Abstract
Understanding the transformation of graphene derivatives by natural amphiphiles is essential for elucidating the biological and environmental implications of this emerging class of engineered nanomaterials. Using rapid discrete-molecular-dynamics simulations, we examined the binding of graphene and graphene oxide with peptides, fatty acids, and cellulose, and complemented our simulations by experimental studies of Raman spectroscopy, FTIR, and UV-Vis spectrophotometry. Specifically, we established a connection between the differential binding and the conformational flexibility, molecular geometry, and hydrocarbon content of the amphiphiles. Importantly, our dynamics simulations revealed a Vroman-like competitive binding of the amphiphiles for the graphene oxide substrate. This study provides a mechanistic basis for addressing the transformation, evolution, transport, biocompatibility, and toxicity of graphene derivatives in living systems and the natural environment.
Due to their unique physical properties1,2,3, graphene and graphene derivatives have emerged as ideal materials for constructing novel nano- and quantum devices. The potential applications of graphene derivatives range from electronic circuits and energy storage to biomedical nanodevices for imaging, sensing, and diagnosis4,5,6,7,8. The increasing application and foreseeable mass-production of graphene derivatives9,10, however, will likely lead to their environmental discharge, while advances in graphene-based nanomedicine will induce biological exposure to such engineered nanostructures. Consequently, it has become increasingly crucial to delineate the transformation, evolution, transport, and biocompatibility of graphene derivatives in the aqueous phase, ranging from biological to environmental systems11.
Once discharged into the environment or introduced to biological systems, graphene derivatives may interact with natural organic matter, biomolecules, and other ionic and molecular complexes through self assembly and chemical reactions. Most of these natural and bio-materials are amphiphilic in nature, and are usually comprised of carbohydrates, peptides, and fatty acids. Since natural amphiphiles may bind with graphene derivatives to render a biocorona12,13, it is conceivable that the fate of graphene derivatives in biological systems and the environment is determined by the entity of the biocorona rather than the nanomaterial substrates alone. Possessing vastly different molecular morphology and physicochemical properties, natural amphiphiles are expected to feature distinct binding affinities for graphene derivatives to ascribe the latter new physical chemical and/or biological identities. Furthermore, differences in the concentration and affinity of natural amphiphiles may lead to their competitive binding for graphene derivatives, similar to the Vroman effect that is exhibited by serum proteins adsorbed onto solid surfaces14.
The binding of small ligands and peptides to graphene derivatives has been a subject of a few recent studies. For example, Dai et al. showed that simple physisorption through π-stacking could be exploited to load doxorubicin -- a commonly used cancer drug -- onto graphene oxide15. It was demonstrated that uniformly dispersed graphene oxide16,17 in a chitosan-ferrocene matrix became positively charged, thereby boosting its capacity to stabilize biomolecules such as glucose oxidase and consequently facilitate the fabrication of a glucose biosensor6. Katoch et al. showed that a dodecamer peptide could bind to graphene by orienting its aromatic residues, such as tryptophan and histidine, parallel to the nanosheet; by comparing the binding affinities of mutant peptides with tryptophans substituted by alanines the researchers identified an essential role of tryptophan in attributing to the strong binding of the peptide and the graphene18. The stability of peptides adsorbed onto a graphene nanosheet was studied through coarse-grained simulations, which underpinned the importance of π-stacking, van der Waals, and hydrophobic interactions in the binding19,20. Density functional theory calculations were employed to study how biomolecular adsorption could affect the density of states of graphene nanosheets21. These studies, however, mostly focused on the binding of graphene derivatives with single-molecular or single-component amphiphiles. A systematic study of the binding of graphene derivatives with a collection of representative natural amphiphiles is essential for elucidating the transformation and dynamics of graphene derivatives in complex biological and environmental media.
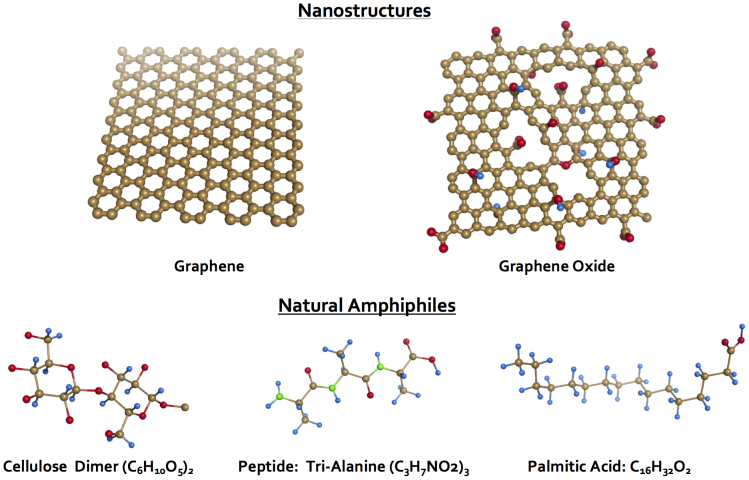
Herein we combined computational modeling and experimental characterizations to examine the binding of graphene and graphene oxide with natural amphiphiles. Specifically for simulations, we adopted cellulose dimers, tri-alanine peptides, and palmitic acids as model amphiphiles (Fig. 1) to represent the sugar, peptide, and fatty acid moieties present in algal exudates used in our experiments, respectively. In addition to being prevalent in aquatic environments, these molecular species are also ubiquitous across the biosphere of living organisms. We performed discrete molecular dynamics (DMD) simulations, a rapid dynamic sampling algorithm22 to characterize the binding between the graphene derivatives and the natural amphiphiles. The molecular systems were modeled using the united atom representation, in which polar hydrogen and heavy atoms were explicitly modeled. The simulations were performed with implicit solvent, and the inter-atomic interactions were modeled by a physical force field adapted from Medusa23, which include van der Waals, solvation24, electrostatic, and hydrogen bond25,26. In our simulations, graphene nanosheet was presented as a two-dimensional honeycomb, where its aromatic carbon atoms featured van der Waals and hydrophobic interactions. In contrast, graphene oxide was modeled by introducing defects, epoxidations, hydroxylations, and carboxylations to its graphene backbone (Fig. 1). These modifications rendered the graphene oxide surface more hydrophilic in physicochemistry and rugged in morphology than graphene.
