Abstract
In the last two decades, new techniques that monitor ionic current modulations as single molecules pass through a nanoscale pore have enabled numerous single-molecule studies. While biological nanopores have recently shown the ability to resolve single nucleotides within individual DNA molecules, similar developments with solid-state nanopores have lagged, due to challenges both in fabricating stable nanopores of similar dimensions as biological nanopores and in achieving sufficiently low-noise and high-bandwidth recordings. Here we show that small silicon nitride nanopores (0.8 to 2-nm-diameter in 5 to 8-nm-thick membranes) can resolve differences between ionic current signals produced by short (30 base) ssDNA homopolymers (poly(dA), poly(dC), poly(dT)), when combined with measurement electronics that allow a signal-to-noise ratio of better than 10 to be achieved at 1 MHz bandwidth. While identifying intramolecular DNA sequences with silicon nitride nanopores will require further improvements in nanopore sensitivity and noise levels, homopolymer differentiation represents an important milestone in the development of solid-state nanopores.
Keywords: Nanopores, silicon nitride, homopolymers, DNA, transmission electron microscopy, sequencing, translocation, single-stranded DNA, polynucleotide
Nanopores are a unique and powerful tool for a variety of single-molecule studies,1–13 including detection of miRNAs,14 discrimination between classes of nucleic acids,14,15 detection of DNA binding,16 measurement of molecular forces,17,18 and detection of anthrax toxin.19 One of the highest-profile potential applications of nanopores is in the field of DNA sequencing.13 Nanopores hold the potential to sequence DNA faster and cheaper than current industrial standards.1,5,10 Although biological nanopore experiments have resolved many details of DNA and RNA structure,10,13,20 the remaining challenges for both biological and solid-state nanopore-based sequencing are in spatial and temporal resolution. To identify random sequences, a nanopore must have a geometry that is sensitive to only a few bases at a time,21,22 and it also must have sufficient temporal resolution to resolve these bases as they passes through the nanopore. Spatial concerns are generally addressed by reducing the length of the active detecting region of the nanopore,14,23–26 while temporal requirements can be satisfied either by slowing down the DNA27–36 or by speeding up signal acquisition.37 By combining ion channel proteins which have detecting regions less than 1.5 nm long with polymerase-based positional control, biological nanopores have recently demonstrated proof-of-principle DNA sequencing.22,38 Proof-of-principle nanopore-based DNA sequencing-by-synthesis,39 based on single molecule mass spectroscopy,40,41 has also been demonstrated recently. Interest remains high to translate these successes to solid-state nanopores that offer the potential for easier manufacturability. The higher signal levels of solid-state nanopores may also make sequencing possible without enzymatic techniques to slow down translocation. This requires scaling the nanopores to sizes comparable to biological nanopores, while significantly improving detection electronics. For solid-state nanopores, previous work with sub-2-nm-diameter nanopores has been largely limited to SiNx membranes with thicknesses ≥ 10 nm.42–45 Only a few biomolecule translocation experiments have been reported on SiNx membranes with thicknesses ≤ 10 nm,14,46,47 but no solid-state nanopores have been reported that combine ultra-thin membranes with nanopore diameters smaller than 2 nm.
In this paper, we measure ionic current signals through 0.8- to 2-nm-diameter nanopores in 5- to 8-nm-thick silicon nitride (SiNx) membranes. ssDNA translocations through nanopores of these dimensions transiently reduce the ionic conductance by up to 70 – 90%, similar to results from biological nanopores.2 The reduced thickness of these nanopores leads to higher ionic conductances, increased bias current,14 and a reduction in the number of DNA bases present in the nanopore constriction to approximately 15 bases for a 5-nm-thick membrane (see Figure 1b). By combining these nanopores with thermoelectric temperature regulation14,48 and a low-noise amplifier that supports signal bandwidths as high as 1 MHz, we demonstrate the ability to differentiate between homopolymer sequences of ssDNA as short as 30 bases with solid-state nanopores.
FIGURE 1.
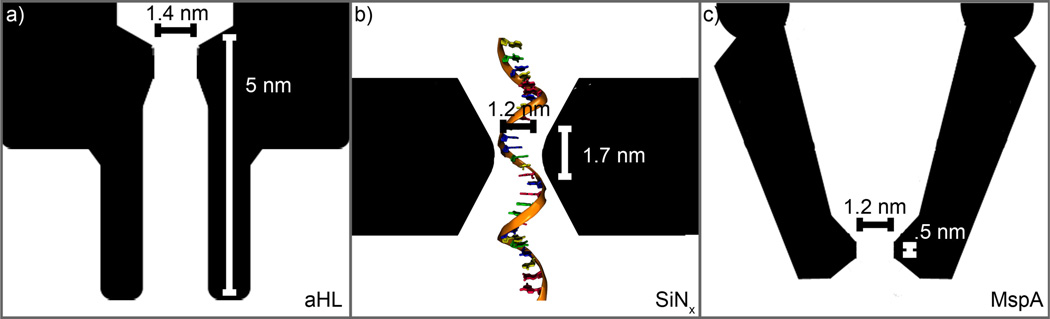
Comparison of the SiNx nanopores presented here to biological nanopores. All images are on the same scale. (a) Biological nanopore α-hemolysin (αHL), which has a thickness of 5 nm and a diameter of 1.4 nm. (b) Representative dimensions of a SiNx nanopore presented here. The nanopore shown is 1.2 nm in diameter and has an effective thickness (heff) of 1.7 nm in a 5-nm-thick membrane. The single-stranded DNA is shown to scale. (c) Biological nanopore MspA, which has a thickness of 0.5 nm and a diameter of 1.2 nm.
