Abstract
Two-pore domain K+ (K2P) channels underlie leak or background potassium conductances in many cells. The Trek subfamily of K2P channels, which includes Trek1/Kcnk2 and Trek2/Kcnk10 and has been implicated in depression, nociception, and cognition, exhibits complex regulation and can modulate cell excitability in response to a wide array of stimuli. While alternative translation initiation and alternative splicing contribute to the structural and functional diversity of Trek1, the impact of post-transcriptional modifications on the expression and function of Trek2 is unclear. Here, we characterized two novel splice isoforms of the mouse Trek2 gene. One variant is a truncated form of Trek2 that possesses two transmembrane segments and one pore domain (Trek2-1p), while the other (Trek2b) differs from two known mouse Trek2 isoforms (Trek2a and Trek2c) at the extreme amino terminus. Both Trek2-1p and Trek2b, and Trek2a and Trek2c, showed prominent expression in the mouse CNS. Expression patterns of the Trek2 variants within the CNS were largely overlapping, though some isoform-specific differences were noted. Heterologous expression of Trek2-1p yielded no novel whole-cell currents in transfected HEK 293 cells. In contrast, expression of Trek2b correlated with robust K+ currents that were ∼5-fold larger than currents measured in cells expressing Trek2a or Trek2c, a difference mirrored by significantly higher levels of Trek2b found at the plasma membrane. This study provides new insights into the molecular diversity of Trek channels and suggests a potential role for the Trek2 amino-terminus in channel trafficking and/or stability.
Two-pore domain K+ (K2P) channels, whose pore-forming subunits contain four membrane-spanning domains, two pore regions, and cytoplasmic amino (N)- and carboxyl (C)-termini, underlie leak or background potassium conductances in many cell types (reviewed in Enyedi and Czirjak, 2010). The K2P family is functionally diverse, with several sub-families exhibiting unique regulatory and biophysical properties that allow them to modulate cell excitability in response to specific stimuli. Members of the Trek subfamily of K2P channels, which includes Trek1/Kcnk2, Trek2/Kcnk10, show perhaps the most complex regulation among K2P channels (Patel and Honore, 2001, Honore, 2007). Trek channels mediate K+ currents sensitive to membrane stretch (Patel et al., 1998, Bang et al., 2000, Lesage et al., 2000), arachidonic acid (Patel et al., 1998, Lesage et al., 2000), temperature (Maingret et al., 2000, Kang et al., 2005), and pH (Maingret et al., 1999, Lesage et al., 2000, Kim et al., 2001). Trek channels are also inhibited by protein kinase A (PKA) and protein kinase C (PKC) phosphorylation (Fink et al., 1996, Patel et al., 1998, Lesage et al., 2000, Maingret et al., 2000, Bockenhauer et al., 2001, Murbartian et al., 2005), which couples channel activity to G protein-dependent signaling cascades involving Gs, Gi/o, and Gq G-proteins.
Trek channels have been implicated in a wide array of physiological and neurobehavioral processes. For example, genetic variation in the TREK1 gene in humans was linked to individual differences in mood and responses to rewarding stimuli (Dillon et al., 2010). Mice lacking the Trek1 gene are more sensitive to painful heat (Alloui et al., 2006), are resistant to the sedative effects of volatile anesthetics (Heurteaux et al., 2004), are more susceptible to ischemia and epilepsy (Heurteaux et al., 2004), and exhibit a depression-resistant phenotype (Heurteaux et al., 2006). While less is known regarding physiological roles for Trek2, recent studies suggest that Trek2 contributes to the resting membrane potential in mouse superior cervical ganglion neurons (Cadaveira-Mosquera et al., 2011), and that Trek2 mediates the postsynaptic inhibitory effects of GABAB and α2 adrenergic receptor activation in neurons of the entorhinal cortex (Deng et al., 2009, Xiao et al., 2009). Moreover, knock-down of Trek2 mRNA in the entorhinal cortex impaired spatial learning in mice (Deng et al., 2009).
Given the contribution of Trek channels to neurophysiology and neuropharmacology, understanding the molecular diversity within this K2P subfamily, particularly within the CNS, is important. Previous studies have documented multiple post-transcriptional modifications that increase structural diversity in the Trek family, and in some instances, these modifications correlate with functional diversity. For example, alternative translation initiation was shown to yield both short and long variants of Trek1 that differ with respect to Na+ permeability (Thomas et al., 2008). At present, the full scope and functional relevance of alternative splicing in the Trek family is unclear, particularly for the Trek2 gene. While some Trek2 splice variants harboring unique N-terminal domains have been characterized (Gu et al., 2002), rigorous comparative assessments of expression or function have not been undertaken. The goal of this study was to identify murine Trek2 splice variants, evaluate their expression patterns, and probe for isoform-dependent differences in channel function.
Experimental Procedures
RT-PCR and Trek isoform expression analysis
Total mouse brain RNA (500 ng, Clontech; Mountain View, CA) was reverse-transcribed using the iScript cDNA synthesis kit according to manufacturer's recommendations (BioRad; Hercules, CA). For subsequent PCR amplification, 2 μL of reverse transcribed cDNA was combined with 0.1 μL Hi-Fidelity Taq (Invitrogen Life Sciences, Carlsbad, CA), 10X Hi-Fidelity Taq buffer, 2.5 μM oligonucleotide mixture, 0.5 μM MgSO4, and 2.5 μM dNTPs in a final volume of 25 μL. Sense oligonucleotides targeting the unique 5′ UTR for Trek2a (5′-gcagagcgagacccaaccactcc-3′), Trek2b (5′-ggctgcaactccaccgagcacg-3′), and Trek2c (5′-ccgttggtctgtttaaccgacgag-3′), were paired with an antisense oligonucleotide targeting either the 3′UTR found in exon 5 (5′-ccaacatggtagcgcacagtcc-3′) to amplify the truncated one-pore (1p) variants, or 3′UTR found in exon 8 (5′-gccactgtctgaaatgaagctcttgc-3′) to amplify full-length Trek2 variants. The thermal cycling protocol consisted of a 2 min denaturation step at 94°C, followed by 25/35/45 cycles of 94°C/30 s, 58°C/30 s, and 68°C/3 min, and a 10 min final extension step at 68°C. Data presented in Fig. 2 derives from experiments involving 35 thermal cycles. While increasing cycle number to 45 increased the intensities of the Trek2 isoform amplicons, no differences in the relative patterns of isoform distribution were observed between experiments involving 35 or 45 cycle numbers. Predicted amplicon sizes were 869 bp (Trek2c-1p), 1871 bp (Trek2a), 1822 bp (Trek2b), 1935 bp (Trek2c), Amplicons were extracted using a gel purification kit (Qiagen, Valencia, CA), inserted into pCR2.1-Topo (Invitrogen Life Sciences), and sequenced. Identical PCR conditions and isoform-specific oligonucleotides were used to evaluate regional differences in Trek2 variant expression in 8 mouse tissues (Clontech; Mountain View, CA) and 15 different brain regions (Zyagen; San Diego, CA). PCR samples were subject to DNA electrophoresis on 1% agarose gels containing ethidium bromide for band visualization.
