Abstract
Applied in tandem, elastin-like polypeptides (ELPs) and the sortase A (SrtA) transpeptidase from Staphylococcus aureus provide a general method for chromatography-free purification of tag-free recombinant proteins and optional, site-specific and homogeneous conjugation of the protein to a small molecule. This system provides an efficient, practical mechanism for generating bioactive proteins and protein-small-molecule combination therapeutics at high yields and purities.
Keywords: Biotechnology, Proteins, Polymers [bio(org.)], Anticancer agents
Column chromatography is the workhorse of protein purification, but the requirements for large volumes of buffers, expensive resins that have limited potential for re-use, and significant operator hours to optimize and execute the separations present major financial and technical hurdles to scaling up production. Additionally, covalent modification of proteins with small molecules and polymers is being routinely attempted, but available conjugation methods frequently produce heterogeneous products as a result of incomplete reactivity or the presence of multiple reaction sites within a protein. These two unit operations currently present major limitations to the production of recombinant proteins at both research and manufacturing scales.
A simple process that would facilitate efficient purification as well as site-specific, covalent conjugation of small molecules to a target protein would have great utility in biopharmaceutical production. Herein, we present a “three-in-one” method that utilizes the transpeptidase activity of Staphylococcus aureus sortase A (SrtA) in tandem with elastin-like polypeptides (ELPs) to enable: (1) recombinant fusion protein purification without chromatography, (2) removal of the ELP fusion tag and facile, chromatography-free recovery of pure target protein, and (3) site-specific covalent coupling of an extrinsic moiety to the purified target protein concurrent with cleavage from its ELP fusion partner. We present two complementary fusion protein designs that achieve these goals. The entire process provides a general platform for the purification and modification of a variety of recombinant proteins that is simple, robust, and scalable.
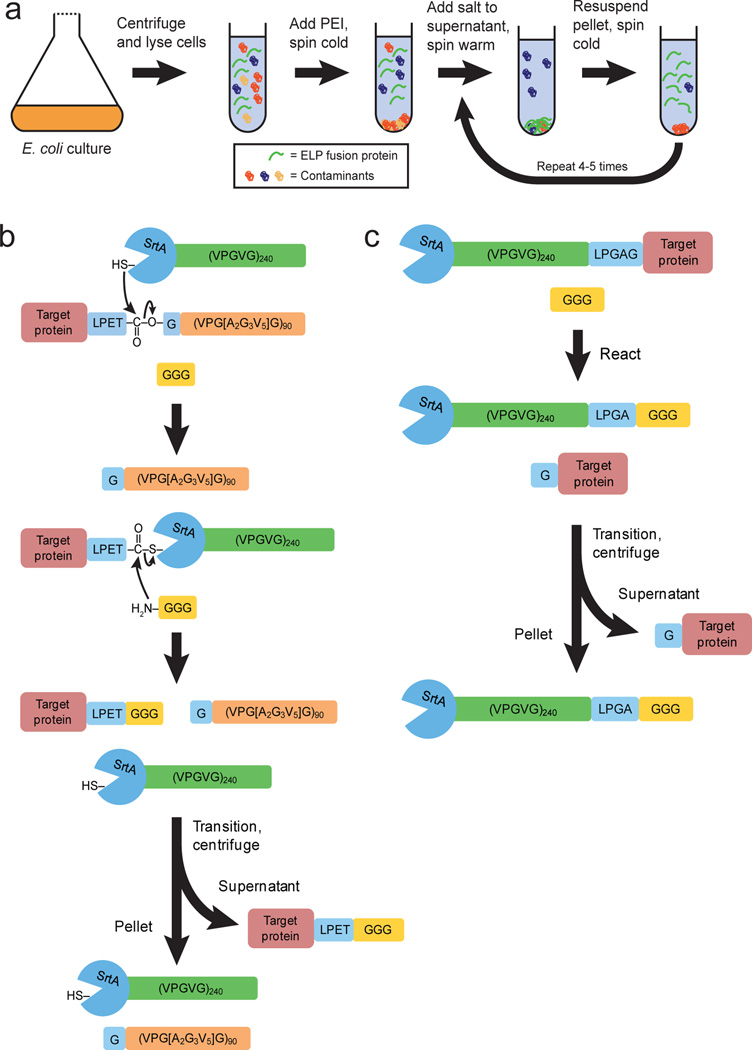
Expressing a protein as a fusion with another protein or a peptide tag is a widely-used strategy for purification by affinity chromatography.[1–4] Elastin-like polypeptides (ELPs) are purification tags that have been developed by our group to provide convenient fusion protein purification without requiring chromatography.[5,6] ELPs are peptide polymers composed of a repeated VPGXG pentapeptide unit, where X is any amino acid except proline. They can be designed to reversibly aggregate above a specific solution temperature – their inverse transition temperature (Tt) – by specifying the ELP amino acid composition, the ELP chain length, and the type and concentration of salt in solution.[7] This reversible phase transition behavior is retained by ELP fusion proteins and forms the basis for their purification by centrifugation rather than chromatography (Scheme 1a).[8,9]
Scheme 1.