Since biological nanopores have been used to differentiate individual DNA bases within a specific DNA sequence or as part of homopolymers,20,22,29,48,49 it is reasonable to expect that SiNx nanopores of similar dimensions may produce comparable results. Figures 1a–c show an illustrated cross-section of a 1.2-nm-diameter nanopore in a 5-nm-thick SiNx membrane (Figure 1b), alongside cross-sections of α-hemolysin (Figure 1a) and MspA (Figure 1c) proteins.50,51 All three are comparable in size, with small differences in diameter and thickness, as detailed in Table 1. The diameters of the solid-state nanopores presented here are comparable to both α-hemolysin (1.4 nm) and MspA (1.2 nm), while the thickness is comparable to α-hemolysin (5 nm) but thicker than MspA (0.5 nm).
TABLE 1.
Comparison of physical properties and experimental results between the SiNx nanopores presented here and published results for the two of the most commonly used biological nanopores: α-hemolysin and MspA. We estimate 10% error in SiNx nanopore diameters from TEM images due to measurement error. For nanopores that were not imaged to avoid damaging them, we estimate our error in nanopore diameter to be larger, ± 0.5 nm, due to the TEM user’s reading error. Error in nanopore diameter may also come from a cleaning step in piranha solution that may slightly change the nanopore size prior to ionic measurements. For the smallest SiNx nanopores, we find that the open pore ionic current is the best measure of their effective size. Because of similar sizes but larger operating voltages in SiNx nanopores, the signal amplitude is about ten times larger and the signal conductance is up to six times larger than in biological nanopores. All comparisons are made for 1M KCl.
| α-hemolysin (α HL) | MspA | Silicon nitride (SiNx) | |
|---|---|---|---|
| Constriction width (nm) | 1.451 | 1.250 | 1–2 (± 10% or 0.5 nm) |
| Constriction height (nm) | 551 | 0.650 | 5–8 (± 3) |
| Conductance (nS) | 12,13 | 1.826 | 3–14 |
| Signal amplitude <ΔI> (nA) | 0.1–0.1052 | 0.15–0.2622,63 | 1–5 |
| Typical operating voltage (mV) | 120 | 180 | up to 1000 |
| Signal conductance <ΔI>/V (nS) | 0.83–0.88 | 0.83–1.4 | 1–5 |
| Percent of pore blocked (%) | 83–952 | 48–8222,26,63 | 30–80 |
| Difference in signal between nucleotides (pA) | 5–152 | 6–1163 | 200–900 |
The geometry of nanopores in solid-state membranes drilled using a transmission electron microscope (TEM) is governed by the interplay between surface tension of the molten SiNx and its ablation kinetics.52 This geometry can be modified by tuning the electron-beam fabrication process.52–54 Based on TEM imaging, ion conductance measurements,53 and annular-dark field scanning TEM (ADF-STEM) studies,14 SiNx nanopore shapes are known to deviate from a perfect cylinder. Electron tomography shows that 7-nm-diameter SiNx nanopores in 50-nm-thick membranes have a truncated double-cone or “hourglass” structure.53 Nevertheless, a simplified geometric model using an equivalent cylinder of reduced effective thickness (heff) is sufficient to quantitatively explain the open and blocked current values measured during DNA translocations.14,53 By fitting both the ionic open-pore and blocked-pore current data for many different-diameter nanopores with the same membrane thickness, heff is estimated to be one-third of the actual membrane thickness (h).14 This implies that TEM drilled nanopores in 5-nm-thick SiNx membranes have heff ~ 1.7 nm. To make a SiNx effective constriction as thin as the constriction in MspA, a nanopore would need to be drilled through a 1.5-nm-thick SiNx membrane , giving heff ~ 0.5 nm, which roughly spans four DNA bases.22
RESULTS AND DISCUSSION
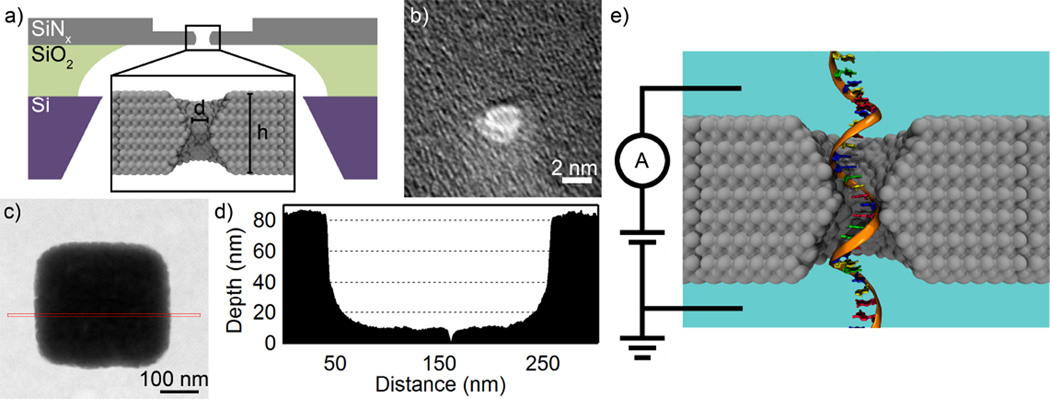
Figure 2a shows the details of the solid-state nanopore design employed in these studies. Our fabrication begins with 10- to 20-µm-square suspended windows of 85-nm-thick low-stress SiNx. In order to reduce the nanopore resistance and increase the nanopore sensitivity, we previously developed a method to obtain sub-10-nm-thick SiNx membranes14 (see Supporting Information for details). A small region (~ 200 × 200 nm2) of a freestanding SiNx membrane is thinned from 85 nm down to the desired thickness. The mass contrast image of one thinned square is shown in Figure 2c; the average line intensity profile (in red) gives the mass-thickness contrast shown in Figure 2d.