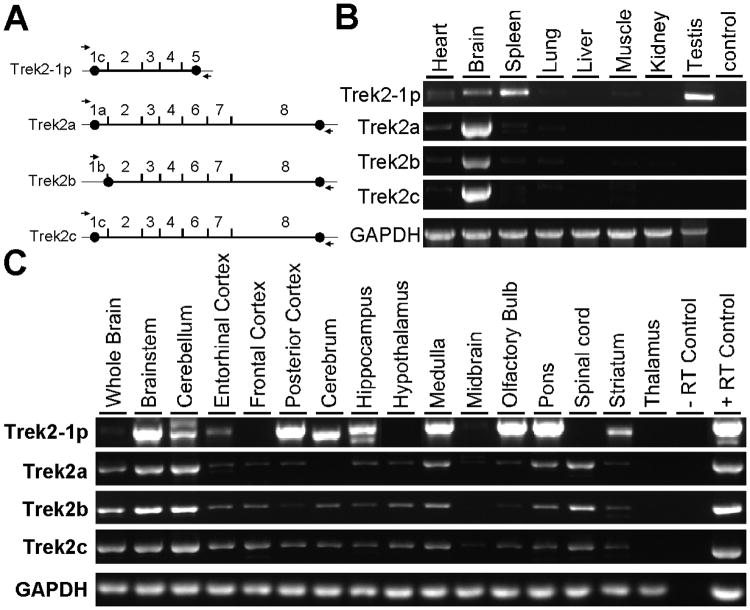
Figure 2. Expression patterns of the mouseTrek2 splice variants.
A) Schematic depiction of mouse Trek2 splice variant mRNAs, showing intron-exon boundaries and locations of isoform-specific oligonucleotides used for analysis of Trek2 variant expression in mouse tissues. Black circles denote the boundaries of Trek2 coding sequence. B) Representative ethidium bromide-stained DNA gel segments showing Trek2 isoform-specific and intron-spanning amplicons in several mouse tissues (n=3 separate experiments per Trek2 isoform). The size and DNA sequence of the primary amplicon corresponded to the intended Trek2 isoform in all cases. Lower intensity bands located near the primary amplicons were seen in some lanes (e.g., Trek2a in spleen). Water was used as a negative control for each unique PCR reaction. A comparable level of GAPDH expression was observed in all mouse tissues. C) Representative ethidium bromide-stained DNA gel segments showing Trek2 splice variant expression in 15 different CNS sub-regions (n=3 separate experiments per isoform). Lanes on the right side of the gel contain results of RT-PCR experiments using isoform-specific PCR primers and total brain mRNA; mRNA samples were treated with (+RT) or without (-RT) reverse-transcriptase. A comparable level of GAPDH expression was observed in all CNS sub-regions.
Generation of Trek2 expression constructs
C-terminal myc-tagged Trek2-1p, Trek2a, Trek2b, and Trek2c expression constructs, were generated by PCR using Hi-Fidelity Taq. Trek2 cDNA (BC132487) was purchased from Open Biosystems (Huntsville, AL) and used as the PCR template. PCR fragments consisting of a Kozak sequence (5′-gccgccacc-3′), Trek2 variant open reading frame, and carboxyl-terminal myc tag sequence (5′-gagcagaaacttagcgaggaggacctg-3′), framed by Nhe1 and HindIII (Trek2a and Trek2c) or Not1 and HindIII (Trek2b) restriction enzyme sites, were then incorporated into the pCR2.1-Topo vector according to manufacturer's protocols (Invitrogen Life Science; Carlsbad, CA). Inserts were sequenced in both directions for accuracy. Amplified fragments were then subcloned into a modified pcDNA3.1- expression plasmid harboring the CAG promoter (pCAG3.1).
Immunoblotting
Human embryonic kidney (HEK) 293FT cells were purchased from Invitrogen Life Sciences (Carlsbad, CA) and cultured according to supplier recommendations. No tests were conducted to rule out the potential contamination of mycoplasma. Cells were plated at 60-70% confluence in 6-well plates, and transfected using the calcium phosphate method with the myc-tagged Trek2 expression constructs (2 μg/well). Two days after transfection, cells were collected in ice-cold PBS and centrifuged at 9200 × g for 1 min. Following centrifugation, the supernatant was aspirated and the pellet was resuspended in 250 μL 2× SDS sample buffer; samples were sonicated briefly. For immunoblotting, 2 μL 1M DTT was added to 10 μL of sample, and the mixture was incubated at 80°C for 10 min. Samples were then loaded onto 12% Bis-Tris gels, and run in a Tris-Glycine buffer under reducing conditions. Samples were transferred using a wet transfer system (Bio-Rad Laboratories, Hercules, CA) onto nitrocellulose membranes (Thermo Fisher Scientific Inc.; Rockford, IL), and probed with either a mouse anti-myc (1:500 dilution, Hoffman-La-Roche, Nutley, NJ) or mouse anti-β-actin (1:10000 dilution, Abcam, Cambridge, MA) antibodies. An IRDye 800CW donkey × mouse secondary antibody (LiCor Biosciences; Lincoln, NE) was used at a dilution of 1:7000. Visualization and quantification of band intensity was performed using the Odyssey Imaging System (LiCor Biosciences; Lincoln, NE). Integrated intensity values were recorded for all lanes and normalized to the intensity of the β-actin band.
