Abstract
Mutationally activated kinases define a clinically validated class of targets for cancer drug therapy1. However, the efficacy of kinase inhibitors in patients whose tumours harbour such alleles is invariably limited by innate or acquired drug resistance2,3. The identification of resistance mechanisms has revealed a recurrent theme—the engagement of survival signals redundant to those transduced by the targeted kinase4. Cancer cells typically express multiple receptor tyrosine kinases (RTKs) that mediate signals that converge on common critical downstream cell-survival effectors—most notably, phosphatidylinositol-3-OH kinase (PI(3)K) and mitogen-activated protein kinase (MAPK)5. Consequently, an increase in RTK-ligand levels, through autocrine tumour-cell production, paracrine contribution from tumour stroma6 or systemic production, could confer resistance to inhibitors of an oncogenic kinase with a similar signalling output. Here, using a panel of kinase-‘addicted’ human cancer cell lines, we found that most cells can be rescued from drug sensitivity by simply exposing them to one or more RTK ligands. Among the findings with clinical implications was the observation that hepatocyte growth factor (HGF) confers resistance to the BRAF inhibitor PLX4032 (vemurafenib) in BRAF-mutant melanoma cells. These observations highlight the extensive redundancy of RTK-transduced signalling in cancer cells and the potentially broad role of widely expressed RTK ligands in innate and acquired resistance to drugs targeting oncogenic kinases.
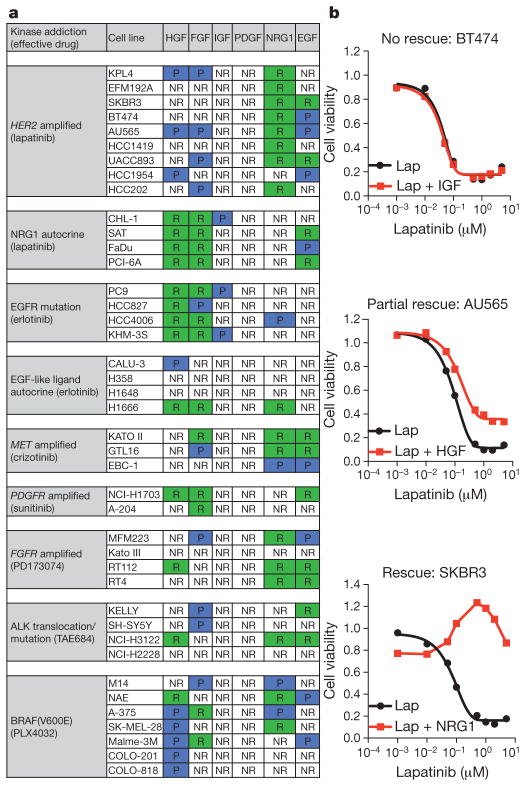
Using 41 human-tumour-derived cell lines with previously defined kinase dependency7–9, we undertook a ‘matrix analysis’ to examine the effects of six different RTK ligands known to be widely expressed in tumours10 (HGF, epidermal growth factor (EGF), fibroblast growth factor (FGF), platelet-derived growth factor (PDGF), neuregulin 1 (NRG1) and insulin-like growth factor (IGF)) on drug response. We quantified the effect of exposing these cell lines to each ligand on the half-maximum inhibitory concentration (IC50) for a kinase inhibitor that otherwise potently suppresses their growth (Supplementary Fig. 1a). Nearly all of the cell lines tested, representing multiple tissue types and distinct kinase dependencies, could be rescued from drug-induced growth inhibition by one or more RTK ligands (Fig. 1a).
Figure 1. RTK ligands attenuate kinase inhibition in oncogene-addicted cancer cell lines.
a, Summary of results from 41 kinase-addicted cancer cell lines co-treated with the appropriate kinase inhibitor and each of six RTK ligands. NR, no rescue; P, partial rescue; R, complete rescue. b, Cell viability assay demonstrating the diversity of ligand effects on drug-treated cell lines (72 h), illustrating examples of no rescue, partial rescue or complete rescue. Lap, lapatinib. Graphs show average values of technical duplicates from one representative experiment out of three independent experiments.