FIGURE 2.
Device fabrication, characterization, and measurement integration. (a) Schematic of the stacked silicon chips used to fabricate nanopores. A silicon chip 500 µm thick is covered with 5 µm of silicon dioxide. 85 nm of SiNx is then deposited. The chips are etched to create a suspended SiNx window. The window is locally thinned, and a nanopore is drilled in it. The nanopore’s height and diameter are defined as shown in the inset. (b) TEM image of a nanopore drilled in a 5-nm-thick membrane. The nanopore’s diameter is measured from the image to be 1.4 nm. (c) STEM mass contrast image of the thinned region of a membrane with a nanopore in it. The red line is integrated over to give a profile of the thinned region (d). The mass contrast data is scaled by the known thickness of the original membrane to give the thickness of the thinned region. (e) Experimental design. The membrane is positioned to separate two chambers of 1 M KCl solution. A bias is applied between the chambers, up to 1V, and the ionic conductance is monitored. As DNA passes through the nanopore, it blocks a considerable fraction of the nanopore volume.
Nanopores are drilled in these thinned membranes on a JEOL 2010F TEM52 set at an accelerating voltage of 200 kV. Figure 2b shows an image of a typical nanopore drilled in TEM mode in a 5-nm-thick membrane (see the Supporting Information for additional discussion and Figure S2 for additional nanopore images). We estimate a 10% error in determining the nanopore diameter from these images; this error takes into account the fact that the shape of the nanopores is more precisely described as an ellipse, rather than a circle (the reported diameter is the average of the major and minor diameters). Most nanopores presented here are not imaged in order to avoid altering the nanopore size with further electron beam exposure.53 Instead, we rely on guides on the TEM phosphor screen and calculations from the open pore conductance53 to determine the nanopore diameter. The ionic conductance has the advantage of incorporating any size changes that may occur during cleaning procedures prior to measurement. We estimate the error in the calculated diameters for the nanopores that were not imaged to be ± 0.5 nm. Figure 2e shows a schematic of the experimental setup for these studies.
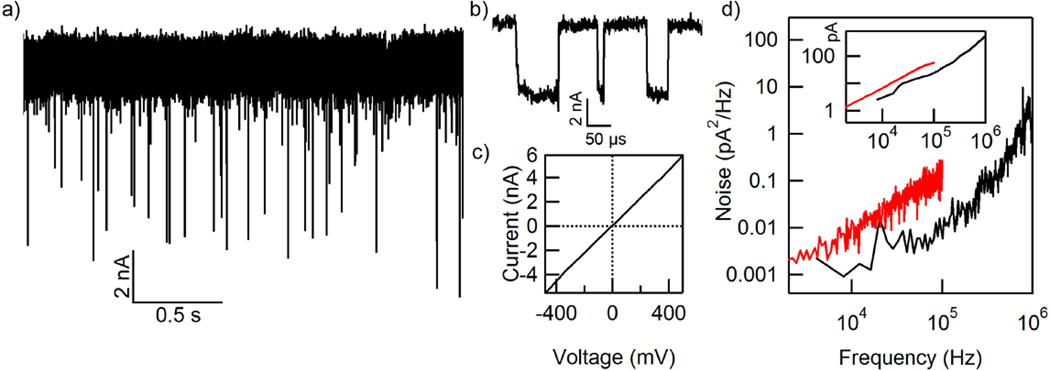
In our experiments, we have translocated short ssDNA molecules from 30 to 180 bases long. Figures 3a–b show the resulting ionic currents through the nanopore for poly(dA)50 molecules translocating through a 2-nm-diameter nanopore in a 5-nm-thick membrane at an applied bias of 1 V with data filtered to a 1 MHz bandwidth. Observed events can be coarsely classified into two groups. We observe shallow events with blocked currents close to the open pore current, which we attribute to DNA deflection events, which have been observed under similar measurement conditions.37 The remaining deeper events correspond to DNA translocations (Figure 3b).37
FIGURE 3.
Characterization of noise in the SiNx nanopores and the experimental setup using two amplifiers. (a) Raw current vs. time trace for 50-mer ssDNA translocating through a 2-nm-diameter nanopore in a 5-nm-thick membrane with an applied bias of 1 V. The trace is digitally low-pass-filtered to B = 1 MHz. (b) Zoomed in events from the trace in (a). Events have minimal attenuation and high signal-to-noise ratios. (c) Current vs. voltage trace for a 2-nm-diameter nanopore in an 8-nm-thick membrane. The slope of this curve yields a conductance of 9.9 nS. (d) Input-referred noise power spectral density for a nanopore measured with an Axopatch 200B (red trace) and a nanopore measured with a Chimera VC100 (black trace). See Figure S4 in the Supporting Information for additional noise spectra. Inset: calculated root-mean-square current noise for both amplifiers, as a function of signal bandwidth.
