nurr1 in the rat hippocampus: implications to spatial discrimination learning and memory. Learn Mem 13: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Crispino M, Tocco G, Feldman JD, Herschman HR, Baudry M (1998) Nurr1 mrna expression in neonatal and adult rat brain following kainic acid-induced seizure activity. Brain Res Mol Brain Res 59: 178–188. [DOI] [PubMed] [Google Scholar]
- 132. Zhang T, Wang P, Ren H, Fan J, Wang G (2009) Ngfi-b nuclear orphan receptor nurr1 interacts with p53 and suppresses its transcriptional activity. Mol Cancer Res 7: 1408–1415. [DOI] [PubMed] [Google Scholar]
- 133. Kim Y, Hong S, Noh MR, Kim SY, Huh PW, et al. (2006) Induction of neuron-derived orphan receptor-1 in the dentate gyrus of the hippocampal formation following transient global ischemia in the rat. Mol Cells 22: 8–12. [PubMed] [Google Scholar]
- 134. Pönniö T, Conneely OM (2004) nor-1 regulates hippocampal axon guidance, pyramidal cell survival, and seizure susceptibility. Mol Cell Biol 24: 9070–9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Bramham CR, Southard T, Sarvey JM, Herkenham M, Brady LS (1996) Unilateral ltp triggers bilateral increases in hippocampal neurotrophin and trk receptor mrna expression in behaving rats: evidence for interhemispheric communication. J Comp Neurol 368: 371–382. [DOI] [PubMed] [Google Scholar]
- 136. Kamei N, Tanaka N, Oishi Y, Hamasaki T, Nakanishi K, et al. (2007) Bdnf, nt-3, and ngf released from transplanted neural progenitor cells promote corticospinal axon growth in organotypic cocultures. Spine (Phila Pa 1976) 32: 1272–1278. [DOI] [PubMed] [Google Scholar]
- 137. Yoo M, Joung I, Han AM, Yoon HH, Kwon YK (2007) Distinct effect of neurotrophins delivered simultaneously by an adenoviral vector on neurite outgrowth of neural precursor cells from different regions of the brain. J Microbiol Biotechnol 17: 2033–2041. [PubMed] [Google Scholar]
- 138. Holm NR, Christophersen P, Olesen SP, Gammeltoft S (1997) Activation of calcium-dependent potassium channels in mouse [correction of rat] brain neurons by neurotrophin-3 and nerve growth factor. Proc Natl Acad Sci U S A 94: 1002–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Cheng B, Mattson MP (1994) Nt-3 and bdnf protect cns neurons against metabolic/excitotoxic insults. Brain Res 640: 56–67. [DOI] [PubMed] [Google Scholar]
- 140. Xu B, Michalski B, Racine RJ, Fahnestock M (2002) Continuous infusion of neurotrophin-3 triggers sprouting, decreases the levels of trka and trkc, and inhibits epileptogenesis and activity-dependent axonal growth in adult rats. Neuroscience 115: 1295–1308. [DOI] [PubMed] [Google Scholar]
- 141. Otal R, Martínez A, Soriano E (2005) Lack of trkb and trkc signaling alters the synaptogenesis and maturation of mossy fiber terminals in the hippocampus. Cell Tissue Res 319: 349–358. [DOI] [PubMed] [Google Scholar]
- 142. Chen G, Kolbeck R, Barde YA, Bonhoeffer T, Kossel A (1999) Relative contribution of endogenous neurotrophins in hippocampal long-term potentiation. J Neurosci 19: 7983–7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Yamagata K, Andreasson KI, Sugiura H, Maru E, Dominique M, et al. (1999) Arcadlin is a neural activity-regulated cadherin involved in long term potentiation. J Biol Chem 274: 19473–11979. [DOI] [PubMed] [Google Scholar]
- 144. Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, et al. (2007) Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering n-cadherin endocytosis via tao2beta and p38 map kinases. Neuron 56: 456–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Lee KJ, Lee Y, Rozeboom A, Lee JY, Udagawa N, et al. (2011) Requirement for plk2 in orchestrated ras and rap signaling, homeostatic structural plasticity, and memory. Neuron 69: 957–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Yang H, Zhang J, Andreasson K, Chen C (2008) Cox-2 oxidative metabolism of endocannabinoids augments hippocampal synaptic plasticity. Mol Cell Neurosci 37: 682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Sang N, Zhang J, Chen C (2007) Cox-2 oxidative metabolite of endocannabinoid 2-ag enhances excitatory glutamatergic synaptic transmission and induces neurotoxicity. J Neurochem 102: 1966–1977. [DOI] [PubMed] [Google Scholar]