(a) Overview of ELP fusion protein purification by inverse transition cycling. (b) Schematic of the reaction catalyzed by SrtA-ELP for recovery of target proteins from binary ELP fusions. (c) Strategy for target recovery by reaction of ternary SrtA-ELP-target protein fusions.
Although purification is initially simplified by fusion of a protein to a tag, in many applications the tag must be removed because it affects the bioactivity of the target protein. This is accomplished by including a protease site or a self-cleaving intein between the target protein and the tag.[10,11] However, the purification handle provided by the tag is lost upon cleavage of the fusion, and isolation of the target protein from the digested product requires additional chromatographic separation steps that must be customized for a given target.
We hypothesized that a SrtA-ELP fusion could be used to cleave other ELP fusion proteins, and that the released target proteins could be easily recovered at high purity by another round of phase transition-mediated purification without chromatography (Scheme 1b). We constructed a gene-level fusion of SrtA lacking its 59 amino-terminal residues – a truncation that has been shown to have no impact on its transpeptidase activity[12] – to an ELP with the sequence (VPGVG)240. A panel of target proteins including thioredoxin (TRX), green fluorescent protein (GFP), soluble murine tumor necrosis factor α (TNFα, amino acids 80–235), and soluble human tumor necrosis factor-related apoptosis-inducing ligand (TRAIL, amino acids 114–281) were fused at the gene level to a diferent ELP, (VPGXG)90, where X represents alanine (A), glycine (G), and valine (V) in a molar ratio of 2:3:5. These target proteins range from well behaved, single sub-unit proteins (TRX and GFP) to more difficult to express, pharmaceutically relevant proteins that form stable quaternary structures (TNFα and TRAIL). The linker between all target proteins and their ELP fusion partners contained the LPETG motif (Supplementary Fig. 1), which was previously demonstrated to be the optimal recognition sequence for SrtA.[13]
We also developed a ternary fusion in which SrtA and the target protein were linked by the ELP (VPGVG)240. This design allowed straightforward purification of the target protein without the need to add extraneous SrtA-ELP (Scheme 1c). Use of the optimal LPETG enzyme recognition site produced no intact ternary fusion due to premature cleavage during expression. However, amino acid substitutions in the LPXTG motif have been demonstrated to lower the SrtA reaction rate,[13] and we identified a variant– LPGAG – that allowed expression of intact ternary fusion when the protein was expressed at low temperature and the purification buffer contained ethylene glycol tetraacetic acid (EGTA) to scavenge the Ca2+ required for SrtA activity[12] (Supplementary Fig. 2).
All fusion proteins were expressed in Escherichia coli (E. coli) and were purified by inverse transition cycling (ITC), a method that we have previously developed.[6,8,9] We designed an ITC protocol that consisted of centrifugation of the cell lysate at 4°C in low-salt buffer where the ELP fusion was below its Tt and soluble, collection of the supernatant, centrifugation at 30°C in 0.3M ammonium sulfate where the ELP fusion was above its Tt and insoluble, followed by collection and solubilization of the pellet – containing the fusion protein – in cold, low-salt buffer. Repeated cycles of cold and warm centrifugation led to enrichment of the ELP fusion protein by eliminating other E. coli proteins that did not exhibit reversible phase transition behavior.
Target protein-ELP fusions and SrtA-ELP were expressed and purified separately, then co-incubated in a buffer that contained Ca2+ and triglycine. For ternary SrtA-ELP-target protein fusions, the cleavage reaction was initiated by transfer of the fusion protein into a buffer that contained Ca2+ and triglycine in the last stage of the ITC protocol. In both systems, the target protein was released by cleavage of the fusion at the SrtA recognition site. Based on scouting studies (Supplementary Fig. 3), we selected overnight incubation at 20°C and a 1:4 enzyme:target ratio (mol/mol) for cleavage of target-protein-ELP fusions by SrtA-ELP.
To isolate the target protein, the phase transitions of all other reaction products – SrtA-ELP, uncleaved target protein-ELP fusions, and free ELP – were triggered by adding sodium chloride to 1M and heating to 40°C. These condtions were selected to ensure aggregation of the SrtA-ELP and unreacted TRX-ELP – one of our more hydrophilic fusions – so that a common protocol could be used for each target protein in our panel (Supplementary Fig. 4 and 5).
