Abstract
RacA is the main Rho GTPase in Aspergillus niger regulating polarity maintenance via controlling actin dynamics. Both deletion and dominant activation of RacA (RacG18V) provoke an actin localization defect and thereby loss of polarized tip extension, resulting in frequent dichotomous branching in the ΔracA strain and an apolar growing phenotype for RacG18V. In the current study the transcriptomics and physiological consequences of these morphological changes were investigated and compared with the data of the morphogenetic network model for the dichotomous branching mutant ramosa-1. This integrated approach revealed that polar tip growth is most likely orchestrated by the concerted activities of phospholipid signaling, sphingolipid signaling, TORC2 signaling, calcium signaling and CWI signaling pathways. The transcriptomic signatures and the reconstructed network model for all three morphology mutants (ΔracA, RacG18V, ramosa-1) imply that these pathways become integrated to bring about different physiological adaptations including changes in sterol, zinc and amino acid metabolism and changes in ion transport and protein trafficking. Finally, the fate of exocytotic (SncA) and endocytotic (AbpA, SlaB) markers in the dichotomous branching mutant ΔracA was followed, demonstrating that hyperbranching does not per se result in increased protein secretion.
Introduction
Filamentous fungi such as Aspergillus niger are widely used in biotechnology for the production of various proteins, enzymes, food ingredients and pharmaceuticals [1]–[4]. During recent years, A. niger became an industrial model fungus, due to its well annotated genome sequence, sophisticated transcriptomics and proteomics technologies and newly established gene transfer systems allowing efficient and targeted genetic and metabolic engineering approaches [3], [5]–[7].
The morphology of filamentous fungi strongly affects the productivity of industrial fermentations [8]–[10]. Basically, Aspergilli and all other filamentous fungi grow either as pellets or as freely dispersed mycelium during submerged growth. Both macromorphologies depend among other things on hyphal branching frequencies – pellets are formed when hyphae branch with a high frequency, dispersed mycelia are a result of low branching frequencies. Whereas the formation of pellets is less desirable because of the high proportion of biomass in a pellet that does not contribute to product formation, long, unbranched hyphae are sensitive to shear forces in a bioreactor. Lysis of hyphae and the subsequent release of intracellular proteases have thus a negative effect on protein production. Hence, from an applied point of view, the preferred fungal macromorphology would consist of dispersed mycelia with short filaments derived from an optimum branching frequency. It is generally accepted that protein secretion occurs mainly at the hyphal apex [11]–[14]. Some studies suggested a positive correlation between the amount of hyphal branches and protein secretion yields [13], [15]–[17], whereas other reports demonstrated no correlation [9], [18]. Therefore, it is still a matter of debate whether a hyperbranching production strain would considerably improve protein secretion rates.
Different mutations can lead to a hyperbranching phenotype in filamentous fungi. For example, dichotomous branching (tip splitting) is a characteristic of the actin (act1) and actinin mutants in Neurospora crassa and A. nidulans [19], [20], a consequence of deleting the formin SepA in A. nidulans [21] or the polarisome component SpaA in A. nidulans and A. niger [22], [23] and a consequence of inactivating the Rho GTPase RacA or the TORC2 complex component RmsA protein in A. niger [24], [25]. Common to these different gene mutations is not only the phenotype they provoke but that they also disturb the dynamics of the actin cytoskeleton. Actin is crucial for polarized hyphal growth in filamentous fungi and controls many cellular processes, including intracellular movement of organelles, protein secretion, endocytosis and cytokinesis [26], [27].
We have recently analyzed the function of all six Rho GTPase encoded in the genome of A. niger (RacA, CftA, RhoA, RhoB, RhoC, RhoD) and uncovered that apical dominance in young germlings and mature hyphae of A. niger is predominantly controlled by RacA [24]. Both RacA and CftA are not essential for A. niger (in contrast to RhoA) but share related functions which are executed in unicellular fungi only by Cdc42p [24]. The data showed that RacA localizes to the apex of actively growing filaments, where it is crucial for actin distribution. Both deletion and dominant activation of RacA (RacAG18V expressed under control of the maltose-inducible glucoamlylase promoter glaA) provoke an actin localization defect and thereby loss of polarized tip extension. In the case of RacA inactivation, actin becomes hyperpolarized, leading to frequent dichotomous branching. Dominant activation of RacA, however, causes actin depolarization, leading to a swollen-tip phenotype and the formation of bulbous lateral branches. Interestingly, the dichotomous branching phenotype suggested that loss of apical dominance in ΔracA can frequently be overcome by the establishment of two new sites of polarized growth. This phenotype resembles the phenotype of the ramosa-1 mutant of A. niger, which harbors an temperature-sensitive mutation in the TORC2 component RmsA causing a transient contraction of the actin cytoskeleton [25], [28], [29].
Altogether, the data supported the model that RacA is important to stabilize polarity axes of A. niger hyphae via controlling actin (de)polymerization at the hyphal apex. The aim of the present study was to unravel the genetic network into which RacA is embedded and which, when disturbed due to deletion or dominant activation of RacA, leads to loss of polarity maintenance and in the case of ΔracA, to reestablishment of two new polarity axes. To determine whether the hyperbranching phenotype of ΔracA leads to an increase in the amount of secreted proteins, the transcriptomes of our previously established RacA mutant strains (ΔracA, PglaA::racAG18V) were compared with the transcriptomes of the respective reference strains (wt, PglaA::racA). By applying defined culture conditions in bioreactor cultivations, branching morphologies as well as physiological parameters including specific growth rate and protein production rate were characterized. Finally, the implication of ΔracA on endocytosis and exocytosis in A. niger was examined by analyzing reporter strains harboring fluorescently tagged SlaB and AbpA (markers for endocytotic actin) and SncA (marker for secretory vesicles). The data obtained were compared with transcriptomic and physiological data of the dichotomous branching mutant ramosa-1 [25], thereby providing new insights into the morphogenetic network of A. niger.
Results
Physiological consequences of RacA inactivation
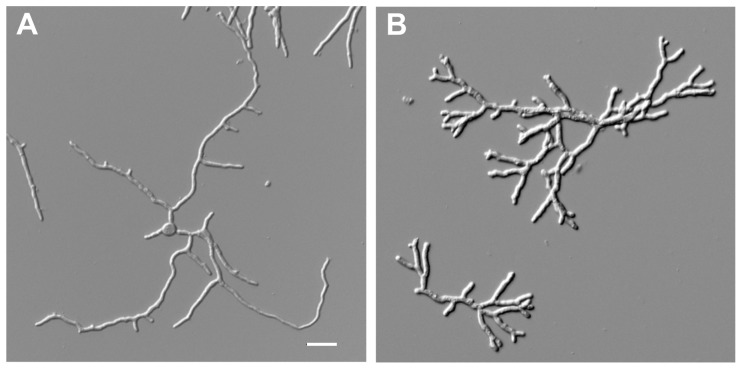
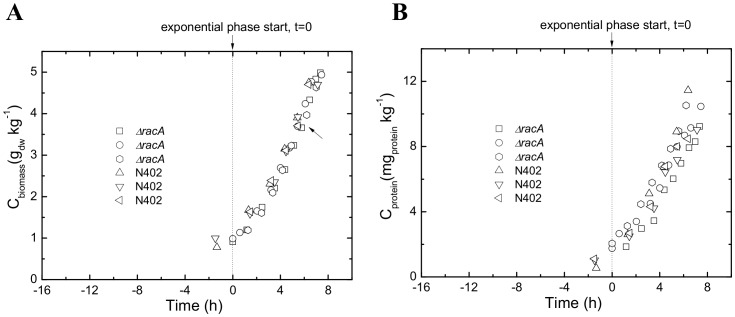
As previously reported, deletion of racA in A. niger provokes hyperbranching germ tubes and hyphae, which are shorter in length but wider in hyphal diameter. This frequent branching results on solid medium in a more compact colony with a reduced diameter due to slower tip extension rates [24]. In order to further characterize the implications of loss of RacA function, the reference strain (wild-type N402) and the ΔracA strain were cultivated in triplicate batch cultures using maltose as growth-limiting carbon source. Propagation of both strains gave rise to homogeneous cultures of dispersed mycelia, whereby loss of RacA resulted in an about 30% higher branching frequency (Fig. 1 and Table 1). Physiological profiles including growth curves, maximum specific growth rates and specific protein secretion rates were obtained with high reproducibility and were nearly identical for both strains despite the significant difference in their morphology (Fig. 2 and Table 2). This result might come with surprise because of the negative effect of the racA deletion on radial colony growth on solid medium [24]. However, growth on solid media can only be assessed based on colony diameter (reflecting tip extension) and not on biomass accumulation (i.e. increase in cell volume per time). During exponential growth, growth yield on substrate (Yx/s) was comparable in both strains; 0.63±0.03 and 0.60±0.02 gbiomass gmaltose −1 for ΔracA and N402, respectively. Notably, the amount of extracellular protein was not altered in ΔracA strain compared to N402 (Table 2). Hence, an increased branching frequency is the only highly significant consequence of racA disruption, which, however, does not per se result in higher protein secretion rates.
Figure 1. Hyphal morphology during dispersed growth.
Mycelial samples of the wild-type strain N402 (A) and the ΔracA mutant strain (B) were taken during the mid-exponential phase when approximately 75% of the carbon source was consumed. Bar, 20 µm.
Table 1. Comparative image analysis of branching morphologies.
| N402 (n = 38) | ΔracA (n = 13) | |
| Mycelium length (µm) | 510±177 | 507±184 |
| No. of hyphal apices | 17±6 | 22±8 |
| Branch length (µm) | 32±6 | 23±2 |
| Central hyphal length (µm) | 257±63 | 162±41 |
Morphological samples were taken from the exponential growth phase and the individual mycelium was randomly selected to measure the length of the mycelium and the number of branching tips using imageJ. Mean values ± standard deviations are given. Bold letters indicate significant differences (two tailed t-test, p<0.01).
Figure 2. Biomass (A) and extracellular protein (B) accumulation for the wild-type strain N402 and the ΔracA strain.
The arrow indicates the time point when biomass samples were harvested for transcriptomics analyses. The graphs represent data for three independent biological replicate cultures per strain.
Table 2. Physiological characterization of N402 and ΔracA strains.
| N402 | ΔracA | |
| Maximum specific growth rate (h−1) | 0.22±0.01 | 0.24±0.01 |
| Yield (gdw gmaltose −1) | 0.60±0.02 | 0.63±0.03 |
| Respiratory quotient (RQ) | 0.97±0.05 | 1.03±0.03 |
| Acidification (mmolbase gdw −1 h−1) | 1.19±0.06 | 1.17±0.02 |
| Specific protein secretion rate (mgprotein gdw −1 h−1) | 0.49±0.07 | 0.53±0.02 |
Biomass samples were taken from triplicate independent batch cultivations using maltose as carbon source (Fig. 2). Mean values ± standard deviations are given. No significant difference was observed with any of the variables (two tailed t-test, p<0.01). RQ, respiratory quotient calculated as the ratio of CO2 production and O2 consumption rates.
Consequences of RacA inactivation on exo- and endocytosis
We previously showed that RacA is important for actin localization at the hyphal tip [24]. As actin is important for both exo- and endocytosis, the consequences of racA deletion on both was assessed in A. niger by following the localization of fluorescently-labelled reporter proteins SncA, AbpA and SlaB, respectively. SncA is the vesicular-SNARE that is specific for the fusion of Golgi derived secretory vesicles with the plasma membrane [30] and used as marker for exocytosis in A. nidulans [31], [32]. Abp1/AbpA and Sla2/SlaB are actin binding proteins and well characterized endocytic markers in yeast and filamentous fungi [31], [33], [34]. Screening of the genome sequence of A. niger [5] predicted for each of the established marker proteins a single orthologue for A. niger: An12g07570 for SncA, An03g06960 for Abp1/AbpA and An11g10320 for Sla2/SlaB.