The consequences of ligand exposure on drug response were categorized as follows: (1) ‘no rescue’, addition of ligand did not affect drug response; (2) ‘partial rescue’, ligand partially abrogated treatment response; or (3) ‘complete rescue’, ligand ‘right-shifted’ the IC50 curve >10-fold, or completely suppressed drug response (Fig. 1b). HGF, FGF and NRG1 were the most broadly active ligands, followed by EGF, whereas IGF and PDGF had relatively little effect, despite activating their corresponding receptors (Supplementary Figs 1b and 5a). Many tested cell lines could be rescued from treatment sensitivity by exposure to as many as four different ligands, highlighting the capacity of such cells to engage redundant survival pathways upon exposure to a variety of ligands. None of the tested ligands could rescue cells from cisplatin sensitivity, suggesting that RTK ligands do not confer broad protection from toxic agents (Supplementary Fig. 1c).
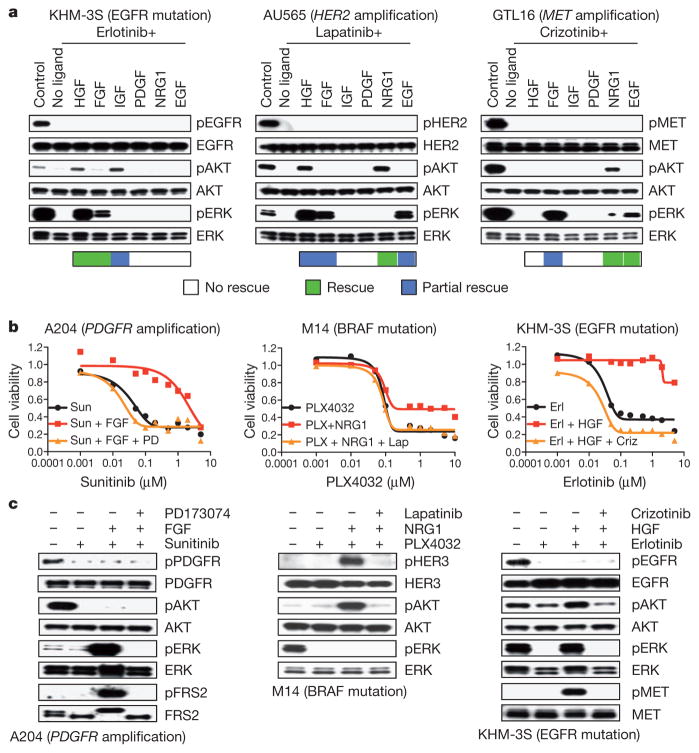
To explore the signalling dynamics associated with ligand-mediated rescue, we assessed the status of two downstream survival signalling pathways commonly engaged by RTKs: the PI(3)K–AKT and MAPK pathways11. Where ligand-mediated rescue was achieved, ligand re-activated at least one of these pathways despite the presence of drug (Fig. 2a). Pathway re-activation was not due to re-activation of the oncogenic kinase, as kinase autophosphorylation remained suppressed after ligand co-treatment. In the tested models, HGF re-activated both PI(3)K and MAPK, IGF and NRG1 only re-activated PI(3)K and FGF, and EGF only re-activated MAPK. Activation of the ‘redundant RTK’ and consequent downstream survival signalling persisted for at least 48 h (Supplementary Fig. 1d). A functional role for re-activation of both PI(3)K and MAPK signalling was observed in lapatinib-treated AU565 cells in the presence of NRG1, FGF or a combination (Supplementary Fig. 2a). However, specifically inhibiting PI(3)K attenuated HGF-promoted drug resistance, which was associated with engagement of both survival pathways (Supplementary Fig. 2b). As expected, ligand rescue was blocked by co-targeting the secondary activated kinase, confirming that effective ligands were acting via their cognate RTKs (Fig. 2b, c and Supplementary Fig. 3a, b). Inhibitors of the ‘secondary’ RTK that mediated ligand rescue had little or no effect as single agents, indicating that the kinase-addicted cells are not initially dependent on multiple different RTKs in the absence of available ligand. Moreover, ligand stimulation had little or no effect on cell proliferation (Figs 1b and 2b).