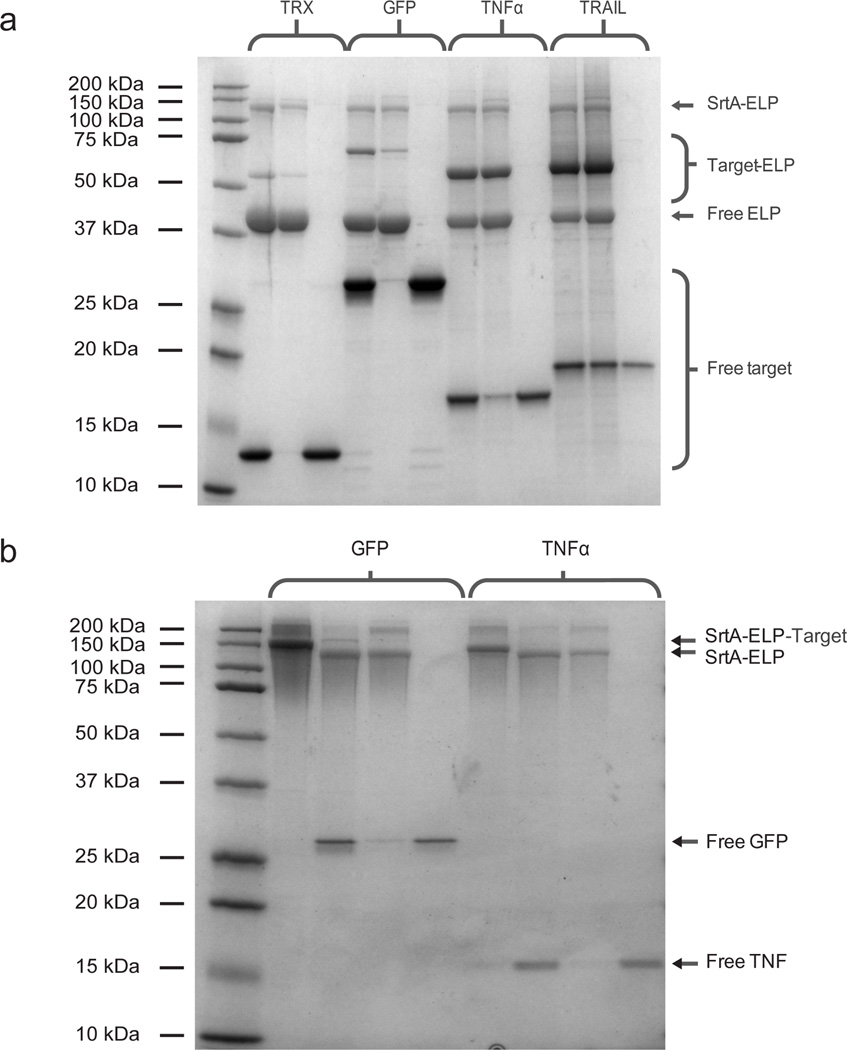
Dialysis or diafiltration of the supernatant removed the remaining triglycine and left behind the purified target protein. Figure 1a shows sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of the unpurified reaction product and centrifugation fractions for the purification of TRX, GFP, TNFα, and TRAIL. Figure 1b shows similar analysis for GFP and TNFα purified from ternary fusion reactions.
Figure 1.
(a) SDS-PAGE of target protein purification by reaction of target protein-ELP and SrtA-ELP fusions. Left to right for each target protein: reaction product, centrifugation pellet, and centrifugation supernatant. (B) SDS-PAGE of ternary fusion reactions. Left to right for each target protein: unreacted ternary fusion, reaction product, centrifugation pellet, and centrifugation supernatant.
We observed excellent cleavage and target protein recovery (Table 1). High-performance liquid chromatography (HPLC) confirmed that purities were greater than 95% (Supplementary Fig. 6) and MALDI-TOF mass spectrometry confirmed the molecular weight of each target protein (Supplementary Figure 7).
Table 1.