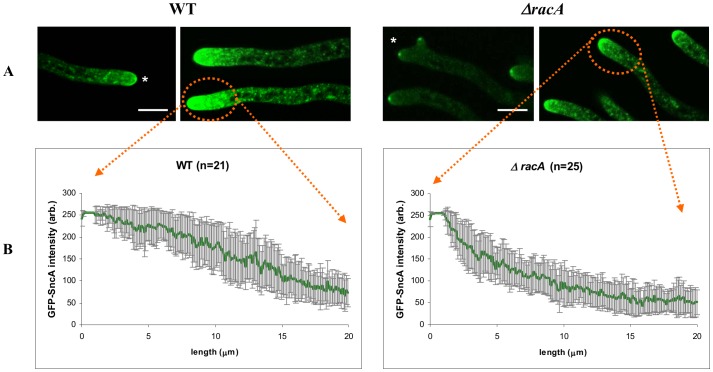
We constructed a reporter strain expressing a fusion of GFP with the v-SNARE SncA as described elsewhere (Kwon et al., manuscript submitted). In brief, physiological expression levels of GFP-SncA was ensured by fusing GFP between the N-terminus and the promoter of sncA and used this cassette to replace it with the endogenous sncA gene (giving strain FG7). As depicted in Figure 3A, GFP-SncA is visible as punctuate intracellular structures representing secretory vesicles. These vesicles accumulate towards the hyphal tip, overlap with the Spitzenkörper and are highest at the extreme apex, which is proposed to be the site of exocytosis in filamentous fungi [31]. This localization of GFP-SncA in A. niger was very similar to the localization previously reported for other filamentous fungi [14], [31], [35]–[37]. Importantly, the amount of secretory vesicles per hyphal tip was affected in the ΔracA strain. Although the localization of GFP-SncA was similar to the wild-type strain, the intensity of the signal was considerably lower. Quantification of the GFP signal intensities in both strains revealed that the tips of wild-type hyphae display a GFP-SncA gradient of ∼20–25 µm but only about ∼10 µm in the racA deletion strain (Fig. 3B). Both strains, however, do not differ in their specific protein secretion rates (Table 2), implying that the total amount of secretory vesicles is the same in both strains. This discrepancy can most easily be explained by the assumption that secretory vesicles in the hyperbranching ΔracA strain are merely distributed to more hyphal tips, which in consequence lowers the amount of vesicles per individual hyphal tip.
Figure 3. Localization of secretory vesicles and fluorescent intensity distributions in the wild-type strain N402 and the ΔracA mutant strain using GFP-SncA as fluorescent marker.
(A) CLSM images showing the localization of GFP-SncA in hyphal tips. The Spitzenkörper is indicated with a star. (B) Fluorescent intensity distributions along hyphal tip compartments (n>20) within a region of 20 µm. Bar, 10 µm.
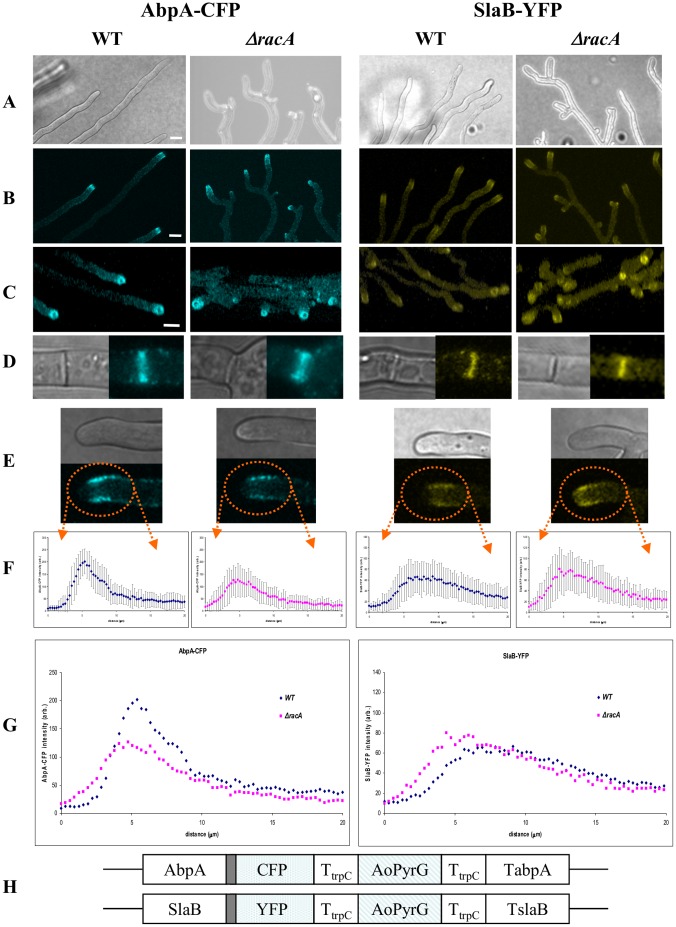
To follow the effect of racA deletion on endocytosis, AbpA and SlaB were labeled using a C-terminal labeling strategy as previously reported for A. nidulans [31]. Importantly, both AbpA-CFP and SlaB-YFP were expressed at physiological levels by using the respective endogenous promoter and by replacing the constructs with the endogenous abpA and slaB gene, respectively (Fig. 4B). AbpA-CFP (strain MK6.1) and SlaB-YFP (strain MK5.1) transformants were phenotypically indistinguishable from the recipient strain, indicating that both AbpA-CFP and SlaB-YFP are functionally expressed (Fig. 4 and data not shown). Although AbpA-CFP and SlaB-YFP fluorescence signals were only weakly detectable (which is a direct consequence of their low endogenous expression level), both proteins were visible in the wild-type background as peripheral punctate structures and formed a subapical ring likely reflecting the endocytic machinery (Fig. 4A–C). Fluorescent signals were excluded from the hyphal apex which is in agreement with previous reports for A. nidulans showing that endocytosis occurs behind the tip [31], [34]. The signal of SlaB-YFP but not AbpA-CFP seemed to be intimately associated with the plasma membrane (data not shown), which would be in agreement with the function of both proteins - Sla2/SlaB is involved in early endocytic site initiation while Abp1/AbpA is important for invagination, scission and release of endocytotic vesicles [38]. SlaB-YFP and AbpA-CFP fluorescence was also occasionally observed at septa or sites destined for septum formation (Fig. 4D), probably suggesting an involvement of endocytotic events at septa as recently reported for A. oryzae [14]. Importantly, the intensity and distribution of SlaB-YFP and AbpA-CFP differed slightly in the wild-type and the ΔracA strain (Fig. 4E–G). The endocytotic actin ring as visualized by AbpA-CFP was sharper formed in the wild-type background but more diffuse in the ΔracA strain, an observation which, however, was not so evident for SlaB-YFP (Fig. 4 F–G). Still, the endocytotic ring formed by both SlaB-YFP and AbpA-CFP seemed to be positioned closer to the hyphal apex in the ΔracA strain (Fig. 4G), suggesting that deletion of RacA affected endocytotic processes and provoked a slight mislocalization of the endocytotic ring.
Figure 4. Localization of endocytic ring structures in the wild-type strain N402 and the ΔracA mutant strain using AbpA-CFP and SlaB-YFP as fluorescent markers.
The cultures were grown for two days at 22°C on MM agar. (A) Transmission light images, (B) two dimensional fluorescent confocal images and (C) three dimensional reconstructions from z-sectional confocal images are shown for both strains. (D, E) depict selected light and fluorescent images at sites of septation (D) and at sites of endocytosis (E). (F, G) Fluorescent intensity distribution of endocytotic ring structures. Fluorescence was measured in at least 25 hyphal tips of each strain. Mean values with (F) or without (G) standard deviation is shown. (H) Schematic representation of the AbpA-CFP and SlaB-YFP constructs designed for integration at the endogenous abpA and slaB loci, respectively. The A. oryzae pyrG gene served as selection marker, a sequence encoding 5× Gly-Ala as peptide linker (grey box) and the 3′ region from the A. nidulans trpC gene as terminator. Bar, 10 µm.
The transcriptomic fingerprint of hyperbranching in ΔracA
To study the transcriptomic consequences due to deletion of racA, RNA samples from triplicate bioreactor cultivations were taken after both wt and ΔracA cultures reached the mid-exponential growth phase (biomass concentration 3.7 gr kg−1). A total of 139 genes out of 14,165 A. niger genes were identified as differentially expressed, 44 of which displayed increased and 95 genes decreased expression levels in ΔracA (FDR<0.05). The complete list of differentially expressed genes, including fold change and statistical significance is given in Tables S1 and S2. Interestingly, the modulated gene set in ΔracA is very small (139 genes, i.e. 1% of all A. niger genes) but in the same range as the differentially expressed gene set in the dichotomous branching mutant ramosa-1 of A. niger (136 genes; [25]. Although the affected gene sets had opposite signs (44 up/95 down in ΔracA, 109 up/27 down in ramosa-1), similar processes were affected in both hyperbranching strains (Table 3): (i) (phospho)lipid signaling, (ii) calcium signaling, (iii) cell wall integrity (CWI) signaling and cell wall remodeling, (iv) γ-aminobutyric acid (GABA) metabolism and (v) transport phenomena. Specific responses for ΔracA but not ramosa-1 included genes related to actin localization and protein trafficking.
Table 3. Selected genes whose expression profile respond to hyperbranching in ΔracA (this work) and ramosa-1 [25]. Genes are ordered into different processes and functions.