The open pore current in our measurements typically drifts by ~ 3%, as seen in Figure 3a. Open pore ionic conductances (Figure 3c) range from 3 nS to 14 nS for nanopores with diameters between 0.8 and 2 nm and membrane thicknesses between 5 and 8 nm, matching theoretical predictions.14,53,55 We did not observe translocation events in pores smaller than 0.9 nm (see Supporting Information).
To support a 1 MHz signal bandwidth, experiments are performed with a VC100 low-noise voltageclamp amplifier (Chimera Instruments, New York, NY). Direct comparison of the noise power spectral densities of measurements with these electronics and the more conventional Axopatch 200B are possible below 100 kHz, as shown in Figure 3d, although there is variance between experiments (see Figure S4 in the Supporting Information). Our traces typically exhibit input-referred noise of 520 pArms at the full bandwidth of 1 MHz, and accordingly less noise at lower bandwidths (see Figure 3d inset). When data are filtered to 100 kHz, a bandwidth closer to many published nanopore recordings,3,10,11 the noise is 24 pArms. The primary source of noise at these frequencies is the interaction between the amplifier’s voltage noise and the capacitance of the solid-state membrane chip, which we estimate to be 50 pF.
For comparisons of ionic currents from different homopolymers, we use short, 30-base ssDNA composed of homopolymers of either adenine (poly(dA)30), thymine (poly(dT)30), or cytosine (poly(dC)30) (Integrated DNA Technologies). Guanine is not included in these experiments due to G-tetrad formation in homopolymers longer than four bases. Longer ssDNA has previously been measured in solid-state nanopores, but observed differences in ionic currents have been attributed to secondary structure in the molecules (i.e., base stacking).8 While ssDNA has a short persistence length (~ 0.3 nm)56 and will coil at shorter lengths than dsDNA, the small diameters of the nanopores prevent the passage of folded ssDNA. In addition, the force on the DNA corresponding to the applied bias (1000 mV) is sufficient to overcome both the entropic and enthalpic barriers from any secondary structure in the homopolymers.57 1 µL of 100 µM solution of one homopolymer is added to the chamber at the lower potential to yield a final concentration of 2 µM. After adding the nucleotides, transient current reductions appear in the ionic-current trace. Between each homopolymer experiment, the nanopore is rinsed thoroughly with deionized water, and a baseline current trace is recorded for five minutes to ensure that no blockades are seen before the next homopolymer is measured.
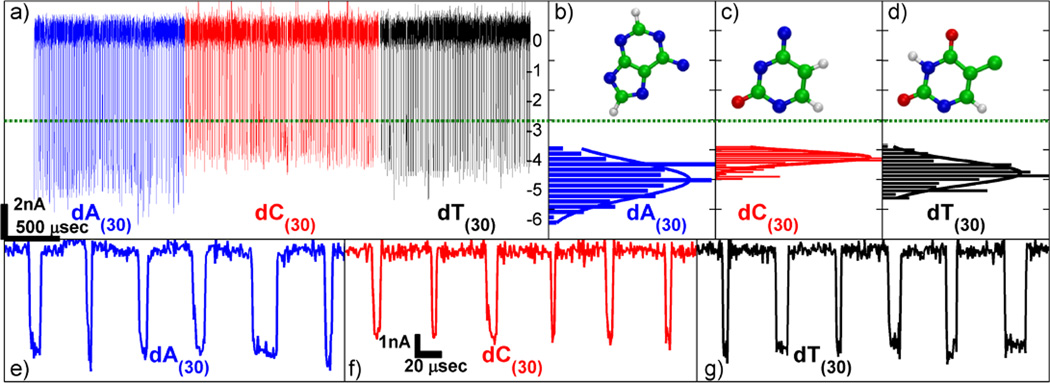
Figure 4a shows concatenated current blockades for each homopolymer, poly(dA)30, poly(dC)30, and poly(dT)30, from a 1.4-nm-diameter nanopore in a 5-nm-thick membrane taken at a 1 V applied bias at a signal bandwidth of 500 kHz (at which a signal-to-noise ratio of better than 10 is achieved). The events are identified using a threshold of 3 nA below the open pore current (green line in Figure 4), which excludes shallower collision events. Figure 4a shows many short, ssDNA translocation events densely packed in time, such that each event in this figure appears as a narrow spike (a magnified view of these events for each homopolymer is shown in Figures 4e–g). The current histograms determined from this data for each homopolymer are shown in Figures 4b–d. Using more than 700 events for each homopolymer, poly(dA)30 gives a mean event depth of <ΔIA> = 5.1 ± 0.4 nA, poly(dC)30 gives a mean event depth of <ΔIC> = 4.2 ± 0.1 nA, and poly(dT)30 gives a mean event depth of <ΔIT> = 4.8 ± 0.2 nA. The mean and error (standard deviation) values for event depth are calculated from Gaussian fits to the histograms. A Welch’s t-test is performed for the difference between the mean depths for each pair of homopolymers, and the p-value is found to be less than 0.0001 in all three cases, indicating that while the distributions overlap, the difference between the means has strong statistical significance.
FIGURE 4.