Summary of reactions of target protein-ELPs and SrtA-ELP for representative batches of each fusion protein. Conversion and recovery percentages were calculated by analysis of SDS-PAGE images and confirmed by quantifying purified reaction product concentrations by spectrophotometry.
| Target protein |
SrtA reaction efficiency (%) |
Target protein recovery (%) |
Target protein purity (% by HPLC) |
Target protein yield (mg/L fermentation) |
|---|---|---|---|---|
| TRX | 88 | 100 | 98 | 35 |
| GFP | 85 | 100 | 100 | 28 |
| TNFα | 54 | 82 | 96 | 16 |
| TRAIL | 46 | 46 | 95 | 9 |
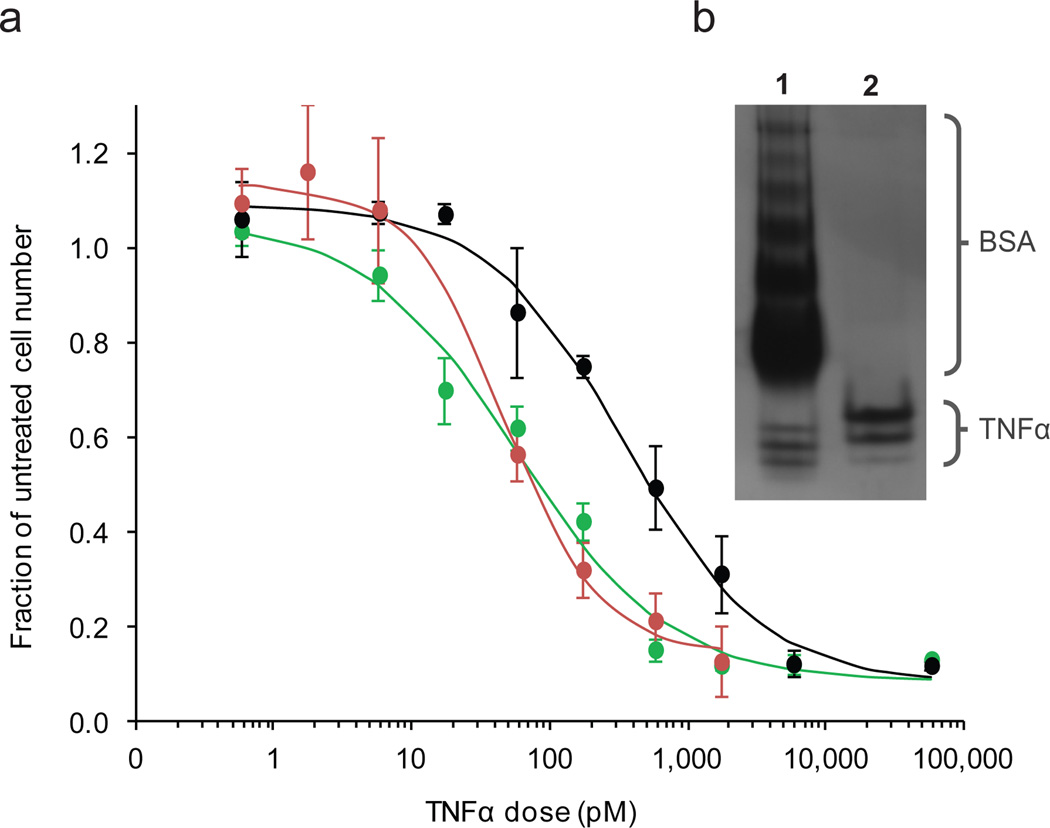
The fluoresence emission spectrum of GFP provided a simple indication that our methodology yielded properly folded, active protein. TNFα was investigated as a more complicated case, as homotrimer assembly is critical for TNFα signaling.[14,15] Non-denaturing, native PAGE was used to evaluate the protein’s quaternary structure against commercially-available TNFα. Protein from both sources ran similarlyand appeared as three distinct bands on the gel, suggestive of monomers, dimers, and trimers (Figure 2b). The commercial preparation contains 0.5% bovine serum albumin, which accounts for the lower mobility bands at the top of the gel (Supplementary Figure 8). Interestingly, matrix assisted laser desorption ionization time-of flight mass spectrometry (MALDI-TOFMS) not only confirmed the mass of monomeric TNFα, but also showed peaks at double and triple the monomer m/z, which provided additional evidence of proper quaternary structure (Supplementary Fig. 7). Static light scattering on TNFα purified from the ternary fusion indicated an average molecular weight of 44.7 kDa (Supplementary Fig. 9), which is consistent with the average molecular weight of 49 kDa reported previously for TNFα.[16]
Figure 2.
(a) Viablility of L929 cells after incubation with TNFα from reaction of TNFα isolated from the binary fusion ( ), from the ternary fusion (
), from the ternary fusion ( ), or from eBioscience (
), or from eBioscience ( ) relative to an untreated control. Error bars indicate standard deviations. (b) Native PAGE of commercial TNFα (lane 1) and TNFα from the ternary fusion (lane 2) are comparable and suggest the presence of monomer, dimer, and trimer populations in the lower (most mobile) bands.
) relative to an untreated control. Error bars indicate standard deviations. (b) Native PAGE of commercial TNFα (lane 1) and TNFα from the ternary fusion (lane 2) are comparable and suggest the presence of monomer, dimer, and trimer populations in the lower (most mobile) bands.