Electrophysiology
HEK 293 cells were cultured according to ATCC specifications, plated on 8-mm glass coverslips (30,000 cells/coverslip), and transfected using Lipofectamine LTX (Life Technologies Corporation; Carlsbad, CA) with an EGFP expression plasmid (0.02 μg/coverslip) and either a Trek2 splice variant or empty vector (0.09 μg/coverslip). One day after transfection, whole-cell currents were measured in EGFP-labeled cells with hardware (Axopatch-200B amplifier, Digidata 1320) and software (pCLAMP v. 9.0) from Molecular Devices, Inc. (Sunnyvale, CA). Borosilicate (3-6 MΩ) pipettes were filled with (in mM): 140 KCl, 2 MgCl2, 1 EGTA, 5 HEPES, pH 7.2 with KOH. The bath solution consisted of (in mM): 136 NaCl, 4 KCl, 1.5 CaCl2, 2 MgCl2, 5 HEPES, pH 7.4 with NaOH. In some experiments, cells were switched from the normal bath solution to solutions containing 20 or 40 mM KCl (osmotically-balanced by equimolar reductions in NaCl) using an SF-77B Perfusion Fast-Step system (Warner Instrument Corp.; Hamden, CT). Upon achieving whole-cell access, cells were held at -70 mV and subjected to either a voltage-ramp (-100 mV to 60 mV in 1 s) or voltage-step (-100 mV to 60 mV in 20 mV increments; 0.5 s/step) protocol. Only experiments with low RA (<20 MΩ) and low CM (<50 pF) were included in the final data set. All measured currents were filtered at 2 kHz and stored directly on hard disk for subsequent analysis.
Surface biotinylation
HEK 293FT cells were plated at 60-70% confluency in 6-well plates, and transfected using the calcium phosphate method with myc-tagged Trek2 expression constructs (2 mg/well). Two days after transfection, cells were washed three times with ice-cold PBS (pH 8.0), and then incubated with 2 mM NHS-PEG4 Biotin (Thermo Fisher Scientific Inc.; Rockford, IL) in ice-cold PBS, with slow shaking at 4°C for 30 min. Cells processed in parallel but not treated with biotin served as negative controls. Biotin was gently removed and cells were washed five times with ice-cold PBS. Cells were collected in PBS and centrifuged at 9200 × g for 1 min. Following centrifugation, supernatants were aspirated and pellets were resuspended in 250 μL Lysis Buffer consisting of PBS, 1% Triton, and a mixture of protease inhibitors (10 μg/ml Pepstatin A, 10 μg/ml Aprotinin, 10 μg/ml PMSF, and 1 μg/ml Leupeptin); Samples were sonicated briefly and incubated with rocking for 30 min at 4°C. Debris was pelleted in a microcentrifuge at 16800 × g at 4°C for 30 min and supernatants were retained. 25 μL of sample (“input”) was removed, and the rest of the sample was mixed by rotation for 30 min at 4°C with 60 μL of a 50% slurry of NeuroAvadin beads (Thermo Fisher Scientific Inc.; Rockford, IL). Beads were washed three times with Lysis Buffer and bound proteins were eluted in 100 μL of 2× SDS sample buffer plus 25 μL 1 M DTT by boiling for 10 min. Immunoblotting for myc-tagged proteins was performed as described above. Visualization and quantification of band intensity was performed using the Odyssey Imaging System (LiCor Biosciences; Lincoln, NE). Integrated intensity values were recorded for all lanes, and normalized to the intensity value of Trek2b.
Data analysis
Data are presented as the mean ± SEM. Statistical analyses were done using Prism v. 11 (GraphPad Software Inc., USA). Whole-cell current amplitudes were analyzed using Kruskal-Wallis or Mann-Whitney tests, as appropriate. Protein levels were analyzed by ANOVA, followed by multiple comparisons with Student-Newman-Keuls test when a significant interaction and/or main effect was found. Differences were considered significant if P<0.05.
Results
A BLAST search was used to identify all mouse cDNAs harboring sequence coding for the first pore domain in Trek2 in the GenBank® database (www.ncbi.nlm.nih.gov/genbank/); Table 1 lists the 8 entries identified using this approach. Five of the 8 entries correspond to a mouse Trek2 variant that shares homology with a human and rat Trek2 isoform termed Trek2a (Gu et al., 2002). Another entry (DQ185134) corresponds to a previously-described mouse Trek2 variant similar to the human and rat Trek2c variant (Gu et al., 2002). In this study, we focused on the 2 remaining Trek2 cDNAs (AK031904 and AK019376), which code for unique Trek2 variants that have not been studied to date.
Table 1. Murine Trek2-related cDNAs identified in the GenBank® database.
| Accession Number | Exon Assembly | Nomenclature | Protein size (AA) |
|---|---|---|---|
| NM_029911 | 1a, 2, 3, 4, 6, 7, 8 | Trek2a | 535 |
| BC137869 | 1a, 2, 3, 4, 6, 7, 8 | Trek2a | 535 |
| BC132487 | 1a, 2, 3, 4, 6, 7, 8 | Trek2a | 535 |
| AK082153 | 1a, 2, 3, 4, 6, 7, 8 | Trek2a | 535 |
| AK036066 | 1a, 2, 3, 4, 6, 7, 8 | Trek2a | 535 |
| AK031904 | 1b, 2, 3, 4, 6, 7, 8 | Trek2b | 520 * |
| DQ185134 | 1c, 2, 3, 4, 6, 7, 8 | Trek2c | 538 |
| AK019376 | 1c, 2, 3, 4, 5 | Trek2c-1p | 241 |
cDNA entries identified from a BLAST search with sequence corresponding to the first pore-domain of Trek2 (taken from NM_029911). Exon assembly, nomenclature and protein size (in number of amino acids, AA) are outlined in the table above.
The AK031904 database entry lacks a thymidine in Exon 2 found in all other Trek2-related cDNAs and the mouse Trek2 gene. The primary amplicon generated by RT-PCR from mouse brain total RNA using Trek2b isoform-specific oligonucleotides contained also this thymidine; as such, the predicted Trek2b protein is corrected to be 520 amino acids.