Figure 1. Simulation scheme of the nanostructures of graphene and graphene oxide nanosheets (top row) and the natural amphiphiles of a cellulose dimer, a tri-alanine peptide, and a palmitic acid (bottom row).
Color schemes: brown - carbon, red - oxygen, blue - hydrogen, green - nitrogen.
Results
Differential binding of nanosheets with single amphiphiles – temperature varying DMD simulations
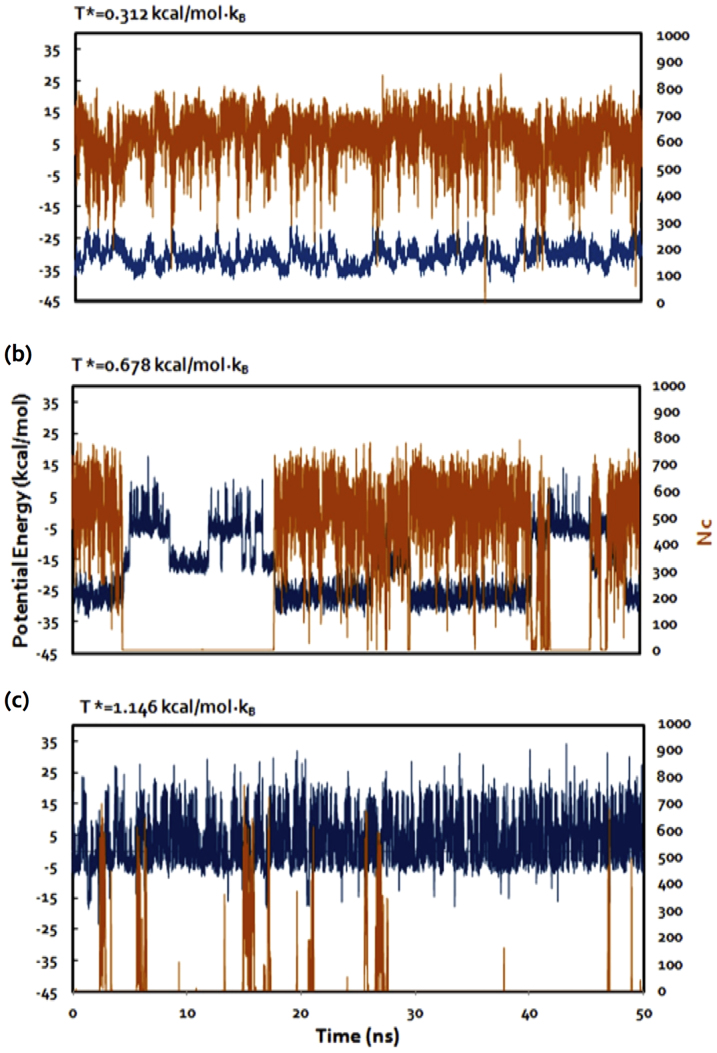
We first characterized the dynamics of single-molecular binding between the nanosheets of graphene derivatives and the amphiphiles. We performed DMD simulations at different temperatures and monitored the binding along the simulation trajectories. For example, in the case of graphene oxide and cellulose27 binding, we observed three different regimes (Fig. 2). At low temperatures (Fig. 2a), the molecular system had low potential energies and the cellulose stayed bound to the nanosheet with a high number of atomic contacts (Nc) occurring between the two species. At high temperatures (Fig. 2c), the cellulose molecule dissociated from the nanosheet with higher potential energies and a low Nc value. The two species only occasionally formed contacts due to thermal fluctuations. Between these two extreme regimes, there existed a mid-point temperature, Tm, where the cellulose had approximately an equal probability of being bound and unbound to graphene oxide (Fig. 2b). Interestingly, in the unbound state, the systems featured an intermediate energy state, which belonged to the excitation of a high-energy normal mode due to harmonic constraint applied to confine the nanosheet (Supplementary Fig. S1). Therefore, the inter-molecular contact, Nc, rather than the potential energy, was a more appropriate parameter to monitor the binding. At Tm, the potential energies and inter-molecular Nc values displayed large and anti-correlated fluctuations, clearly resulting from the interplay of enthalpy and entropy. Here the contributions of entropy include freedoms in both translation and configuration. The values of Tm were used to quantify the binding affinities between the different amphiphiles and the nanosheets.
Figure 2. Binding simulation trajectories at different temperatures.
Both potential energy (E, blue, left vertical axis) and number of atomic contacts (Nc, orange, right vertical axis) are shown for DMD simulations of cellulose-graphene oxide binding. Simulations at three different temperatures are shown: T* = 0.312 (a), 0.678 (b), and 1.146 (c), where the temperature unit is kcal/mol·kB.