We examined the bioactivity of TNFα by measuring its effect on the L929 mouse fibrosarcoma cell line, which is known to be sensitive to cytolysis by TNFα at picomolar concentrations.[17,18] Cell number relative to an untreated group was assessed for cells treated with SrtA-purified or commercial TNFα (Figure 2a). ED50 values were 48 pM, 350 pM, and 57 pM, for TNFα isolated from cleavage of TNFα-ELP by SrtA-ELP, TNFα isolated from the ternary fusion, and commercial protein, respectively. The similar bioactivity of the commercial and SrtA-purified TNFα further reinforced the proper folding and multimerization suggested by native PAGE, static light scattering, and MALDI-TOFMS.
SrtA, unlike proteases or self-splicing inteins that simply cleave a target protein from a tag, also enables site-specific, covalent attachment of other molecules to the target protein. Our protein purification reactions were designed to mimic a typical protease digestion by using synthetic triglycine as a nucleophile. However, triglycine-modified lipids and small molecules, as well as proteins with amino-terminal glycine residues have also been used effectively as nucleophiles in transpeptidation reactions catalyzed by sortase A.[19–23] In principle, our system is flexible in that it can be easily extended to accomodate these nucleophiles. Moreover, this conjugation approach has the attractive feature that only one molecule is installed specifically at the C-terminus of the protein.
To demonstrate the flexibiity of our system, we conjugated the chemotherapeutic camptothecin (CPT) to TRAIL, generating a hybrid anticancer agent. We chose CPT for its potent antitumor activity and because it is a chromophore with an extinction peak at 365 nm, which allowed the product to be tracked spectrophotometrically. The drug was chemically modified such that the hydroxyl group on the E-ring was coupled to the carboxyl terminus of triglycine (Supplementary Fig. 10).
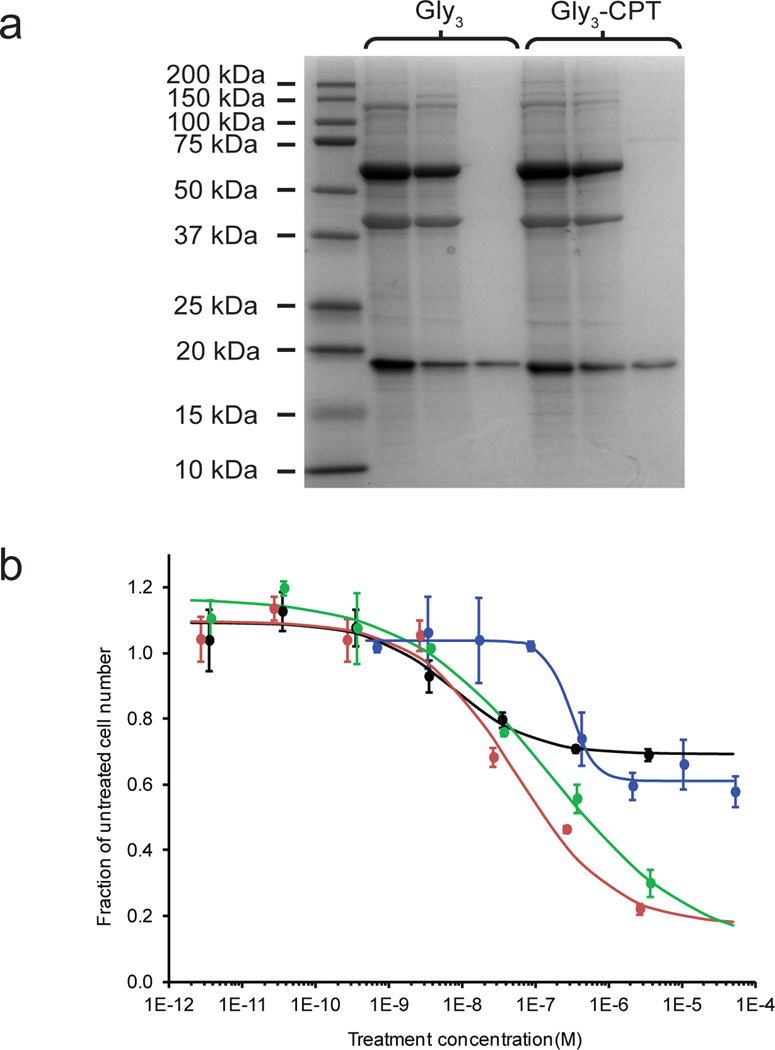
The conditions used for the TRAIL-CPT conjugation reaction and product recovery were the same as those used for the purification of unlabeled protein, except that Gly3-CPT was substituted for triglycine. SDS-PAGE of parallel reactions with triglycine or Gly3-CPT (Figure 3a) indicated significant conversion in both cases, suggesting that Gly3-CPT was an effective nucleophile. Based on analysis of SDS-PAGE images, we determined that the reaction conversion using Gly3-CPT was approximately 80% of that for a reaction using Gly3. Similarly, overall recovery after reaction purification for Gly3-CPT reactions were consistently 65% of recoveries for reactions using Gly3. We attribute the reductions in yield and recovery to the reduced solubility of the CPT moiety, which likely lowered the concentration driving force for reaction and possibly contributed to some nonspecific aggregtion of the product.