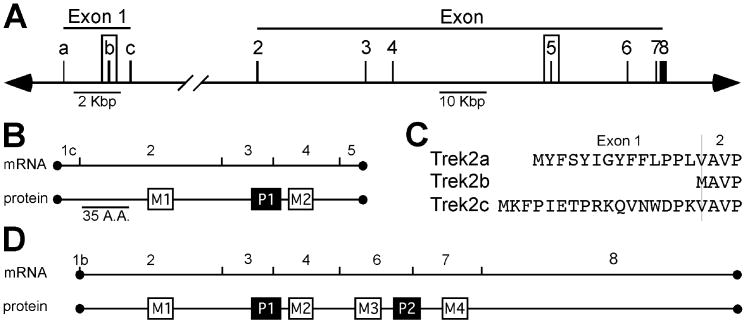
cDNA AK019376 codes for a C-terminal truncation of Trek2. The cDNA sequence was aligned with the mouse Trek2/Kcnk10 gene (MGI:1919508), found on chromosome 12 (99672204-99816150: Mouse Genome Database). Five exons were identified in this splice variant (exons 1c, 2, 3, 4, 5), the first four of which were known previously (Fig. 1A,B). Exon 5 contains a short stretch (60 bases) of coding sequence followed by an in-frame stop codon and 188 bp of 3′ UTR. This variant generates a protein with only two transmembrane segments (M1 and M2) and one pore (P1) domain (Fig. 1B), for a total of 241 amino acids; we termed this a one-pore domain-containing Trek2 (Trek2-1p) variant.
Figure 1. Structure and splicing of the mouseTrek2 gene.
A) Trek2 intron-exon structure and nomenclature. Exons 1b and 5 are previously-unreported exons. Note the different scales on the left and right side of the gene depiction. B) Linear alignments of Trek2-1p mRNA and protein. Numbers above the mRNA refer to the corresponding Trek2 gene exons. M1 and M2 denote the location of the two transmembrane segments, whereas P1 denotes the location of the pore domain. Scale bar = 35 amino acids (A.A.). C) Sequence alignment of the unique N-terminal domains in Trek2a, Trek2b, and Trek2c. The thin vertical line denotes the boundary between corresponding coding sequence in exon 1(a/b/c) and exon 2. D) Linear alignments of Trek2b mRNA and protein. Numbers above the mRNA refer to the corresponding Trek2 gene exons. M1-M4 denote the locations of the four transmembrane segments, and P1-2 denote the location of the 2 pore domains. The scale is the same as that used in panel B.
cDNA AK031904 codes for an N-terminal Trek2 splice variant (Fig. 1A,C,D). Alignment of this cDNA with the mouse Trek2/Kcnk10 gene revealed a previously-unrecognized exon that we termed exon 1b based on its location between exon 1a and 1c. In total, 7 exons contribute to this splice variant (exons 1b, 2, 3, 4, 6, 7, 8), which we termed Trek2b (Fig. 1D). It should be noted that the database sequence for AK031904 (Trek2b) is lacking a thymidine in exon 2 (between bases 181 and 182 of coding sequence) that is found in the mouse Trek2a and Trek2c variants, as well as the Trek2 gene. Absence of this thymidine introduces a frame shift leading to premature termination of Trek2 translation. To determine whether the database sequence was in error, we designed intron-spanning oligonucleotides targeting the unique 5′UTR in exon 1b along with the 3′UTR from exon 8 for use in RT-PCR experiments in whole brain total RNA (Fig. 2). We were able to amplify a product of the predicted size, and DNA sequencing demonstrated that the amplicon did contain the missing thymidine.
The translation initiation codon for the Trek2b isoform spans two exons, with the adenosine found at the 3′ exon-intron boundary of exon 1b, and the thymidine and guanosine found at the 5′ intron-exon boundary of exon 2. As such, Trek2b is slightly shorter (520 amino acids) than the two other N-terminal splice variants Trek2a (535 amino acids) and Trek2c (538 amino acids). Trek2a, Trek2b, and Trek2c share coding sequence contributed by exons 2, 3, 4, 6, 7, and 8, and differ only at their N-termini (Fig. 1C).
We next examined the tissue distribution of Trek2-1p and Trek2b, along with Trek2a and Trek2c, using a multi-tissue mouse cDNA panel and a PCR strategy involving intron-spanning, isoform-specific oligonucleotides (Fig. 2A,B). Trek2-1p expression was detected in the mouse brain, spleen and testis (Fig. 2B), with lower levels observed in heart, lung, and muscle; no expression of Trek2-1p was detected in liver or kidney. Trek2a, Trek2b, and Trek2c were detected in brain, with lower levels observed in heart, spleen, and lung. Little or no expression of Trek2a, Trek2b, or Trek2c was detected in muscle, kidney, liver or testis.
We next sought insight into the regional distribution of the Trek2 splice variants in the CNS. Expression of the four Trek2 splice variants was evaluated in a cDNA panel containing 15 different CNS structures (Fig. 2A,C). While all four variants showed broad distribution in the CNS, Trek2-1p exhibited the most restricted expression pattern of the splice variants tested. Robust signals corresponding to Trek2-1p were observed in brainstem, cerebellum, posterior cortex, cerebrum, hippocampus, medulla, olfactory bulb and pons. Lower intensity signals were observed in entorhinal cortex and striatum, whereas little or no expression was detected in frontal cortex, hypothalamus, midbrain, spinal cord, and thalamus. Trek2a, Trek2b, and Trek2c were broadly expressed in the CNS, with the most robust signals observed in the brainstem and cerebellum. None of the 3 N-terminal Trek2 variants was detected in the thalamus, despite the clear presence of the positive control transcript (GAPDH). While the regional expression patterns for Trek2 splice variants were largely overlapping, some isoform-dependent differences were observed. For example, Trek2b and Trek2c were detected in the cerebrum, while Trek2a expression was not. Similarly, expression of Trek2a and Trek2c, but not Trek2b, was observed in the midbrain.