Results for poly(dA)30, poly(dC)30, and poly(dT)30. (a) Concatenated events from each homopolymer. The green line is the threshold for defining events. (b) Normalized histogram of event depths for poly(dA)30. The mean value is 5.1 ± 0.4 nA. (c) Normalized histogram of event depths for poly(dC)30. The mean value is 4.2 ± 0.1 nA. (d) Normalized histogram of event depths for poly(dT)30. The mean value is 4.8 ± 0.2 nA. Mean values and errors are calculated from Gaussian fits to the histograms. Insets in (b)–(d) are diagrams of the base corresponding to the histogram. (e)–(g) Sample events from the data set shown in (a). The left trace (blue) shows events from poly(dA)30, the middle trace (red) shows events from poly(dC)30, and the right trace (black) shows events from poly(dT)30. This data is low-pass filtered to a bandwidth of 500 kHz.
We find the ratios of mean event depths between homopolymers (e.g., <ΔIA> / <ΔIC>) to be constant across the range of nanopore dimensions considered. In particular, in three experiments on different nanopores with nanopore diameters between 1 and 2 nm and membrane thicknesses between 5 and 8 nm, the ratio of mean event depths of adenine to cytosine is <ΔIA> / <ΔIC= = 1.25 ± 0.05, and the ratio of mean event depths of thymine to cytosine is <ΔIT> / <ΔIC> = 1.16 ± 0.02. The current blocked by a homopolymer in the nanopore is given by <ΔI> ~ S/h, where S is the cross-sectional area of the homopolymer, and h is the thickness of the nanopore. Therefore, the ratio of mean currents for two homopolymers, e.g., poly(dA)30 and poly(dC)30, is then equal to the ratio of homopolymer cross-sectional areas, SA and SC, and independent of nanopore thickness, <ΔIA> / <ΔIC> = SA/SC. In contrast, other measurements, such as the mean current difference (e.g., <ΔIA> − <ΔIC> ~ (SA−SC)/h), vary for different nanopores. It is promising that homopolymer differentiation was achieved using a range of nanopore diameters and membrane thicknesses, as this suggests that some geometric variability in solid-state nanopores may be tolerable in future nanopore DNA sequencing systems.
Since all three homopolymers have the same length (~ 10 nm), we would expect them to produce similar blockade durations unless they have different interactions with the nanopore surfaces. Event durations for data in Figure 4 range from 4 µs to 200 µs, with characteristic durations of 18 µs for poly(dA)30, 33 µs for poly(dC)30, and 22 µs for poly(dT)30. Histograms of event durations for these homopolymers exhibit an exponential form, exp(−t/τ) (see Figure S5 in the Supporting Information),58,59 which has also been reported for thin SiNx nanopores14,37 and graphene nanopores.23 These timescales correspond to a typical DNA velocity between 0.3 – 0.6 µs per base. The event durations measured are also of the same order of magnitude as previous experimental results of 20 µs for dsDNA of similar length (25 base pairs).14 Event duration was not used as a basis for homopolymer differentiation.
These results qualitatively agree with a model of blockade levels that depends predominantly on the physical size of each base. Adenine, the largest base, blocks the most ionic current, while the two smaller bases (cytosine and thymine) block the least ionic current. The inset of Figures 4b–d shows the atomic structure of adenine, cytosine, and thymine. Additionally, as observed in Figures 4b–d, the width of the current distribution for poly(dA)30 is larger than the width of the current distributions for poly(dC)30 and poly(dT)30. This may be due to the effects of base orientation; the larger adenine base can have more conformations, possibly leading to a wider spread of current blockades compared to cytosine and thymine. For a range of thin nanopores with 0.9 to 2-nm-diameters, similar blocked currents, 30 – 80% of the open pore current, are observed for various homopolymers (see Figure S6 in the Supporting Information), and current differences between pairs of homopolymers are similar to those shown in Figures 4b–d. Further studies should attempt to quantitatively explain the observed differences in the magnitudes of blocked currents and optimize parameters to narrow these distributions. This could be achieved by lowering the system noise and performing systematic studies of the effects of DNA length, nanopore dimensions, applied voltage, and salt concentration. We note that standard deviations in the blockade histograms (Figures 4b–d) are comparable to the baseline current noise amplitudes, which suggests that further reduction of measurement noise may reduce the overlap of current distributions between homopolymers.
CONCLUSION
In this work, we fabricated solid-state nanopores having diameters of 1 to 2 nm in membranes as thin as approximately 5 nm, comparable to the dimensions of commonly-used biological nanopores. We measured the ionic current with reduced electronic noise and improved temporal resolution, which allowed us to demonstrate proof-of-principle differentiation of ssDNA homopolymers. The mean homopolymer current signals differed by 200 – 900 pA in 1 M KCl solution at applied voltages between 600 mV and 1 V. We anticipate that future studies will further address the relationship between individual nucleotides’ properties and observed solid-state nanopore current signals, as well as improve experimental parameters to achieve differentiation of individual bases within a single DNA molecule.
METHODS
The SiNx is supported by a 5 × 5 mm2 Si chip. Sandwiched between the Si and the SiNx is a 5-µm-thick, thermally-grown SiO2 layer, which reduces the membrane capacitance and electrical noise.14,60,61 The nanopore device is cleaned using hot piranha solution followed by repeated water rinsing. The nanopore is then assembled in a fluoropolymer cell using a homemade, quick-cure silicone gasket that divides the cell into two chambers containing a salt solution composed of 1 M KCl + 1 mM EDTA buffered to pH 8 using 10 mM Tris-HCl. Our fluoropolymer cell accommodates volumes of 10–50 µL and features temperature regulation using a thermoelectric device connected to a copper block that houses the cell. Data are obtained with the copper block cooled to 2°C. Bias potentials between 600 mV and 1V are applied across the nanopore through Ag/AgCl electrodes, and ionic current is monitored as a function of time. These high voltage biases are used to increase the signal and the resulting signal-to-noise ratio.