Figure 3.
(a) SDS-PAGE analysis of TRAIL-ELP reactions run in parallel with triglycine (Gly3) or triglycine-modified camptothecin (Gly3-CPT). Shown left to right for each reaction are the raw product, centrifugation pellet, and centrifugation supernatant. (b) Cell death in MDA-MB-231 cells treated with TRAIL ( ), Gly3-CPT (
), Gly3-CPT ( ), TRAIL-CPT conjugate (
), TRAIL-CPT conjugate ( ), or a mixture of Gly3-CPT and TRAIL at a 0.8:1 molar ratio. (
), or a mixture of Gly3-CPT and TRAIL at a 0.8:1 molar ratio. ( ). For combination treatments, the TRAIL concentration is indicated. The viable cell number is reported relative to an untreated group. Error bars indicate standard deviations.
). For combination treatments, the TRAIL concentration is indicated. The viable cell number is reported relative to an untreated group. Error bars indicate standard deviations.
ESI-MS confirmed that the m/z ratios of TRAIL and the TRAIL-CPT conjugate differed by the mass of a single molecule of CPT (Table 2 and Supplementary Figure 11). HPLC in 70/30 (v/v) Milli-Q water/acetonitrile indicated that the conjugated and non-conjugated compounds eluted differently, and that excess triglycine-CPT was successfully removed by diafiltration of the recovered reaction product (Supplementary Fig. 12).
Table 2.
Summary of ESI mass spectrometry for TRAIL and TRAIL-CPT purified from TRAIL-ELP by reaction with SrtA-ELP. Molecular weights are reported as the average of 14 and 11 charge states for TRAIL-Gly3 and TRAIL-CPT, respectively.
| Species | Predicted Mw (Da) | ESI-MS Mw (Da) |
|---|---|---|
| TRAIL-Gly3-CPT | 20893.2 | 20896.2 |
| TRAIL-Gly3 | 20563.9 | 20564.3 |
| Difference = CPT | 329.3 | 331.9 |
Notably, in our reaction design the unreacted protein retained the ELP tag, so that homogeneous conjugate could be purified from unreacted protein by a single centrifugation regardless of the extent of reaction conversion. Based on a calibration curve for Gly3-CPT (Supplementary Fig. 13), we consistently obtained conjugation ratios greater than 0.8:1 (mol CPT/mol TRAIL). The controlled stoichiometry and homogeneous product produced in our conjugation protocol, as well as the ability to easily separate the conjugate from unreacted protein are especially important for pharmaceutical applications, where non-specific or incomplete reactivity must be controlled precisely to achieve batch-to-batch consistency for regulatory approval and patient safety.
Bioactivity of the TRAIL-CPT conjugate was assessed by measuring apoptosis in the TRAIL-sensitive human breast adenocarcinoma cell line MDA-MB-231 (Figure 3b). Viable cell number relative to an untreated control was assayed after incubation with TRAIL, Gly3-CPT, TRAIL-CPT conjugate, or TRAIL and Gly3-CPT in a molar ratio equivalent to that of the conjugate. High concentrations of Gly3-CPT alone killed 40% of the cells, whereas TRAIL killed a maximum of 30% of the cells and activated caspases 3 and 7 in a dose-dependent manner (Supplementary Fig. 14). Notably, the IC50 for the caspase activation assay confirmed that determined by the MTS assay.
Dosing TRAIL and CPT as a conjugate reduced the viable cell number by a maximum of 75%. Though improved cell killing was anticipated using a combination of CPT and TRAIL, it is noteworthy that the TRAIL-CPT conjugate had the same potency and efficacy as the combination of non-conjugated Gly3-CPT and TRAIL in an equivalent molar ratio, which suggested that each molecule within the conjugate retained its activity. Interestingly, both conjugated and non-conjugated drug combinations showed an additive effect (Supplementary Fig. 15). We also tested the bioactivity of the TRAIL-CPT conjugate on the TRAIL-insensitive human prostate adenocarcinoma cell line PC3 (Supplementary Figure 16). In this case, both TRAIL-CPT and a combination of Gly3-CPT and TRAIL killed cells in a manner equivalent to Gly3-CPT alone over the concentration range tested, suggesting that these cells were not sensitized to TRAIL by co-treatment with CPT. Though the particular combination of TRAIL and CPT does not offer an advantage in our cytotoxicity assays, it nonetheless provides a proof-of-concept example of the ease with which bioactive combination therapeutics can be produced using our system.