Figure 2. Pro-survival pathway re-activation correlates with RTK-ligand rescue.
a, Immunoblots showing effects of ligand (50 ng ml−1) on RTK, AKT and extracellular signal-regulated kinase (ERK) phosphorylation (p) after kinase inhibition (1 μM, 2 h). Ligand rescue is indicated: green squares, complete rescue; blue squares, partial rescue. b, Drug-induced suppression of viability in three kinase-addicted cell lines (72 h). Cells were co-treated with ligand and an appropriate secondary kinase inhibitor (0.5 μM). Graphs show average values of technical duplicates from one representative experiment out of three independent experiments. Criz; crizotinib; Erl, erlotinib; Lap, lapatinib; PD, PD173074; PLX, PLX4032; Sun, sunitinib. c, Immunoblots showing effect of kinase inhibition (1 μM) ± ligands (2 h) on RTK, AKT and ERK phosphorylation. Cells were co-treated with secondary kinase inhibitor (0.5 μM).
Analysis of baseline RTK expression confirmed that the tested lines express multiple RTKs. Ligand-induced rescue was well correlated with expression of certain RTKs in some cases (Supplementary Fig. 4), suggesting that the RTK profile of tumours before treatment could inform an optimal treatment strategy that anticipates the need to co-target two or more kinases. In some cases, ligands were unable to rescue cells from drug effects despite expression of the corresponding RTK. In a few cases, the RTK ligand activated its receptor; however, engagement of PI(3)K or MAPK was not observed (Supplementary Fig. 5a). In other cases, the ligand activated its receptor as well as at least one downstream effector; however, that was not sufficient for rescue (Supplementary Fig. 5b, c). This was observed, for example, with H2228 and H358 cells exposed to HGF, or with COLO-201 cells exposed to NRG1. However, H2228 and H358 cells are rescued by HGF after longer-term drug treatment, implicating the presence of a subpopulation of HGF-responsive cells selected over time in the presence of an inhibitory kinase (Supplementary Fig. 6c, d).
There were several findings with clinical implications. For example, one of two tested non-small-cell lung carcinoma (NSCLC) cell lines harbouring an anaplastic lymphoma kinase (ALK) translocation (NCI-H3122 cell line), and exhibiting ALK addiction, was rescued from ALK inhibition by brief exposure to HGF (Supplementary Fig. 6a, b). In these HGF receptor (MET)-expressing cells, HGF activates extracellular signal-regulated kinase (ERK) and AKT even in the presence of the ALK-selective inhibitor TAE684. In contrast, survival of these cells was disrupted even in the presence of HGF by crizotinib, a dual ALK/MET kinase inhibitor approved for treatment of ALK-translocated NSCLCs12. The second ALK-translocated NSCLC line, NCI-H2228, also expresses MET, but was not rescued from ALK inhibition by HGF at the 72-h time point. However, HGF re-activated AKT and ERK in the presence of TAE684 (Supplementary Fig. 5b), and longer-term TAE684 treatment in the presence of HGF yielded TAE684-resistant cells (Supplementary Fig. 6c). The relatively durable clinical responses observed in ALK-translocated NSCLC patients might be attributed in part to the dual ALK/MET inhibitory activity of crizotinib.
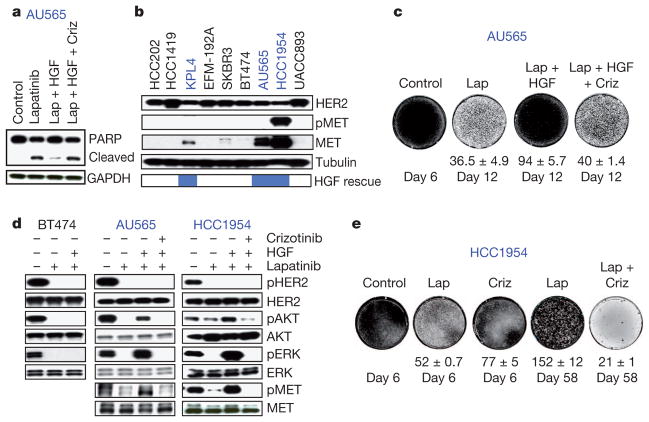
The ability of HGF to rescue three of nine tested human epidermal growth factor receptor 2 (HER2)-amplified breast cancer cell lines from growth inhibition by the HER2 kinase inhibitor lapatinib was also unexpected (Fig. 3a, b). These three cell lines express MET, which was correlated with the ability of HGF to attenuate lapatinib response (Fig. 3b). As with NCI-H228 cells, longer-term co-treatment (12 days) of the partially HGF-rescued AU565 cells revealed that HGF rapidly promoted lapatinib resistance, potentially by driving selection of a subpopulation of MET-expressing cells (Fig. 3c). Indeed, 9-day lapatinib/HGF co-treatment yielded a population of cells with increased MET (Supplementary Fig. 7a). HGF re-activated PI(3)K and MAPK signalling specifically in MET-positive cells (Fig. 3d).