Accurate estimation of Tm requires sufficient sampling of the conformational space. We therefore applied replica exchange DMD simulations25 (Methods) to enhance the sampling, where multiple simulations were running in parallel at different temperatures and the replica temperatures were subject to exchange periodically according to the Metropolis criteria28. A system in a kinetically trapped state has a chance to escape the local minimum by running at a higher temperature, thus enhancing the sampling. Based on the replica exchange simulations, we computed the thermodynamic parameters using the weighted histogram method (Methods)29.
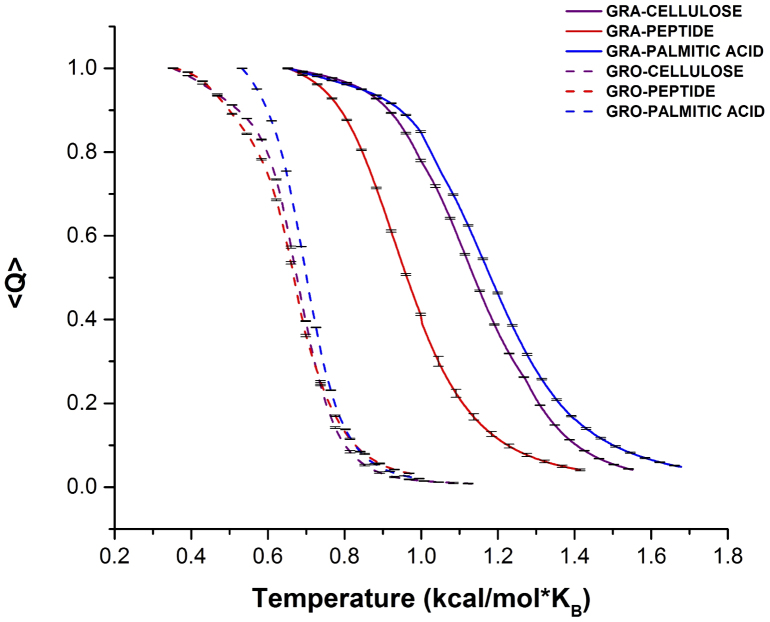
For both graphene and graphene oxide, we performed independent replica exchange DMD simulations of their binding to cellulose, tri-alanine, and palmitic acid. We computed the average Nc as a function of temperature (Fig. 3a). For comparison between different molecular systems, we normalized the average Nc by its maximum value at low temperature to obtain the Q-value, which quantified the fraction of inter-molecular contacts. The amphiphiles showed a lower Tm when bound to graphene oxide than graphene, indicating a weaker binding associated with graphene oxide due to its various surface modifications that compromised inter-molecular hydrophobic interaction while encouraged electrostatic repulsion.
Figure 3. Differential binding affinities of natural amphiphiles with graphene derivatives.
The average value of normalized fraction of contacts, <Q>, was computed as the function of temperatures (T*) using the weighted histogram analysis method with replica exchange DMD simulations (Methods). The error bars were computed as the statistical uncertainty47. The temperature dependence of <Q> values illustrates the melting. For each of the three amphiphile species, the melting curves of both graphene oxide (“GRO”, dashed) and graphene (“GRA”, solid) are shown.
Interestingly, for both graphene and graphene oxide, palmitic acid displayed the strongest binding while tri-alanine showed the weakest (Fig. 3). The strong binding of palmitic acid to the nanosheets correlated with its longer molecular chain that consisted of a higher number of hydrocarbons and hence a greater degree of chain flexibility. The conformational flexibility of palmitic acid allowed its long hydrophobic tail to pack tightly against the nanosheet, taking advantage of contributions from van der Waals and hydrophobic interactions. Both the cellulose and the peptide were more rigid and hydrophilic compared to the palmitic acid tail. In addition, the higher melting temperature for cellulose on graphene, compared to that for peptide, can be attributed to stacking. In contrast to the ring-like structure of the cellulose, the peptide backbone of tri-alanine was unable to form many contacts with the nanosheet. This is in agreement with the molecular dynamics study by Katoch et al., in which a lower binding affinity was observed when tryptophan residues were replaced by alanine. In the case of graphene oxide, the melting curves for cellulose and peptide were closer to each other (Fig. 3), suggesting that stacking was compromised by the functional groups of the nanosheet to shield its aromatic structure.