The combination of SrtA and ELPs represents a powerful and flexible system for purification and site-specific chemical modification of proteins. As with inteins, our ternary fusion provides a straightforward, all-in-one system that is subject to premature cleavage during expresion. However, our binary fusion system provides an alternative with excellent control over reactivity, high product yields, and virtually no increase in complexity.
Using our reaction strategies, significant quantities of high-purity, bioactive recombinant proteins can be purified without column chromatography by a protocol that is practical and applicable to a variety of target proteins. The ease with which our protocol is executed and the flexibility to perform an optional, one-step site-specific conjugation reaction that homogeneously labels the product provide a valuable, “three-in-one” set of tools for the production of therapeutic proteins and protein-small molecule conjugates.
Experimental Section
Fusion protein genes: The SrtA gene was cloned from S. aureus and the amino-terminal 59 amino acids were removed by polymerase chain reaction. DNA coding for the ELPs (VPGVG)240, and (VPGXG)90, where X represents A, G, and V in a molar ratio of 2:3:5 were constructed previously in our lab. The genes for thioredoxin (TRX) and green fluorescent protein (GFP) were available from previous studies. DNA coding for soluble murine tumor necrosis factor α (TNFα, amino acids 80–235), and soluble human tumor necrosis factor-related apoptosis-inducing ligand (TRAIL, amino acids 114–281) were designed for E. coli codon usage and purchased from Life Technologies (Carlsbad, CA).
Fusion protein expression and purification: Expression vectors were transformed into E. coli strain BL21 (DE3). Frozen stocks were used to inoculate a starter culture that was grown overnight at 37°C with orbital shaking at 250 rpm. Starters were centrifuged, resuspended in fresh media, and used to inoculate 4L shake flasks containing 1L terrific broth with appropriate antibiotic. 2% inoculums were used. All cultures were allowed to grow for 6–8 hours with 200 rpm orbital shaking at 25°C. Protein expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to 0.5 mM final concentration. Induction was allowed to proceed overnight at 16°C. Cells were lysed by sonication and fusion proteins were recovered by inverse transition cycling.
Sortase reactions: SrtA-ELP and target protein-ELP were combined to achieve an enymze:substrate molar ratio of approximately 1:4. Protein concentrations were determined by the Beer-Lambert Law using calculated extinction coefficients[24] and the absorbance at 280 nm measured by a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE). Reaction buffer was added to a final concentration of 50 mM Tris-HCl, 150 mM NaCl, 10 mM CaCl2, pH 7.5. Synthetic triglycine peptide was added to 10–20 molar excess over the target protein-ELP fusion and the reaction was allowed to proceed for approximately 18 hours at 20°C.
Cleaved target proteins in both reaction designs were purified by centrifugation at 16.1 rcf in a fixed-angle benchtop centrifuge with temperature controlled at 40°C for 15 minutes. Prior to centrifugation, 1M sodium chloride was added to reactions where the target protein was originally fused to ELP (VPGXG)90 [X=V5A2G3].
Labeling reactions were identical to the reaction protocol for target protein purification using SrtA-ELP, except that triglycine was replaced with camptothecin modified by the Duke University Small Molecule Synthesis Facility (Durham, NC) to contain triglycine covalently linked to the E-ring hydroxyl group.
Cytotoxicity assays: L929 cells were cultured in 96-well culture dishes at an initial density of 50,000 cells/mL and treated with a range of concentrations of TNFα purified by cleavage of TNFα-ELP, from the ternary fusion, or purchased from eBioscience (San Diego, CA). Negative control groups were included that were untreated. Cultures were incubated for 36 hours.
MDA-MB-231 cells were cultured in 96-well culture dishes at an initial density of 100,000 cells/mL and incubated for approximately 18 hours. The growth media was replaced with serum-free media and TRAIL, Gly3-CPT, TRAIL-CPT conjugate, or non-conjugated CPT and TRAIL (0.8:1 mol/mol).
Growth inhibition versus untreated controls was assessed using a CellTiter 96 One Solution MTS/PMS viability assay purchased from Promega (Madison, WI). Absorbance was measured at 490 nm in a multi-well spectrophotometer. All treatment concentrations were repeated in triplicate, and the means for each group were normalized to that of the untreated group. All samples were corrected for background absorbance by measuring the 490 nm absorbance of the MTS/PMS reagent added to cell-free media.