Figure 3. HGF promotes lapatinib resistance in HER2-amplified breast cancer cell lines.
a, Immunoblots showing apoptosis in AU565 cells after lapatanib (Lap; 1 μM), HGF or crizotinib (Criz; 0.5 μM) treatment. Blue font indicates partially rescued cell lines. b, Immunoblots showing phospho-MET (pMET) and MET in HER2-amplified breast cancer cell lines with partial HGF rescue indicated. c, Syto 60 cell staining of AU565 cells treated with lapatinib (1 μM), HGF or crizotinib (0.5 μM). d, Immunoblots showing AKT and ERK re-activation in MET-positive (blue) and MET-negative (black) cells. Cells were treated with lapatinib (1 μM), HGF or crizotinib (0.5 μM) (2 h). e, Syto 60 staining of HCC1954 cells treated with lapatinib (5 μM) or crizotinib (1 μM). Images are representative of three biological replicates and values indicate mean ± s.d.
A subset of tested HER2-positive primary breast tumours express MET protein (Supplementary Fig. 7b). One HER2-amplified breast cancer cell line (HCC1954) showed elevated phospho-MET in the absence of exogenous HGF, implicating an autocrine mechanism (Fig. 3b), and MET inhibition in these cells delayed the emergence of lapatinib resistance (Fig. 3e). Collectively, these results suggest that MET-expressing HER2-positive breast tumours might evade HER2 inhibition by engaging MET in a subpopulation of ‘primed’ tumour cells, depending on the availability of HGF. Additionally, eight of nine tested HER2-amplified breast cell lines were rescued from lapatinib sensitivity by exposure to the HER3 ligand NRG1 (Fig. 1a and Supplementary Fig. 7c), possibly implicating NRG1 levels in the tumour microenvironment in the variable response to HER2 kinase inhibition.
Another observation with clinical implications was the unexpected finding that HGF attenuated the response to the BRAF kinase inhibitor PLX4032 (vemurafenib) in BRAF-mutant cell lines. PLX4032 recently demonstrated remarkable efficacy in BRAF-mutant melanoma, leading to clinical approval13. Notably, among 446 tested secreted factors, HGF was among a very small number that could rescue BRAF-mutant melanoma cells from PLX4032 sensitivity (Supplementary Fig. 8).
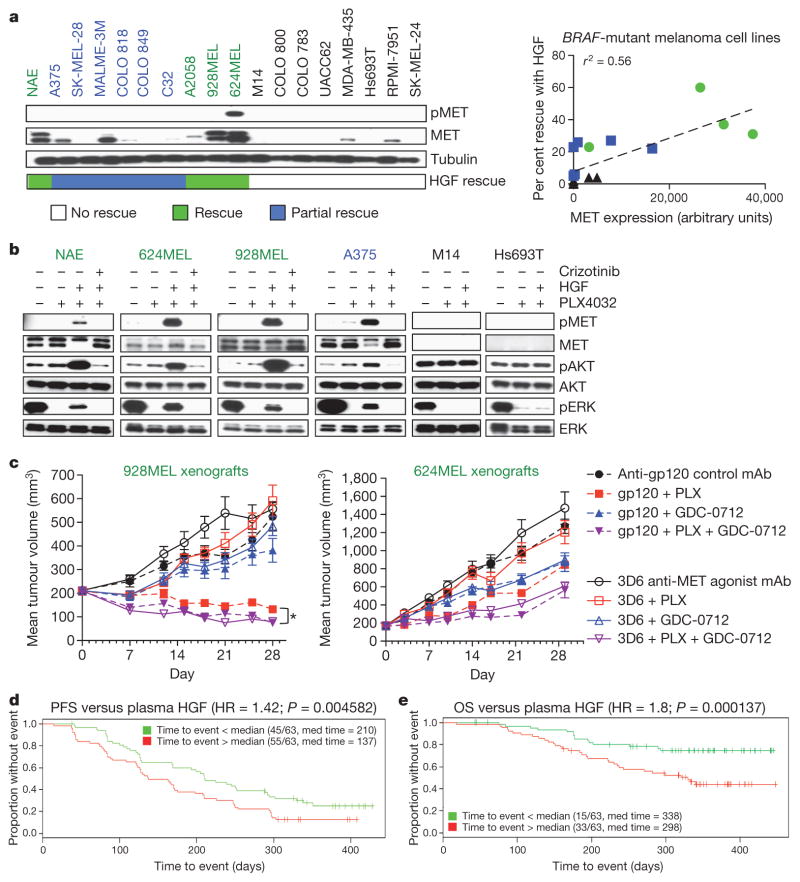
Among 12 additional BRAF-mutant melanoma cell lines tested, HGF significantly attenuated PLX4032 sensitivity in 5 lines (Fig. 4a). Eight of ten HGF-rescued cell lines expressed detectable MET, whereas MET was undetectable or barely detectable in the non-rescued cells. MET expression was correlated with HGF rescue (Fig. 4a), and HGF re-activated MAPK in lines rescued by HGF, but not in MET-negative HGF-non-rescued cells (Fig. 4b). As anticipated, rescue by HGF was blocked when MET was inhibited by crizotinib (Fig. 4b and Supplementary Fig. 9a). One BRAF-mutant cell line (624MEL) showed elevated phospho-MET in the absence of exogenous HGF, consistent with an autocrine mechanism (Fig. 4a), and MET inhibition in these cells delayed PLX4032 resistance (Supplementary Fig. 9b). Crizotinib co-treatment also prevented resistance to PLX4032 in two cell lines (A375 and 928MEL) with undetectable phospho-MET, further supporting a role for HGF-activated MET in PLX4032 resistance (Supplementary Fig. 9b).