| Predicted protein function* | ΔracA versus wt | ramosa-1 versus wt | ||||
| Open reading frame code | Up/Down | Closest S. cerevisiae ortholog | Open reading frame code | Up/Down | Closest S. cerevisiae ortholog | |
| (Phospho)lipid metabolism and signaling | ||||||
| phosphatidyl synthase, synthesis of phosphatidyl alcohols | An02g08050 | ↑ | ||||
| sterol 24-C-methyltransferase, ergosterol synthesis | An04g04210 | ↑ | Erg6 | |||
| C-14 sterol reductase, ergosterol synthesis | An01g07000 | ↓ | Erg24 | An01g07000 | ↑ | Erg24 |
| inositol hexaki-/heptaki-phosphate kinase, synthesis of IP6, IP7 | An14g04590 | ↓ | Kcs1 | An16g05020 | ↑ | Vip1 |
| plasma membrane protein promoting PI4P synthesis | An18g06410 | ↑ | Sfk1 | |||
| phospholipase B, synthesis of glycerophosphocholine | An18g01090 | ↓ | Plb3 | |||
| phospholipase B, synthesis of glycerophosphocholine | An02g13220 | ↓ | Plb1 | |||
| phospholipase D, synthesis of phosphatidic acid | An15g07040 | ↑ | Spo14 | |||
| diacylglycerol pyrophosphate phosphatase, synthesis of DAG | An04g03870 | ↓ | Dpp1 | An04g03870 | ↑ | Dpp1 |
| diacylglycerol pyrophosphate phosphatase, synthesis of DAG | An02g01180 | ↓ | Dpp1 | An11g05330 | ↑ | |
| choline/ethanolamine permease | An16g01200 | ↑ | Hnm1 | |||
| choline/ethanolamine permease | An01g13290 | ↓ | Hnm1 | |||
| transcription factor important for sterol uptake | An02g09780 | ↓ | Upc2 | |||
| transcription factor important for sterol uptake | An12g00680 | ↓ | Upc2 | |||
| mannosyl-inositol phosphorylceramide (MIPC) synthase | An05g02310 | ↓ | Sur1 | |||
| Calcium homeostasis and signaling | ||||||
| Ca2+/calmodulin dependent protein kinase | An02g05490 | ↑ | Cmk2 | |||
| Ca2+/calmodulin dependent protein kinase | An16g03050 | ↑ | Cmk2 | |||
| vacuolar Ca2+/H+ exchanger | An01g03100 | ↓ | Vcx1 | An01g03100 | ↑ | Vcx1 |
| vacuolar Ca2+/H+ exchanger | An05g00170 | ↓ | Vcx1 | An05g00170 | ↑ | Vcx1 |
| vacuolar Ca2+/H+ exchanger | An14g02010 | ↓ | Vcx1 | |||
| Ca2+ transporting ATPase | An19g00350 | ↓ | Pmc1 | An02g06350 | ↑ | Pmc1 |
| Ca2+/phospholipid-transporting ATPase | An04g06840 | ↑ | Drs2 | |||
| Cell wall remodeling and integrity | ||||||
| membrane receptor, CWI signaling | An01g14820 | ↑ | Wsc2 | |||
| MAP kinase kinase, CWI signaling, MkkA | An18g03740 | ↑ | Mkk1/2 | |||
| plasma protein responding to CWI signaling | An07g08960 | ↓ | Pun1 | |||
| plasma protein responding to CWI signaling | An08g01170 | ↓ | Pun1 | |||
| α-1,3-glucanase | An04g03680 | ↑ | An08g09610 | ↑ | ||
| β-1,3-glucanosyltransferase (GPI-anchored) | An16g06120 | ↑ | Gas1 | |||
| β-1,4-glucanase | An03g05530 | ↓ | An03g05530 | ↑ | ||
| chitin synthase class II, similar to ChsA of A. nidulans | An07g05570 | ↑ | ||||
| chitin transglycosidase (GPI-anchored) | An07g01160 | ↓ | Crh2 | An13g02510 | ↑ | Crh1 |
| chitinase (GPI-anchored), similar to ChiA of A. nidulans | An09g06400 | ↓ | Cts1 | |||
| β-mannosidase | An11g06540 | ↑ | ||||
| endo-mannanase (GPI-anchored), DfgE | An16g08090 | ↓ | Dfg1 | An16g08090 | ↑ | Dfg1 |
| α-1,2-mannosyltransferase | An14g03910 | ↑ | Kre2 | |||
| α-1,2-mannosyltransferase | An18g05910 | ↑ | Kre2 | |||
| α-1,3-mannosyltransferase | An15g04810 | ↓ | Mnt2 | |||
| α-1,6-mannosyltransferase | An05g02320 | ↓ | ||||
| cell wall protein | An14g01840 | ↓ | Tir3 | An04g05550 | ↑ | Flo11 |
| cell wall protein | An11g01190 | ↓ | Sps22 | An11g01190 | ↑ | Sps22 |
| cell wall protein | An03g05560 | ↑ | ||||
| cell wall protein | An04g03830 | ↑ | ||||
| cell wall protein | An02g11620 | ↓ | ||||
| plasma membrane protein | An02g08030 | ↓ | Pmp3 | |||
| GABA metabolism | ||||||
| glutaminase | An11g07960 | ↑ | ||||
| γ-aminobutyrate transaminase | An17g00910 | ↑ | Uga1 | |||
| NAD(+)-dependent glutamate dehydrogenase | An02g14590 | ↑ | Gdh2 | |||
| GABA permease | An16g01920 | ↑ | ||||
| transcription factor for GABA genes | An02g07950 | ↓ | ||||
| Transporter | ||||||
| mitochondrial phosphate translocator | An02g04160 | ↑ | Mir1 | |||
| mitochondrial ABC transporter during oxidative stress | An12g03150 | ↓ | Mdl1 | |||
| vacuolar glutathione S-conjugate ABC-transporter | An13g02320 | ↑ | Ycf1 | |||
| plasma membrane Na+/K+-exchanging ATPase alpha-1 chain | An09g00930 | ↓ | An09g00930 | ↑ | ||
| plasma membrane K+ transporter | An03g02700 | ↓ | Trk1 | |||
| multidrug transporter | An01g05830 | ↓ | Qdr1 | An02g01480 | ↑ | |
| low-affinity Fe(II) transporter of the plasma membrane | An16g06300 | ↓ | Fet4 | An16g06300 | ↑ | Fet4 |
| vacuolar H+-ATPase subunit, required for copper and iron homeostasis | An10g00680 | ↓ | Vma3 | |||
| siderophore-iron transporter | An12g05510 | ↓ | Taf1 | |||
| mitochondrial carrier protein | An14g01860 | ↓ | Rim2 | |||
| vacuolar zinc transporter | An15g03900 | ↓ | Zrc1 | |||
| allantoin permease | An18g01220 | ↓ | Dal5 | |||
| oligopeptide transporter | An13g01760 | ↑ | Opt1 | An15g07460 | ↓ | Opt1 |
| oligopeptide transporter | An11g01040 | ↓ | Opt1 | |||
| hexose/glucose transporter | An06g00560 | ↑ | Hxt13 | An09g04810 | ↓ | |
| amino acid transporter | An03g00430 | ↑ | ||||
| galactose transporter | An01g10970 | ↓ | Gal2 | |||
| FAD transporter into endoplasmatic reticulum, CWI-related | An01g09050 | ↓ | Flc2 | |||
| Protein trafficking | ||||||
| GTPase activating protein involved in protein trafficking | An01g02860 | ↓ | Gyp8 | An15g01560 | ↑ | Gyp7 |
| t-SNARE protein important for fusion of secretory vesicles with the plasma membrane | An02g05390 | ↓ | Sec9 | |||
| vacuolar protein important for endosomal-vacuolar trafficking pathway | An11g01810 | ↓ | Rcr2 | |||
| Actin localisation | ||||||
| polarisome component SpaA | An07g08290 | ↓ | Spa2 | |||
| actin-binding protein involved in endocytosis | An03g01160 | ↓ | Lsb4 | |||
| protein required for normal localization of actin patches | An16g02680 | ↓ | Apd1 | |||
| Amphysin-like protein required for actin polarization | An17g01945 | ↓ | Rvs161 | |||
| Amphysin-like protein required for actin polarization | An09g04300 | ↓ | Rvs167 | |||
| Other signaling processes | ||||||
| Ser/Thr protein kinase important for K+ uptake | An17g01925 | ↓ | Sat4 | |||
| transcription factor for RNA polymerase II | An16g07220 | ↓ | Tfg2 | |||
| negative regulator of Cdc42 | An12g04710 | ↓ | Vtc1 | |||
| putative C2H2 zinc-finger transcription factor | An04g01500 | ↑ | ||||
| SUN family protein involved in replication | An08g07090 | ↓ | Sim1 | |||
| similar to A. nidulans transcription factor RosA | An16g07890 | ↓ | Ume6 | |||
| Others | ||||||
| hypothetical aspergillosis allergen rAsp | An03g00770 | ↓ | An03g00770 | ↑ | ||
Genes up-regulated are indicated with ↑, genes down-regulated with ↓. Differential gene expression was evaluated by moderated t-statistics using the Limma package [82] with a FDR threshold at 0.05 [83]. Identical ORFs which are differentially expressed in both ΔracA and ramosa-1 are indicated in bold. Fold changes and statistical significance is given in Additional file 1 and 2.
: Protein functions were predicted based on information inferred from the Saccharomyces genome data base SGD (http://www.yeastgenome.org/) and the Aspergillus genome database AspGD (http://www.aspergillusgenome.org/).
In the case of (phosho)lipid signaling, genes encoding enzymes for the synthesis of the key regulatory lipid molecules diacylglycerol (DAG) and inositolpyrophosphates (IP6 and IP7) were differentially expressed in both ΔracA and ramosa-1. These molecules play important roles in the regulation of actin polarisation, CWI and calcium signaling in lower and higher eukaryotes (see discussion). Notably, expression of An04g03870 predicted as ortholog of the S. cerevisiae Dpp1p (DAG pyrophosphate phosphatase) is affected in both strains; however, down-regulated in ΔracA but up-regulated in ramosa-1. The same opposite response was observed for two A. niger ORFs predicted to encode inositol hexaki-/heptaki-phosphate kinases synthesizing the signaling molecules inositol pyrophosphates IP6 or IP7: An14g04590 (Ksc1p ortholog) is down-regulated in ΔracA, whereas An16g05020 (Vip1p ortholog) is up-regulated in ramosa-1. Inositol polyphosphates IP4-IP6 are known to bind to the C2B domain of the calcium sensor protein synaptotagmin [39]. This binding inhibits exocytosis of secretory vesicles, whereas binding of calcium to the C2A domain of synaptotagmin activates exocytosis [40]. In the ΔracA strain, four calcium transporters are down-regulated compared to the wild-type situation, two of which (An01g03100, An05g00170) code for the ortholog of the S. cerevisiae Vcx1p protein, which is also differentially expressed under hyperbranching conditions in ramosa-1 (Table 3). This observation hints at the possibility that reduced GFP-SncA fluorescence at ΔracA hyphal tips is somehow linked with changes in IP6/IP7 and calcium levels in ΔracA, which would be in agreement with a recent report which demonstrated that calcium spikes accompany hyphal branching in Fusarium and Magnaporthe hyphae [41]. Changes in the intracellular calcium distribution also affect the homeostasis of other ions. Congruently, 12 genes putatively encoding transport proteins for ions (Na+, K+, Fe2+) and small molecules (phospholipids, amino acids, peptides, hexoses) displayed differential transcription in ΔracA as also observed for ramosa-1 (Table 3), suggesting that ion homeostatic and/or metabolic control systems are also important to maintain polar growth. Reduced exocytotic GFP-SncA signals at hyphal tips of the ΔracA strain would imply that less cell wall biosynthetic/remodeling enzymes are transported to the tip. Indeed, expression of ten ORFs encoding cell membrane and cell wall genes and three ORFs involved in protein trafficking were down-regulated in ΔracA (Table 3). Whatever the consequences of reduced expression of cell wall or ion homeostasis genes are, none of these changes led to increased sensitivity of the ΔracA strain towards cell wall stress agents (calcofluor white), different salts (MgCl2, KCl, NaCl) or oxidative conditions (H2O2, menadion; data not shown), suggesting that the integrity of the cell wall or cell membrane is not disturbed in the racA strain.
Inactivation of RacA, however, has considerable consequences on actin localization as previously reported [23]. Congruently, five ORFs involved in actin polarization were down-regulated in the ΔracA strain (Table 3). Of special importance are the polarisomal component SpaA, whose deletion has been shown to cause a hyperbranching phenotype and reduced growth speed in A. niger [23] and two ORFs which are homologous to the S. cerevisiae amphyphysin-like proteins Rvs161p and Rvs167p. The latter function as heterodimer in S. cerevisiae, bind to phospholipid membranes and have established roles in organization of the actin cytoskeleton, endocytosis and cell wall integrity [42].
The transcriptomic fingerprint of apolar growth in RacAG18V
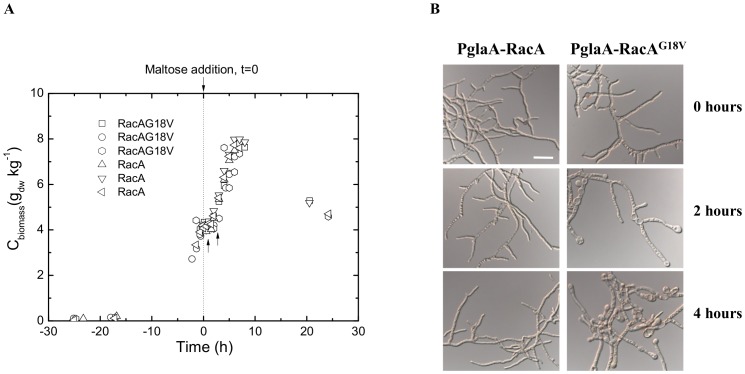
Next, we wished to dissect the transcriptomic adaptation of A. niger to dominant activation of RacA. Batch cultures of our previously established RacA mutant strains PglaA::racAG18V and PglaA::racA (reference strain) were started with xylose (0.75%) as repressing carbon source. After the cultures reached the exponential growth phase and xylose was consumed, maltose (0.75%) was added to induce expression of genes under control of PglaA. Hypothetically, a so far unknown RacA-dependent GAP ensures that the activity of RacA is spatially restricted to the hyphal apex in the PglaA-racA strain thereby maintaining a stable polarity axes even under inducing conditions (Fig. 5). However, this control mechanism is leveraged off in the PglaA-racAG18V mutant strain, as the GTPase-negative G18V mutation traps RacA in its active, GTP-bound form [24]. Hence, the switch from xylose to maltose leads to a loss of polarity maintenance in the PglaA-racAG18V strain and the formation of clavate-shaped hyphal tips and bulbous lateral branches (Fig. 5). RNA samples were extracted from duplicate cultures 2 and 4 h after the maltose shift and used for transcriptomic comparison. Expression of 3,757 (506) genes was modulated after 2 h (4 h) of induction, 1,906 (282) of which showed increased and 1,851 (224) decreased expression levels in the PglaA-racAG18V strain when compared to the PglaA-racA reference strain (FDR<0.05). The complete list of differentially expressed genes, including fold change and statistical significance is given in Tables S1 and S2. GO enrichment analysis using the FetGOat tool [43] discovered that most of these genes belong to primary metabolism, suggesting that both strains differed in their ability to quickly adapt to the new carbon source (Table S3).
Figure 5. Biomass accumulation and morphology during submerged cultivation of PglaA-RacAG18V and PglaA-RacA mutant strains.
(A) Growth curve for both mutant strains. The dashed line indicates the time point when the inducing carbon source maltose was added. The two arrows indicate the time points at which samples for transcriptome analysis were taken. (B) Dispersed hyphal morphology at the time point of maltose addition as well as 2 and 4 h after maltose addition. Bar, 20 µm.