To probe the impact of alternative splicing on Trek2 channel function, we next measured whole-cell currents in HEK 293 cells expressing individual Trek2 splice variants. Expression constructs for each variant were generated with sequence for the myc epitope tag engineered into the C-terminus. All three N-terminal Trek2 splice variants yielded novel currents exhibiting outward rectification (under conditions of physiological internal and external K+ concentrations) that were distinct from currents measured in mock-transfected control cells (Fig. 3A,D). While considerable cell-to-cell variability in whole-cell current amplitudes was observed, clear group differences were found in cells transfected with Trek2a, Trek2b, and Trek2c. Cells expressing Trek2b exhibited large currents (3-20 nA at +60 mV), whereas currents measured in cells expressing Trek2a and Trek2c were markedly smaller (0.5-5 nA at 60 mV) (Fig. 3A,D).
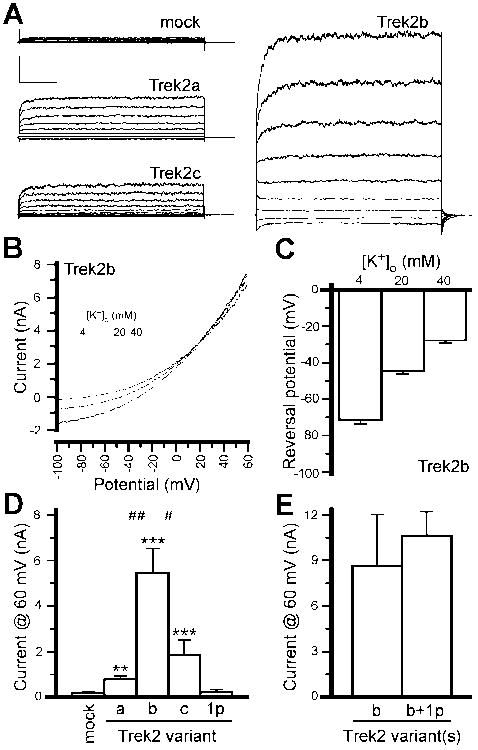
Figure 3. Functional characterization of Trek2 splice variants.
A) Sample traces showing whole-cell currents evoked by a voltage step protocol (-100 to +60 mV, in 20 mV increments; 500 ms step durations) in HEK 293 cells expressing myc-tagged Trek2a, Trek2b, or Trek2c variants. Currents observed in mock-transfected cells were negligible. Scale bars = 1 nA, 100 ms. B) Sample traces showing the effect of external K+ concentration ([K+]o; 4, 20, 40 mM) on the whole-cell current evoked by a ramp protocol (-100 to 60 mV in 1 s) in a cell expressing Trek2b. Note the rightward shift in reversal potential accompanying the increase in [K+]o., and the increased inward current flow at higher [K+]o, consistent with previous analyses of the Trek2 rectification profile. C) Summary plot of current reversal potentials in Trek2b-expressing cells as a function of [K+]o (n=5). D) Summary of outward currents (measured at 60 mV) in cells expressing Trek2a, Trek2b, Trek2C, and Trek2-1p, as well as mock-transfected controls. The results of a Kruskal-Wallis test were significant (H=55.3, 4 df, P<0.0001), indicating that mean current amplitudes were significantly different across groups. Symbols: **,*** P<0.01 and 0.001, respectively, vs. mock-transfected; #,## P<0.05 and 0.01, respectively, vs. Trek2b. E) Summary of outward currents (measured at 60 mV) in cells expressing Trek2b in the absence or presence of Trek2-1p (n=7 per group). The results of a Mann-Whitney test were not significant (U=11.0, P=0.1), indicating that mean current amplitudes were not significantly different between the 2 groups.
Reversal potentials measured for cells expressing Trek2a (-60±6 mV; n=18), Trek2b (-70±2 mV; n=21), and Trek2c (-62±3 mV; n=27) were comparable, and significantly more negative than those measured in mock-transfected control cells (-24±4 mV; n=12, P<0.001). These observations are expected for cells over-expressing K+-selective ion channels. Indeed, when K+ concentrations in the bath were increased from 4 mM to either 20 or 40 mM, the reversal potential of the whole-cell current measured in cells expressing the novel Trek2b variant shifted toward more depolarized membrane potentials, consistent with expectations for a K+-selective ion channel (Fig. 3B,C). Moreover, the slope of the line fitting the plot of whole-cell current reversal potential versus the natural log of [K+]o/[K+]i was 20±1 mV, close to the predicted value of 25.3 mV for a perfectly-selective K+ channel measured at room temperature.
Cells transfected with Trek2-1p showed no novel currents (Fig. 3D), despite the appearance of recombinant protein (not shown). As a truncated C-terminal splice variant of Trek1 has been reported and shown to exert a dominant negative influence on the functional expression of Trek1 (Fink et al., 1998, Veale et al., 2010), we asked whether Trek2-1p acts as a dominant negative influence on Trek2. Cells were co-transfected with 1:1 mixtures of Trek2b and either empty vector or Trek2-1p. We did not detect any difference in current amplitudes between cells expressing Trek2b in the absence or presence of Trek2-1p (Fig. 3E).
Previous comparative functional assessments of recombinant human TREK2A and TREK2C splice variants revealed no isoform-dependent differences in channel selectivity, single channel conductance, or mean open time, arguing that the short unique N-terminal domains do not influence fundamental biophysical properties of Trek2 channels (Gu et al., 2002, Simkin et al., 2008). Indeed, a mutant version of rat Trek2c lacking the first 21 amino acids (i.e., a mutant with a N-terminus virtually identical to the murine Trek2b variant described herein) exhibited functional properties indistinguishable from full-length Trek2c (Simkin et al., 2008). Accordingly, we surmised that the elevated whole-cell currents seen in cells expressing Trek2b reflected isoform-dependent differences in channel expression and/or distribution on the membrane surface. Indeed, levels of total Trek2b protein were significantly greater than levels of either Trek2a or Trek2c in whole-cell extracts from transfected HEK 293 cells (Fig. 4A,C).