Figure 4. HGF promotes PLX4032 resistance in BRAF-mutant melanoma cell lines.
a, Left, immunoblots showing MET status in melanoma cells, with HGF rescue indicated. Right, correlation between MET expression and HGF rescue in PLX4032 (1 μM)-treated cells (72 h). Black font indicates no rescue; blue, partial rescue; green, complete rescue. b, Immunoblots showing ERK reactivation in MET-positive (green/blue) and MET-negative (black) cells treated with PLX4032 (1 μM), HGF or crizotinib (0.5 μM) (2 h). c, Effect of activating MET (3D6) on tumour growth inhibition by PLX4032 in two xenografts (n =10 per group). mAb, monoclonal antibody; PLX, PLX4032. Differences between PLX4032-treated and PLX4032- and GDC-0712 (MET inhibitor)-treated control antibody (gp120) groups (*P =0.0008). Error bars represent mean ± s.e.m. (biological replicates). d, e, PFS (d) and OS (e) in PLX4032-treated melanoma patients stratified based on plasma HGF (green < median HGF; red > median HGF).
To confirm these findings in vivo, we performed xenograft studies with BRAF-mutant 928MEL and 624MEL melanoma cells. Activation of MET in tumours using the MET-agonistic antibody 3D6 strongly abrogated the growth-suppressive effects of PLX4032 (Fig. 4c). The relevance of MET activation by 3D6 in attenuating the PLX4032 response was verified by co-treating with a MET kinase inhibitor. Inhibiting MET enhanced the effect of PLX4032 on tumour regression (Fig. 4c), with more partial responses observed in the 928MEL model (Supplementary Fig. 10).
To extend these findings in a clinical context, we tested the hypothesis that circulating HGF in BRAF-mutant melanoma patients could contribute to clinical outcome. Pre-treatment plasma HGF levels were measured from 126 of the 132 patients enrolled onto the BRIM2 clinical trial (BRAF-mutant metastatic melanoma patients treated with PLX4032). HGF levels ranged from 33–7,200 pg ml−1 with a median level of 334 pg ml−1 (Supplementary Fig. 11a). Increased plasma HGF was associated with worse outcome as measured by progression-free survival (PFS; hazard ratio, 1.42; P <0.005) and overall survival (OS; hazard ratio, 1.8; P <0.001; Fig. 4d, e). Segregating patients into tertiles revealed a continuous relationship between HGF level and outcome, rather than a threshold effect (Supplementary Fig. 11b). As BRIM2 was a single-arm study in which all patients received PLX4032, it is not possible to determine whether higher HGF levels confer drug resistance; however, this study implicates HGF–MET signalling in disease progression and overall survival, and together with related findings14, suggests a potential role for HGF in the response to BRAF inhibition in BRAF-mutant melanoma.