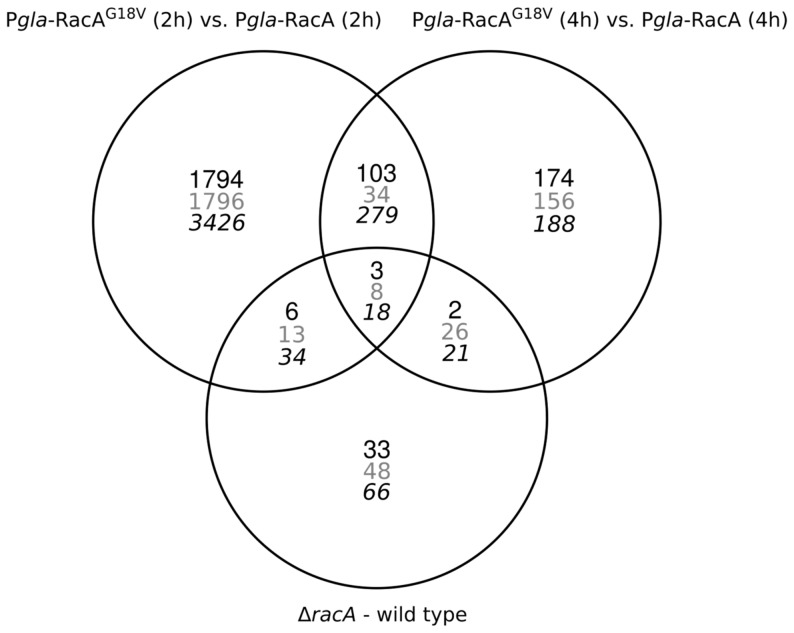
To identify those genes which only relate to the difference between polar to apolar morphology in PglaA-racAG18V but are independent from time and carbon source, a Venn diagram was constructed (Fig. 6) and the intersection determined representing genes which are differently expressed after both 2 and 4 h in PglaA-racAG18V when compared to the 2 h and 4 h data sets of the PglaA-racA reference strain. Overall, 148 genes showed different expression, 106 of which were up-regulated and 42 of which were down-regulated in PglaA-racAG18V (Table S4). Again, only a small set of genes (about 1% of the A. niger genome) show different expression levels during polar and apolar growth. Table 4 highlights the most interesting genes of this compilation, which could be grouped into several regulatory processes including (i) (phospho)lipid signaling, (ii) calcium signaling, (iii) CWI signaling and (iv) nitrogen signaling. In addition, metabolic processes including primary metabolism (amino acid biosynthesis) and secondary metabolism (polyketide synthesis, non-ribosomal peptide synthesis) were affected as well.
Figure 6. Venn diagrams of induced (black numbers), repressed (grey numbers) and up- or down-regulated (italics numbers) genes for the PglaA-RacAG18V/PglaA-RacA and ΔracA/N402 comparisons.
Table 4. Selected genes whose expression profiles differ between polar and apolar growth in PglaA-racAG18V versus PglaA-racA in a time- and carbon source-independent manner. Genes are ordered into different processes and functions.
| Predicted protein function* | Open reading frame code | Up/Down | Closest S. cerevisiae ortholog |
| (Phospho)lipid metabolism and signaling | |||
| dehydrogenase involved in sphingosin 1-phosphate breakdown | An01g09260 | ↑ | Hfd1 |
| Lysophospholipase, synthesis of glycerophosphocholine | An16g01880 | ↑ | |
| plasma membrane flippase transporting sphingoid long chain bases | An02g06440 | ↑ | Rsb1 |
| glycerophosphocholine phosphodiesterase, synthesis of phosphocholine | An18g03170 | ↑ | Gde1 |
| lanosterol 14A-demethylase | An11g02230 | ↓ | Erg11 |
| C-14 sterol reductase, ergosterol synthesis | An01g07000 | ↓ | Erg24 |
| choline/ethanolamine permease | An01g13290 | ↓ | Hnm1 |
| Calcium homeostasis and signaling | |||
| Ca2+/calmodulin dependent protein kinase | An02g05490 | ↓ | Cmk2 |
| Cell wall remodeling and integrity | |||
| endo-glucanase EglA | An14g02760 | ↑ | |
| endo-glucanase EglB | An16g06800 | ↑ | |
| endo-glucanase similar to Trichoderma reesei egl4 | An14g02670 | ↑ | |
| alpha-glucanosyltransferase AgtA (GPI-anchored) | An09g03100 | ↑ | |
| chitin synthase ChsL | An02g02340 | ↑ | Chs3 |
| chitin transglycosidase | An16g02850 | ↑ | Crh1 |
| chitinase | An01g05360 | ↑ | Cts2 |
| cell wall protein similar to A. nidulans PhiA | An14g01820 | ↑ | |
| cell wall protein with internal repeats | An12g10200 | ↑ | |
| cell wall protein (flocculin) | An12g00140 | ↑ | Flo11 |
| protein involved in β-1,3 glucan synthesis | An05g00130 | ↓ | Knh1 |
| α-1,2-mannosyltransferase | An04g06730 | ↓ | Mnn2 |
| Primary metabolism | |||
| isocitrate lyase AcuD | An01g09270 | ↑ | Icl1 |
| citrate lyase | An11g00510 | ↑ | |
| citrate lyase | An11g00530 | ↑ | |
| succinate dehydrogenase | An16g07150 | ↑ | Osm1 |
| aspartate transaminase, synthesis of glutamate | An16g05570 | ↑ | |
| acetyl-CoA carboxylase, synthesis of fatty acids | An12g04020 | ↑ | |
| homo-isocitrate dehydrogenase, synthesis of lysine | An15g02490 | ↓ | Lys12 |
| arginosuccinate synthetase, synthesis of arginine | An15g02340 | ↓ | Arg1 |
| acetylornithine aminotransferase, synthesis of arginine | An15g02360 | ↓ | Arg8 |
| arginyl-tRNA synthetase | An02g04880 | ↓ | |
| aspartic beta semi-aldehyde dehydrogenase, synthesis of threonine and methionine | An11g09510 | ↓ | Hom2 |
| homoserine kinase, synthesis of threonine | An17g02090 | ↓ | Thr1 |
| threonine synthase | An16g02520 | ↓ | Thr4 |
| phosphoribosylglycinamide formyltransferase, synthesis of purines | An02g02700 | ↓ | Ade8 |
| Secondary metabolism | |||
| polyketide synthase | An11g07310 | ↑ | |
| similar to plant zeaxanthin epoxidase ABA2 | An03g06500 | ↑ | |
| similar to enniatin synthase esyn1 of Fusarium scirpi | An13g03040 | ↑ | |
| similar to enoyl reductase LovC of the lovastatin biosynthesis A. terreus | An13g02940 | ↑ | |
| similar to enoyl reductase LovC of the lovastatin biosynthesis A. terreus | An09g01880 | ↑ | |
| similar to HC-toxin peptide synthase HTS of Cochliobolus carbonum | An16g06720 | ↑ | |
| Transporter | |||
| polyamine transporter | An11g07300 | ↑ | Tpo3 |
| polyamine transporter | An12g07400 | ↑ | Tpo3 |
| polyamine transporter | An13g03220 | ↓ | Tpo1 |
| vacuolar basic amino acid transporter | An06g00770 | ↑ | Vba5 |
| oligopeptide transporter | An11g01040 | ↓ | Opt1 |
| hexose transporter | An02g07610 | ↑ | Hxt5 |
| galactose transporter | An01g10970 | ↓ | Gal2 |
| low-affinity Fe(II) transporter of the plasma membrane | An16g06300 | ↓ | Fet4 |
| siderophore transporter | An03g03560 | ↓ | Arn1 |
| plasma membrane multidrug transporter | An07g01250 | ↑ | Pdr5 |
| multidrug transporter | An13g03060 | ↑ | Snq2 |
| Protein trafficking | |||
| protein kinase involved in exocytosis | An08g03360 | ↑ | Kin1 |
| Other signaling processes | |||
| zinc finger transcriptional repressor | An04g08620 | ↑ | Oaf3 |
| protein recruiting the SAGA complex to promoters | An07g04540 | ↑ | Cti6 |
| histidine kinase osmosensor | An14g02970 | ↑ | Sln1 |
| transcriptional regulator involved in nitrogen repression | An02g11830 | ↑ | Ure2 |
| transcription factor similar to A. nidulans MedA | An02g02150 | ↑ | |
| transcription factor | An02g06180 | ↑ | |
| transcription factor important for salt stress resistance | An12g09020 | ↑ | Hal9 |
| DNA damage checkpoint protein during replication | An03g06930 | ↓ | Rad24 |
| SUN family protein involved in replication | An08g07090 | ↓ | Sim1 |
| transcription factor important for Zn2+ homeostasis | An08g01860 | ↓ | Zap1 |
| alpha subunit of the translation initiation factor | An18g04650 | ↓ | Gcn3 |
| Others | |||
| pathogenesis-related protein | An08g05010 | ↑ | Pry1 |
| hypothetical aspergillosis allergen rAsp | An03g00770 | ↓ |
Genes up-regulated are indicated with ↑, genes down-regulated with ↓. Differential gene expression was evaluated by moderated t-statistics using the Limma package [82] with a FDR threshold at 0.05 [83]. Identical ORFs which are differentially expressed in PglaA-racAG18V and ΔracA are indicated in bold. Fold changes and statistical significance is given in Additional file 1 and 2.
: Protein functions were predicted based on information inferred from the Saccharomyces genome data base SGD (http://www.yeastgenome.org/) and the Aspergillus genome database AspGD (http://www.aspergillusgenome.org/).
The transcriptomic fingerprint indicated that the turgor pressure is increased during apolar growth and thereby an osmotic and cell wall stress is sensed in the PglaA-racAG18V strain. An14g02970, an ORF with strong similarity to the Sln1p histidine kinase osmosensor of S. cerevisiae which forms a phosphorelay system to activate the Hog1 MAP kinase cascade [44], showed increased expression. In agreement, some cell wall genes which have been shown to respond to caspofungin-induced cell wall stress in A. niger [45] were up-regulated as well: agtA, a GPI-anchored alpha-glucanosyltransferase and two putative cell wall protein encoding genes phiA and An12g10200. In addition, other proteins, known to be up-regulated under cell wall and osmotic stress in S. cerevisiae showed enhanced expression in PglaA-racAG18V: An16g02850 (ortholog of the chitin transglycosylase Crh1p, [46], An02g05490 (Ca2+/calmodulin dependent protein kinase Cmk2p, [47] and An07g01250 (ortholog of the multidrug transporter Pdr5p, [48]. Furthermore, increased expression was also observed for An03g06500 encoding an ortholog of the plant zeaxanthin epoxidase, which catalyses one step in the biosynthetic pathway of the plant hormone abscisic acid, known to protect plant cells against dehydration under high-salinity stress [49].
The expression of many transporters and permeases (iron, hexoses, amino acids and peptides) was also modulated in PglaA-racAG18V as well as the expression of several amino acid biosynthetic genes, most of which were down-regulated (lysine, arginine, threonine, methionine; Table 4). As also some ORFs predicted to function as important activators in replication (An03g06930, An08g07090) and translation (An18g04650) displayed decreased expression suggests that reduced tip extension during apolar growth slows down basic cellular processes. In this context, it is interesting to note that An01g09260 predicted to break down sphingosin 1-phosphate (S1P) showed increased expression during apolar growth. S1P is a sphingolipid acting as second messenger in lower and higher eukaryotes regulating respiration, cell cycle and translation [50] and is also important for the sole sphingolipid-to-glycerolipid metabolic pathway [51]. As also expression of An02g06440, a predicted ortholog of the S. cerevisiae sphingosin flippase Rsb1p which extrudes sphingosin into the extracytoplasmic side of the plasma membrane, was increased during apolar growth might suggest that reduced levels of S1P are present in apolar growing hyphal tips (see discussion).