Differential binding of nanosheets with algal exudates – experiments
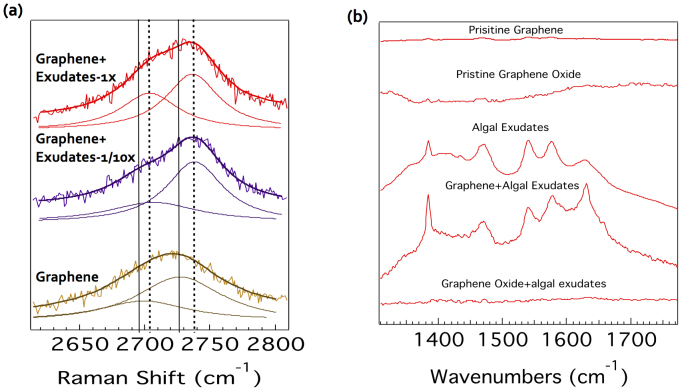
To complement the simulations, algal exudates were acquired from freshly cultured Chlorella sp. following a protocol developed in our lab30. The algal exudates were used to mimic the natural amphiphiles of cellulose, peptides, and fatty acids in the simulations. Graphene was synthesized using previously described chemical vapor deposition method31 while graphene oxide was produced using the modified Hummer's method32. Graphene-based materials exhibited strong Raman spectra due to the double resonance phenomenon33. Importantly, the overtone of disorder band (referred to as the 2D-band) exhibited two peaks at ~2690 cm−1 and ~2725 cm−1 (see Fig. 4a). This band is highly sensitive to charge transfer from substrates, dopants or any adsorbents. We observed that the 2D-band in graphene upshifted upon exposure to the algal exudates, suggesting a possible charge transfer from graphene to algal exudates. However, no such charge transfer was observed in the case of graphene oxide, implying a weaker interaction occurring between the graphene oxide and algal exudates (Supplementary Fig. S2), in agreement with our simulations (Fig. 3). Furthermore, our Fourier transform infrared (FTIR) spectra (Fig. 4b) showed that the exudate peaks were present and absent in the graphene and graphene oxide incubated with algal exudates and after washing with de-ionized water, consistent with both the Raman spectra and the simulations.
Figure 4. Raman (a) and FTIR (b) spectra of algal exudate-graphene/graphene oxide system.
1×: stock, 1/10×: ten-time diluted.
To further examine the binding kinetics of graphene and graphene oxide with algal exudates, we incubated the nanosheets with exudates in water and observed their precipitation at different temperatures (Methods). The absorbance peak of the algal exudates at 205 nm was monitored over time for both graphene and graphene oxide (Figs. 5a, b). The normalized absorbance value corresponded to the total fraction of exudates and graphene (graphene oxide) still present in solution at a given time. This process was performed with fresh suspensions at both 30°C and 35°C. We also performed a control experiment of graphene and graphene oxide in the absence of algal exudates at both temperatures, and did not identify significant temperature dependence of the control precipitation over the temperature range examined.
Figure 5. UV-V is absorbance spectra of algal exudates mixed with graphene (a) and graphene oxide (b) measured at 30°C (blue) and 35°C (maroon).
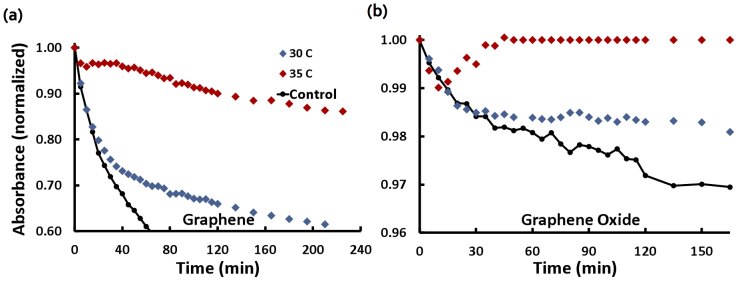
Control kinetics performed in the absence of exudates at 30°C (with no significant difference seen at 35°C) at concentrations equal to test samples.
For both graphene and graphene oxide, algal exudates slowed the rate of precipitation at both temperatures. This general behavior is indicative of exudates binding with graphene and graphene oxide to render both types of nanosheets more water-soluble. The binding with algal exudates should also discourage π-stacking of the nanosheets, further slowing their rate of precipitation. The temperature-dependent behavior of graphene and graphene oxide with algal exudates displayed qualitatively the same trends – the overall difference in precipitation rate is due to the stark difference in hydrophobicity between graphene and graphene oxide. During the initial precipitation at 30°C, both graphene and graphene oxide with exudates followed the control precipitation closely, indicating that under this temperature stacking of the nanosheets dominated exudate-nanosheet interactions to favor precipitation. The suspensions at 35°C showed less pronounced precipitations, reaching a complete suspension after 50 min incubation with the graphene oxide. At 240-min incubation with the graphene, while the suspension reached 60% of normalized absorbance at 30°C, it registered 86% of normalized absorbance at 35°C in the same time period. This behavior clearly confirmed that significant binding occurred between algal exudates and both graphene and graphene oxide, and this binding was further enhanced by elevated temperature.