Overall, the findings highlight the extensive nature of signal crosstalk among RTKs that are co-expressed in most tumour cells, and the potentially broad role of RTK ligands in innate and acquired resistance to kinase inhibitors. Such ligands could be produced by tumour cells themselves to drive autocrine survival mechanisms, by tumour stroma, or systemically to affect drug response15,16.
The increasingly appreciated heterogeneity of human tumours complicates the elucidation of drug resistance mechanisms17–19. In the context of our findings, we imagine distinct mechanisms by which such heterogeneity could contribute to acquired resistance. Thus, a subpopulation of tumour cells present before therapy capable of responding to a survival-promoting RTK ligand might be expanded through the selective pressure of drug treatment if such a ligand becomes available within the tumour microenvironment. Indeed, analysis of MET expression in BRAF-mutant melanoma cells revealed a heterogeneous cell population (Supplementary Fig. 12). Similarly, in EGFR-mutant NSCLC, a subpopulation of MET-driven tumour cells may emerge upon exposure to HGF during EGFR tyrosine kinase inhibitor treatment20. Activation of multiple RTKs has been reported in glioblastoma, and suppression of pro-survival signals and consequent cell death was only observed after co-targeting multiple activated RTKs21. A subpopulation of RTK-ligand-producing tumour cells may be selected during treatment. In various pre-clinical models, the observed acquired resistance mechanism involved a ‘switch’ to a new RTK dependency22–27, which in some cases could be attributed to increased RTK-ligand production.
Although genomic biomarkers have been critical in identifying the patients most likely to benefit from therapy, there is a largely unexplained wide range of clinical benefit among such patients in terms of magnitude and duration of response12,13. Our findings support a potentially broad role for RTK ligands in the overall clinical benefit from such therapies, and provide a foundation for the use of biomarkers based on the expression of RTKs and their ligands to inform treatment strategies that anticipate both innate and acquired resistance mechanisms.
METHODS
Cell lines
Human cancer cell lines were obtained and tested for sensitivity using an automated platform as previously described7. Cell lines were maintained at 37 °C in a humidified atmosphere at 5% CO2 and grown in RPMI 1640 or DMEM/F12 growth media (GIBCO) supplemented with 10% fetal bovine serum (GIBCO), 50 units ml−1 penicillin and 50 μg ml−1 streptomycin (GIBCO).
Reagents
Lapatinib, sunitinib and erlotinib were purchased from LC Laboratories. Crizotinib, TAE684, AZD6244 and BEZ235 were purchased from Selleck Chemicals. PD173074 was purchased from Tocris Bioscience. PLX4032 was purchased from Active Biochem. Recombinant human HGF, EGF, FGF-basic, IGF-1 and PDGF-AB were purchased from Peprotech. Recombinant human NRG1-β1 was purchased from R&D Systems. For in vivo studies, 3D6 anti-MET agonist antibody, PLX4032 and GDC-0712 were generated at Genentech. GDC-0712 was used in xenograft experiments as it has a similar kinase profile as crizotinib28 (Supplementary Fig. 13) and was available in quantities sufficient for in vivo study. See Supplementary Methods for synthesis protocol.
Immunoblot analysis
Cell lysates were collected using Nonidet-P40 lysis buffer, supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Scientific) and immunodetection of proteins was carried out using standard protocols. The phospho-HER2 (Y1248; catalogue no. 2247), HER2 (no. 2242), phospho-HER3 (Y1289; no. 4791), phospho-MET (Y1234/5; no. 3126), PDGFRα (no. 5241), phospho-FRS2α (Y196; no. 3864), IGF-1Rβ (no. 3027), phospho-ALK (Y1604; no. 3341), AKT (no. 9272), phospho-ERK (T202/Y204; no. 9101), ERK (no. 9102), GAPDH (no. 2118) and β-tubulin (no. 2146) antibodies were purchased from Cell Signaling Technologies. Antibodies to HER3 (SC-285), MET (SC-10), phospho-PDGFRα (SC-12911), FRS2α (SC-8318), FGFR1 (SC-7945), FGFR2 (SC-122), FGFR3 (SC-13121) and ALK (SC-25447) were purchased from Santa Cruz Biotechnologies. Phospho-AKT (S473; no. 44-621G) antibody was purchased from Invitrogen. Phospho-EGFR (Y1068; ab5644) antibody was purchased from Abcam. EGFR (no. 610017) antibody was purchased from BD Biosciences. PARP (no. 14-6666-92) antibody was purchased from eBioscience. Densitometry was carried out using ImageJ software.