Interestingly, none of the extracted 148 genes (Table S4) showed any link to actin filament organization, although actin polarization is lost in the PglaA-racAG18V strain and actin patches are randomly scattered intracellularly or at the cell periphery [24]. One explanation might be that transcription of actin-related genes was immediately altered after maltose addition which induced the switch from polar to apolar growth in PglaA-racAG18V. To still get a glimpse on genes involved in actin patch formation, the 2 h dataset of PglaA-racAG18V versus PglaA-racA was screened for the presence of enriched GO terms related to actin (Table S3). Using this approach, 10 actin-related genes were extracted which are summarized in Table 5. Most interestingly, An18g04590, an ortholog of the S. cerevisiae Rho GDP dissociation inhibitor Rdi1p displayed increased expression. Rdi1p regulates the Rho GTPases Cdc42p, Rho1p and Rho4p, localizes to polarized growth sites at specific times of the cell cycle and extracts all three proteins from the plasma membrane to keep them in an inactive cytosolic state [52]–[54]. Overexpression of Rdi1p causes slightly rounder cell morphology in S. cerevisiae [55]. Another interesting actin-related gene showing increased expression in strain PglaA-racAG18V is An11g02840, the predicted homolog of the S. cerevisiae Slm2 protein. Slm2p binds to the second messenger phosphatidylinositol-4,5-bisphosphate (PIP2) and to the TORC2 signaling complex and integrates inputs from both signaling pathways to control polarized actin assembly and cell growth [56]. In addition, it is also a target of sphingolipid and calcium signaling during heat stress response in S. cerevisiae and promotes cell survival by coordinating cell growth and actin polarization [57]. Both An18g04590 (Rdi1p ortholog) and An11g02840 (Slm2p ortholog) might thus be two key proteins important to sustain tip growth and proper actin polarization in A. niger. The other eight GO enriched proteins of strain PglaA-racAG18V are orthologs of S. cerevisiae proteins with a function in cortical actin patch formation (Table 5). For example, subunits of the Arp2/3 complex which is required for the motility and integrity of cortical actin patches are up-regulated in apolar growing hyphal tips of strain PglaA-racAG18V, one of which (An18g06590, Arc40p ortholog) also responds to caspofungin-induced loss of cell polarity in A. niger [45]. Another interesting gene showing increased expression was An04g09020, the ortholog of twinfilin. Twf1p has been shown to localize to cortical actin patches in S. cerevisiae, forms a complex with the capping protein Cap2 (An01g05290, up-regulated in PglaA-racAG18V), sequesters actin monomers to sites of actin filament assembly and is regulated by PIP2 [58], providing an additional hint that re-structuring of the actin cytoskeleton in PglaA-racAG18V might be orchestrated by PIP2 signaling. Taken together, the transcriptomic fingerprint of A. niger hyphae expressing dominant active RacA suggests that several signaling pathways and secondary messengers might orchestrate the morphological switch from polar to apolar growth.
Table 5. GO term enriched actin-related genes whose expression responds to the switch from polar to apolar growth in PglaA-racAG18V.
| Predicted protein function* | Open reading frame code | Up/Down | Closest S. cerevisiae ortholog |
| Rho GDP dissociation inhibitor | An18g04590 | ↑ | Rdi1 |
| TORC2 and phosphoinositide PI4,5P(2) binding protein | An11g02840 | ↑ | Slm2 |
| Arp2/3 complex subunit | An02g06360 | ↑ | Arc15 |
| Arp2/3 complex subunit | An01g05510 | ↑ | Arc35 |
| Arp2/3 complex subunit | An18g06590 | ↑ | Arc40 |
| tropomyosin 1 | An13g00760 | ↑ | Tpm1 |
| actin cortical patch component | An02g14620 | ↑ | Aip1 |
| twinfilin | An04g09020 | ↑ | Twf1 |
| actin-capping protein | An01g05290 | ↑ | Cap2 |
| protein recruiting actin polymerization machinery | An10g00370 | ↑ | Bzz1 |
Genes up-regulated are indicated with ↑. Differential gene expression was evaluated by moderated t-statistics using the Limma package [82] with a FDR threshold at 0.05 [83]. Fold changes and statistical significance is given in Additional file 1 and 2.
: Protein functions were predicted based on information inferred from the Saccharomyces genome data base SGD (http://www.yeastgenome.org/) and the Aspergillus genome database AspGD (http://www.aspergillusgenome.org/).
The RacA effector gene set
We finally compared the transcriptomic dataset of ΔracA versus wt with the dataset of PglaA-racAG18V versus PglaA-racA (4 h after maltose addition) to identify those genes whose transcription is generally affected by morphological changes independently whether provoked by RacA inactivation or by RacA hyperactivation. Overall, 38 genes fulfill this criterion (Fig. 6, Table S4) and are summarized in Table 6. The affected gene list covered processes such as (i) (phospho)lipid signaling, (ii) CWI and remodeling, (iii) actin localization, (iv) transport phenomena and (v) protein trafficking. Most interestingly, 12 out of the 38 genes were also differentially expressed during apical branching in ramosa-1 [25], including two orthologs of diacylglycerol pyrophosphate phosphatase (Dpp1p), which synthesizes the secondary messenger diacylglycerol (DAG), the activator of mammalian and fungal protein kinase C, which in fungi is a component of the CWI pathway localized upstream of the MAP kinase kinase Mkk1/2 (MkkA in A. niger; for review see [59]). Targets of the CWI signaling pathway are cell wall remodeling genes, five of which were differentially expressed in RacA hyper- or inactivation strains (Table 6). From these five genes, three are of special importance as these were also effector genes in the hyperbranching mutant ramosa-1 [25]. Although calcium signaling genes seemed not be among the extracted 38 genes, its indirect involvement might be conceivable. For example, An18g01090 encoding the predicted ortholog of the S. cerevisiae phospholipase B (Plb3p) is among this gene set. Plb3p is activated at high concentrations of Ca2+ and specifically accepts phosphatidylinositol as a substrate to keep its concentration on the outer membrane leaflet low [60]. Finally, 17 RacA effector genes encode proteins of unknown function, most of which have no predicted orthologs in S. cerevisiae. As their function, however, seems to be important for morphological changes in A. niger, they are highly interesting candidate genes for future analyses.
Table 6. Complete list of genes whose expression respond to hyperbranching in ΔracA versus wild-type and to the switch from polar and apolar growth in PglaA-racAG18V versus PglaA-racA (4 h after induction).
| Predicted protein function* | Open reading frame code | Up/Down in PglaA-racAG18V versus PglaA-racA (4 h) | Up/Down in ΔracA versus wt | Closest S. cerevisiae ortholog |
| (Phospho)lipid metabolism and signaling | ||||
| phospholipase B, synthesis of glycerophosphocholine | An18g01090 | ↓ | ↓ | Plb3 |
| diacylglycerol pyrophosphate phosphatase, synthesis of DAG | An02g01180 | ↓ | ↓ | Dpp1 |
| diacylglycerol pyrophosphate phosphatase, synthesis of DAG | An04g03870 | ↓ | ↓ | Dpp1 |
| sterol 24-C-methyltransferase, ergosterol synthesis | An04g04210 | ↑ | ↑ | Erg6 |
| C-14 sterol reductase, ergosterol synthesis | An01g07000 | ↓ | ↓ | Erg24 |
| transcription factor important for sterol uptake | An02g07950 | ↓ | ↓ | Upc2 |
| transcription factor important for sterol uptake | An12g00680 | ↓ | ↓ | Upc2 |
| Cell wall remodeling and integrity | ||||
| α-1,3-mannosyltransferase | An15g04810 | ↓ | ↓ | Mnt2 |
| endo-mannanase (GPI-anchored), DfgE | An16g08090 | ↓ | ↓ | Dfg1 |
| β-1,4-glucanase | An03g05530 | ↓ | ↓ | |
| cell wall protein | An11g01190 | ↓ | ↓ | Sps22 |
| plasma membrane protein | An02g08030 | ↓ | ↓ | Pmp3 |
| Actin localization | ||||
| amphysin-like protein required for actin polarization | An17g01945 | ↓ | ↓ | Rvs161 |
| actin-binding protein involved in endocytosis | An03g01160 | ↓ | ↓ | Lsb4 |
| Transporter | ||||
| choline/ethanolamine permease | An01g13290 | ↓ | ↓ | Hnm1 |
| low-affinity Fe(II) transporter | An16g06300 | ↓ | ↓ | Fet4 |
| oligopeptide transporter | An11g01040 | ↓ | ↓ | Opt1 |
| galactose/glucose permease | An01g10970 | ↓ | ↓ | Gal2 |
| Protein trafficking | ||||
| GTPase activating protein involved in protein trafficking | An01g02860 | ↓ | ↓ | Gyp8 |
| protein important for endosomal-vacuolar trafficking | An11g01810 | ↓ | ↓ | Rcr2 |
| peptidase | An03g02530 | ↓ | ↓ | |
| Others | ||||
| hypothetical aspergillosis allergen rAsp | An03g00770 | ↓ | ↓ | |
| cytochrome P450 protein | An11g02990 | ↓ | ↓ | Dit2 |
| isoamyl alcohol oxidase | An03g06270 | ↓ | ↓ | |
| protein with strong similarity to penicillin V amidohydrolase | An12g04630 | ↓ | ↓ | |
| oxidoreductase | An03g00280 | ↑ | ↑ | |
| protein with nucleotide binding domain | An01g08150 | ↑ | ↑ | Irc24 |
| protein with methyltransferase domain | An09g00160 | ↑ | ↑ | |
| protein with unknown function | An15g03880 | ↓ | ↓ | |
| protein with unknown function | An01g10900 | ↓ | ↓ | |
| protein with unknown function | An07g05820 | ↓ | ↓ | |
| protein with unknown function | An18g00810 | ↓ | ↓ | |
| protein with unknown function | An04g04630 | ↓ | ↓ | |
| protein with unknown function | An06g00320 | ↓ | ↓ | |
| protein with unknown function | An07g04900 | ↓ | ↓ | |
| protein with unknown function | An01g13320 | ↓ | ↓ | |
| protein with unknown function | An16g07920 | ↑ | ↑ | |
| protein with unknown function | An01g03780 | ↓ | ↓ |
Genes up-regulated are indicated with ↑, genes down-regulated with ↓. Differential gene expression was evaluated by moderated t-statistics using the Limma package [82] with a FDR threshold at 0.05 [83]. Identical ORFs or proteins with predicted similar function being also differentially expressed in ramosa-1 are indicated in bold. Fold changes and statistical significance is given in Additional file 1 and 2.
: Protein functions were predicted based on information inferred from the Saccharomyces genome data base SGD (http://www.yeastgenome.org/) and the Aspergillus genome database AspGD (http://www.aspergillusgenome.org/).
Discussion
The fungal actin cytoskeleton is highly dynamic and fulfils multiple functions important for cell polarity regulation, endocytosis, exocytosis and septation. Central regulators of actin polymerization and depolymerization are Rho GTPases whose activity is regulated by their membrane-cytoplasmatic shuttling which itself is modulated by external or internal morphogenetic signals. Actin dynamics is thus controlled by a network of signaling pathways that sense and integrate different stimuli [61]. We have recently proposed that the A. niger GTPases RacA and CftA (Cdc42p) can substitute each other with respect to actin assembly but that actin disassembly is mainly under control of RacA. A racA deletion mutant is thus not affected in actin polymerization (because is secured by CftA) but impaired in actin disassembly. In consequence, maintenance of apical dominance can become frequently lost in the racA deletion strain resulting in a hyperbranching phenotype. In contrast, RacA trapped in its active, GTP-bound form (RacAG18V) provokes the formation of higher-order actin structures, i.e. actin patches, which cause loss of polarity maintenance and the formation of round, apolar growing cells [24]. The purpose of the current study was to identify the transcriptional signature associated with morphological changes in hyphal tip growth of A. niger. The transcriptional response of A. niger provoked by inactivation and hyperactivation of RacA, respectively, was determined and compared with the transcriptomic fingerprint of the apical branching transcriptome of the ramosa-1 mutant [25]. The data obtained allowed us to reconstruct the transcriptomic network that helps A. niger to adapt to abnormal morphologies and to secure the integrity of its cell wall.