Experiments are carried out using the VC100 high-bandwidth, low-noise voltage-clamp amplifier (Chimera Instruments, New York, NY) to apply a voltage bias and measure the current through the nanopore. This amplifier extends a traditional patch-clamp circuit topology62 to support higher signal bandwidths. While its noise floor is not as low as a fully-integrated design37, the new instrument is largely interchangeable with an Axopatch 200B (Molecular Devices), and it co-exists easily with the temperature-controlled fluid cell. The amplifier includes a fourth-order Bessel low-pass filter at 1 MHz, and signals are digitized at 4 – 6 MS/s. Acquired data are digitally low-pass filtered to the desired signal bandwidth before analysis in Matlab (MathWorks, Natick, MA). Not all of the datasets required the full amplifier bandwidth, but higher sample rates ensure many data points per translocation event and provide flexibility during data analysis. We have previously shown that the limited bandwidth of popular patch-clamp amplifiers can lead to attenuation by as much as ~ 20% of solid-state nanopore blockades briefer than 16 µs.14 In Figure 4, we consider events that are unimodal, i.e., events with no obvious intra-event structure37 and whose blockade currents have a standard deviation similar to that of the open pore current (300 pArms at a 500 kHz bandwidth).
Supplementary Material
Acknowledgments
The authors thank Dr. Matthew Johnston for instrumentation assistance, Dr. Robert Johnson for graphics, and Dr. Aleksei Aksimentiev and Dr. Meni Wanunu for useful comments. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship (K.V.) under Grant No. DGE-0822. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. M.P. acknowledges funding from the NSF-IGERT program (Grant DGE-0221664). This work was supported in part by the MRSEC NSF grants DMR-0520020 and DMR-1120901, NIH Grants R21HG004767 and R01HG006879, and by the Nano/Bio Interface Center through the National Science Foundation NSEC DMR08-32802. We gratefully acknowledge use of the TEM in the NSF-MRSEC electron microscopy facility.
Footnotes
Supporting Information Available. Details of membrane thinning and characterization, details of nanopore fabrication, discussion and ionic-current trace for a nanopore too small to allow translocations, more TEM and STEM images of nanopores, additional power spectral density graphs for the Axopatch 200B and Chimera VC100 amplifier, event duration histograms, the percent of the open pore current blocked by ssDNA translocations through additional sub-2-nm-diameter nanopores, and additional experimental data. This material is available free of charge via the Internet at http://pubs.acs.org.
Competing financial interests. KS and JKR are principals in Chimera Instruments. The VC100 from Chimera Instruments was employed in these studies.
REFERENCES
- 1.Deamer DW, Akeson M. Nanopores and Nucleic Acids: Prospects for Ultrarapid Sequencing. Trends Biotechnol. 2000;18:147–151. doi: 10.1016/s0167-7799(00)01426-8. [DOI] [PubMed] [Google Scholar]
- 2.Deamer DW, Branton D. Characterization of Nucleic Acids by Nanopore Analysis. Acc Chem Res. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- 3.Healy K, Schiedt B, Morrison AP. Solid-State Nanopore Technologies for Nanopore-Based DNA Analysis. Nanomedicine (London, U. K) 2007;2:875–897. doi: 10.2217/17435889.2.6.875. [DOI] [PubMed] [Google Scholar]
- 4.Dekker C. Solid-State Nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 5.Branton D, Deamer DW, Marziali A, Bayley H, Benner S, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, et al. The Potential and Challenges of Nanopore Sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasianowicz JJ, Robertson JWF, Chan ER, Reiner JE, Stanford VM. Nanoscopic Porous Sensors. Annu Rev Anal Chem. 2008;1:737–766. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 7.Howorka S, Siwy Z. Nanopore Analytics: Sensing of Single Molecules. Chem Soc Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 8.Keyser UF. Controlling Molecular Transport through Nanopores. J R Soc, Interface. 2011;8:1369–1378. doi: 10.1098/rsif.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatesan BM, Bashir R. Nanopore Sensors for Nucleic Acid Analysis. Nat Nanotechnol. 2011;6:615–624. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 10.Wanunu M. Nanopores: A Journey towards DNA Sequencing. Phys Life Rev. 2012;9:125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiner JE, Balijepalli A, Robertson JWF, Campbell J, Suehle J, Kasianowicz J. Disease Detection and Management via Single Nanopore-Based Sensors. Chem Rev. 2012;112:6431–6451. doi: 10.1021/cr300381m. [DOI] [PubMed] [Google Scholar]
- 12.Miles BN, Ivanov AP, Wilson KA, Dogan F, Japrung D, Edel JB. Single Molecule Sensing with Solid-State Nanopores: Novel Materials, Methods, and Applications. Chem Soc Rev. 2013;42:15–28. doi: 10.1039/c2cs35286a. [DOI] [PubMed] [Google Scholar]