Differential binding of nanosheets with multiple amphiphile species – DMD simulation
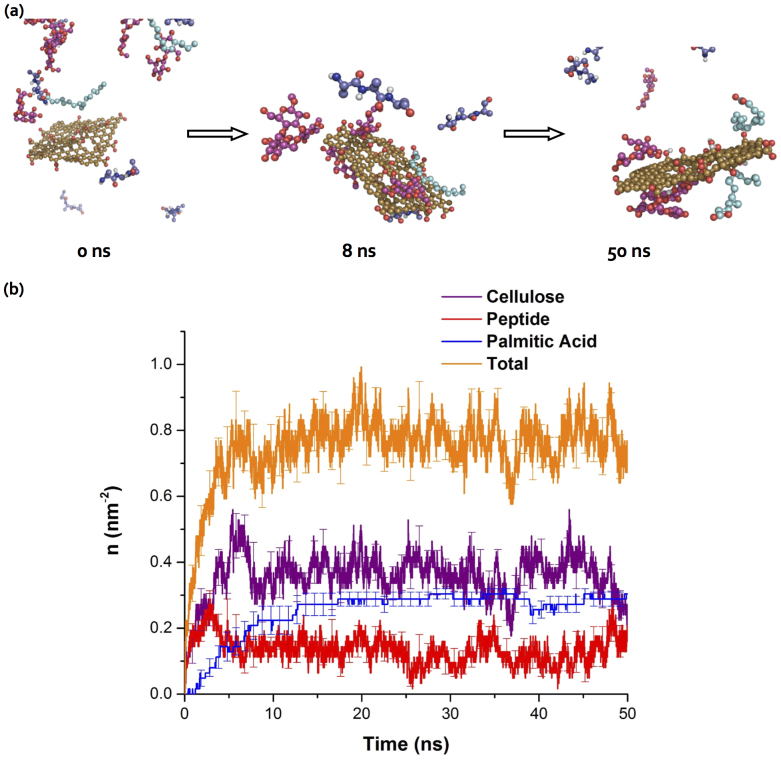
To model the binding of graphene derivatives with a collection of natural amphiphiles in biological and environmental media, we performed a constant-temperature DMD simulation of a graphene oxide nanosheet mixed with the three amphiphilie species simultaneously (Methods). We used the relative ratios of glucose to peptide to palmitic acid as found in algal exudates34, 7:3:1, and accordingly we included 14 cellulose, 6 peptide, and 2 palmitic acid molecules. The amphiphilic molecules were initially positioned away from the nanosheet (Fig. 6a). We chose a simulation temperature T ≈ Tm of tri-alanine binding. This temperature allowed rapid equilibration while all molecules were able to bind to the graphene oxide nanosheet.
Figure 6. Vroman-like competitive binding of amphiphile mixture with graphene oxide.
(a) The snapshots of the binding between graphene oxide and amphiphile mixture in a typical DMD simulation: t = 0 ns, 8 ns, and 50 ns. (b) The average number density of molecules, n, bound to the nanosheet is shown as a function of temperature. The averages and standard deviations (error bars) are calculated from 10 independent simulations with different initial configurations.
We then monitored the number density of molecules bound to the nanosheet as a function of the simulation time (Fig. 6b). The averages and standard deviations (error bars in Fig. 6b) were computed over ten independent simulations with different initial conditions. Due to their high concentrations, peptides and celluloses rapidly covered the nanosheet to form a nanosheet-amphiphile biocorona (Figs. 6a), which hindered the binding of palmitic acids. However, due to their relatively weak binding affinity, the peptides and celluloses on the nanosheet underwent rapid exchange with the molecules in solution to assume a “soft” biocorona35. Despite having the lowest concentration in the simulation, palmitic acids occasionally interacted with the dynamic biocorona under diffusion. The hydrophobic palmitic acids could also bind to themselves before interacting with the nanosheet (Supplementary Fig. S3). Once the nanosheet surface was available due to dynamic fluctuations, palmitic acids bound to the surface and remained attached as evidenced by a monotonic increase and small error bars of the corresponding number density (Fig. 6b). As a result, the biocorona became “hardened” as the weakly-bound peptides and celluloses were replaced by the palmitic acids. In the case of higher stoichiometric ratios of amphiphiles to the nanosheet, we expect a complete coverage of the nanosheet by strong binders like the palmitic acids to render a “hard” biocorona. Our results illustrate the general applicability of the Vroman effect for describing the binding kinetics of biomolecular species competing for graphene derivatives.
Discussion
We systematically studied the interactions of graphene derivatives with natural amphiphiles to elucidate the general fate of graphene nanostructures in biological and environmental milieu. Our computational studies showed that both graphene and graphene oxide could bind to the amphiphiles, although graphene oxide displayed a weaker binding capacity owing to its surface charge and surface functionalizations. Using algal exudates as a model system, our experimental characterizations confirmed the differential binding of graphene and graphene oxide for natural amphiphiles. Furthermore, our simulations revealed that natural amphiphiles of cellulose, peptide and palmitic acid -- owing to their differences in hydrocarbon content, conformational flexibility, and molecular geometry -- displayed distinct binding affinities for the graphene derivatives. Specifically, we were able to directly observe in our simulations, for the first time, a Vroman-like binding during which amphiphiles of different abundance and binding affinity rapidly competed for the graphene nanosheet surface; here amphiphiles of high abundance but low binding affinity readily covered the surface of the nanosheet to initiate a “soft” biocorona, while amphiphiles of low abundance but high affinity eventually took over to render a “hard” biocorona. This study offers a mechanistic basis for our understanding of the physicochemical properties and the fate and implications of graphene derivatives in biological and environmental matrices.
Methods
Construction of the model systems
The graphene nanosheet was prepared using the VMD36 carbon nanostructure builder plugin (www.ks.uiuc.edu/Research/vmd/plugins/nanotube). The dimension of the nanosheet was set as 25 × 25 Å2. The structure of graphene oxide was obtained by chemically editing the graphene nanosheet using Avogadro, a cross-platform molecule editor (avogadro.openmolecules.net)37. To mimic the experimentally-observed structural defects in graphene oxide38,39,40, we included random vacancies in the graphene oxide matrix. We then introduced epoxide, hydroxyl, and carboxyl groups to the lattice in a stochastic manner while maintaining the valencies of the composing carbon atoms. After the introduction of defects and chemical modifications, we performed energy minimization using the MMFF94s force field41. Because of the introduction of sp3 carbons on the planner carbon latter, the graphene oxide nanosheet became non-planner after the energy minimization (Fig. 1).