- 148. Slanina KA, Roberto M, Schweitzer P (2005) Endocannabinoids restrict hippocampal long-term potentiation via cb1. Neuropharmacology 49: 660–668. [DOI] [PubMed] [Google Scholar]
- 149. Chen C, Magee JC, Bazan NG (2002) Cyclooxygenase-2 regulates prostaglandin e2 signaling in hippocampal long-term synaptic plasticity. J Neurophysiol 87: 2851–2857. [DOI] [PubMed] [Google Scholar]
- 150. Lee J, Kosaras B, Aleyasin H, Han JA, Park DS, et al. (2006) Role of cyclooxygenase-2 induction by transcription factor sp1 and sp3 in neuronal oxidative and dna damage response. FASEB J 20: 2375–2377. [DOI] [PubMed] [Google Scholar]
- 151. Yang H, Chen C (2008) Cyclooxygenase-2 in synaptic signaling. Curr Pharm Des 14: 1443–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Stolle K, Schnoor M, Fuellen G, Spitzer M, Cullen P, et al. (2007) Cloning, genomic organization, and tissue-specific expression of the rasl11b gene. Biochim Biophys Acta 1769: 514–524. [DOI] [PubMed] [Google Scholar]
- 153. Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, et al. (2000) Regulation of t cell activation, anxiety, and male aggression by rgs2. Proc Natl Acad Sci U S A 97: 12272–12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Hutchison RM, Chidiac P, Leung LS (2009) Hippocampal long-term potentiation is enhanced in urethane-anesthetized rgs2 knockout mice. Hippocampus 19: 687–691. [DOI] [PubMed] [Google Scholar]
- 155. Han J, Mark MD, Li X, Xie M, Waka S, et al. (2006) Rgs2 determines short-term synaptic plasticity in hippocampal neurons by regulating gi/o-mediated inhibition of presynaptic ca2+ channels. Neuron 51: 575–586. [DOI] [PubMed] [Google Scholar]
- 156. Seredenina T, Gokce O, Luthi-Carter R (2011) Decreased striatal rgs2 expression is neuroprotective in huntington's disease (hd) and exemplifies a compensatory aspect of hd-induced gene regulation. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Heidenreich O, Neininger A, Schratt G, Zinck R, Cahill MA, et al. (1999) Mapkap kinase 2 phosphorylates serum response factor in vitro and in vivo. J Biol Chem 274: 14434–14443. [DOI] [PubMed] [Google Scholar]
- 158. Pintchovski SA, Peebles CL, Kim HJ, Verdin E, Finkbeiner S (2009) The serum response factor and a putative novel transcription factor regulate expression of the immediate-early gene arc/arg3.1 in neurons. J Neurosci 29: 1525–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Johnson AW, Crombag HS, Smith DR, Ramanan N (2011) Effects of serum response factor (srf) deletion on conditioned reinforcement. Behav Brain Res 220: 312–318. [DOI] [PubMed] [Google Scholar]
- 160. Ramanan N, Shen Y, Sarsfield S, Lemberger T, Schütz G, et al. (2005) Srf mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci 8: 759–767. [DOI] [PubMed] [Google Scholar]
- 161. Etkin A, Alarcón JM, Weisberg SP, Touzani K, Huang YY, et al. (2006) A role in learning for srf: deletion in the adult forebrain disrupts ltd and the formation of an immediate memory of a novel context. Neuron 50: 127–143. [DOI] [PubMed] [Google Scholar]
- 162. Chang LF, Lin PC, Ho LI, Liu PY, Wu WC, et al. (2011) Overexpression of the orphan receptor nur77 and its translocation induced by pch4 may inhibit malignant glioma cell growth and induce cell apoptosis. J Surg Oncol 103: 442–450. [DOI] [PubMed] [Google Scholar]
- 163. Kawaai K, Tominaga-Yoshino K, Urakubo T, Taniguchi N, Kondoh Y, et al. (2010) Analysis of gene expression changes associated with long-lasting synaptic enhancement in hippocampal slice cultures after repetitive exposures to glutamate. J Neurosci Res 88: 2911–2922. [DOI] [PubMed] [Google Scholar]
- 164. Dijkmans TF, van Hooijdonk LW, Schouten TG, Kamphorst JT, Fitzsimons CP, et al. (2009) Identification of new nerve growth factor-responsive immediate-early genes. Brain Res 1249: 19–33. [DOI] [PubMed] [Google Scholar]