A transcriptomic perspective on the morphogenetic network of A. niger
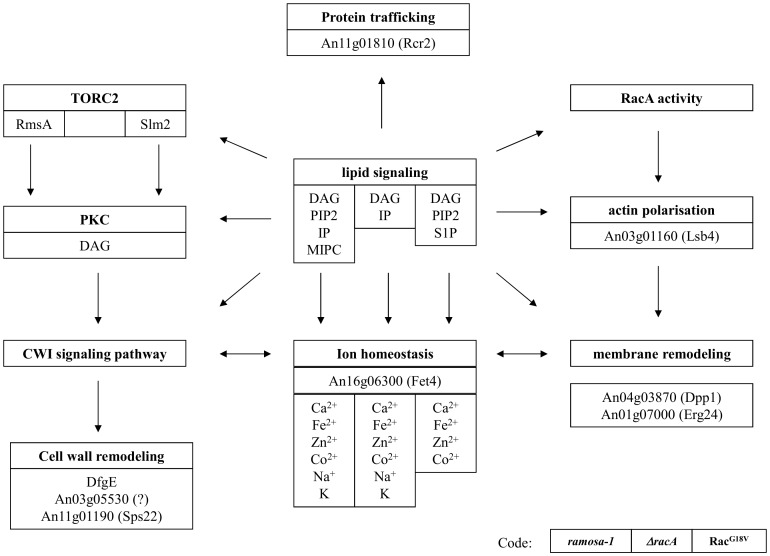
A central result of our comparative transcriptomics approach is the finding that several lipid molecules are likely involved in the maintenance of polar growth in A. niger (Fig. 7). The synthesis of important phospho- and sphingolipid molecules (phosphatidic acid, DAG, PIP2, inositolphosphates (IP), glycerophosphocholine, mannose-inositol-phosphoceramide (MIPC) and S1P seem to be modulated during apical branching (ΔracA, ramosa-1) and apolar growth (PglaA-racAG18V), as genes encoding respective synthetic or degrading enzymes showed differential expression in comparison to the wild-type (Tables 3, 4 and 6). Many of these molecules function as secondary messengers in eukaryotes (DAG, PA, IP, PIP2, S1P), others are essential components of fungal membranes (plasma membrane, organelles, lipid droplets), whereby sphingolipids (e.g. MIPC) and ergosterol are worth highlighting as they concentrate to form lipid rafts in plasma membranes which organize and regulate signaling cascades involved in polar growth control of S. cerevisiae [62]. Lipid rafts have been shown to form ordered subdomains of eukaryotic plasma membranes into which monomeric and trimeric G proteins associate in a dynamic and selective manner to organize signal transduction complexes [63]. It is therefore intriguing that expression of An01g07000, the ortholog of the ergosterol synthesizing enzyme Erg24p, is modulated in all three strains ΔracA, ramosa-1 and PglaA-racAG18V, and being also among the cell wall stress responsive genes when A. niger is exposed to caspofungin or fenpropimorph [45]. This suggests that ergosterol metabolism is of main importance for polarized growth and cell wall integrity in A. niger.
Figure 7. A reconstructed model for the morphogenetic network of A. niger based on the transcriptomic fingerprints determined for the apical branching mutant ramosa-1 [25], the apical branching mutant ΔracA (this work) and the apolar growing mutant PglaA-RacAG18V (this work).
The model also rests on cell biological and phenotypic data obtained for all three strains (this work and [25]) as well as on literature data for conserved mechanisms from yeast to humans (see discussion for references). Indicated are signalling and metabolic processes, which showed transcriptional responses in all three strains, some deduced key players and the hypothetical connection of these processes.
Unfortunately, data on fungal lipid signaling networks are sparse. So far, it is known that sphingolipids play a key role in pathogenicity in Cryptococcus neoformans, that the quorum sensing molecule farnesol is involved in mycelial growth, biofilm formation and stress response of Candida albicans, that both sphingolipids and farnesol are important for maintaining cell wall integrity and virulence of A. fumigatus (for review see [59]) and that the activity of two ceramide synthases is important for the formation of a stable polarity axis in A. nidulans [64] (Li et al.). In Schizosaccharomyces pombe, MIPC was shown to be required for endocytosis of a plasma-membrane-localized transporter and for protein sorting into the vacuole [65]. As ΔracA is affected in endocytosis (see below) and the MIPC synthesizing enzyme Sur1p (An05g02310) is down-regulated as well might suggest that MIPC has a similar function in A. niger. Notably, the sphingolipid synthesizing protein inositol-phosphoryl ceramide synthase (Ipc1) plays a major role in both establishment and maintenance of cell polarity in A. nidulans by regulating actin dynamics [66], [67]. However, it is not known whether this is mediated by the sphingolipid inositol-phosphoryl ceramide (IPC) or by other products of the ceramide synthetic pathway such as DAG, MIPC or sphingosines. Anyhow, inhibition of sphingolipid synthesis in A. nidulans caused wider hyphal cells, abnormal branching and tip splitting and is not suppressible by the addition of sorbitol [66], [67] - observations which also hold true for ΔracA and ramosa-1 [24], [25], suggesting that sphingolipid mediated control of hyphal cell polarity is not mediated by the CWI pathway in Aspergillus. Still, S. cerevisiae strains defective in CWI signaling (e.g. pkc1Δ, mpk1Δ) also exhibit severe defects in lipid metabolism, including accumulation of phosphatidylcholine, DAG, triacylglycerol, and free sterols as well as aberrant turnover of phosphatidylcholine, suggesting that CWI signaling and lipid homeostasis are nevertheless closely linked in fungi [68].
A second important outcome of this study is that not only calcium signaling seems to be of utmost importance for morphological decisions in all three mutant strains ΔracA, ramosa-1 and PglaA-racAG18V, but ion homeostasis in general (Fig. 7). Many ion transport proteins are differentially expressed in all three strains when compared to the wild-type situation including transport proteins for Na+, K+, Ca2+, Fe2+, Zn2+ and Co2+. Of special importance is An16g06300, a predicted Fe(II) transporter, homologous to the S. cerevisiae plasma membrane transporter Fet4p, whose transcriptional regulation is affected in all three strains. Fet4p is a low-affinity Fe(II) transporter also transporting Zn2+ and Co2+ and is under combinatorial control of iron (Atf1p transcriptional activator), zinc (Zap1p transcriptional factor) and oxygen (Rox1p repressor) [69], being for example important for S. cerevisiae to tolerate alkaline pH [70]. It has been postulated that changes in the phospholipid composition govern the function of membrane-associated zinc transporters such as Fet4p [71]. Vice versa, the transcriptional factor Zap1p controls not only expression of zinc-related transporters but also expression of the DAG pyrophosphate phosphatase Dpp1p [71]. This is a very interesting observation in view of the fact that one predicted Dpp1p ortholog (An04g03870) shows differential expression in all three morphological mutant strains of A. niger, an analogy which might suggest that polarized growth of A. niger might be orchestrated by (phospho)lipid signaling which is somehow interconnected with zinc metabolism.
Finally, our transcriptomics comparison uncovered that endocytotic processes are likely to be involved in the morphogenetic network of A. niger. In all three strains, ΔracA, ramosa-1 and PglaA-racAG18V, expression of An03g01160, a predicted ortholog of the S. cerevisiae Lsb4p was modulated (Table 6, Fig. 7). Lsb4p is an actin-binding protein, conserved from yeast to humans, binds to actin patches and promotes actin polymerization together with the WASP protein Lsb17 in an Arp2/3-independent pathway thereby mediating inward movement of vesicles during endocytosis [72]. Lsb4p is also a PIP binding protein due to a phosphoinositide-binding domain (SYLF), which is highly conserved from bacteria to humans. The human homolog of Lsb4p (SH3YL1) binds to PIP3 and couples the synthesis of PIP2 with endocytotic membrane remodeling, whereas Lsb4p binds directly to PIP2 (note that PIP3 is believed to be absent in yeast). Thus, Lsb4p homologs are predicted to couple PIP2 with actin polymerization to regulate actin and membrane dynamics involved in membrane ruffling during endocytosis [73]. Beside An03g01160 (Lsb4p ortholog), An17g01945 is worth highlighting in this context as well. An17g01945 encodes an ortholog of the amphysin-like lipid raft protein Rvs161p and is differentially expressed in both ΔracA and PglaA-racAG18V. Rvs161p affects the membrane curvature in S. cerevisiae and mediates in conjunction with Rvs167 and PIP2 membrane scission at sites of endocytosis [74].
Taken together, the transcriptomic signature of the three morphological mutants predicts that the morphological changes are brought about the interconnection of several signaling and metabolic pathways. Remarkably, the responding gene set in ΔracA and ramosa-1 seems to be, although substantially overlapping, oppositely regulated. One explanation might be that inactivation of RacA and TORC2 induces dichotomous branching in different manners. As the subcellular distribution of actin is different in both strains (the ramosa-1 mutant shows scattered actin patches at hyphal tips, whereas actin is concentrated at hyphal apices in the ΔracA mutant) might suggest that different causes (loss of actin polarization/actin hyperpolarization) provoke different responding transcriptional changes, which, however, eventually result in the same phenotypic response, namely tip splitting. Similarly puzzling is the observation that the core set of 38 genes which are responsive in both ΔracA and PglaA-racAG18V respond in the same direction (Table 6), although they are associated with excessive polar growth (hyperbranching) in the ΔracA strain but with the absence of polar growth (tip swelling) in strain PglaA-racAG18V. A plausible explanation might be that loss of polarity maintenance in both strains is connected with similar transcriptional changes controlling actin dynamics and vesicle flow, but that reestablishment of polar growth in the racA deletion strain requires genes which are not important for tip swelling in the PglaA-racAG18V strain.
Does a hyperbranching strain secrete more proteins?
In filamentous fungi, it is believed that protein secretion occurs at the hyphal tip. This holds true for glucoamylase (GLA), the most abundant secreted enzyme in A. niger [11], [12]. Another example is α-amylase, the major secretory protein of A. oryzae [14]. Hence, one might expect that a higher branching frequency would result in higher secretion yields. However, our study demonstrated that more tips in the ΔracA strain do not necessarily increase protein secretion; instead, protein yields were the same in both mutant and wild-type (Table 2). The most logical explanation is that the same amount of secretory vesicles is merely distributed to more tips in the ΔracA strain, resulting in less secretory vesicles per individual tip. The quantitative data obtained for the exocytotic marker GFP-SncA and the endocytotic marker AbpA-CFP and SlaB-YFP clearly demonstrate that fewer vesicles are transported to the apex of an individual tip (Fig. 3) and that endocytosis is slowed down as well – the endocytotic ring seems to be less well defined and the fluorescence intensity of both endocytotic markers is decreased (Fig. 4). This data is corroborated by the transcriptomic fingerprint of the ΔracA strain. The transcription of genes predicted to function in protein trafficking and actin localization is down-regulated as well as expression of genes governing phospholipid signaling and cell wall remodeling. Remarkably, biomass formation is the same in both ΔracA and wt. This suggests that the amount of secreted vesicles is adjusted in both strains just to ensure hyphal tip growth but that the capacity of a hyphal tip growing apparatus to accommodate vesicles is much higher (at least in ΔracA). Hence, challenging the ΔracA strain to overexpress a certain protein of interest might increase the number of secretory vesicles thus resulting in higher secretion yields. We currently run respective experiments to test this hypothesis. In any case, the hyperbranching ΔracA mutant could already be of value for high-density cultivation during industrial processes: it forms a less shear stress-sensitive, compact macromorphology but does not form pellets. It thus exhibits improved rheological properties without any apparent disadvantages with respect to growth rate and physiology.
Conclusions
The transcriptomic signature of the three individual mutants ΔracA, ramosa-1 and PglaA-racAG18V uncovered specific and overlapping responses to the morphological changes induced and suggests the participation as well as interconnectedness of several regulatory and metabolic pathways in these processes. The data obtained predict a role for different signaling pathways including phospholipid signaling, sphingolipid signaling, TORC2 signaling, calcium signaling and CWI signaling in the morphogenetic network of A. niger. These pathways likely induce different physiological adaptations including changes in sterol, zinc and amino acid metabolism and changes in ion transport and protein trafficking. Central to the morphological flexibility of A. niger is the actin cytoskeleton whose dynamics can be precisely controlled in these mutants. Future attempts are necessary to address important issues which cannot be resolved by transcriptomics. For example, how is the lipid composition in apical and subapical regions of A. niger hyphae? Which morphogenetic proteins are also parts of the network whose expression is regulated post-transcriptionally and can thus not be detected by transcriptomics approaches? Where are they localized during (a)polar growth? What are the metabolic prerequisites to sustain fast polar growth coupled with high secretion rates? Clearly, a comprehensive understanding of the morphogenetic network of A. niger will require and integrated systems biology approach where transcriptomics analyses will be combined with proteomics, metabolomics and lipidomics approaches and linked with cell biological studies.
Materials and Methods
Strains, culture conditions and molecular techniques
Aspergillus strains used in this study are given in Table 7. Strains were grown on minimal medium (MM) [75] containing 1% (w v−1) glucose and 0.1% (w v−1) casamino acids or on complete medium (CM), containing 0.5% (w v−1) yeast extract in addition to MM. When required, plates were supplemented with uridine (10 mM). Transformation of A. niger and fungal chromosomal DNA isolation was performed as described [76]. All molecular techniques were carried out as described earlier [77].