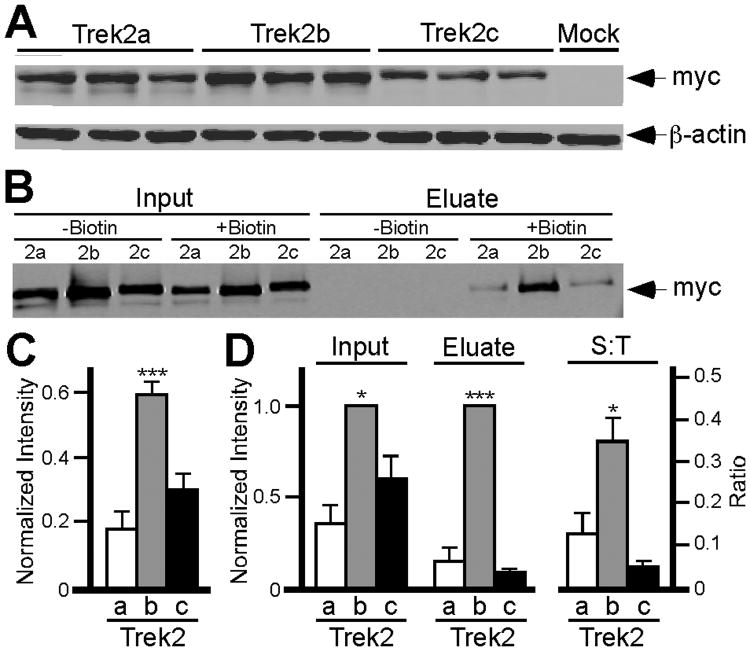
Figure 4. Whole-cell and plasma membrane expression levels of Trek2 splice variants.
A) Representative immunoblot of total Trek2 protein from HEK 293 cells expressing Trek2a, Trek2b, or Trek2c. Three separate transfections per Trek2 variant were performed, and the levels of both the Trek2 variant and β-actin were determined in each sample. B) Representative immunoblot of Trek2 splice variants found in samples of biotinylated surface proteins. C) Summary plot depicting the level of total Trek2 protein expressed in HEK293 cells, normalized to the level of β-actin (N=10). ANOVA revealed a statistically greater amount of Trek2b total protein compared to both Trek2a and Trek2c (n=10, F(2,27)=21.45: P<0.001). D) Summary graph depicting total Trek2 protein (Input), total Trek2 biotinylated protein (Eluate), and surface-to-total protein ratio (S:T). Three independent experiments are summarized. Isoform-dependent differences in the levels of Trek2 in input (F(2,7)=18.7; P<0.05) and eluate (F(2,7)=131.964; P<0.001) samples were observed, as well as in the S-T ratio (F(2,7)=9.5; P<0.05). Symbols: *,*** P<0.05 and 0.001, respectively, vs. Trek2a and Trek2c.
Plasma membrane levels of the N-terminal Trek2 splice variants were determined using a surface biotinylation approach (Materials and Methods). In brief, membrane proteins were tagged with a membrane-impermeable biotin reagent, and biotinylated proteins were then enriched using streptavidin precipitation, followed by quantitative immunoblotting for the myc-tagged Trek2 variants. The level of Trek2b at the plasma membrane of transfected cells was significantly greater than surface levels of either Trek2a or Trek2c (Fig. 4B, D). Moreover, the ratio of surface-to-total Trek2b protein was significantly higher than that measured for either Trek2a or Trek2c (Fig. 4D, right plot).
Discussion
Most previous studies involving rodent Trek2 expression patterns or function have not differentiated between the Trek2a and Trek2c variants, or have involved only a specific (often un-specified) Trek2 isoform (Bang et al., 2000, Lesage et al., 2000, Kim et al., 2001, Kang et al., 2005, Kim et al., 2005, Sandoz et al., 2006, Kang et al., 2007, Kreneisz et al., 2009). In addition, most effort to understand the structural influences on Trek channel function have centered on the intracellular C-terminus, which has been implicated in channel modulation by phosphorylation, temperature, arachidonic acid, and volatile anesthetics, and on protein-protein interactions involving the A kinase anchoring protein (AKAP-150) and microtubule associated protein-2 (mTap2) (Sandoz et al., 2006, Sandoz et al., 2008, Kreneisz et al., 2009, Enyedi and Czirjak, 2010). Accordingly, one goal of the present study was to evaluate possible distinctions in the expression patterns or function of the known N-terminal splice variants of Trek2, Trek2a and Trek2c. In addition, we describe two novel variants of the mouse Trek2 gene, including a third N-terminal splice variant (Trek2b).
While tissue and CNS distributions of the three N-terminal splice variants were comparable, whole-cell currents seen in cells expressing Trek2b were significantly larger that those measured in cells expressing Trek2a or Trek2c. Previous work has ruled out a contribution of the extreme N-terminus to mean open time, single channel conductance, and ion selectivity, as a truncated mutant of rat Trek2c lacking the first 37 N-terminal amino acids behaved identically to the full length protein (Simkin et al. 2008). While the different current amplitudes observed in cells expressing Trek2a, Trek2b, and Trek2c could be explained by differences in the activation properties of these channels, the correlation between current and surface protein levels suggests a simpler explanation. As Trek2b lacks the unique and short N-terminal domains found on Trek2a and Trek2c, we infer that the Trek2 N-terminus regulates channel trafficking and/or stability. Indeed, the ratio of surface-to-total Trek2b was greater than that determined for Trek2a or Trek2c, arguing that Trek2b is either more efficiently targeted to the plasma membrane, or is less efficiently internalized or degraded, or both. Interestingly, the N-terminus has been implicated in the regulation of surface trafficking for Trek1 through an interaction with β-COP (Kim et al., 2010).
N-terminal Trek2 splice variants in humans and rats corresponding to mouse Trek2a (human: NM_021161; rat: EDL81687) and Trek2c (human: NM_138317; rat: NM_023096) have been reported (Gu et al., 2002). Interestingly, sequence homology is relatively low for the unique N-terminal domain in the Trek2a variant across the three species (not shown). As key functional domains are typically conserved across species, this could argue that the unique N-terminus of Trek2a serves no important role in channel function or regulation. Alternatively, given the relative hydrophobicity of this domain, it may serve as a signal or leader sequence that targets Trek2a to a particular cellular sub-domain. In contrast to Trek2a, the unique N-terminal domain in Trek2c is perfectly conserved across mice, rats, and humans, and it possesses some intriguing elements. For example, the lysine residue found at the N-terminus of Trek2c is predicted to be a destabilizing influence on protein stability, as per the N-end rule (Bachmair et al., 1986). Moreover, the Trek2c N-terminal domain consists of a combination of hydrophobic and hydrophilic residues, a signature of proteins that undergo Hsc70-mediated degradation (Majeski and Dice, 2004). The lack of specific targeting sequences and/or trafficking signals in Trek2b may explain the robust expression levels observed for this isoform in transfected cells.