The molecular structures of cellulose dimer, tri-alanine and palmitic acid were generated and minimized using open babel (openbabel.org)42. We adapted the MedusaScore force field43, an extended Medusa force field44 for small-molecule ligands, to model the inter-atomic interactions of biomolecules beyond proteins45.
Simulation setup
The simulation was setup in a cubic box with each dimension of 100 Å. In our simulations, we applied harmonic constraints to the edge carbon atoms of graphene and graphene oxide with a weak spring constant k = 0.1 kcal/mol·Å2. The rest of nanosheet atoms were free to move. For each of the molecular system, we first performed equilibration simulations of 5 ns, and then production simulations of at least 50 ns.
In DMD simulations, the temperature unit is kcal/mol·kB. Here, kB is the Boltzmann constant. For the replica exchange simulations, we used 14 replicas with different temperatures to sample the conformational space. Ranges of temperatures used in replica exchange were from 0.65 to 1.55 for graphene-cellulose, 0.65 to 1.4 for graphene-peptide, 0.65 to 1.65 for graphene-palmitic acid, and from 0.35 to 1.2 for graphene oxide-cellulose, 0.35 to 1.0 for graphene oxide-peptide and 0.5 to 1.0 for graphene oxide-palmitic acid. The temperatures were adjusted to ensure sufficient exchange between replicas with neighboring temperatures and that the exchange rates were approximately equal.
To study competitive binding of the amphiphile mixtures, we placed a graphene oxide nanosheet together with 14 cellulose, 6 peptide and 2 palmitic acid molecules in a cubic box with the linear dimension of 110 Å. We followed previous report of algae exudate composition34 to set up the molecular system. We performed the constant-temperature DMD simulation at T = 0.67 kcal/mol·Å2 for 50 ns.
Inter-molecular contacts
We monitored inter-molecule interactions by measuring the number of atomic contacts between two molecules, Nc. Two atoms were counted as in contact if the inter-atomic distance was smaller than 6.5 Å. The fraction of inter-molecular contacts, Q, corresponded to Nc normalized by the maximum number of atomic contacts when two molecules were bound at low temperature.
Normal modes of the nanosheet
In our simulations, we constrained the nanosheets with harmonic constraints in order to direct monitor amphiphile binding without re-centering the molecular system around the graphene sheet. As a result, the constrained nanosheet can undergo collective motions, i.e. normal modes, which feature different frequencies and corresponding energies. At low temperatures, only the low-energy modes that the atoms moved along approximately the same direction were observed. As the temperature increased, higher-energy normal modes could be excited (Supplementary Fig. S1).
Weighted histogram analysis method (WHAM)
The WHAM method utilizes multiple simulation trajectories with overlapping sampling along the reaction coordinates to self-consistently compute the density of states ρ(E) by combining histograms from different simulation trajectories46. Given the density of states, the partition function can be computed: 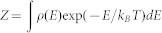 . To compute any physical parameter as a function of temperature, we calculated the conditional probability P(A | E) of observing a structure with the parameter of A at given energy E, evaluated from all simulation trajectories. The average RMSD as a function of temperature can be computed as
. To compute any physical parameter as a function of temperature, we calculated the conditional probability P(A | E) of observing a structure with the parameter of A at given energy E, evaluated from all simulation trajectories. The average RMSD as a function of temperature can be computed as 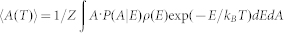 .
.
Syntheses of graphene derivatives
Few-layer graphene samples were prepared using chemical vapor deposition technique. Briefly, 25 μm Ni foils were placed away from the center of tube furnace (diameter: 24 mm), which was maintained at 900°C under a flow of Ar (230 sccm) and H2 (120 sccm). After 60 min, Ni foils were moved to the center and graphene was synthesized by decomposing methane (10 sccm) for 10 min at a reduced temperature (850°C). Subsequently, methane flow was shut off and the samples were moved away from the center. The furnace temperature was ramped down to 400°C at 5°C/min and was maintained at 400°C for 90 min. The H2 flow was shut off immediately upon reaching 400°C, and the samples were cooled to room temperature under Ar flow.
For solvent exfoliation of graphene, bulk graphite (~1 g) was dispersed in 100 mL of N-methyl-2-pyrrolidinone (NMP) and sonicated using 1/8” tip sonicator (Branson 250) at 100 W for 2 h. The resulting dispersion was filtered through a 0.45 μm nylon filter and re-suspended in 100 mL of fresh NMP. Subsequently, the solution was bath sonicated for 6 h and centrifuged at 500 rpm for 45 min. The supernatant was vacuum filtered using a 0.45 μm nylon filter. Finally, the filtered powder was washed several times using deionized water to remove residual NMP. We used modified Hummer's method to prepare graphene oxide.
Briefly, exfoliated graphene (2 g) was dispersed in concentrated H2SO4 (46 mL). KMnO4 (6 g) was added gradually with stirring in an ice bath. The mixture was then stirred at 35°C for 2 h, and deionized water (100 mL) was added. In 1 h, the reaction was terminated by the addition of a large amount of deionized water (300 mL) and 30% H2O2 solution (5 mL), causing violent effervescence and an increase in temperature to 100°C, after which the color of the suspension changed to bright yellow. The suspension was washed with 1:10 HCl solution (500 mL) in order to remove metal ions by filter paper and funnel. The paste collected from the filter paper was dried at 60°C, until it became agglomerated. The agglomeration was washed several times with deionized water and air-dried to obtain graphene oxide samples.