- 165. Park CS, Gong R, Stuart J, Tang SJ (2006) Molecular network and chromosomal clustering of genes involved in synaptic plasticity in the hippocampus. J Biol Chem 281: 30195–30211. [DOI] [PubMed] [Google Scholar]
- 166. Do JH, Choi DK (2008) Clustering approaches to identifying gene expression patterns from dna microarray data. Mol Cells 25: 279–288. [PubMed] [Google Scholar]
- 167. Daliri MR, Torre V (2006) Unsupervised clustering of shapes. Lecture Notes in Computer Science (LNCS) 4291: 712–720, 2006, Springer Publisher. 4291. [Google Scholar]
- 168. Penke Z, Chagneau C, Laroche S (2011) Contribution of egr1/zif268 to activity-dependent arc/arg3.1 transcription in the dentate gyrus and area ca1 of the hippocampus. Front Behav Neurosci 5: 48–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Yilmaz-Rastoder E, Miyamae T, Braun AE, Thiels E (2011) Ltp- and ltd-inducing stimulations cause opposite changes in arc/arg3.1 mrna level in hippocampal area ca1 in vivo. Hippocampus 21: 1290–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Kelly MP, Deadwyler SA (2003) Experience-dependent regulation of the immediate-early gene arc differs across brain regions. J Neurosci 23: 6443–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Lockhart DJ, Winzeler EA (2000) Genomics, gene expression and dna arrays. Nature 405: 827–836. [DOI] [PubMed] [Google Scholar]
- 172. Turrigiano G (2012) Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Turrigiano G (2007) Homeostatic signaling: the positive side of negative feedback. Curr Opin Neurobiol 17: 318–324. [DOI] [PubMed] [Google Scholar]
- 174. Kucharska A, Rushworth LK, Staples C, Morrice NA, Keyse SM (2009) Regulation of the inducible nuclear dual-specificity phosphatase dusp5 by erk mapk. Cell Signal 21: 1794–1805. [DOI] [PubMed] [Google Scholar]
- 175. Marie-Claire C, Benturquia N, Lundqvist A, Courtin C, Noble F (2008) Characteristics of dual specificity phosphatases mrna regulation by 3,4-methylenedioxymethamphetamine acute treatment in mice striatum. Brain Res 1239: 42–48. [DOI] [PubMed] [Google Scholar]
- 176. Nunes-Xavier CE, Tárrega C, Cejudo-Marín R, Frijhoff J, Sandin A, et al. (2010) Differential up-regulation of map kinase phosphatases mkp3/dusp6 and dusp5 by ets2 and c-jun converge in the control of the growth arrest versus proliferation response of mcf-7 breast cancer cells to phorbol ester. J Biol Chem 285: 26417–26430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Ma'ayan A, Lipshtat A, Iyengar R, Sontag ED (2008) Proximity of intracellular regulatory networks to monotone systems. IET Syst Biol 2: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Ma'ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, et al. (2005) Formation of regulatory patterns during signal propagation in a mammalian cellular network. Science 309: 1078–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Lindecke A, Korte M, Zagrebelsky M, Horejschi V, Elvers M, et al. (2006) Long-term depression activates transcription of immediate early transcription factor genes: involvement of serum response factor/elk-1. Eur J Neurosci 24: 555–563. [DOI] [PubMed] [Google Scholar]
- 180. Benito E, Valor LM, Jimenez-Minchan M, Huber W, Barco A (2011) camp response element-binding protein is a primary hub of activity-driven neuronal gene expression. J Neurosci 31: 18237–18250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Tullai JW, Schaffer ME, Mullenbrock S, Kasif S, Cooper GM (2004) Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and mek/erk signaling pathways. J Biol Chem 279: 20167–20177. [DOI] [PubMed] [Google Scholar]
- 182. O'Sullivan NC, Pickering M, Di Giacomo D, Loscher JS, Murphy KJ (2010) Mkl transcription cofactors regulate structural plasticity in hippocampal neurons. Cereb Cortex 20: 1915–1925. [DOI] [PubMed] [Google Scholar]