- 13.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of Individual Polynucleotide Molecules using a Membrane Channel. Proc Natl Acad Sci U. S. A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndić M. Rapid Electronic Detection of Probe-Specific microRNAs using Thin Nanopore Sensors. Nat Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 15.Fologea D, Gershow M, Ledden B, McNabb DS, Golovchenko J, Li J. Detecting Single Stranded DNA with a Solid State Nanopore. Nano Lett. 2005;5:1905–1909. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalczyk SW, Hall AR, Dekker C. Detection of Local Protein Structures along DNA Using Solid-State Nanopores. Nano Lett. 2010;10:324–328. doi: 10.1021/nl903631m. [DOI] [PubMed] [Google Scholar]
- 17.Keyser UF, Koeleman BN, Van Dorp S, Krapf D, Smeets RMM, Lemay SG, Dekker NH, Dekker C. Direct Force Measurements on DNA in a Solid-State Nanopore. Nat Phys. 2006;2:473–477. [Google Scholar]
- 18.Dudko OK, Mathe J, Szabo A, Meller A, Hummer G. Extracting Kinetics from Single-Molecule Force Spectroscopy: Nanopore Unzipping of DNA Hairpins. Biophys J. 2007;92:4188–4195. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halverson KM, Panchal RG, Nguyen TL, Gussio R, Little SF, Misakian M, Bavari S, Kasianowicz JJ. Anthrax Biosensor, Protective Antigen Ion Channel Asymmetric Blockade. J Biol Chem. 2005;280:34056–34062. doi: 10.1074/jbc.M507928200. [DOI] [PubMed] [Google Scholar]
- 20.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond Time-Scale Discrimination among Polycytidylic Acid, Polyadenylic Acid, and Polyuridylic Acid as Homopolymers or as Segments within Single RNA Molecules. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timp W, Comer J, Aksimentiev A. DNA Base-Calling from a Nanopore Using a Viterbi Algorithm. Biophys J. 2012;102:L37–L39. doi: 10.1016/j.bpj.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok N, Niederweis M, Gundlach JH. Reading DNA at Single-Nucleotide Resolution with a Mutant MspA Nanopore and phi29 DNA Polymerase. Nat Biotechnol. 2012;30 doi: 10.1038/nbt.2171. 349-U174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, Fischbein MD, Venta K, Luo Z, Johnson ATC, et al. DNA Translocation through Graphene Nanopores. Nano Lett. 2010;10:2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 24.Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LMK, Dekker C. DNA Translocation through Graphene Nanopores. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 25.Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Graphene as a Subnanometre Trans-Electrode Membrane. Nature. 2010;467 doi: 10.1038/nature09379. 190-U173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-Molecule DNA Detection with an Engineered MspA Protein Nanopore. Proc Natl Acad Sci U. S. A. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Slowing Down DNA Translocation through a Nanopore in Lithium Chloride. Nano Lett. 2012;12:1038–1044. doi: 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic Focusing of Unlabelled DNA into Nanoscale Pores using a Salt Gradient. Nat Nanotechnol. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieberman KR, Cherf GM, Doody MJ, Olasagasti F, Kolodji Y, Akeson M. Processive Replication of Single DNA Molecules in a Nanopore Catalyzed by phi29 DNA Polymerase. J Am Chem Soc. 2010;132:17961–17972. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeh LH, Zhang MK, Joo SW, Qian SZ. Slowing Down DNA Translocation through a Nanopore by Lowering Fluid Temperature. Electrophoresis. 2012;33:3458–3465. doi: 10.1002/elps.201200142. [DOI] [PubMed] [Google Scholar]
- 31.Wei RS, Martin TG, Rant U, Dietz H. DNA Origami Gatekeepers for Solid-State Nanopores. Angew Chem Int Ed. 2012;51:4864–4867. doi: 10.1002/anie.201200688. [DOI] [PubMed] [Google Scholar]
- 32.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science. 2012;338:932–936. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirsaidov U, Comer J, Dimitrov V, Aksimentiev A, Timp G. Slowing the Translocation of Double-Stranded DNA using a Nanopore Smaller than the Double Helix. Nanotechnology. 2010;21:1–10. doi: 10.1088/0957-4484/21/39/395501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luan BQ, Wang D, Zhou R, Harrer S, Peng H, Stolovitzky G. Dynamics of DNA Translocation in a Solid-State Nanopore immersed in Aqueous Glycerol. Nanotechnology. 2012;23:1–7. doi: 10.1088/0957-4484/23/45/455102. [DOI] [PubMed] [Google Scholar]
- 35.Kawano R, Schibel AEP, Cauley C, White HS. Controlling the Translocation of Single-Stranded DNA through alpha-Hemolysin Ion Channels Using Viscosity. Langmuir. 2009;25:1233–1237. doi: 10.1021/la803556p. [DOI] [PubMed] [Google Scholar]
- 36.He Y, TsuTsui M, Scheicher RH, Bai F, Taniguchi M, Kawai T. Thermophoretic Manipulation of DNA Translocation through Nanopores. Acs Nano. 2012;7:538–546. doi: 10.1021/nn304914j. [DOI] [PubMed] [Google Scholar]