For more detailed experimental methods, please see the Supporting Information section.
Supplementary Material
Acknowledgments
The authors thank Wafa Hassouneh for performing light scattering data collection and analysis and Kate Clancy for assistance with determining initial reaction conditions. This work was supported by a University Scholars Fellowship awarded by the Graduate School at Duke University and by the National Institutes of Health through grants 5T32 GM008487, R01 GM061232, and R01 AI46611.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Joseph J. Bellucci, Department of Biomedical Engineering, Duke University, Durham, North Carolina 27708 (USA)
Miriam Amiram, Department of Biomedical Engineering, Duke University, Durham, North Carolina 27708 (USA).
Jayanta Bhattacharyya, Department of Biomedical Engineering, Duke University, Durham, North Carolina 27708 (USA).
Dewey McCafferty, Department of Chemistry and Biochemistry, Duke University, Durham, North Carolina 27708 (USA).
Ashutosh Chilkoti, Department of Biomedical Engineering, Duke University, Durham, North Carolina 27708 (USA), chilkoti@duke.edu.
References
- 1.Braun P, et al. Proc. Natl. Acad. Sci. USA. 2002;99:2654–2659. doi: 10.1073/pnas.042684199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hang Q, Woods L, Feiss M, Catalano CE. J. Biol. Chem. 1999;274:15305–15314. doi: 10.1074/jbc.274.22.15305. [DOI] [PubMed] [Google Scholar]
- 3.de Boer E, et al. Proc. Natl. Acad. Sci. USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rigaut G, et al. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 5.Wu WY, Mee C, Califano F, Banki R, Wood DW. Nat. Protoc. 2006;1:2257–2262. doi: 10.1038/nprot.2006.314. [DOI] [PubMed] [Google Scholar]
- 6.Meyer D, Chilkoti A. Nat. Biotech. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 7.Cho Y, et al. J. Phys. Chem. B. 2008;112:13765–13771. doi: 10.1021/jp8062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreher MR, et al. J. Am. Chem. Soc. 2008;130:687–694. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassouneh W, Christensen T, Chilkoti A. Curr. Protein Sci. 2010;61:6.11.1–6.11.16. doi: 10.1002/0471140864.ps0611s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge X, et al. J. Am. Chem. Soc. 2005;127:11228–11229. doi: 10.1021/ja0531125. [DOI] [PubMed] [Google Scholar]
- 11.Banki MR, Feng L, Wood DW. Nat. Methods. 2005;2:659–661. doi: 10.1038/nmeth787. [DOI] [PubMed] [Google Scholar]
- 12.Ilangovan U, Ton-That H, Iwahara J, Schneewind I, Clubb RT. Proc. Natl. Acad. Sci. USA. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruger RG, et al. Biochemistry. 2004;43:1541–1551. doi: 10.1021/bi035920j. [DOI] [PubMed] [Google Scholar]
- 14.Ameloot P, Declercg W, Fiers W, Vandenabeele P, Brouckaert P. J. Biol. Chem. 2001;276:27098–27103. doi: 10.1074/jbc.M104486200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang XM, Weber I, Chen MJ. J. Biol. Chem. 1992;267:24069–24075. [PubMed] [Google Scholar]
- 16.Schoenfeld HJ, et al. J. Biol. Chem. 1991;266:3863–3869. [PubMed] [Google Scholar]
- 17.Flick DA, Gifford GE. J. Immunol. Methods. 1984;68:167–175. doi: 10.1016/0022-1759(84)90147-9. [DOI] [PubMed] [Google Scholar]
- 18.Liddil JD, Dorr RT, Scuderi P. Cancer Res. 1989;49:2722–2728. [PubMed] [Google Scholar]
- 19.Mao H, Hart SA, Schink A, Pollok BA. J. Am. Chem. Soc. 2004;126:2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- 20.Antos JM, Miller GM, Grotenbreg GM, Ploegh HL. J. Am. Chem. Soc. 2008;130:16338–16343. doi: 10.1021/ja806779e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antos JM, et al. J. Am. Chem. Soc. 2009;131:10800–10801. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z, Guo X, Wang Q, Swarts BM, Guo Z. J. Am. Chem. Soc. 2010;132:1567–1571. doi: 10.1021/ja906611x. [DOI] [PubMed] [Google Scholar]
- 23.Popp MW, Dougan SK, Chuang TY, Spooner E, Ploegh HL. Proc. Natl. Acad. Sci. USA. 2011;108:3169–3174. doi: 10.1073/pnas.1016863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill SC, von Hippel PH. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.