Tissue samples
Primary breast tumour samples with appropriate Institutional Review Board (IRB) approval and informed patient consent were obtained from the following sources: Cureline, ILSbio and the Cooperative Human Tissue Network of the National Cancer Institute. Metastatic melanoma tumour samples with appropriate IRB approval and informed patient consent were obtained from the BRIM2 trial. The human tissue samples used in the study were de-identified (double-coded) before their use and thus the study using these samples is not considered human subject research under the US Department of Human and Health Services regulations and related guidance (45 CFR, Part 46). Immunohistochemistry for MET was performed on formalin-fixed paraffin-embedded sections cut at a thickness of 4 μm on to positively charged glass slides. The staining was performed on a Discovery XT autostainer with Ultraview detection (VMSI) using the MET rabbit monoclonal antibody SP44 (Spring BioScience; no. M3441) and CC1 standard antigen retrieval. Sections were counterstained with haematoxylin and specific membranous staining for MET was scored on a scale from 0 (no staining) to 3+ (strong staining).
HGF ELISA
Plasma was obtained from a metastatic melanoma patient’s pre-dose PLX4032 cycle one and the concentration of HGF in patient-derived plasma was quantitatively measured using a sandwich ELISA as previously described29.
Xenograft studies
All procedures were approved by and conformed to the guidelines and principles set by the Institutional Animal Care and Use Committee of Genentech and were carried out in an Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facility. Ten million 928MEL or 624MEL BRAF-mutant melanoma cells (suspended in a 1:1 mixture of HBSS/Matrigel) were inoculated in the right flank of CRL C.B-17 SCID.bg mice (Charles River Laboratories). When tumours reached an average volume of 200 mm3, mice (10 per group) were treated with either control antibody (anti-gp120; 10 mg kg−1 once per week; intraperitoneal), 3D6 (anti-MET agonistic antibody; 10 mg kg−1 once per week; intraperitoneal), PLX4032 (50 mg kg−1 twice daily; periocular), GDC-0712 (MET small molecular inhibitor28, previously named GNE-A; 100 mg kg−1 every day; periocular) as indicated for 4 weeks. Tumours were measured twice weekly using digital calipers (Fred V. Fowler Company) and tumour volumes were calculated using the formula (L×(W×W))/2. A partial response was defined as a reduction in tumour volume greater than 50% but less than 100%. A complete response was defined as 100% reduction in tumour volume. Differences between the PLX4032-treated and the PLX4032- and GDC-0712-treated control antibody groups were determined using two-way ANOVA (P =0.0008).
Secreted factor screen
Recombinant purified secreted factors were purchased from Peprotech and R&D Systems as indicated, and were reconstituted in PBS/ 0.1% BSA (Supplementary Table 1). Secreted factors were transferred into 96-well plates at a concentration of 1 μg ml−1, and subsequently diluted to 100 ng ml−1 in media containing either no drug or 5 μM PLX4032. Equal volumes of diluted factor (final concentration 50 ng ml−1) were arrayed into the 384 well plates pre-seeded with SK-MEL-28 cells (500 cells per wells seeded the day before) using an Oasis liquid handler. After 72 h incubation, cell viability was determined using Cell Titer Glo (Promega).
Statistics
Cell viability assays were carried out in duplicate wells within an individual experiment (technical replicates) and carried out multiple times (biological replicates) as indicated. Correlation of receptor expression with cognate ligand rescue was carried out using a 2×2 contingency table with the following groups: receptor positive, RTK-ligand rescued; receptor positive, RTK-ligand non-rescued; receptor negative, RTK-ligand rescued; receptor negative, RTK-ligand non-rescued. Significance was determined using a two-tailed Fisher exact probability test.