Table 7. Strains used in this work.
| Strain | Relevant genotype | Source |
| N402 | cspA1 (derivative of ATCC9029) | [86] |
| AB4.1 | pyrG− | [87] |
| MA70.15 | ΔkusA pyrG− (derivative of AB4.1) | [88] |
| MA80.1 | ΔkusA, ΔracA::AopyrG | [24] |
| FG7 | ΔkusA pyrG+ egfp::sncA (derivative of MA70.15) | Kwon et al, submitted |
| MA1.8 | PglaA::racA (derivative of AB4.1) | [24] |
| MA60.15 | PglaA::racAG18V (derivative of AB4.1) | [24] |
| MK5.1 | ΔkusA, slaB::eyfp (derivative of MA70.15) | This work |
| MK6.1 | ΔkusA, abpA::ecfp (derivative of MA70.15) | This work |
| MK7.1 | ΔkusA, ΔracA, slaB::eyfp (derivative of MA80.1) | This work |
| MK8.1 | ΔkusA, ΔracA, abpA::ecfp (derivative of MA80.1) | This work |
Bioreactor cultivation conditions
Maltose-limited batch cultivation was initiated by inoculation of 5 L (kg) ammonium based minimal medium with conidial suspension to give 109 conidia L−1. Maltose was sterilized separately from the MM and final concentration was 0.8% (w/v). Temperature of 30°C and pH 3 were kept constant, the latter by computer controlled addition of 2 M NaOH or 1 M HCl, respectively. Acidification of the culture broth was used as an indirect growth measurement [78]. Submerged cultivation was performed with 6.6 L BioFlo3000 bioreactors (New Brunswick Scientific, NJ, USA). A more detailed description of the fermentation medium and cultivation is given in [79]. Batch cultivation for PglaA-RacAG18V or PglaA-RacA were run similarly as the maltose-limited batch cultivations of ΔracA cultures except that 0.75% xylose was used as a initial carbon source instead of maltose. When the exponential growth phase was over (indicated by a sharp rise of the dissolved oxygen tension and the pH value), 0.75% maltose was added to induce expression of PglaA-RacAG18V or PglaA-RacA, respectively. Samples for the analysis of morphological characteristics, biomass formation, protein yield and RNA were taken every hour.
Analysis of culture broth
Dry weight biomass concentration was determined by weighing lyophilized mycelium separated from a known mass of culture broth. Culture broth was filtered through GF/C glass microfiber filters (Whatman). The filtrate was collected and frozen for use in solute analyses. The mycelium was washed with demineralised water, rapidly frozen in liquid nitrogen and lyophilized. Glucose concentration was measured as previously described [80] with slight modifications: 250 mM triethanolamine (TEA) was used as buffer (pH 7.5). Extracellular protein concentration was determined using the Quick Start Bradford Protein Assay (Bio-Rad) using BSA as standard. The total organic carbon in the culture filtrate was measured with a Total Organic Carbon Analyzer (TOC-Vcsn; Shimadzu, Japan) using glucose as standard.
Microarray analysis
Total RNA extraction, RNA quality control, labeling, Affymetrix microarray chip hybridization and scanning were performed as previously described [25]. Background correction, normalization and probe summarization steps were performed according to the default setting of the robust multi-array analysis (RMA) package as recently described [81]. Differential gene expression was evaluated by moderated t-statistics using the Limma package [82] with a threshold of the Benjamini and Hochberg False Discovery Rate (FDR) of 0.05 [83]. Fold change of gene expression from different samples was calculated from normalized expression values. Geometric means of the expression values as well as fold change for all strains and comparisons are summarized in Tables S1 and S2 and have been deposited at the GEO repository (http://www.ncbi.nlm.nih.gov/geo/info/linking.html) under the accession number GSE42258. Transcriptomic data for the exponential growth phase of the reference strain N402 was published recently [84].
Gene Ontology (GO) and enrichment analysis
Over-represented GO terms in sets of differentially expressed genes were determined by Fisher's exact test [85] as implemented in FetGOat [43] using a FDR of q<0.05. An improved GO annotation for A. niger CBS513.88 was applied based on ontology mappings from A.nidulans FGSCA4 [43].
Construction of AbpA-CFP and SlaB-YFP expression cassettes
Standard PCR and cloning procedures were used for the generation of the constructs [77]. All PCR amplified DNA sequences and cloned fragments were confirmed by DNA sequencing (Macrogene). Primers used in this study are listed in Table S5. Correct integrations of constructs in A. niger were verified by Southern analysis [77].The expression vectors, AbpA-CFP and SlaB-YFP were constructed using the fusion PCR approach as described previously [23] with slight modifications. Plasmid pVM3-1 [23] harboring the GA5 peptide linker followed by the CFP, TtrpC and the selection marker pyrG from A. oryzae was used as starting point. A second TtrpC terminator sequence was generated by PCR and ligated via a SalI restriction site into pVM3-1 which would later allow looping out of the pyrG marker by FOA counter-selection [76]. The resulting plasmid was named pMK3. For the fusion PCR, three separate fragments were amplified by PCR: the C-terminal part of abpA ORF (∼1 kb), the module containing CFP-Ttrpc-AopyrG (∼3.5 kb) and the terminator region of abpA (∼1 kb). Subsequently, the three individual fragments were fused together by a fusion PCR and the resulting amplicon (∼5.6 kb) was cloned into pJET (Fermentas) to give plasmid pMK5. SlaB-YFP was also constructed in a similar way and the final plasmid was named pMK6.
Microscopy
Light microscopic pictures were captured using an Axioplan 2 (Zeiss) equipped with a DKC-5000 digital camera (Sony). For light and fluorescence images for SlaB-YFP and AbpA-CFP transformants, pictures were captured with 40× C-apochromatic objective on an inverted LSM5 microscope equipped with a laser scanning confocal system (Zeiss Observer). The observation conditions for the life-imaging of hyphae were described previously [24]. To determine branching frequencies, the lengths of hyphae and branches were measured and evaluated using the program Image J. For the quantification of GFP-SncA, AbpA-CFP and SlaB-YFP signals, a single section of individual hyphal tip was captured (n>20).
Supporting Information
Complete transcriptome data containing RMA expression values (log2 scale), mean expression values, p-values, q-values and fold changes.
(XLSX)
Subset of the transcriptome data containing up-down-regulated gene sets.
(XLSX)
ZIP archive file containing FetGOat enrichment results for the up- and down-regulated gene sets of all six comparisons.
(ZIP)
Subset of the transcriptome data for selected intersection of the Venn diagram.
(XLSX)
Primers used in this study.
(DOCX)
Acknowledgments
This project was carried out within the research programme of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Funding Statement
This project was carried out within the research programme of the Kluyver Centre for Genomics of Industrial Fermentation which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fleissner A, Dersch P (2010) Expression and export: recombinant protein production systems for Aspergillus . Appl Microbiol Biotechnol 87: 1255–1270. [DOI] [PubMed] [Google Scholar]
- 2. Meyer V (2008) Genetic engineering of filamentous fungi–progress, obstacles and future trends. Biotechnol Adv 26: 177–185. [DOI] [PubMed] [Google Scholar]
- 3. Meyer V, Wu B, Ram AF (2010) Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnol Lett 33: 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saloheimo M, Pakula TM (2012) The cargo and the transport system: secreted proteins and protein secretion in Trichoderma reesei (Hypocrea jecorina). Microbiology 158: 46–57. [DOI] [PubMed] [Google Scholar]
- 5. Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, et al. (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25: 221–231. [DOI] [PubMed] [Google Scholar]
- 6. Carvalho ND, Arentshorst M, Kwon MJ, Meyer V, Ram AF (2010) Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl Microbiol Biotechnol 87: 1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jacobs DI, Olsthoorn MM, Maillet I, Akeroyd M, Breestraat S, et al. (2009) Effective lead selection for improved protein production in Aspergillus niger based on integrated genomics. Fungal Genet Biol 46 Suppl 1: S141–152. [DOI] [PubMed] [Google Scholar]
- 8. Grimm LH, Kelly S, Krull R, Hempel DC (2005) Morphology and productivity of filamentous fungi. Appl Microbiol Biotechnol 69: 375–384. [DOI] [PubMed] [Google Scholar]
- 9. McIntyre M, Muller C, Dynesen J, Nielsen J (2001) Metabolic engineering of the morphology of Aspergillus . Adv Biochem Eng Biotechnol 73: 103–128. [DOI] [PubMed] [Google Scholar]
- 10. Papagianni M (2004) Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol Adv 22: 189–259. [DOI] [PubMed] [Google Scholar]
- 11. Wösten HA, Moukha SM, Sietsma JH, Wessels JG (1991) Localization of growth and secretion of proteins in Aspergillus niger . J Gen Microbiol 137: 2017–2023. [DOI] [PubMed] [Google Scholar]
- 12. Gordon CL, Khalaj V, Ram AF, Archer DB, Brookman JL, et al. (2000) green fluorescent protein fusions to monitor protein secretion in Aspergillus niger . Microbiology 146 Pt 2: 415–426. [DOI] [PubMed] [Google Scholar]
- 13. Muller C, McIntyre M, Hansen K, Nielsen J (2002) Metabolic engineering of the morphology of Aspergillus oryzae by altering chitin synthesis. Appl Environ Microbiol 68: 1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hayakawa Y, Ishikawa E, Shoji JY, Nakano H, Kitamoto K (2011) Septum-directed secretion in the filamentous fungus Aspergillus oryzae . Mol Microbiol 81: 40–55. [DOI] [PubMed] [Google Scholar]
- 15. te Biesebeke R, Record E, van Biezen N, Heerikhuisen M, Franken A, et al. (2005) Branching mutants of Aspergillus oryzae with improved amylase and protease production on solid substrates. Appl Microbiol Biotechnol 69: 44–50. [DOI] [PubMed] [Google Scholar]
- 16. Amanullah A, Christensen LH, Hansen K, Nienow AW, Thomas CR (2002) Dependence of morphology on agitation intensity in fed-batch cultures of Aspergillus oryzae and its implications for recombinant protein production. Biotechnol Bioeng 77: 815–826. [DOI] [PubMed] [Google Scholar]
- 17. Wongwicharn A, McNeil B, Harvey LM (1999) Effect of oxygen enrichment on morphology, growth, and heterologous protein production in chemostat cultures of Aspergillus niger B1-D. Biotechnol Bioeng 65: 416–424. [DOI] [PubMed] [Google Scholar]
- 18. Bocking SP, Wiebe MG, Robson GD, Hansen K, Christiansen LH, et al. (1999) Effect of branch frequency in Aspergillus oryzae on protein secretion and culture viscosity. Biotechnol Bioeng 65: 638–648. [DOI] [PubMed] [Google Scholar]
- 19. Virag A, Griffiths AJ (2004) A mutation in the Neurospora crassa actin gene results in multiple defects in tip growth and branching. Fungal Genet Biol 41: 213–225. [DOI] [PubMed] [Google Scholar]
- 20. Wang J, Hu H, Wang S, Shi J, Chen S, et al. (2009) The important role of actinin-like protein (AcnA) in cytokinesis and apical dominance of hyphal cells in Aspergillus nidulans . Microbiology 155: 2714–2725. [DOI] [PubMed] [Google Scholar]
- 21. Harris SD, Morrell JL, Hamer JE (1994) Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136: 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Virag A, Harris SD (2006) Functional characterization of Aspergillus nidulans homologues of Saccharomyces cerevisiae Spa2 and Bud6. Eukaryot Cell 5: 881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meyer V, Arentshorst M, van den Hondel CA, Ram AF (2008) The polarisome component SpaA localises to hyphal tips of Aspergillus niger and is important for polar growth. Fungal Genet Biol 45: 152–164. [DOI] [PubMed] [Google Scholar]
- 24. Kwon MJ, Arentshorst M, Roos ED, van den Hondel CA, Meyer V, et al. (2011) Functional characterization of Rho GTPases in Aspergillus niger uncovers conserved and diverged roles of Rho proteins within filamentous fungi. Mol Microbiol 79: 1151–1167. [DOI] [PubMed] [Google Scholar]
- 25. Meyer V, Arentshorst M, Flitter SJ, Nitsche BM, Kwon MJ, et al. (2009) Reconstruction of signaling networks regulating fungal morphogenesis by transcriptomics. Eukaryot Cell 8: 1677–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berepiki A, Lichius A, Read ND (2011) Actin organization and dynamics in filamentous fungi. Nat Rev Microbiol 9: 876–887. [DOI] [PubMed] [Google Scholar]
- 27. Lanzetti L (2007) Actin in membrane trafficking. Curr Opin Cell Biol 19: 453–458. [DOI] [PubMed] [Google Scholar]