Micro-Raman and FTIR measurement
Micro-Raman spectra were collected using a Dilor XY triple grating spectrometer equipped with TE-cooled CCD coupled to an Ar+ laser excitation at 514.5 nm. The Raman spectrum of graphene oxide exhibited a weaker 2D-band compared to graphene. Nonetheless, we observed that the 2D-band for graphene oxide samples did not upshift (unlike graphene) upon incubation with algal exudates (see Supplementary Fig. S2). For FTIR measurements, the samples were incubated with algal exudates for 12 h. Subsequently, the samples were washed using de-ionized water to remove any unadsorbed exudates. For graphene oxide, the samples were centrifuged at 3,000 rpm for 2 h and the obtained pellets were washed in deionized water. Finally, the sample was air-dried overnight for performing FTIR studies. The dried samples were encased in a KBr matrix and their FTIR spectra were measured using a Bruker IFS v66 spectrometer.
UV-Vis measurement
For UV-Vis kinetics measurements, graphene was suspended in water from dry state and both graphene and graphene oxide (both 10 μg/mL) were bath sonicated for 5 min. Algal exudates, prepared as described previously30, were then added and the resulting suspensions were immediately placed in a temperature-controlled UV-Vis spectrophotometer (Cary 300-Bio, Varian Instruments). The maximum temperature fluctuation of the device was ±0.02°C.
Author Contributions
S.R. performed the simulations, analyzed the results and wrote the paper. N.G. performed the UV-Vis measurement and wrote the relevant Methods and experimental results. R.P. synthesized graphene derivatives, conducted the Raman and FTIR studies, and wrote the relevant Methods. A.K. and P.C. prepared algal exudates and contributed to the analysis. P.C.K. and F.D. conceived and initiated the study, contributed to the analysis and edited the paper.
Supplementary Material
Supplementary Figures
Acknowledgments
The work was supported by an NSF Grant CBET-1232724 (to P.C.K.), the startup funds from Clemson University (to F.D.), and the DoRa program with the Foundation Archimedes, Estonia (to A.K.). The simulations were performed on the Palmetto high performance cluster, which is managed and maintained by Clemson University CCIT.
References
- Novoselov K. S. et al. Electric field effect in atomically thin carbon films. Science 306, 666–669 (2004). [DOI] [PubMed] [Google Scholar]
- Huang X. et al. Graphene-based materials: synthesis, characterization, properties, and applications. Small 7, 1876–1902 (2011). [DOI] [PubMed] [Google Scholar]
- Geim A. K. Graphene: status and prospects. Science 324, 1530–1534 (2009). [DOI] [PubMed] [Google Scholar]
- He Q., Wu S., Yin Z. & Zhang H. Graphene-based electronic sensors. Chem. Sci. 3, 1764–1772 (2012). [Google Scholar]
- Yang W. et al. Carbon Nanomaterials in biosensors: should you use nanotubes or graphene? Angew. Chem.-Int. Edit. 49, 2114–2138 (2010). [DOI] [PubMed] [Google Scholar]
- Qiu J.-D., Huang J. & Liang R.-P. Nanocomposite film based on graphene oxide for high performance flexible glucose biosensor. Sens. Actuator B-Chem. 160, 287–294 (2011). [Google Scholar]
- Morales-Narvaez E. & Merkoci A. Graphene oxide as an optical biosensing platform. Adv. Mater. 24, 3298–3308 (2012). [DOI] [PubMed] [Google Scholar]
- Yang K., Feng L., Shi X. & Liu Z. Nano-graphene in biomedicine: theranostic applications. Chem. Soc. Rev. 42, 530–547 (2013). [DOI] [PubMed] [Google Scholar]
- Sivudu K. S. & Mahajan Y. Mass production of high quality graphene: An analysis of worldwide patents, http://www.nanowerk.com/spotlight/spotid=25744.php (2012).
- The Global Market for Graphene to 2020, Future Markets, Inc., http://www.researchandmarkets.com/reports/2492343 (2013).