There is currently no homolog of Trek2b reported in either human or rat. Alignment of mouse exon 1b with the human TREK2 gene suggests a possible human homolog. In rat, however, we were unable to detect a possible Trek2b variant. As coding sequence in exon 1b is restricted to a single adenosine at the intron-exon boundary, the only sequence that can be used for alignment purposes is 5′UTR, which may be less conserved across species. Mouse Trek2b was so-named as its first exon is located between the exons that are utilized to generate Trek2a and Trek2c. While a human TREK2B variant has been reported (NP_612191; Gu et al., 2002) it is clearly distinct from the mouse Trek2b variant characterized herein. The human TREK2B variant exhibits a unique 18 amino-acid N-terminal domain and is expressed in pancreas and kidney, but not brain (Gu et al., 2002). Moreover, the first exon of human TREK2B is located downstream of human exon 1c. Alignment of human exon 1b with the mouse Trek2 gene yielded no obvious homology. Thus, species differences exist with respect to Trek2 gene structure and alternative splicing, producing multiple distinct and species-specific Trek2 variants.
The Trek2-1p variant evaluated in this study, which utilizes exon 1c and exon 5, resembles an inwardly-rectifying K+ (KIR) channel in terms of predicted membrane topology, and is expressed in multiple tissues and brain regions. We have also detected expression of Trek2-1p variants utilizing exons 1a and 1b in the mouse CNS (not shown). To date, we have been unable to detect novel currents resulting from heterologous over-expression of Trek2a/b/c-1p. While a C-terminal splice variant of Trek1 exhibiting only a single transmembrane segment was shown recently to act as a dominant negative modulator of Trek1 by limiting its surface delivery (Veale et al., 2010), we observed no impact of Trek2-1p expression on current amplitudes in cells expressing Trek2b. It is possible that Trek2-1p variants require heteroassembly with another channel subunit, or a binding partner, to form functional channels. Alternatively, these variants might be regulated in a distinct manner, such that our recording conditions were not suitable to observe substantial channel activity Finally, these variants may be subject to nonsense-mediated decay, where the splicing event serves to down-regulate Trek2 activity.
In summary, we identified two novel alternative splice variants of the mouse Trek2 gene that exhibit widespread distribution in the mouse CNS. Our findings demonstrate that N-terminal variation can influence current amplitude and surface level of Trek2 channels. Accordingly, differential and dynamic regulation of Trek2 isoform expression may mediate changes in the intrinsic excitability of neurons.
Acknowledgments
The authors would like to thank members of the Wickman laboratory for providing helpful comments on this manuscript. This work was supported by NIH grants RO1 MH061933, P50 DA011806, R21 DA029343 (KW), and T32 DA07234 (KM).
Footnotes
Disclosures: The authors have approved the final article and declare no conflicts of interest, financial or otherwise.
References
- Alloui A, Zimmermann K, Mamet J, Duprat F, Noel J, Chemin J, Guy N, Blondeau N, Voilley N, Rubat-Coudert C, Borsotto M, Romey G, Heurteaux C, Reeh P, Eschalier A, Lazdunski M. TREK-1, a K+ channel involved in polymodal pain perception. Embo J. 2006;25:2368–2376. doi: 10.1038/sj.emboj.7601116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bang H, Kim Y, Kim D. TREK-2, a new member of the mechanosensitive tandem-pore K+ channel family. J Biol Chem. 2000;275:17412–17419. doi: 10.1074/jbc.M000445200. [DOI] [PubMed] [Google Scholar]
- Bockenhauer D, Zilberberg N, Goldstein SA. KCNK2: reversible conversion of a hippocampal potassium leak into a voltage-dependent channel. Nat Neurosci. 2001;4:486–491. doi: 10.1038/87434. [DOI] [PubMed] [Google Scholar]
- Cadaveira-Mosquera A, Ribeiro SJ, Reboreda A, Perez M, Lamas JA. Activation of TREK currents by the neuroprotective agent riluzole in mouse sympathetic neurons. J Neurosci. 2011;31:1375–1385. doi: 10.1523/JNEUROSCI.2791-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P, Xiao Z, Yang C, Rojanathammanee L, Grisanti L, Watt J, Geiger J, Liu R, Porter J, Lei S. GABA(B) receptor activation inhibits neuronal excitability and spatial learning in the entorhinal cortex by activating TREK-2 K+ channels. Neuron. 2009;63:230–243. doi: 10.1016/j.neuron.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon DG, Bogdan R, Fagerness J, Holmes AJ, Perlis RH, Pizzagalli DA. Variation in TREK1 gene linked to depression-resistant phenotype is associated with potentiated neural responses to rewards in humans. Hum Brain Mapp. 2010;31:210–221. doi: 10.1002/hbm.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi P, Czirjak G. Molecular background of leak K+ currents: two-pore domain potassium channels. Physiol Rev. 2010;90:559–605. doi: 10.1152/physrev.00029.2009. [DOI] [PubMed] [Google Scholar]
- Fink M, Duprat F, Lesage F, Reyes R, Romey G, Heurteaux C, Lazdunski M. Cloning, functional expression and brain localization of a novel unconventional outward rectifier K+ channel. Embo J. 1996;15:6854–6862. [PMC free article] [PubMed] [Google Scholar]