- 183. Kumar V, Fahey PG, Jong YJ, Ramanan N, O'Malley KL (2011) Activation of the intracellular metabotropic glutamate receptor 5 in striatal neurons leads to upregulation of genes associated with sustained synaptic transmission including arc/arg3.1. J Biol Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Kalita K, Kharebava G, Zheng JJ, Hetman M (2006) Role of megakaryoblastic acute leukemia-1 in erk1/2-dependent stimulation of serum response factor-driven transcription by bdnf or increased synaptic activity. J Neurosci 26: 10020–10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Papadia S, Stevenson P, Hardingham NR, Bading H, Hardingham GE (2005) Nuclear ca2+ and the camp response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J Neurosci 25: 4279–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Zhang SJ, Steijaert MN, Lau D, Schütz G, Delucinge-Vivier C, et al. (2007) Decoding nmda receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron 53: 549–562. [DOI] [PubMed] [Google Scholar]
- 187. Broccard FD, Pegoraro S, Ruaro ME, Altafini C, Torre V (2009) Characterization of the time course of changes of the evoked electrical activity in a model of a chemically-induced neuronal plasticity. BMC Res Notes 2: 13–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Ryser S, Tortola S, van Haasteren G, Muda M, Li S, et al. (2001) Map kinase phosphatase-1 gene transcription in rat neuroendocrine cells is modulated by a calcium-sensitive block to elongation in the first exon. J Biol Chem 276: 33319–33327. [DOI] [PubMed] [Google Scholar]
- 189. Ryser S, Massiha A, Piuz I, Schlegel W (2004) Stimulated initiation of mitogen-activated protein kinase phosphatase-1 (mkp-1) gene transcription involves the synergistic action of multiple cis-acting elements in the proximal promoter. Biochem J 378: 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Williams GT, Lau LF (1993) Activation of the inducible orphan receptor gene nur77 by serum growth factors: dissociation of immediate-early and delayed-early responses. Mol Cell Biol 13: 6124–6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, et al. (2008) Genome-wide analysis of mef2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Lam BY, Zhang W, Ng DC, Maruthappu M, Roderick HL, et al. (2010) Creb-dependent nur77 induction following depolarization in pc12 cells and neurons is modulated by mef2 transcription factors. J Neurochem 112: 1065–1073. [DOI] [PubMed] [Google Scholar]
- 193. Lemberger T, Parkitna JR, Chai M, Schütz G, Engblom D (2008) Creb has a context-dependent role in activity-regulated transcription and maintains neuronal cholesterol homeostasis. FASEB J 22: 2872–2879. [DOI] [PubMed] [Google Scholar]
- 194. Gähwiler BH (1981) Organotypic monolayer cultures of nervous tissue. J Neurosci Methods 4: 329–342. [DOI] [PubMed] [Google Scholar]
- 195. Uchida I, Cestari IN, Yang J (1996) The differential antagonism by bicuculline and sr95531 of pentobarbitone-induced currents in cultured hippocampal neurons. Eur J Pharmacol 307: 89–96. [DOI] [PubMed] [Google Scholar]
- 196. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, et al. (2003) Summaries of affymetrix genechip probe level data. Nucleic Acids Res 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Huang daW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 198. Rozen S, Skaletsky H (2000) Primer3 on the www for general users and for biologist programmers. Methods Mol Biol 132: 365–386. [DOI] [PubMed] [Google Scholar]
- 199. Odenwald WF, Rasband W, Kuzin A, Brody T (2005) Evoprinter, a multigenomic comparative tool for rapid identification of functionally important dna. Proc Natl Acad Sci U S A 102: 14700–14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, et al. (2010) Jaspar 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38: 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables. - Supplementary Table A: The complete list of microarray probes/genes. - Supplementary Table B: List of primers used for the time course analysis. - Supplementary Table C: RT-PCR validation data.
(DOCX)
Supplementary notes.
(DOCX)