Statistical analysis of BRIM2 clinical samples
HGF levels were log transformed, and the Kolmogorov–Smirnoff test was used to test the resulting distribution for departure from the Gaussian distribution. The Cox-proportional model was used to test the log-transformed HGF levels for association with the PFS and OS. Association between the response and HGF levels was tested using the Wilcoxon rank-sum test. Kaplan–Meier curves were used to show the relationship between the HGF levels and the time-to-event outcomes (PFS and OS). The number of events/patients and medium time to event is shown for each group. The Cox-proportional model of the outcome as the function of the continuous HGF level was used to calculate the hazard ratio and corresponding P-value.
Supplementary Material
Acknowledgments
We thank members of the Settleman laboratory, G. Bray and C. Bowdoin for helpful discussions, K. Trunzer and B. Nelson for assistance with access to clinical samples.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions T.R.W. and J.S. designed the study, analysed data, discussed results and co-wrote the paper. T.R.W. performed in vitro experiments. T.R.W., E.L. and R.N. designed and performed the 446 soluble factor screen. H.K. performed immunohistochemistry analysis. E.C., J.P. and M.M. designed and performed in vivo experiments. E.P., L.B., Y.W. and Y.Y. assessed BRIM2 study material, including HGF enzyme-linked immunosorbent assay (ELISA) from plasma. J.F. carried out the biostatistical analysis. J.L. carried out efficacy and safety analyses on the BRIM2 study. J.S. and A.R. were clinical investigators on the BRIM2 study. J.M. and D.P.S. characterized GDC-0712.
Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 2.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrett JT, Arteaga CL. Resistance to HER2-directed antibodies and tyrosine kinase inhibitors: mechanisms and clinical implications. Cancer Biol Ther. 2011;11:793–800. doi: 10.4161/cbt.11.9.15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–79. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Moritz A, et al. Akt–RSK–S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal. 2010;3:ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, Huang P. Cancer-stromal interactions: role in cell survival, metabolism and drug sensitivity. Cancer Biol Ther. 2011;11:150–156. doi: 10.4161/cbt.11.2.14623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDermott U, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci USA. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson TR, Lee DY, Berry L, Shames DS, Settleman J. Neuregulin-1-mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer Cell. 2011;20:158–172. doi: 10.1016/j.ccr.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Yonesaka K, et al. Autocrine production of amphiregulin predicts sensitivity to both gefitinib and cetuximab in EGFR wild-type cancers. Clin Cancer Res. 2008;14:6963–6973. doi: 10.1158/1078-0432.CCR-08-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller MM, Fusenig NE. Friends or foes — bipolar effects of the tumour stroma in cancer. Nature Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 11.Grant S, Qiao L, Dent P. Roles of ERBB family receptor tyrosine kinases, and downstream signaling pathways, in the control of cell growth and survival. Front Biosci. 2002;7:d376–d389. doi: 10.2741/grant. [DOI] [PubMed] [Google Scholar]
- 12.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012 doi: 10.1038/nature11183. http://dx.doi.org/10.1038/nature11183. [DOI] [PMC free article] [PubMed]
- 15.Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–366. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang K, et al. Recombinant human erythropoietin antagonizes trastuzumab treatment of breast cancer cells via Jak2-mediated Src activation and PTEN inactivation. Cancer Cell. 2010;18:423–435. doi: 10.1016/j.ccr.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintana E, et al. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calbo J, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. 2011;19:244–256. doi: 10.1016/j.ccr.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 22.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guix M, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res. 2010;70:1625–1634. doi: 10.1158/0008-5472.CAN-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, et al. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Res. 2009;69:6871–6878. doi: 10.1158/0008-5472.CAN-08-4490. [DOI] [PubMed] [Google Scholar]
- 27.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 28.Liederer BM, et al. Preclinical absorption, distribution, metabolism, excretion, and pharmacokinetic-pharmacodynamic modelling of N-(4-(3-((3S,4R)-1-ethyl-3-fluoropiperidine-4-ylamino)-1H-pyrazolo[3,4-b]p yridin-4-yloxy)-3-fluorophenyl)-2-(4-fluorophenyl)-3-oxo-2,3-dihydropyrida zine-4-carboxamide, a novel MET kinase inhibitor. Xenobiotica. 2011;41:327–339. doi: 10.3109/00498254.2010.542500. [DOI] [PubMed] [Google Scholar]
- 29.Catenacci DVT, et al. Durable complete response of metastatic gastric cancer with anti-met therapy followed by resistance at recurrence. Cancer Discovery. 2011;1:573–579. doi: 10.1158/2159-8290.CD-11-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.