- Fink M, Lesage F, Duprat F, Heurteaux C, Reyes R, Fosset M, Lazdunski M. A neuronal two P domain K+ channel stimulated by arachidonic acid and polyunsaturated fatty acids. Embo J. 1998;17:3297–3308. doi: 10.1093/emboj/17.12.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Schlichthorl G, Hirsch JR, Engels H, Karschin C, Karschin A, Derst C, Steinlein OK, Daut J. Expression pattern and functional characteristics of two novel splice variants of the two-pore-domain potassium channel TREK-2. J Physiol. 2002;539:657–668. doi: 10.1113/jphysiol.2001.013432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurteaux C, Guy N, Laigle C, Blondeau N, Duprat F, Mazzuca M, Lang-Lazdunski L, Widmann C, Zanzouri M, Romey G, Lazdunski M. TREK-1, a K+ channel involved in neuroprotection and general anesthesia. Embo J. 2004;23:2684–2695. doi: 10.1038/sj.emboj.7600234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurteaux C, Lucas G, Guy N, El Yacoubi M, Thummler S, Peng XD, Noble F, Blondeau N, Widmann C, Borsotto M, Gobbi G, Vaugeois JM, Debonnel G, Lazdunski M. Deletion of the background potassium channel TREK-1 results in a depression-resistant phenotype. Nat Neurosci. 2006;9:1134–1141. doi: 10.1038/nn1749. [DOI] [PubMed] [Google Scholar]
- Honore E. The neuronal background K2P channels: focus on TREK1. Nat Rev Neurosci. 2007;8:251–261. doi: 10.1038/nrn2117. [DOI] [PubMed] [Google Scholar]
- Kang D, Choe C, Cavanaugh E, Kim D. Properties of single two-pore domain TREK-2 channels expressed in mammalian cells. J Physiol. 2007;583:57–69. doi: 10.1113/jphysiol.2007.136150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Choe C, Kim D. Thermosensitivity of the two-pore domain K+ channels TREK-2 and TRAAK. J Physiol. 2005;564:103–116. doi: 10.1113/jphysiol.2004.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Hwang EM, Yarishkin O, Yoo JC, Kim D, Cho M, Lee YS, Choong-Hyun S, Yi GS, Yoo J, Kang D, Han J, Hong SG, Park JY. Enhancement of TREK1 channel surface expression by protein-protein interactions with Beta-COP. Biochemical and Biophysical Research Communications. 2010;395:244–250. doi: 10.1016/j.bbrc.2010.03.171. [DOI] [PubMed] [Google Scholar]
- Kim JS, Park JY, Kang HW, Lee EJ, Bang H, Lee JH. Zinc Activates TREK-2 Potassium Channel Activity. JPET. 2005;314:618–625. doi: 10.1124/jpet.105.084418. [DOI] [PubMed] [Google Scholar]
- Kim Y, Gnatenco C, Bang H, Kim D. Localization of TREK-2 K+ channel domains that regulate channel kinetics and sensitivity to pressure, fatty acids and pHi. Pflugers Arch. 2001;442:952–960. doi: 10.1007/s004240100626. [DOI] [PubMed] [Google Scholar]
- Kreneisz O, Benoit JP, Bayliss DA, Mulkey DK. AMP-activated protein kinase inhibits TREK channels. J Physiol. 2009;587(24):5819–5830. doi: 10.1113/jphysiol.2009.180372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage F, Terrenoire C, Romey G, Lazdunski M. Human TREK2, a 2P domain mechano-sensitive K+ channel with multiple regulations by polyunsaturated fatty acids, lysophospholipids, and Gs, Gi, and Gq protein-coupled receptors. J Biol Chem. 2000;275:28398–28405. doi: 10.1074/jbc.M002822200. [DOI] [PubMed] [Google Scholar]
- Maingret F, Lauritzen I, Patel AJ, Heurteaux C, Reyes R, Lesage F, Lazdunski M, Honore E. TREK-1 is a heat-activated background K+ channel. Embo J. 2000;19:2483–2491. doi: 10.1093/emboj/19.11.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maingret F, Patel AJ, Lesage F, Lazdunski M, Honore E. Mechano- or acid stimulation, two interactive modes of activation of the TREK-1 potassium channel. J Biol Chem. 1999;274:26691–26696. doi: 10.1074/jbc.274.38.26691. [DOI] [PubMed] [Google Scholar]
- Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. International Journal of Biochemistry and Cell Biology. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Murbartian J, Lei Q, Sando JJ, Bayliss DA. Sequential phosphorylation mediates receptor- and kinase-induced inhibition of TREK-1 background potassium channels. J Biol Chem. 2005;280:30175–30184. doi: 10.1074/jbc.M503862200. [DOI] [PubMed] [Google Scholar]
- Patel AJ, Honore E. Properties and modulation of mammalian 2P domain K+ channels. Trends Neurosci. 2001;24:339–346. doi: 10.1016/s0166-2236(00)01810-5. [DOI] [PubMed] [Google Scholar]
- Patel AJ, Honore E, Maingret F, Lesage F, Fink M, Duprat F, Lazdunski M. A mammalian two pore domain mechano-gated S-like K+ channel. Embo J. 1998;17:4283–4290. doi: 10.1093/emboj/17.15.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz G, Tardy MP, Thummler S, Feliciangeli S, Lazdunski M, Lesage F. Mtap2 is a constituent of the protein network that regulates twik-related K+ channel expression and trafficking. J Neurosci. 2008;28:8545–8552. doi: 10.1523/JNEUROSCI.1962-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz G, Thummler S, Duprat F, Feliciangeli S, Vinh J, Escoubas P, Guy N, Lazdunski M, Lesage F. AKAP150, a switch to convert mechano-, pH- and arachidonic acid-sensitive TREK K+ channels into open leak channels. Embo J. 2006;25:5864–5872. doi: 10.1038/sj.emboj.7601437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin D, Cavanaugh EJ, Kim D. Control of the singe channel conductance of K2P10.1 (TREK-2) by the amino-terminus: role of alternative translation initiation. Journal of Physiology. 2008;586:5651–5663. doi: 10.1113/jphysiol.2008.161927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Plant LD, Wilkens CM, McCrossan ZA, Goldstein SA. Alternative translation initiation in rat brain yields K2P2.1 potassium channels permeable to sodium. Neuron. 2008;58:859–870. doi: 10.1016/j.neuron.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veale EL, Rees KA, Mathie A, Trapp S. Dominant negative effects of a non-conducting TREK1 splice variant expressed in brain. J Biol Chem. 2010;285:29295–29304. doi: 10.1074/jbc.M110.108423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Deng PY, Rojanathammanee L, Yang C, Grisanti L, Permpoonputtana K, Weinshenker D, Doze VA, Porter JE, Lei S. Noradrenergic depression of neuronal excitability in the entorhinal cortex via activation of TREK-2 K+ channels. J Biol Chem. 2009;284:10980–10991. doi: 10.1074/jbc.M806760200. [DOI] [PMC free article] [PubMed] [Google Scholar]