- 37.Rosenstein JK, Wanunu M, Merchant CA, Drndić M, Shepard KL. Integrated Nanopore Sensing Platform with Sub-Microsecond Temporal Resolution. Nat Methods. 2012;9 doi: 10.1038/nmeth.1932. 487-U112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Automated Forward and Reverse Ratcheting of DNA in a Nanopore at 5-Angstrom Precision. Nat Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Tao C, Chien M, Hellner B, Balijepalli A, Robertson JWF, Li Z, Russo JJ, Reiner JE, Kasianowicz JJ, et al. PEG-Labeled Nucleotides and Nanopore Detection for Single Molecule DNA Sequencing by Synthesis. Sci Rep U. K. 2012;2:1–8. doi: 10.1038/srep00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson JWF, Rodrigues CJ, Stanford VM, Rubinson KA, Krasilnikov OV, Kasianowicz JJ. Single-Molecule Mass Spectrometry in Solution using a Solitary Nanopore. Proc Natl Acad Sci U. S. A. 2007;104:8207–8211. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reiner JE, Kasianowicz JJ, Nablo BJ, Robertson JWF. Theory for Polymer Analysis using Nanopore-Based Single-Molecule Mass Spectrometry. Proc Natl Acad Sci U. S. A. 2010;107:12080–12085. doi: 10.1073/pnas.1002194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Q, Comer J, Dimitrov V, Yemenicioglu S, Aksimentiev A, Timp G. Stretching and Unzipping Nucleic Acid Hairpins using a Synthetic Nanopore. Nucleic Acids Res. 2008;36:1532–1541. doi: 10.1093/nar/gkm1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Comer J, Dimitrov V, Zhao Q, Timp G, Aksimentiev A. Microscopic Mechanics of Hairpin DNA Translocation through Synthetic Nanopores. Biophys J. 2009;96:593–608. doi: 10.1016/j.bpj.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. Stretching DNA using the Electric Field in a Synthetic Nanopore. Nano Lett. 2005;5:1883–1888. doi: 10.1021/nl0510816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho C, Qiao R, Heng JB, Chatterjee A, Timp RJ, Aluru NR, Timp G. Electrolytic Transport through a Synthetic Nanometer-Diameter Pore. Proc Natl Acad Sci U. S. A. 2005;102:10445–10450. doi: 10.1073/pnas.0500796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wanunu M, Bhattacharya S, Xie Y, Tor Y, Aksimentiev A, Drndić M. Nanopore Analysis of Individual RNA/Antibiotic Complexes. Acs Nano. 2011;5:9345–9353. doi: 10.1021/nn203764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niedzwiecki DJ, Iyer R, Borer PN, Movileanu L. Sampling a Biomarker of the Human Immunodeficiency Virus across a Synthetic Nanopore. Acs Nano. 2013 doi: 10.1021/nn400125c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid Nanopore Discrimination between Single Polynucleotide Molecules. Proc Natl Acad Sci U. S. A. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lieberman KR, Dahl JM, Mai AH, Akeson M, Wang HY. Dynamics of the Translocation Step Measured in Individual DNA Polymerase Complexes. J Am Chem Soc. 2012;134:18816–18823. doi: 10.1021/ja3090302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faller M, Niederweis M, Schulz GE. The Structure of a Mycobacterial Outer-Membrane Channel. Science. 2004;303:1189–1192. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- 51.Song LZ, Hobaugh MR, Shustack C, Cheley S, Bayley H, Gouaux JE. Structure of Staphylococcal Alpha-Hemolysin, a Heptameric Transmembrane Pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 52.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Fabrication of Solid-State Nanopores with Single-Nanometre Precision. Nat Mater. 2003;2:537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 53.Kim MJ, Wanunu M, Bell DC, Meller A. Rapid Fabrication of Uniformly Sized Nanopores and Nanopore Arrays for Parallel DNA Analysis. Adv Mater. 2006;18:3149–3153. [Google Scholar]
- 54.van den Hout M, Hall AR, Wu MY, Zandbergen HW, Dekker C, Dekker NH. Controlling Nanopore Size, Shape and Stability. Nanotechnology. 2010;21:1–6. doi: 10.1088/0957-4484/21/11/115304. [DOI] [PubMed] [Google Scholar]
- 55.Kowalczyk SW, Grosberg AY, Rabin Y, Dekker C. Modeling the Conductance and DNA Blockade of Solid-State Nanopores. Nanotechnology. 2011;22:1–5. doi: 10.1088/0957-4484/22/31/315101. [DOI] [PubMed] [Google Scholar]
- 56.Tinland B, Pluen A, Sturm J, Weill G. Persistence Length of Single-Stranded DNA. Macromolecules. 1997;30:5763–5765. [Google Scholar]
- 57.Meller A, Nivon L, Branton D. Voltage-Driven DNA Translocations through a Nanopore. Phys Rev Lett. 2001;86:3435–3438. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 58.Meller A, Branton D. Single Molecule Measurements of DNA Transport through a Nanopore. Electrophoresis. 2002;23:2583–2591. doi: 10.1002/1522-2683(200208)23:16<2583::AID-ELPS2583>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 59.Wanunu M, Sutin J, McNally B, Chow A, Meller A. DNA Translocation Governed by Interactions with Solid-State Nanopores. Biophys J. 2008;95:4716–4725. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Healy K, Ray V, Willis LJ, Peterman N, Bartel J, Drndić M. Fabrication and Characterization of Nanopores with Insulated Transverse Nanoelectrodes for DNA Sensing in Salt Solution. Electrophoresis. 2012;33:3488–3496. doi: 10.1002/elps.201200350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fischbein MD, Drndić M. Nanogaps by Direct Lithography for High-Resolution Imaging and Electronic Characterization of Nanostructures. Appl Phys Lett. 2006;88:1–3. [Google Scholar]
- 62.Sigworth FJ. Electronic Design of the Patch Clamp. In: Sakmann B, Neher E, editors. Single Channel Recording. Vol. 209. New York: Springer; pp. 95–106. [Google Scholar]
- 63.Derrington IM, Butler TZ, Collins MD, Manrao E, Pavlenok M, Niederweis M, Gundlach JH. Nanopore DNA Sequencing with MspA. Proc Natl Acad Sci U. S. A. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.