- 28. Reynaga-Pena CG, Bartnicki-Garcia S (1997) Apical branching in a temperature sensitive mutant of Aspergillus niger . Fungal Genet Biol 22: 153–167. [DOI] [PubMed] [Google Scholar]
- 29. Reynaga-Pena CG, Bartnicki-Garcia S (2005) Cytoplasmic contractions in growing fungal hyphae and their morphogenetic consequences. Arch Microbiol 183: 292–300. [DOI] [PubMed] [Google Scholar]
- 30. Chen YA, Scheller RH (2001) SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol 2: 98–106. [DOI] [PubMed] [Google Scholar]
- 31. Taheri-Talesh N, Horio T, Araujo-Bazan L, Dou X, Espeso EA, et al. (2008) The tip growth apparatus of Aspergillus nidulans . Mol Biol Cell 19: 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taheri-Talesh N, Xiong Y, Oakley BR (2012) The functions of myosin II and myosin V homologs in tip growth and septation in Aspergillus nidulans . PLoS One 7: e31218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quintero-Monzon O, Rodal AA, Strokopytov B, Almo SC, Goode BL (2005) Structural and functional dissection of the Abp1 ADFH actin-binding domain reveals versatile in vivo adapter functions. Mol Biol Cell 16: 3128–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Araujo-Bazan L, Penalva MA, Espeso EA (2008) Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in Aspergillus nidulans . Mol Microbiol 67: 891–905. [DOI] [PubMed] [Google Scholar]
- 35. Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K (2007) Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol Biol Cell 18: 295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valkonen M, Kalkman ER, Saloheimo M, Penttila M, Read ND, et al. (2007) Spatially segregated SNARE protein interactions in living fungal cells. J Biol Chem 282: 22775–22785. [DOI] [PubMed] [Google Scholar]
- 37. Kuratsu M, Taura A, Shoji JY, Kikuchi S, Arioka M, et al. (2007) Systematic analysis of SNARE localization in the filamentous fungus Aspergillus oryzae . Fungal Genet Biol 44: 1310–1323. [DOI] [PubMed] [Google Scholar]
- 38. Kaksonen M, Toret CP, Drubin DG (2006) Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 7: 404–414. [DOI] [PubMed] [Google Scholar]
- 39. Joung MJ, Mohan SK, Yu C (2012) Molecular level interaction of inositol hexaphosphate with the C2B domain of human synaptotagmin I. Biochemistry 51: 3675–3683. [DOI] [PubMed] [Google Scholar]
- 40. Mikoshiba K, Fukuda M, Ibata K, Kabayama H, Mizutani A (1999) Role of synaptotagmin, a Ca2+ and inositol polyphosphate binding protein, in neurotransmitter release and neurite outgrowth. Chem Phys Lipids 98: 59–67. [DOI] [PubMed] [Google Scholar]
- 41. Kim HS, Czymmek KJ, Patel A, Modla S, Nohe A, et al. (2012) Expression of the Cameleon calcium biosensor in fungi reveals distinct Ca(2+) signatures associated with polarized growth, development, and pathogenesis. Fungal Genet Biol 49: 589–601. [DOI] [PubMed] [Google Scholar]
- 42. Friesen H, Humphries C, Ho Y, Schub O, Colwill K, et al. (2006) Characterization of the yeast amphiphysins Rvs161p and Rvs167p reveals roles for the Rvs heterodimer in vivo. Mol Biol Cell 17: 1306–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nitsche BM, Crabtree J, Cerqueira GC, Meyer V, Ram AF, et al. (2011) New resources for functional analysis of omics data for the genus Aspergillus . BMC Genomics 12: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, et al. (1996) Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86: 865–875. [DOI] [PubMed] [Google Scholar]
- 45. Meyer V, Damveld RA, Arentshorst M, Stahl U, van den Hondel CA, et al. (2007) Survival in the presence of antifungals: genome-wide expression profiling of Aspergillus niger in response to sublethal concentrations of caspofungin and fenpropimorph. J Biol Chem 282: 32935–32948. [DOI] [PubMed] [Google Scholar]
- 46. Bermejo C, Rodriguez E, Garcia R, Rodriguez-Pena JM, Rodriguez de la Concepcion ML, et al. (2008) The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol Biol Cell 19: 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dudgeon DD, Zhang N, Ositelu OO, Kim H, Cunningham KW (2008) Nonapoptotic death of Saccharomyces cerevisiae cells that is stimulated by Hsp90 and inhibited by calcineurin and Cmk2 in response to endoplasmic reticulum stresses. Eukaryot Cell 7: 2037–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ernst R, Klemm R, Schmitt L, Kuchler K (2005) Yeast ATP-binding cassette transporters: cellular cleaning pumps. Methods Enzymol 400: 460–484. [DOI] [PubMed] [Google Scholar]
- 49. Xiong L, Lee H, Ishitani M, Zhu JK (2002) Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis . J Biol Chem 277: 8588–8596. [DOI] [PubMed] [Google Scholar]
- 50. Cowart LA, Shotwell M, Worley ML, Richards AJ, Montefusco DJ, et al. (2010) Revealing a signaling role of phytosphingosine-1-phosphate in yeast. Mol Syst Biol 6: 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakahara K, Ohkuni A, Kitamura T, Abe K, Naganuma T, et al. (2012) The Sjogren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol Cell 46: 461–471. [DOI] [PubMed] [Google Scholar]
- 52. Richman TJ, Toenjes KA, Morales SE, Cole KC, Wasserman BT, et al. (2004) Analysis of cell-cycle specific localization of the Rdi1p RhoGDI and the structural determinants required for Cdc42p membrane localization and clustering at sites of polarized growth. Curr Genet 45: 339–349. [DOI] [PubMed] [Google Scholar]
- 53. Tiedje C, Sakwa I, Just U, Hofken T (2008) The Rho GDI Rdi1 regulates Rho GTPases by distinct mechanisms. Mol Biol Cell 19: 2885–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Johnson JL, Erickson JW, Cerione RA (2009) New insights into how the Rho guanine nucleotide dissociation inhibitor regulates the interaction of Cdc42 with membranes. J Biol Chem 284: 23860–23871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tcheperegine SE, Gao XD, Bi E (2005) Regulation of cell polarity by interactions of Msb3 and Msb4 with Cdc42 and polarisome components. Mol Cell Biol 25: 8567–8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fadri M, Daquinag A, Wang S, Xue T, Kunz J (2005) The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol Biol Cell 16: 1883–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Daquinag A, Fadri M, Jung SY, Qin J, Kunz J (2007) The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol Cell Biol 27: 633–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P (2001) Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol 155: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Singh A, Del Poeta M (2010) Lipid signalling in pathogenic fungi. Cell Microbiol 13: 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Merkel O, Oskolkova OV, Raab F, El-Toukhy R, Paltauf F (2005) Regulation of activity in vitro and in vivo of three phospholipases B from Saccharomyces cerevisiae . Biochem J 387: 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lichius A, Berepiki A, Read ND (2011) Form follows function – the versatile fungal cytoskeleton. Fungal Biol 115: 518–540. [DOI] [PubMed] [Google Scholar]
- 62. Wachtler V, Balasubramanian MK (2006) Yeast lipid rafts?–an emerging view. Trends Cell Biol 16: 1–4. [DOI] [PubMed] [Google Scholar]
- 63. Moffett S, Brown DA, Linder ME (2000) Lipid-dependent targeting of G proteins into rafts. J Biol Chem 275: 2191–2198. [DOI] [PubMed] [Google Scholar]
- 64. Li S, Du L, Yuen G, Harris SD (2006) Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans . Mol Biol Cell 17: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nakase M, Tani M, Morita T, Kitamoto HK, Kashiwazaki J, et al. (2010) Mannosylinositol phosphorylceramide is a major sphingolipid component and is required for proper localization of plasma-membrane proteins in Schizosaccharomyces pombe . J Cell Sci 123: 1578–1587. [DOI] [PubMed] [Google Scholar]
- 66. Cheng J, Park TS, Chio LC, Fischl AS, Ye XS (2003) Induction of apoptosis by sphingoid long-chain bases in Aspergillus nidulans . Mol Cell Biol 23: 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cheng J, Park TS, Fischl AS, Ye XS (2001) Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans . Mol Cell Biol 21: 6198–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nunez LR, Jesch SA, Gaspar ML, Almaguer C, Villa-Garcia M, et al. (2008) Cell wall integrity MAPK pathway is essential for lipid homeostasis. J Biol Chem 283: 34204–34217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Waters BM, Eide DJ (2002) Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J Biol Chem 277: 33749–33757. [DOI] [PubMed] [Google Scholar]
- 70. Serrano R, Bernal D, Simon E, Arino J (2004) Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J Biol Chem 279: 19698–19704. [DOI] [PubMed] [Google Scholar]
- 71. Carman GM, Han GS (2007) Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion. Biochim Biophys Acta 1771: 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Robertson AS, Allwood EG, Smith AP, Gardiner FC, Costa R, et al. (2009) The WASP homologue Las17 activates the novel actin-regulatory activity of Ysc84 to promote endocytosis in yeast. Mol Biol Cell 20: 1618–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hasegawa J, Tokuda E, Tenno T, Tsujita K, Sawai H, et al. (2011) SH3YL1 regulates dorsal ruffle formation by a novel phosphoinositide-binding domain. J Cell Biol 193: 901–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Youn JY, Friesen H, Kishimoto T, Henne WM, Kurat CF, et al. (2010) Dissecting BAR domain function in the yeast Amphiphysins Rvs161 and Rvs167 during endocytosis. Mol Biol Cell 21: 3054–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bennett JW, Lasure LL (1991) More gene manipulations in fungi. New York: Academic Press. 441–457 p.
- 76.Meyer V, Ram AF, Punt PJ (2010) Genetics, genetic manipulation and approaches to strain improvement of filamentous fungi. In: AL D, J D, editors. Manual of industrial microbiology and biotechnology. 3rd ed. NY: Wiley. pp. 318–329.
- 77.Sambrook J, Russel D (2001) Molecular cloning: A laboratory manual. New York: Cold Spring Habour Laboratory Press.
- 78. Iversen JJL, Thomsen JK, Cox RP (1994) On Line Growth Measurements in Bioreactors by Titrating Metabolic Proton-Exchange. Applied Microbiology and Biotechnology 42: 256–262. [Google Scholar]
- 79. Jørgensen TR, Nitsche BM, Lamers GE, Arentshorst M, van den Hondel CA, et al. (2010) Transcriptomic insights into the physiology of Aspergillus niger approaching a specific growth rate of zero. Appl Environ Microbiol 76: 5344–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bergmeyer H, Bernt E, Schmidt F, Stork H (1974) Glucose. In: Bergmeyer H, editor. Methods of Enzymatic Analysis. New York: Academic Press. pp. 1196–1201.
- 81. Nitsche BM, Ram AF, Meyer V (2012) The use of open source bioinformatics tools to dissect transcriptomic data. Methods Mol Biol 835: 311–331. [DOI] [PubMed] [Google Scholar]
- 82. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 83. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Stat Methodol 57: 289–300. [Google Scholar]
- 84. Nitsche BM, Jorgensen TR, Akeroyd M, Meyer V, Ram AF (2012) The carbon starvation response of Aspergillus niger during submerged cultivation: insights from the transcriptome and secretome. BMC Genomics 13: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fisher RA (1922) On the Interpretation of X2 from Contingency Tables, and the Calculation of P. J R Stat Soc 85: 87–94. [Google Scholar]
- 86. Bos CJ, Debets AJ, Swart K, Huybers A, Kobus G, et al. (1988) Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger . Curr Genet 14: 437–443. [DOI] [PubMed] [Google Scholar]
- 87. van Hartingsveldt W, Mattern IE, van Zeijl CM, Pouwels PH, van den Hondel CA (1987) Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol Gen Genet 206: 71–75. [DOI] [PubMed] [Google Scholar]
- 88. Meyer V, Arentshorst M, El-Ghezal A, Drews AC, Kooistra R, et al. (2007) Highly efficient gene targeting in the Aspergillus niger kusA mutant. J Biotechnol 128: 770–775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete transcriptome data containing RMA expression values (log2 scale), mean expression values, p-values, q-values and fold changes.
(XLSX)
Subset of the transcriptome data containing up-down-regulated gene sets.
(XLSX)
ZIP archive file containing FetGOat enrichment results for the up- and down-regulated gene sets of all six comparisons.
(ZIP)
Subset of the transcriptome data for selected intersection of the Venn diagram.
(XLSX)
Primers used in this study.
(DOCX)