- Wang K. et al. Biocompatibility of graphene oxide. Nanoscale Research Letters 6, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedervall T. et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 104, 2050–2055 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke P. C. Fiddling the string of carbon nanotubes with amphiphiles. Phys. Chem. Chem. Phys. 9, 439–447 (2007). [DOI] [PubMed] [Google Scholar]
- Hlady V. & Buijs J. Protein adsorption on solid surfaces. Curr. Opin. Biotechnol. 7, 72–77 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Robinson J. T., Sun X. & Dai H. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J. Am. Chem. Soc. 130, 10876–10877 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson K. et al. Determination of the local chemical structure of graphene oxide and reduced graphene oxide. Adv. Mater. 22, 4467–4472 (2010). [DOI] [PubMed] [Google Scholar]
- Pacilé D. et al. Electronic properties and atomic structure of graphene oxide membranes. Carbon 49, 966–972 (2011). [Google Scholar]
- Katoch J. et al. Structure of a peptide adsorbed on graphene and graphite. Nano Lett. 12, 2342–2346 (2012). [DOI] [PubMed] [Google Scholar]
- Kim S. N. et al. Preferential binding of peptides to graphene edges and planes. J. Am. Chem. Soc. 133, 14480–14483 (2011). [DOI] [PubMed] [Google Scholar]
- Pandey R. B., Kuang Z., Farmer B. L., Kim S. S. & Naik R. R. Stability of peptide (P1 and P2) binding to a graphene sheet via an all-atom to all-residue coarse-grained approach. Soft Matter 8, 9101–9109 (2012). [Google Scholar]
- Qin W., Li X., Bian W.-W., Fan X.-J. & Qi J.-Y. Density functional theory calculations and molecular dynamics simulations of the adsorption of biomolecules on graphene surfaces. Biomaterials 31, 1007–1016 (2010). [DOI] [PubMed] [Google Scholar]
- Rapaport D. C. The art of molecular dynamics simulation (Cambridge University Press, 2004). [Google Scholar]
- Ding F. & Dokholyan N. V. Emergence of protein fold families through rational design. PLoS Comput. Biol. 2, e85 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis T. & Karplus M. Effective energy functions for protein structure prediction. Curr. Opin. Struct. Biol. 10, 139–145 (2000). [DOI] [PubMed] [Google Scholar]
- Ding F., Tsao D., Nie H. & Dokholyan N. V. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure 16, 1010–1018 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F., Furukawa Y., Nukina N. & Dokholyan N. V. Local unfolding of Cu, Zn superoxide dismutase monomer determines the morphology of fibrillar aggregates. J. Mol. Biol. 421, 548–560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y., Sugiyama J., Chanzy H. & Langan P. Crystal structure and hydrogen bonding system in cellulose Iα from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 125, 14300–14306 (2003). [DOI] [PubMed] [Google Scholar]
- Metropolis N. & Ulam S. The Monte Carlo method. J. Am. Stat. Assoc. 44, 335–341 (1949). [DOI] [PubMed] [Google Scholar]
- Chodera J. D., Swope W. C., Pitera J. W., Seok C. & Dill K. A. Use of the weighted histogram analysis method for the analysis of simulated and parallel tempering simulations. J. Chem. Theory Comput. 3, 26–41 (2007). [DOI] [PubMed] [Google Scholar]
- Chen P., Powell B. A., Mortimer M. & Ke P. C. Adaptive interactions between zinc oxide nanoparticles and Chlorella sp. Environ. Sci. Technol. 46, 12178–12185 (2012). [DOI] [PubMed] [Google Scholar]
- Podila R., Vedantam P., Ke P. C., Brown J. M. & Rao A. M. Evidences for charge transfer-induced conformational changes in carbon nanostructure-protein corona. J. Phys. Chem. C Nanomater Interfaces 116, 22098–22103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. & Ruoff R. S. Chemical methods for the production of graphenes. Nature Nanotech. 4, 217–224 (2009). [DOI] [PubMed] [Google Scholar]
- Podila R., Rao R., Tsuchikawa R., Ishigami M. & Rao A. M. Raman spectroscopy of folded and scrolled graphene. ACS Nano 6, 5784–5790 (2012). [DOI] [PubMed] [Google Scholar]
- Haas A. F. & Wild C. Composition analysis of organic matter released by cosmopolitan coral reef-associated green algae. Aquat. Biol. 10, 131–138 (2010). [Google Scholar]
- Milani S., Bombelli F. B., Pitek A. S., Dawson K. A. & Raedler J. Reversible versus irreversible binding of transferrin to polystyrene nanoparticles: soft and hard corona. ACS Nano 6, 2532–2541 (2012). [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A. & Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- Hanwell M. D., Curtis D. E., Lonie D. C., Vandermeersch T., Zurek E. & Hutchison G. R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 4, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K. P., Bao Q. L., Eda G. & Chhowalla M. Graphene oxide as a chemically tunable platform for optical applications. Nature Chem. 2, 1015–1024 (2012). [DOI] [PubMed] [Google Scholar]
- Bagri A., Mattevi C., Acik M., Chabal Y. J., Chhowalla M. & Shenoy V. B. Structural evolution during the reduction of chemically derived graphene oxide. Nature Chem. 2, 581–587 (2010). [DOI] [PubMed] [Google Scholar]
- Dreyer D. R., Park S., Bielawski C. W. & Ruoff R. S. The chemistry of graphene oxide. Chem. Soc. Rev. 39, 228–240 (2010). [DOI] [PubMed] [Google Scholar]
- Halgren T. A. MMFF VI. MMFF94s option for energy minimization studies. J. Comput. Chem. 20, 720–729 (1999). [DOI] [PubMed] [Google Scholar]
- O'Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T. & Hutchison G. R. Open babel: an open chemical toolbox. J. Cheminformatics 3, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S., Biedermannova L., Vondrasek J. & Dokholyan N. V. MedusaScore: an accurate force-field based scoring function for virtual drug screening. J. Chem. Inf. Model 48, 1656–1662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F. & Dokholyan N. V. Emergence of protein fold families through rational design. PLoS Comput. Biol. 2, e85 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor E. A., Ding F. & Dokholyan N. V. Structural and thermodynamic effects of post-translational modifications in mutant and wild type Cu, Zn superoxide dismutase. J. Mol. Biol. 408, 555–567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Rosenberg J. M., Bouzida D., Swendsen R. H. & Kollman P. A. The weighted histogram analysis method for free energy calculations on biomolecules. I. The method. J. Comput. Chem. 13, 1011–1021 (1992). [Google Scholar]
- Chodera J. D., Swope W. C., Pitera J. W., Seok C. & Dill K. A. Use of the weighted histogram analysis method for the analysis of simulated and parallel tempering simulations. J. Chem. Theory Comput. 3, 26–41 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures