Background: The PIM-2 kinase is a potent survival factor; its role in the DNA damage response has never been addressed.
Results: PIM-2 promotes DNA lesions repair in an E2F-1 and ATM-dependent manner; Pim-2-silenced cells are more susceptible to UV damage.
Conclusion: PIM-2 is an upstream activator of E2F-1 and ATM in the UV damage response.
Significance: PIM-2 is an essential component of the UV damage response.
Keywords: Apoptosis, Base Excision Repair, Cancer Biology, Cell Signaling, DNA Damage Response, ATM, E2F-1, PIM-2
Abstract
The oncogenic nature ascribed to the PIM-2 kinase relies mostly on phosphorylation of substrates that act as pro-survival/anti-apoptotic factors. Nevertheless, pro-survival effects can also result from activating DNA repair mechanisms following damage. In this study, we addressed the possibility that PIM-2 plays a role in the cellular response to UV damage, an issue that has never been addressed before. We found that in U2OS cells, PIM-2 expression and activity increased upon exposure to UVC radiation (2–50 mJ/cm2), and Pim-2-silenced cells were significantly more sensitive to UV radiation. Overexpression of PIM-2 accelerated removal of UV-induced DNA lesions over time, reduced γH2AX accumulation in damaged cells, and rendered these cells significantly more viable following UV radiation. The protective effect of PIM-2 was mediated by increased E2F-1 and activated ATM levels. Silencing E2F-1 reduced the protective effect of PIM-2, whereas inhibiting ATM activity abrogated this protective effect, irrespective of E2F-1 levels. The results obtained in this study place PIM-2 upstream to E2F-1 and ATM in the UV-induced DNA damage response.
Introduction
The DNA damage response (DDR)4 is a complex cellular mechanism aimed to preserve DNA integrity after it has been damaged. DNA double-strand breaks (DSB) are considered the most deleterious damage that must be rapidly repaired through either the nonhomologous end joining or the homologous recombination pathways. Failure to properly repair DSBs leads to apoptosis, but it might also lead to cancer. One of the earliest events in the cell response to DSB is activation of the phosphatidylinositol 3-kinase like protein kinases: ATM (ataxia-telangiectasia mutated), ATR (ataxia-telangiectasia and Rad3 related), and DNA-PKcs (DNA-dependent protein kinase catalytic subunit), which phosphorylate a wide range of checkpoint and repair proteins (reviewed by Shiloh (1)). Among these kinases, ATM is primarily activated after ionizing radiation or treatment with radiomimetic drugs (1–5). ATR, on the other hand, is the primary kinase activated after replication-dependent DSB/replication stress (6, 7). One of the first proteins to be phosphorylated upon DSBs is the histone variant H2AX (8), and the appearance of phosphorylated H2AX on Ser-139 (γH2AX) became a common measure for detecting DSBs (reviewed by Takahashi and Ohnishi (9)). Another biologically important damaging agent is UV radiation. Although UVA radiation (320–400 nm) can mainly cause DNA damage by inducing reactive oxygen species, UVB (280–320 nm) and UVC (200–280 nm) can directly target nuclear DNA by formation of DNA lesions such as cyclobutane-pyrimidine dimers (CPDs) and TC(6–4) photoproducts (10–14). These lesions can inhibit DNA replication, as well as transcription, and may cause chromosomal breakage, mutations, and cell death (reviewed by Batista et al. (15)). UV-induced DNA damage is mainly repaired by the nucleotide excision repair (NER) mechanism. NER-deficient cells are generally hypersensitive to the cytotoxic and mutagenic effects of UV light (10, 16). UVB and UVC can indirectly promote DNA DSBs through the collapse of replication forks upon collision with UV-induced DNA photoproducts (17, 18). As in the case of DSBs, the UV-induced DNA damage (UVB and UVC) is also characterized by nuclear accumulation of γH2AX, although the kinetics of this accumulation is different from that seen after ionizing radiation-induced DSB (17, 19–21).
Members of the PIM serine/threonine kinase family, Pim-1–3, are generally regarded as proto-oncogenes and are overexpressed in a range of hematopoietic malignancies and solid cancers (22–30). Transgenic mice overexpressing Pim-1 or Pim-2, together with c-Myc, tend to developed lymphomas with high frequency (31, 32), whereas Eμ-Myc transgenic mice (overexpressing c-Myc alone) that were deficient for both Pim-1 and Pim-2 exhibited delayed lymphoma development (33).
The human Pim-2 gene encodes for two isoforms, of 34 and 41 kDa, that share an identical kinase domain but differ at their N terminus, with the 41-kDa isoform having an extended N terminus because of an alternative translation initiation site (25). Both isoforms share a panel of substrates, with increased phosphorylation efficiency reported for the 34-kDa isoform (34). During the past decade, PIM-2 has been shown to directly contribute to cell survival. It was found to phosphorylate BAD on Ser-112, thus preventing it from binding to and activating the pro-apoptotic Bcl-Xl protein (34, 35). Other phosphorylation targets include: COT, the IkB kinase activator, which upon phosphorylation leads to NF-κB-dependent pro-survival effect (36); 4E-BP1, which upon phosphorylation promotes release of the pro-survival eukaryotic translation initiation factor eIF-4e (35); and API-5, an apoptotic inhibitor that is stabilized by phosphorylation by PIM-2 (37). PIM-2 is involved also in regulation of cell cycle progression and cell proliferation. PIM-2 can promote G1 arrest either by directly phosphorylating and thus stabilizing the cell cycle inhibitor p21Cip1/WAF1 (p21) (38) or by promoting proteasome-dependent down-regulation of CDC25A (39). On the other hand, PIM-2 can promote cell cycle progression by phosphorylating the cell cycle inhibitor p27Kip1, leading to its nuclear export and proteasome-dependent degradation (40). In addition, PIM-2 can phosphorylate c-Myc on Ser-329, thus stabilizing it and enhancing its transcriptional activity (32).
Although cell survival effects have been ascribed to PIM-2, its role in the cellular response to UV damage has never been reported. The aim of this study was to address this issue. We found that expression and activity of PIM-2 increased upon exposure to UVC radiation and that Pim-2-silenced cells were significantly more sensitive to UV radiation. Overexpression of PIM-2 in U2OS cells accelerated reduction of UV-induced DNA lesions over time, reduced γH2AX accumulation in damaged cells, and rendered these cells less sensitive to the UV radiation. The protective effect of PIM-2 was mediated by increased E2F-1 levels and an increase in the activating phosphorylation of ATM on Ser-1981 (pATM). Silencing E2F-1 reduced the protective effect of PIM-2, whereas inhibiting ATM activity abrogated the protective effect of PIM-2, irrespective of E2F-1 levels. The results obtained in this study place PIM-2 upstream to E2F-1 and ATM in the UV-induced DNA damage response.
EXPERIMENTAL PROCEDURES
Cell Culture
U2OS cells (human osteosarcoma) were cultured in DMEM, supplemented with 10% fetal bovine serum, l-glutamine (2 mm), and 1% penicillin-streptomycin-nystatin (Invitrogen). The cells were maintained at 37 °C with 5% CO2 in a humidified chamber. For synchronization of cells to the G1 phase, the double thymidine block method was applied. To this end, cells were grown with DMEM without serum for 24 h, washed with PBS, and kept growing with DMEM containing 10% FBS and 2.5 mm thymidine (diluted to 250 mm, in PBS) for 18 h (first block). Next, the cells were washed twice with PBS and allowed to grow with fresh DMEM containing 10% FBS for 9 h before thymidine was added again to growing medium (final concentration, 2.5 mm) for 18 h (second block). At this stage, the cells were taken for FACS analysis to verify G1 arrest. For the S phase, the cells were washed twice with PBS and allowed to grow for 6 h with fresh DMEM containing 10% FBS. FACS analysis was performed to verify synchronization at the S phase.
Real Time PCR Analysis
RNA was isolated using EZ-RNA II kit (Biological Industries, Kibbutz Beit-Haemek, Israel). 2 μg of total RNA were taken for cDNA preparation, using the RevertAid Moloney murine leukemia virus reverse transcriptase kit (Fermentas). cDNA was diluted 1:4 in HPLC-purified H2O. Each reaction (10 μl) contained 5 μl of fast SYBR green (Ready mix with Teq-pol) (Applied Biosystems), 1 μl of diluted cDNA, and 0.3 pm from each primer (forward and reverse). Real time PCR and data analysis were performed on the StepOnePlus real time PCR system (Applied Biosystems). The primers used were: Pim-2 forward, 5′-ccctgcttcatgatgaaccctacact-3′; Pim-2 reverse, 5′-ccatgtcatagaggaggatgcccag-3′; S12 forward, 5′-ggaaggcattgctgctgg-3′; and s12 reverse, 5′-cctcaatgacatccttgg-3′.
Protein Extraction and Western Blot Analysis
Cells were harvested using lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40) containing protease inhibitor complete EDTA free (Roche Applied Science), and 1 mm PSMF. Protein concentration was determined using the Bradford method, and 40 μg of protein were used for Western analysis (7–15% polyacrylamide gels), according to standard procedure. Signal was visualized by chemiluminescence detection reagents (Pierce). The following primary antibodies were used: anti-PIM-2-Self-made (25), anti-E2F-1 (Santa Cruz), anti-actin (Santa Cruz), anti-ATM (Epitomics), anti-pAMT (Ser-1981) (Epitomics), anti-γH2AX (Trevigen), anti-tubulin (Sigma), anti-pKap1 (Ser-824) (Bethyl), anti-p53 (Santa Cruz), anti-pp53 (Ser-15) (Cell Signaling), anti-CHK1 (Santa Cruz), anti-pCHK1 (Ser-345) (Cell Signaling), anti-FLAG (Sigma), and anti-H3K9 acetylated (Cell Signaling). The secondary antibodies used were: goat anti-mouse IgG HRP-conjugated (Jackson Laboratories) and donkey anti-goat IgG HRP-conjugated (Jackson Laboratories). The fluorescence secondary antibody used was: Alexa 594 (Invitrogen).
Inducible Gene Expression
The Complete Control Inducible Mammalian Expression System (Stratagene) was used to generate a stable U2OS cell line expressing a FLAG-tagged 41-kDa isoform of PIM-2 in a PON-A-dependent inducible manner. The Tet-On Gene Expression System (Clontech) was used to generate a stable U2OS cell line expressing a HA-tagged 34-kDa isoform of PIM-2 in a doxycycline (DOX)-dependent inducible manner. If not otherwise stated, activation of the FLAG-tagged 41-kDa isoform of PIM-2 was induced by PON-A 10 μm for 48 h, and activation of the HA-tagged 34 kDa isoform was induced by DOX 1 μg/ml for 48 h.
Transfections, Infections, and Silencing
Transfections of equal amounts of cells (4 × 105 cells/10-cm plate) were carried out using JetPEI transfection (Biomol) or polyjet reagent (SignaGen Laboratories) according to the manufacturer's instructions. For E2F-1 silencing, each experiment was done with at least two different sets of siRNA (Sigma-MISSION predesigned siRNA), with the same results. Key data are shown with both sets. siRNAs were introduced to U2OS cells (5–20 pmol/ml), using the PepMute siRNA transfection reagent (SignaGen Laboratories), according to the manufacturer's protocol. The cells were harvested and analyzed 48–96 h post-transfection. For Pim-2 silencing, each targeting experiment was performed with shRNA and an unrelated siRNA (Sigma), with similar results. Key data are shown with both targeting approaches. For siRNA, the cells were treated as described above. For shRNA, retroviral vectors were generated in 293T cells by co-transfection of ψ-ecotropic packaging plasmid (10 μg), pSV-ψ-E-MLV (which provides packaging helper function), and 10 μg of the pSUPER retro plasmid containing Pim-2 shRNA. Polyjet transfection reagent was used for transfection. After 8 h, post-transfection medium was replaced with fresh DMEM supplemented with 10% FBS, and 5-ml aliquots of the retrovirus-containing supernatant were collected thereafter at 6-h intervals. Five collections were pooled together and frozen in aliquots. To infect cells, they were incubated in 4.5 ml of retroviral supernatant supplemented with 8 μg/ml polybrene (Sigma) for 5 h at 37 °C. Then 5.5 ml of medium was added, and 24 h later the medium was replaced by fresh medium, containing 2 μg/ml puromycin (Sigma). The Pim-2 shRNA primers were: forward, 5′-gatccccaacatcctgatagacctacgcttcaagagagcgtaggtctatcaggatgtttttttggaaa-3′; and reverse, 5′-agcttttccaaaaaaacatcctgatagacctacgctctcttgaagcgtaggtctatcaggatgttggg-3′.
UV Irradiation
The cells were washed once with PBS, which was removed from the dish prior to exposure to UVC irradiation (254 nm) at intensities of 2–50 mJ/cm2. Medium was added to the culture immediately after irradiation.
PIM-2 in Vitro Kinase Assay
PIM-2 in vitro kinase assay was performed on Ser-75 (Ser-112 in mouse) of its substrate BAD (34, 35). 105 cells were irradiated at 8 mJ/cm2 and harvested 10 and 20 min postirradiation (unirradiated cells were used as control). PIM-2 was immunoprecipitated from total protein extracts of these cells, using PIM-2 antibodies and agarose A/G beads (Santa Cruz Biotechnology). 1 μg of recombinant BAD (Santa Cruz Biotechnology) was added to 47 μl of immunoprecipitated PIM-2 beads or to 0.1 μg of recombinant PIM-2 (Abgen) in 47 μl of kinase assay buffer, together with 3 μl of a [γ-32P]ATP solution (3 μl of [γ-32P]ATP (250 μCi) + 3 μl of cold ATP (1 mm) + 24 μl of H2O). The reaction was incubated 1 h at room temperature (gently resuspending the beads every 10 min). Following incubation, the samples were boiled for 5 min in sample buffer and separated in 10% SDS-PAGE. The gels were then dried in a gel drier and exposed overnight to a Fujifilm imaging plate, and signal was detected by the BAS-MP phosphorimaging device (Fujifilm).
Monitoring DNA Damage by Alkaline Unwinding Flow Cytometry
We have adopted the flow cytometry acridine orange assay previously described (41), with slight modifications. U2OS cells were UV irradiated (8 mJ/cm2) and trypsinized (1 ml) 5, 30, or 60 min post-UV irradiation (control cells were not irradiated). The cells were washed once with culture medium and again with cold PBS, and the cell pellet was resuspended in 300 μl of cold PBS followed by 1 h of fixation by dropwise addition of 3 ml of ice-cold 70% ethanol. After fixation, the cells were centrifuged (230 g) and resuspended in 35 μl of cold solution of 20 mm Tris, 1 mm EDTA, 100 mm NaCl, pH 7.4, and warmed slowly to 37 °C. The cells were then embedded in 85 μl of 2.25% ultra-low temperature melting agarose (Sigma) in PBS at 40 °C (on the wall of an Eppendorf tube), left at room temperature for few minutes, and then left on ice (or overnight at 4 °C) until solidification of agar. The following steps were carried under subdued light. The cells were treated with 1 ml of alkaline buffer (300 mm NaOH, 300 mm KCl, 50 μm EDTA, pH 13) for 1 min, immediately washed three times with 1 ml of neutralization buffer (150 mm Tris, 150 mm NaCl, pH 7.4) followed by 1 h of incubation in neutralization buffer at room temperature (or overnight at 4 °C). Next, we aspirated the buffer and warmed the agarose to 60 °C until melting, cooled to 40 °C, and mixed with 250 μl of prewarmed (37 °C) solution of 0.1% Triton X-100, 80 mm HCl, 150 mm NaCl, pH 1.3, for 30 s. Then cells were stained by adding 900 μl of freshly made acridine orange buffer (126 mm Na2HPO4, 37 mm citric acid, 150 mm NaCl, 1 mm EDTA, 10 μg/ml RNase A, 22.1 μm acridine orange). The samples were analyzed 15 min post-acridine orange staining using FACStar plus flow cytometer (Becton Dickinson, San Jose, CA). At least 104 cells were collected. Acridine orange green florescent was monitored at 500–530 nm (dsDNA) and acridine orange red florescent was monitored at 645 nm (ssDNA).
CPD Quantitation
U2OS cells were irradiated at 8 mJ/cm2 and trypsin harvested 5, 30, and 60 min post-UV irradiation (control cells were not radiated). The cells were washed with 10 ml of culture medium and centrifuged (230 × g), and DNA was extracted using a genomic DNA miniprep kit (Sigma). CPD quantification was carried out using the STA-322 DNA damage ELISA kit (Cell Biolabs) according to company's protocol. Briefly, DNA was diluted to 0.5 μg/ml, denatured at 95 °C for 10 min, and rapidly cooled on ice for 10 min. 100 μl of DNA was added to each well in the DNA high binding plate and incubated at 37 °C for 2 h. The wells were washed twice with PBS and blocked with 150 μl of diluent buffer for 1 h at room temperature. The buffer was removed, and 100 μl of anti-CPD antibody was added to each well for 1 h at room temperature on an orbital shaker. The plate was washed (three times) with 250 μl of washing buffer, blocked with 150 μl of blocking reagent (1 h at room temperature on an orbital shaker), and washed again (three times) with washing buffer before 100 μl of secondary antibody was added to each well (1 h at room temperature on an orbital shaker). The plate was washed again, and 100 μl of substrate solution was added to each well. Approximately 15 min later, the reaction was stopped using 100 μl of stop solution and immediately read at 450 nm.
Fluorescence-activated Cell Sorting Analysis
The cells were trypsinized and peeled off the plates into a 15-ml tube containing 10 ml of medium, centrifuged for 5 min (210 × g at 4 °C), and pellet was resuspended in 1 ml of PBS. 250 μl were taken for analysis. The cells were incubated 20 min in PI buffer (PBS containing 50 μg/ml PI, 0.1% Triton X-100, 0.07% sodium citrate, and 1 mg/ml RNase A). Fluorescence intensity was analyzed by a FACStar plus flow cytometer (Becton Dickinson, San Jose, CA), using the Quest or ModFit software for analysis. For annexin V/PI assay, the cells were harvested as described and stained with annexin V and PI using the MEBCYT apoptosis kit (MBL International), according to the company's protocol. Importantly, apoptotic rates determined by the sub-G1 phase of the cell cycle or by the annexin V/PI assay gave rather similar results.
MTT Assay for Cell Survival
U2OS Tet-on cells were plated in 96-wells plate (2 × 103 cells/well), and 24 h later DOX was added to the cultures to a final concentration of 1 μg/ml for 48 h. UV irradiation was carried out as described above, with the addition of DOX-containing medium after irradiation. 24 h later, the medium was replaced by 1 mg/ml MTT solution in PBS (pH 7.4) for 2 h at 37 °C. The MTT solution was then removed, and Me2SO was added to dissolve the insoluble formazan product. The absorbance of the colored solution was read by a spectrophotometer at 550 nm.
Immunocytochemistry
The cells were grown on a cover glass in a 35-mm culture dish. At designated times after UV radiation, the cells were fixed (20 min in 4% paraformaldehyde) and then maintain in blocking buffer (PBS containing 20% fetal bovine serum and 0.5% Triton X-100) for 30 min. Next, the cells were incubated at room temperature with anti-γH2AX antibodies diluted 1:100 in blocking buffer, followed by incubation with the secondary antibody Alexa 596 goat anti-rabbit diluted 1:400 in blocking buffer. Chromatin was stained using Hoechst. The cells were mounted with mounting buffer (for 50 ml buffer, 44 ml of glycerol containing 5 ml of 0.2 m Tris, pH 8.5, and 2.5 g of n-propyl gallate) and further analyzed using Axio imager fluorescent microscope.
ATM Inhibition
The cells were grown in the presence of 5 μm KU60019 (Tocris) 30 min prior to UV irradiation. After UV radiation KU60019 (5 μm) was added again to the medium until the cells were harvested. A similar inhibition protocol was used for the KU55399 inhibitor (Tocris) but with a concentration of 10 μm.
Statistical Analysis
Throughout this study, Student's t test was applied to determine the statistical significance of differences between groups.
RESULTS
PIM-2 Protects Cells from UV-induced Damage
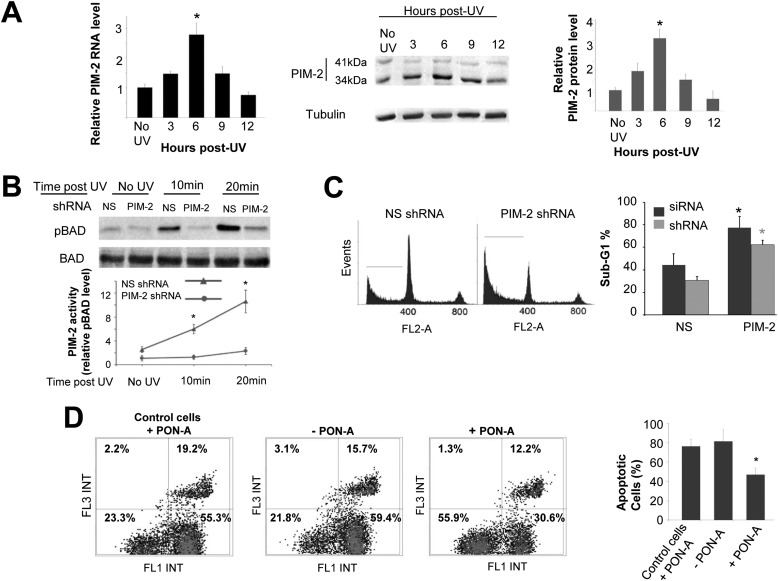
To get an idea of whether Pim-2 is involved in the cell response to genotoxic damage, we first followed the expression level of endogenous Pim-2 in U2OS cells, at both the RNA and protein levels, in response to UVC (50 mJ/cm2) or etoposide (50 μm/24 h) treatment. We found that endogenous Pim-2 was significantly up-regulated, at both the RNA and protein levels (mainly the 34-kDa isoform), in response to UVC irradiation (Fig. 1A). In fact, a prominent increase in the activity of PIM-2, judged by increased phosphorylation of PIM-2 substrate BAD on Ser-75 (Ser-112 in mouse), was already evident 10–20-min postradiation (Fig. 1B). This increase in BAD phosphorylation (pBAD) could not be seen in Pim-2-silenced cells (Fig. 1B and supplemental Fig. S1A), suggesting that the increase in pBAD is indeed PIM-2-dependent. A PIM-2 in vitro kinase assay that was performed on protein extracts from UV-irradiated cells further substantiated the increased activity of PIM-2 10–20 min following radiation, an activity that was associated with an increase in the level of PIM-2 (supplemental Fig. S1, B and C). Only a minor change was evident in PIM-2 protein levels following exposure to etoposide (supplemental Fig. S1D). To determine whether the increased expression of PIM-2 was specifically due to the rather high UV radiation intensity of 50 mJ/cm2, we repeated these experiments with various intensities from 2 to 50 mJ/cm2 and found that UV radiation-dependent elevation in PIM-2 levels was evident at all intensities, with a more prominent increase at 8, 15, and 50 mJ/cm2 (supplemental Fig. S1E). To further assess the importance of PIM-2 to the cell response to UV damage, we silenced Pim-2 in U2OS cells and compared apoptotic rates, 24 h after UV radiation, to cells treated with nonspecific (NS) shRNA or siRNA. As can be seen in Fig. 1C, Pim-2-silenced cells exhibited significantly higher apoptotic rates relative to NS control cells. Similar results were obtained using both Pim-2-targeting agents (shRNA and siRNA), suggesting that PIM-2 is indeed important for the UV damage response of the cell. If this is the case, one could expect that overexpression of Pim-2 would render the cells more stable to UV radiation. To test this hypothesis, we developed two U2OS cell lines overexpressing Pim-2 in a stable and inducible manner. In the first line, a FLAG-tagged 41-kDa isoform of PIM-2 was induced by PON-A in a dose-dependent manner (supplemental Fig. S1F), whereas in the second line, a HA-tagged 34-kDa isoform of PIM-2 was induced by DOX, in a dose-dependent manner (supplemental Fig. S1G). We found that upon overexpression of either isoform of PIM-2 prior to UV radiation, the percentage of apoptotic cells 24 h after radiation was significantly reduced compared with control cells (Fig. 1D and supplemental Fig. S2A). These results were further substantiated by monitoring cell survival using the MTT assay (supplemental Fig. S2B). It is noteworthy that the survival assays corresponded to the apoptotic rates throughout this study. Importantly, the protective effect of PIM-2 was kinase activity-dependent because overexpression of a kinase-dead form of PIM-2 (PIM-2KD, 39) did not reveal any protection (supplemental Fig. S2C). Interestingly, Pim-2 overexpression exerted only minor protection against etoposide treatment (∼5–10% decrease in apoptotic rates relative to control cells; supplemental Fig. S3, A and B), suggesting a differential role for PIM-2 in the cell's response to different DNA damage inducers and implying that PIM-2 has a significant role in the UV-induced DNA damage response.
FIGURE 1.
Involvement of PIM-2 in the UV-induced DNA damage response. A, up-regulation of PIM-2 following exposure to UVC radiation (254 nm). Cells were irradiated at 50 mJ/cm2 and harvested at the indicated times postirradiation (control cells were not radiated). Left panel, real time PCR analysis of total RNA from the irradiated cells, using Pim-2-specific primers. Primers for the S12 gene were used for endogenous control. Middle panel, Western analysis of proteins extracted from the irradiated cells (40 μg) using antibodies against the PIM-2 protein. Anti-tubulin antibodies were used for equal loading control and normalization. Right panel, densitometric analysis of PIM-2 signal (34-kDa isoform) from gels such as that depicted in the middle panel. Signal intensity with no UV was determined as 1. B, increased activity of PIM-2 in UV-irradiated cells, judged by increased phosphorylation of BAD on Ser-75 (pBAD). Upper panel, Western analysis of pBAD in extracts of naïve U2OS cells treated with nonspecific shRNA (NS), or Pim-2-silenced cells (PIM-2), at the indicated postirradiation times. Anti-BAD antibodies were used for equal loading control and normalization. Lower panel, graphic representation of relative intensity of pBAD obtained by densitometric analysis of pBAD signal from gels such as that depicted in the upper panel. Signal intensity in Pim-2-silenced cell (Pim-2 shRNA) with of no UV was determined as 1. C, Pim-2 knockdown cells are more sensitive to UVC-induced apoptosis. Left panel, cell cycle FACS analysis of PI-stained cells, either silenced for Pim-2 or transfected with NS shRNA, 24 h postirradiation with UVC (50 mJ/cm2). The horizontal line in each pattern indicates the channels included in calculation of the sub-G1 phase. Right panel, percentage of cells at the sub-G1 phase, calculated from cell cycle patterns such as that presented in the left panel. Results of both shRNA and siRNA are shown. D, cells overexpressing PIM-2 exhibit better resistance to UVC induce apoptosis. Left panel, annexin V/PI FACS assay for cells either induced to overexpress the 41-kDa isoform of PIM-2 (+PON-A, 10 μm for 24 h) or not (−PON-A), 24 h postirradiation with UVC (50 mJ/cm2). Control cell cannot induce PIM-2 expression upon treatment with PON-A. Right panel, percentage of apoptotic cells (early + late apoptosis). In all panels, the histograms summarize data from three independent experiments. *, statistically significant differences, p < 0.05.
Accelerated Repair of UV-induced DNA Lesions in PIM-2-expressing Cells
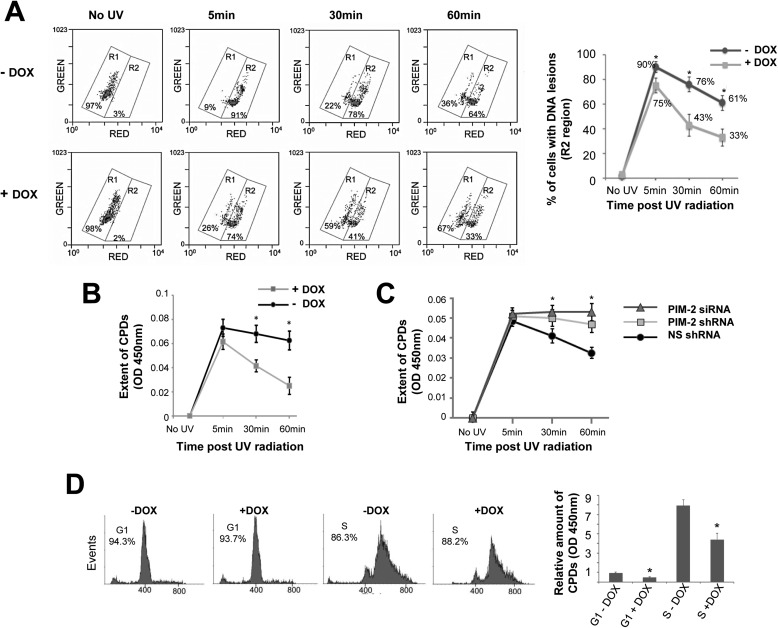
The protective effect exerted by PIM-2 could result from either reduced amount of DNA lesions or accelerated and more efficient repair of the DNA lesions. To differentiate between these options we used two independent approaches to assess the extent of DNA lesions at different time intervals following exposure to UV radiation (8 mJ/cm2). The first approach, monitoring DNA damage by alkaline unwinding flow cytometry, is based on differences in emitted fluorescence from single versus double-stranded acridine orange-stained nucleic acids, red and green, respectively. A direct positive correlation has been reported between the red fluorescence emitted from a cell, after alkaline unwinding of the DNA, and the amount of DNA lesions it contains (41, 42). In our assay, a significant difference (p < 0.05) was evident in the percentage of cells exhibiting DNA lesions (increased red fluorescence) between cells overexpressing PIM-2 (+DOX) and control cells (−DOX), as soon as 5 min after radiation (75% versus 90%, respectively), and this difference became even more apparent 30 min (43% versus 76%, respectively) or 60 min (33% versus 61%, respectively) after radiation (Fig. 2A). No difference was observed in unirradiated controls, either treated or not with DOX. Similar differences between PIM-2-overexpressing cells and control cells were obtained using an additional approach of a direct ELISA assay for UV-induced CPDs, using anti-CPD antibodies. As can be seen in Fig. 2B, 5 min after radiation, the amount of CPDs in PIM-2-overexpressing cells is reduced compared with control cells, and as time passes the difference becomes significant (p < 0.04 and p < 0.03 for 30 and 60 min, respectively). Moreover, the kinetics of removal of CPDs was slower upon silencing Pim-2 (with both shRNA and siRNA), compared with nonrelevant shRNA/siRNA control, reaching a statistically significant difference (p < 0.05) 30 min after radiation (Fig. 2C). These differences seem not to be related to a specific cell cycle stage because the cultures in these experiments were not synchronized, and the cell cycle distribution did not diverge significantly between samples. Nonetheless, given that cells at the S phase of the cell cycle are known to be more sensitive to UV radiation, we repeated the later set of experiments in synchronized cell populations to determine any possible cell cycle effects on our results. To this end, cells overexpressing PIM-2 (+DOX) and control cells (−DOX) were synchronized to either the G1 or S phase, using the double thymidine block method (Fig. 2D), before being exposed to UV radiation (8 mJ/cm2), and 45 min postirradiation the relative amount of CPDs was assessed as described above. As expected, the relative amount of legions was much higher in the S phase population, but in both the G1 and S phase cell populations, PIM-2-expressing cells exhibited significantly reduced CPDs compared with control cells (p < 0.05; Fig. 2D), excluding a cell cycle effect on the protective properties of PIM-2. We concluded that PIM-2 plays a role in the removal of the DNA lesions and that overexpression of PIM-2 results in accelerated and more efficient repair of the DNA lesions.
FIGURE 2.
Accelerated repair of UV-induced DNA lesions in PIM-2-expressing cells. A, monitoring DNA damage in PIM-2-overexpressing cells (+DOX) versus control cells (−DOX), exposed to UVC radiation (8 mJ/cm2), by alkaline unwinding flow cytometry. At the indicated time intervals postirradiation, cells were fixed, embedded in low temperature melting agarose, treated with alkaline buffer to unwind damaged DNA, stained with acridine orange, and analyzed by FACS for green and red fluorescence emitted from double- and single-stranded DNA, respectively. Left panel, region R1 represents distribution of undamaged cells, and region R2 represents distribution of damaged cells. The percentage of cells within each region is indicated. Right panel, percentage of cells with DNA lesions, calculated from FACS analyzes such as that depicted in the left panel. *, statistically significant differences, p < 0.05. B, direct ELISA assay for UV-induced CPDs in PIM-2-overexpressing cells (+DOX) versus control cells (−DOX), exposed to UVC radiation (8 mJ/cm2), using anti-CPD antibodies. *, statistically significant differences, p < 0.05. C, direct ELISA assay for UV-induced CPDs in Pim-2-silenced cells (Pim-2 shRNA and siRNA) versus control cells treated with NS shRNA, exposed to UVC radiation (4 mJ/cm2), using anti-CPD antibodies. D, the protective effect of PIM-2 overexpression is not cell cycle-dependent. Left panel, cells overexpressing PIM-2 (+DOX) and control cells (−DOX) were synchronized to either the G1 or S phase, using the double thymidine block method. Percentage of cells at either G1 or S phase is indicated. Right panel, relative amount of CPDs 45 min after UV radiation (8 mJ/cm2). CPDs in G1 synchronized control cells (G1-DOX) were determined as 1. *, statistically significant difference, p < 0.05.
E2F-1 Mediates PIM-2-stimulated Protection against UV-induced DNA Damage
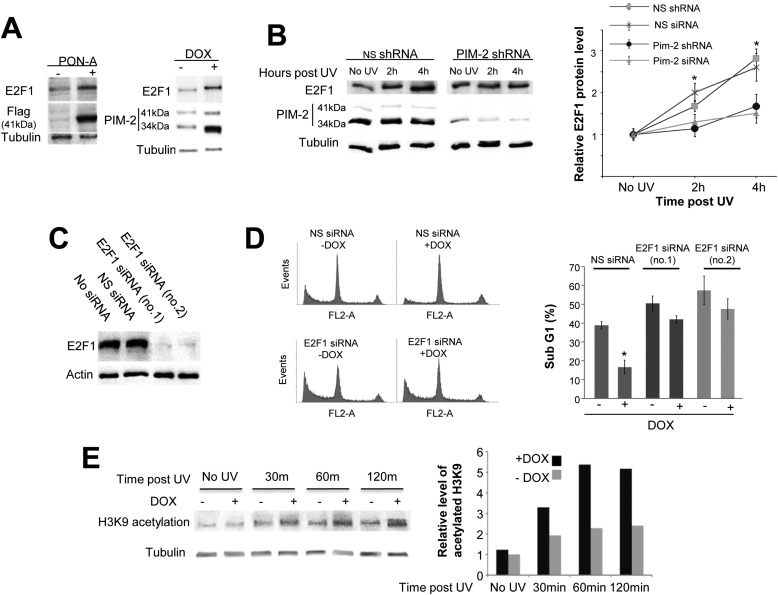
It has recently been reported that E2F-1 plays an important role in the UV-induced DNA damage response (43, 44). Given these reports and given our previous observation that E2F-1 is up-regulated upon overexpression of PIM-2 in HeLa cells (39), we hypothesized that PIM-2-dependent up-regulation of E2F-1 might contribute to the protective effect of PIM-2 following UV radiation. To address this possibility, we first determined E2F-1 levels following induction of expression of either PIM-2 isoform (41- and 34-kDa isoforms) in our U2OS system. We found that E2F-1 levels indeed increased upon induction of expression of either PIM-2 variant (Fig. 3A). We also found that upon exposure to UV radiation, E2F-1 levels were elevated in U2OS cells, as expected (Fig. 3B). However, when Pim-2 was silenced, E2F-1 levels were only moderately increased, if at all, relative to cells treated with nonspecific shRNA/siRNA (Fig. 3B), indicating that PIM-2 contributes to the UV-related increase in E2F-1 levels. Furthermore, we monitored apoptotic rates 24 h post-UVC radiation (50 mJ/cm2) under conditions of E2F-1 knockdown (Fig. 3C), with and without PIM-2 (34-kDa isoform) overexpression. As expected, in E2F-1 unsilenced cells (NS-siRNA), PIM-2 overexpression revealed ∼2-fold decrease in apoptotic levels, relative to −DOX control cells (Fig. 3D). However, for E2F-1 knockdown cells, the percentage of apoptotic cells was consistently higher compared with the NS-siRNA control, with only mild (but consistent and statistically significant) protection upon PIM-2 overexpression (Fig. 3D). Furthermore, it has recently been reported that E2F-1 stimulates NER by promoting GCN5 acetyltransferase-dependent acetylation of histone H3 Lys-9 (H3K9) (44). Therefore, to further substantiate the role of E2F-1 in the PIM-2-dependent accelerated repair of the UV-induced DNA legions, we tested whether PIM-2 also promotes H3K9 acetylation. We found that indeed PIM-2-overexpressing cells (+DOX) exhibit increased acetylation at all post-UV time intervals tested, compared with control cells (Fig. 3E). These results, together with the fact that E2F-1 silencing does not affect the increase in PIM-2 levels upon UV radiation (supplemental Fig. S4A), imply that PIM-2 acts upstream to E2F-1 and that E2F-1 mediates the protective effect of PIM-2 against UV-induced damage. However, our results also suggest the existence of an alternative route through which PIM-2 supports resistance to UV damage.
FIGURE 3.
Involvement of E2F-1 in the PIM-2-dependent protection effect against UV-induced DNA damage. A, Western blot analysis (40 μg of total cell extracts) of E2F-1 levels in cells overexpressing either the 41-kDa isoform of PIM-2 (PON-A) or the 34-kDa isoform (DOX), using anti-E2F-1 antibodies. For detection of overexpressed PIM-2, anti-FLAG and anti-PIM-2 antibodies were used for the 41- and 34-kDa isoforms, respectively. Anti-tubulin antibodies were used for equal loading control and normalization. B, up-regulation of E2F-1 following exposure to UVC is delayed in Pim-2-silenced cells. Left panel, Western blot analysis of E2F-1 levels in cells either silenced for Pim-2 expression (Pim-2 shRNA) or cells transfected with NS shRNA as control, 2 and 4 h post-UVC irradiation (50 mJ/cm2). Anti-tubulin antibodies were used for equal loading control and normalization. Right panel, graphic representation of relative E2F-1 levels, obtained from densitometric analysis of gels such as that depicted in the left panel for Pim-2-silenced cells with either shRNA and siRNA. E2F-1 levels in nonirradiated cells were determined as 1. C, E2F-1 silencing using two unrelated sets of E2F-1-specific siRNAs (no.1 and no.2). Nonspecific siRNA was used for control. Anti-actin antibodies were used for equal loading control. D, left panel, cell cycle FACS analysis of PI-stained cells, under conditions of E2F-1 knockdown (E2F-1 siRNA), with (+DOX) and without (−DOX) PIM-2 overexpression, 24 h postirradiation with UVC (50 mJ/cm2). Nonspecific siRNA (NS siRNA) was used as control. Right panel, percentage of cells at the sub-G1 phase, calculated from cell cycle patterns such as those presented in the left panel, for E2F-1-silenced cells with both sets of siRNA. Graphic representations in B and D summarize data from three independent experiments. *, statistically significant differences, p < 0.05. E, left panel, Western bolt analysis of histone H3 Lys-9 (H3K9) acetylation in cells overexpressing the 34-kDa isoform of PIM-2 (+DOX), versus control cell (−DOX), at the indicated time intervals postradiation (50 mJ/cm2). Anti-tubulin antibodies were used for equal loading control and normalization. Right panel, graphic representation of relative acetylated H3K9 levels at different time intervals following radiation, obtained by densitometric analysis of signals in the blot presented in the left panel. Level of acetylated H3K9 in unirradiated control cells (−DOX) was determined as 1.
PIM-2 Activates ATM and ATR
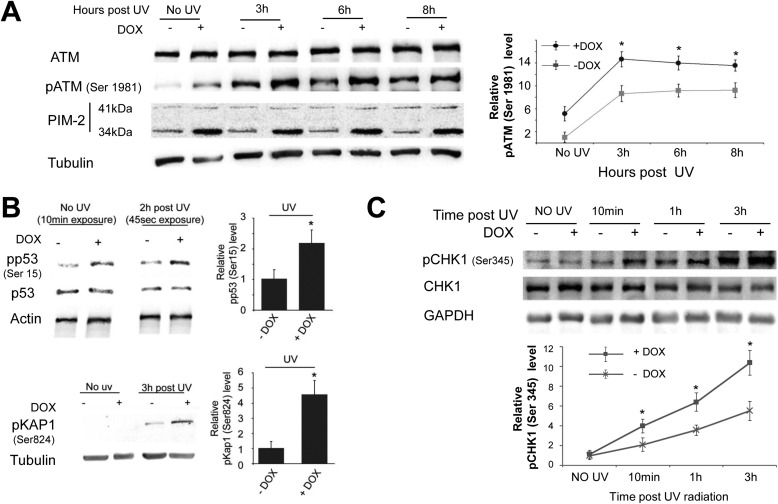
Both ATM and ATR were reported to be activated and to play a role in the UV-induced DDR (45, 46), thus also being potential mediators of the PIM-2-dependent protection. In undamaged cells ATM exists as an inactive dimer, which upon damage undergoes autophosphorylation on Ser-1981 and dimer dissociation, rendering it active to phosphorylate various substrates needed for the damage response (47). We therefore examined the levels of ATM and phospho-ATM (Ser(P)-1981, pATM) in response to PIM-2 overexpression. Cells overexpressing the 34-kDa isoform of PIM-2 (+DOX) were exposed to UVC radiation (15 mJ/cm2) and harvested 3, 6, or 8 h after exposure to radiation before ATM and pATM levels were compared with −DOX control cells. As can be seen in Fig. 4A, pATM levels were considerably higher in all PIM-2-expressing cells, including unirradiated cells, whereas ATM levels were unchanged. No such increase in pATM could be obtained in control U2OS cells treated with DOX (data not shown), indicating that DOX by itself cannot affect the level of pATM. To further verify that PIM-2 indeed activated ATM, we tested the phosphorylative status of both p53 (on Ser-15, pp53) and Kap1 (on Ser-824, pKap1), known substrates of ATM (48, 49). We found that upon overexpression of PIM-2, an increase in the level of pp53 was evident, even in unirradiated cells, supporting the notion that ATM was indeed activated (Fig. 4B). As to phospho-Kap1, a clear difference between PIM-2-overexpressing cells and control cells could be seen following radiation (Fig. 4B). However, no signal of pKap1 could be detected 6 h postirradiation in PIM-2-overexpressing cells (+DOX) in the presence of the ATM-specific inhibitor KU60019 (supplemental Fig. S4B), verifying that phosphorylation of Kap1 is indeed ATM-dependent. Likewise, no signal of pKap1 could be detected in unirradiated cells (Fig. 4B); possibly implying that the extent to which ATM was activated in unirradiated cells was beneath the threshold needed to detect Kap1 phosphorylation. Alternatively, it might imply that Kap1 becomes phosphorylation-competent only after damage. These results indicate that PIM-2 can activate ATM and that this activation might not be restricted to cells with actually damaged DNA.
FIGURE 4.
PIM-2-dependent activation of ATM and ATR. A, left panel, Western blot analysis (40 μg of total cell extracts) of ATM and phosphorylated ATM on Ser-1981 (pATM) in cells overexpressing the 34-kDa isoform of PIM-2 (+DOX), compared with (−DOX) control cells, 3, 6, and 8 h post-UVC irradiation (15 mJ/cm2). Anti-PIM-2 antibodies were used for detection of PIM-2 overexpression. Anti-tubulin antibodies were used for equal loading control and normalization. Right panel, graphic representation of relative pATM levels, obtained from densitometric analysis of gels such as that depicted in the left panel. pATM levels in nonirradiated −DOX control cells were determined as 1. Graph summarizes data from three independent experiments. *, statistically significant differences, p < 0.05. B, left upper panel, Western blot analysis (40 μg of total cell extracts) of phosphorylated p53 on Ser-15 (pp53). Left lower panel, phosphorylated Kap1 on Ser-824 (pKap1), as indicators of active ATM, in cells overexpressing the 34-kDa isoform of PIM-2 (+DOX), compared with (−DOX) control cells, 2 and 3 h post-UVC irradiation (15 mJ/cm2), respectively. Anti-actin and anti-tubulin antibodies were used for equal loading control and normalization. Note the different exposure times of pp53 blots in unirradiated and irradiated cells. Right panel, relative pp53 and pKap levels, obtained from densitometric analysis of gels such as those depicted in the left panel. pp53 and pKap1 levels in −DOX cells were determined as 1. *, statistically significant differences, p < 0.05. C, ATR increased activation in PIM-2-overexpressing cells. Upper panel, Western blot analysis (40 μg of total cell extracts) of phosphorylated CHK1 on Ser-345 (pCHK1), in cells overexpressing the 34-kDa isoform of PIM-2 (+DOX), compared with (−DOX) control cells, at the indicated times post-UVC irradiation (15 mJ/cm2). Anti-GAPDH antibodies were used for equal loading control and normalization. Lower panel, graphic representation of relative pCHK1 levels, obtained from densitometric analysis of gels such as that depicted in the upper panel. pCHK1 levels in nonirradiated (−DOX) control cells were determined as 1. Graph summarizes data from three independent experiments. *, statistically significant differences, p < 0.05.
Stiff et al. (45) have reported that following UV treatment ATM activation is ATR-dependent. To test whether ATR is also activated upon PIM-2 overexpression, we analyzed phosphorylation of Chk1 on Ser-345 (pChk1), a known ATR-dependent phosphorylation site, at different time intervals postirradiation. Only a mild signal of Chk1 phosphorylation could be detected in unirradiated cells (Fig. 4C), suggesting that ATR was not significantly active before UV treatment. Nonetheless, after exposure to UV radiation PIM-2-overexpressing cells consistently exhibited significantly increased pChk1 levels, compared with −DOX control cells (Fig. 4C), at all time intervals tested (p < 0.05), suggesting increased activation of ATR. We concluded that PIM-2 amplifies not only the activation of ATM but that of ATR as well, following UV-induced damage.
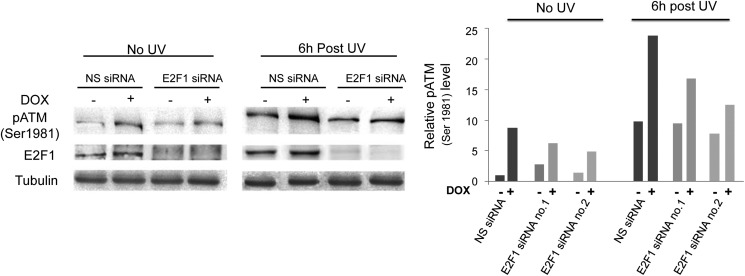
Does ATM activation by PIM-2 depend on E2F-1? To address this question we analyzed pATM levels in E2F-1 knockdown cells versus cells treated with NS-siRNA, in unirradiated cells as well as 6 h after UVC radiation (15 mJ/cm2). We found that E2F-1 silencing (with either siRNA) reduced the ATM activation by PIM-2 but did not block it completely (Fig. 5), suggesting that E2F-1 plays some role in PIM-2-dependent activation of ATM. It also suggested the existence of alternative routes through which PIM-2 can activate ATM.
FIGURE 5.
Knockdown of E2F-1 reduces PIM-2-dependent activation of ATM. Left panel, Western blot analysis (40 μg of total cell extracts) of pATM in cells overexpressing the 34-kDa isoform of PIM-2 (+DOX), compared with control cells (−DOX), under conditions of E2F-1 silencing or transfection with NS siRNA, with or without UVC irradiation (15 mJ/cm2). Tubulin antibodies were used for equal loading control and normalization. Right panel, relative pATM levels, obtained from densitometric analysis of gels such as that depicted in the left panel using both sets of siRNA (no.1 and no.2). pATM level in unirradiated −DOX cells treated with NS siRNA was determined as 1.
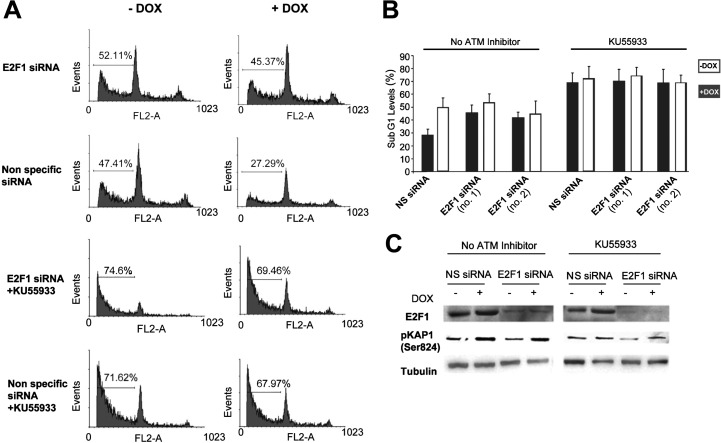
Is ATM activation important for the PIM-2-induced protection effect following UV radiation? To address this issue we monitored the apoptotic rates 6 h after exposure to UV radiation (15 mJ/cm2) in cells overexpressing PIM-2 and in −DOX control cells under conditions of E2F-1 silencing or nonspecific siRNA, with or without the ATM inhibitor KU55933. As expected, the lowest rate of apoptotic cells was observed in nonsilenced E2F-1 cells overexpressing PIM-2 with no inhibition of ATM (Fig. 6, A and B). Likewise, silencing of E2F-1 significantly reduced the protective effect of PIM-2, consistent with the above described results. However, when the ATM inhibitor was added, apoptotic rates were much higher, irrespective of whether E2F-1 was silenced or not (Fig. 6, A and B). The efficiency of E2F-1 silencing and ATM inhibition were verified as depicted in Fig. 6C. These results, together with the fact that inhibition of ATM does not affect the increase in PIM-2 levels upon UV radiation (supplemental Fig. S4C), indicated that ATM indeed acts downstream to PIM-2 and is indispensable for the PIM-2 protective effect during the UV damage response.
FIGURE 6.
ATM inhibition abolishes the PIM-2 protective effect regardless of E2F-1 expression. A, cell cycle FACS analysis (propidium iodide-stained cells) of cells overexpressing PIM-2 (+DOX), compared with (−DOX) control cells, under conditions of E2F-1 silencing (E2F-1 siRNA no.1) or nonspecific siRNA, with or without the ATM inhibitor KU55933 (10 μm), 6 h post-UVC radiation (15 mJ/cm2). The horizontal line in each pattern indicates the channels included in calculation of the sub-G1 phase. The percentage of sub-G1 cells is written above the horizontal line. B, percentage of cells at the sub-G1 phase, calculated from cell cycle patterns such as that presented in A with both E2F-1 siRNA sets (no.1 and no.2). C, Western blot analysis (40 μg of total protein extract) for E2F-1 and phosphorylated Kap1 on Ser-824 (pKap1) levels, in cells treated as described in A. Anti-tubulin was used for equal loading control and normalization.
Reduced γH2AX Accumulation Following Exposure to UV Irradiation in PIM-2 -overexpressing Cells
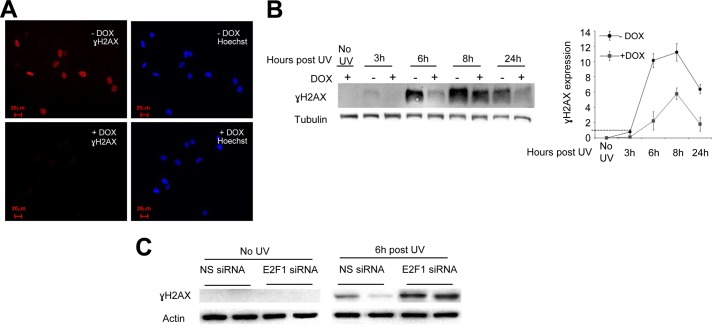
It is well documented that in cells exposed to UVB or UVC radiation, unrepaired DNA lesions, such as CPDs, cause DSBs during the S phase because of collapse of the replication fork. We assumed that if PIM-2-overexpressing cells indeed remove the UV-induced DNA lesions more efficiently, the extent of DSB accumulation will also be reduced. To address this issue, we monitored the accumulation and disappearance of γH2AX following UVC radiation in UV-irradiated (8 mJ/cm2) PIM-2-overexpressing cells (+DOX) compared with control cells (−DOX). The cells were fixed 3, 6, 8, and 24 h post-UV radiation, and γH2AX levels were determined immunocytochemically or by Western blot analysis. In the immunocytochemical analysis, a most distinct difference was observed 6 h postirradiation where cells overexpressing PIM-2 clearly showed reduced levels of γH2AX compared with control cells (Fig. 7A and supplemental Fig. S5). For Western blot analysis, PIM-2-expressing cells (34-kDa isoform) were irradiated (15 mJ/cm2), and proteins were extracted from cells before treatment, as well as 3, 6, 8, and 24 h post-UV radiation. No γH2AX could be detected in DOX-treated, unirradiated cells, indicating that neither DOX nor PIM-2 induce γH2AX accumulation (Fig. 7B). γH2AX started to accumulate 3 h postirradiation in −DOX control cells, reaching a maximum level at 8 h postirradiation and with substantial residual levels 24 h postirradiation. In PIM-2-overexpressing cells, however, significantly lower levels of γH2AX were apparent for all time points tested (Fig. 7B). Importantly, the reduced γH2AX levels were E2F-1-dependent because no such reduction was detected in E2F-1-silenced cells overexpressing PIM-2 (Fig. 7C). On the contrary, γH2AX levels were increased relative to control cells, upon E2F-1 silencing, regardless of DOX stimulation. We therefore concluded that γH2AX accumulation upon UV irradiation is significantly reduced in PIM-2-overexpressing cells, indicating reduced amounts of DSBs.
FIGURE 7.
Reduced γH2AX accumulation following UVC in PIM-2-overexpressing cells. A, immunocytochemical staining of γH2AX in cells overexpressing the 34 kDa isoform of PIM-2 (+DOX), compared with (−DOX) control cells, 6 h post-UVC radiation (8 mJ/cm2). Rabbit anti-γH2AX antibodies were used as primary antibody and Alexa 596 goat anti-rabbit as a secondary antibody (red). The nuclei were stained with Hoechst (blue). The bar represents 50 μm. B, left panel, Western blot analysis (40 μg of total cell extracts) of γH2AX in cells overexpressing the 34-kDa isoform of PIM-2 (+DOX), compared with (−DOX) control cells, at the indicated times post-UVC radiation (8 mJ/cm2). Anti-tubulin antibodies were used for equal loading control and normalization. Right panel, graphic representation of relative γH2AX levels, obtained from densitometric analysis of gels such as that depicted in the left panel. γH2AX levels in −DOX control cells 3 h post-UV were determined as 1. Graph summarizes data from three independent experiments. *, statistically significant differences in γH2AX levels between PIM-2-expressing cells and control cells (p < 0.05) at 6, 8, and 24 h. C, Western blot analysis (40 μg of total cell extracts) of γH2AX in cells overexpressing the 34-kDa isoform of PIM-2 (+DOX), compared with control cells (−DOX), under conditions of E2F-1 silencing (E2F1 siRNA) or NS siRNA. Actin antibodies were used for equal loading control and normalization.
DISCUSSION
The oncogenic nature ascribed to PIM-2 relies mostly on the pro-survival effect exerted by phosphorylation of substrates that upon phosphorylation act as pro-survival/anti-apoptotic factors. Nevertheless, pro-survival effects can also result from activating DNA repair mechanisms following damage. The possibility that PIM-2 is involved in the DDR has never been addressed. In this study, we found that endogenous PIM-2 expression (mainly the 34-kDa isoform) and activity are up-regulated soon after exposure to UV radiation but much less so after treatment with etoposide, suggesting that PIM-2 plays an intrinsic role in the cell response to UV-induced damage. This conclusion was further verified by the dramatic increase in the sensitivity of Pim-2-silenced cells to UV radiation and by the fact that both PIM-2 isoforms had a significant protective effect against UV-derived apoptosis. This protective effect could reflect reduced amount of UV-induced DNA lesions, as has been reported in pluripotent cells compared with differentiated fibroblasts exposed to equal intensities of UVC radiation (50), but it could also imply increased efficiency of the repair process in these cells. We showed that it is the kinetics of removal of the UV-induced DNA lesions that was faster in PIM-2-overexpressing cell versus control cell and in control cells versus Pim-2-silenced cells, suggesting that PIM-2 contributes to a more efficient removal of the lesions. This conclusion was further supported by the reduced γH2AX accumulation over time in PIM-2-overexpressing cells, suggesting reduced amounts of DSB. It is noteworthy, however, that the mechanisms by which Pim-2 expression and activity are up-regulated upon UV radiation remain an open question that needs further investigation. A clue to the answer of this question might be two consensus NF-κB binding sites in the promoter region of Pim-2 (−270 and −595 from the transcription start site), because it has been reported that transcription factor NF-κB is involved in mediating the UV response in mammalian cells (51).
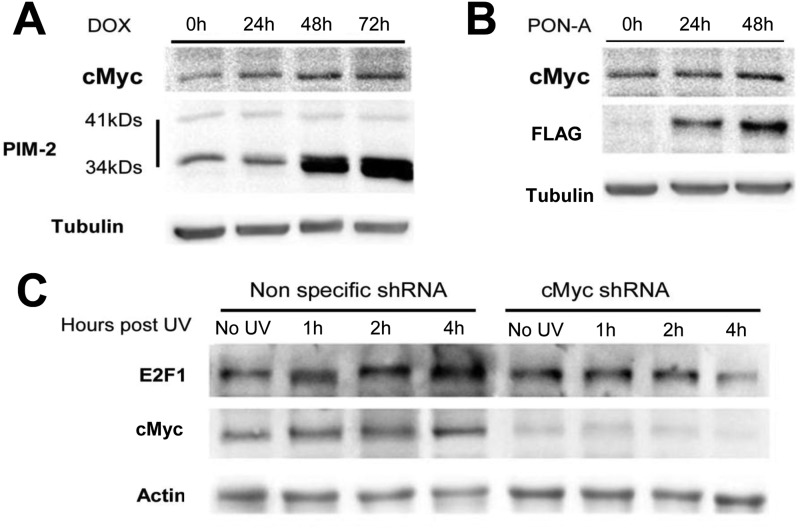
Our results suggest that PIM-2 exerts its protective effect through up-regulation of E2F-1, because the protective effect was greatly diminished in E2F-1-silenced cells. Previous reports have tied E2F-1 to the UV damage response. Epidermic cells from E2F-1 knock-out mice exhibit increased apoptosis upon exposure to UVB radiation, and apoptosis was suppressed in keratinocytes from transgenic mice overexpressing E2F-1, following UVB (43). E2F-1 levels were reported to rise following UV radiation and to be recruited to the UV-induced DNA damage sites, independent of the ability of E2F-1 to regulate transcription (44). Moreover, E2F-1 seems to be required for the recruitment of NER factors to the sites of UV damage because depletion of E2F-1 impairs the recruitment of these factors, leading to reduced efficiency of DNA repair (44). Thus, the unadvanced increase in E2F-1 levels shown in this study caused by PIM-2 overexpression, can, of course, contribute to faster recruitment of NER factors to the sites of UV damage and hence to a more efficient repair. What, then, regulates the increased levels of E2F-1 upon exposure to UV radiation? E2F-1 can be phosphorylated by ATM and ATR on Ser-31, phosphorylation that stabilizes it and can definitely contribute to its up-regulation (52, 53). This requires active ATM and/or ATR prior to E2F-1 up-regulation, and it is possible, therefore, that PIM-2-dependent activation of ATM and ATR (as shown in this study) contributes to E2F-1 up-regulation. However, we have shown in this study that PIM-2-mediated ATM activation depends, at least in part, on E2F-1, suggesting an additional mechanism for PIM-2-dependent up-regulation of E2F-1. An alternative explanation could be that PIM-2 activates transcription of E2F-1 by stabilizing c-Myc, a known positive regulator of E2F-1. Zhang et al. (32) have shown that PIM-2 phosphorylates c-Myc on Ser-329, leading to its stabilization and increasing its transcriptional activity. It was also reported that c-Myc is up-regulated upon exposure to UVC (54). These reports, together with our finding showing c-Myc up-regulation following PIM-2 overexpression or exposure to UVC radiation in our U2OS system (Fig. 8) and showing that E2F-1 is not up-regulated upon exposure to UVC in c-Myc-silenced cells (Fig. 8C), support the notion that c-Myc is a PIM-2 target upon exposure to UV radiation, leading to transcriptional activation of E2F-1.
FIGURE 8.
Increased c-Myc levels upon PIM-2 overexpression and UV radiation. A and B, Western blot analysis (40 μg of total cell extracts) of c-Myc levels in cells overexpressing either the 34-kDa isoform (1 μg/ml DOX for the indicated times) (A) or the 41-kDa isoform of PIM-2 (10 μm PON-A for the indicated times) (B). Anti-PIM-2 and anti-FLAG antibodies were used for detection of the 34- and 41-kDa isoforms, respectively. Anti-tubulin antibodies were used for equal loading control and normalization. C, Western blot analysis (40 μg of total cell extracts) of c-Myc and E2F-1 levels in cells either silenced for c-Myc (cMyc shRNA) or transfected with nonspecific shRNA as control, 1, 2, and 4 h post-UVC irradiation (50 mJ/cm2). Anti-actin antibodies were used for equal loading control and normalization.
Guo et al. (44) reported that recruitment of E2F-1 to UV damage sites is ATR-dependent. Stiff et al. (45) have reported that ATM activation following UV treatment does not rely on autophosphorylation of Ser-1981, but rather it is ATR-dependent. However, Yajima et al. (46) showed that ATM is important for the UV-induced damage response, mainly in later stages when DSB are formed because of collapse of the replication fork, and that ATM was activated in an ATR-independent manner. Our results reported herein demonstrate that in PIM-2-overexpressing cells activation of both ATM and ATR is intensified upon exposure to UV radiation and suggest that ATM plays a crucial role in the PIM-2-induced stability after exposure to UV. We do not know yet how exactly PIM-2 activates ATM and ATR. An intriguing possibility, based on our results indicating reduced ATM activation upon E2F-1 silencing, suggests that E2F-1 contributes to ATM activation. E2F-1 was reported to promote p53 phosphorylation and apoptosis in primary human fibroblasts through stimulating ATM activation (55). Moreover, E2F-1 was reported to be required for ATM activation in the DDR induced upon infection of cells by the human cytomegalovirus (56), supporting a role for E2F-1 in ATM activation in some contexts. As to ATR, in our PIM-2 overexpression experimental system it seems not to be required for ATM initial activation because ATM was preactivated even in unirradiated cells, whereas ATR was not. In fact, we could show that in PIM-2-overexpressing cells ATR intensified activation after UV radiation was eliminated in the presence of ATM inhibitors (not shown), suggesting that ATM contributes to ATR activation in this case. Thus understanding the mutual relationship between ATR and ATM in response to UV radiation needs further investigation.
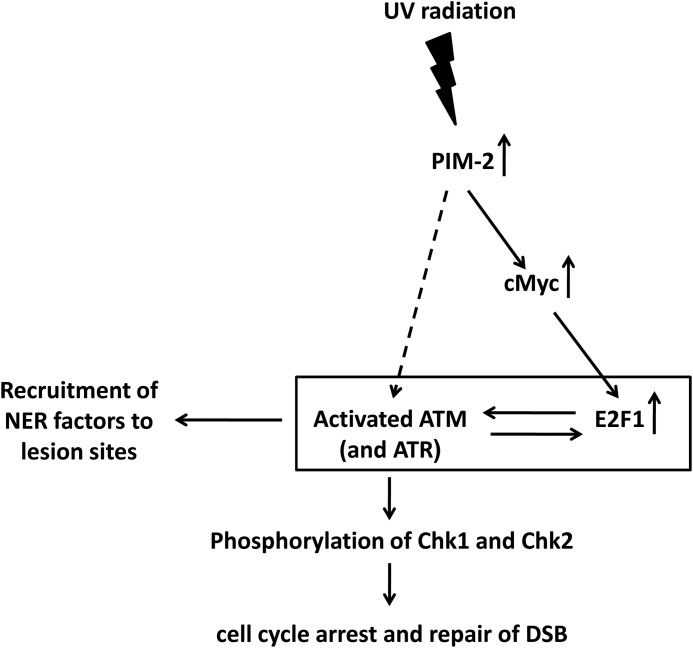
In conclusion, this study attributes an important role for PIM-2 in the cell response to UV-induced DNA damage and places it upstream to E2F-1 and ATM. Prealerting the response mechanism by overexpressing PIM-2 prior to UV radiation results in a more efficient removal of the induced DNA lesions and exertion of a protection effect. We propose a model in which PIM-2 induces activation of ATM, either by elevation of E2F-1 through c-Myc or by an as yet unidentified E2F-1-independent pathway, leading also to activation of ATR. Once ATM and ATR are activated, an amplification cycle is initiated where ATM and ATR can phosphorylate E2F-1 on Ser-31 and stabilize it, thus enabling its further contribution to the amplification cycle. This culminates in ATR- and E2F-1-dependent recruitment of NER factors to the damage sites, ATM- and ATR-dependent activation of the cell cycle checkpoints through Chk1 and Chk2, respectively, and repair of the late appearing DSB (Fig. 9). Finally, it seems that PIM-2 is not the only member of the PIM family that is involved in the DDR. Hsu et al. (57) have recently reported that knockdown of PIM-1, or inhibition of its activity, in human prostate cancer cells resulted in disruption of the nonhomologous end joining repair of DSBs, including disrupted activation of ATM.
FIGURE 9.
A proposed schematic model describing the E2F-1- and ATM-mediated involvement of PIM-2 in the UV-induced DDR.
Supplementary Material
Acknowledgments
We are grateful to Prof. Doron Ginsberg (Bar-Ilan University) for fruitful discussions and for providing aliquots of siRNAs targeting E2F-1. We thank Prof. Uri Nir (Bar Ilan University) and Prof. Yossi Shiloh (Tel Aviv University) for providing aliquots of the ATM-specific inhibitor KU60019 and anti-pKap1 antibodies, respectively. We also thank Dr. Yuval Ebenstein, Dr. Adar Makovski, Dana Bashari, and Orit Feldstein for fruitful discussions and technical help.
This study was supported by Grant 3-00000-5049 from the Weinkselbaum family medical research fund administered by the Israeli Ministry of Health Chief Scientist Research Fund.

This article contains supplemental Figs. S1–S5.
- DDR
- DNA damage response
- DSB
- double-strand break
- CPD
- cyclobutane-pyrimidine dimer
- NER
- nucleotide excision repair
- BAD
- BCL2-associated agonist of cell death
- DOX
- doxycycline
- PON-A
- ponasterone A
- PI
- propidium iodide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NS
- nonspecific.
REFERENCES
- 1. Shiloh Y. (2006) The ATM-mediated DNA-damage response. Taking shape. Trends Biochem. Sci. 31, 402–410 [DOI] [PubMed] [Google Scholar]
- 2. Downs J. A., Côté J. (2005) Dynamics of chromatin during the repair of DNA double-strand breaks. Cell Cycle 4, 1373–1376 [DOI] [PubMed] [Google Scholar]
- 3. Kitagawa R., Kastan M. B. (2005) The ATM-dependent DNA damage signaling pathway. Cold Spring Harb. Symp. Quant. Biol. 70, 99–109 [DOI] [PubMed] [Google Scholar]
- 4. Lavin M. F. (2007) ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26, 7749–7758 [DOI] [PubMed] [Google Scholar]
- 5. Paull T. T., Lee J. H. (2005) The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle 4, 737–740 [DOI] [PubMed] [Google Scholar]
- 6. Furuta T., Takemura H., Liao Z. Y., Aune G. J., Redon C., Sedelnikova O. A., Pilch D. R., Rogakou E. P., Celeste A., Chen H. T., Nussenzweig A., Aladjem M. I., Bonner W. M., Pommier Y. (2003) Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278, 20303–20312 [DOI] [PubMed] [Google Scholar]
- 7. Yajima H., Lee K. J., Chen B. P. (2006) ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell. Biol. 26, 7520–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 9. Takahashi A., Ohnishi T. (2005) Does gamma H2AX foci formation depend on the presence of DNA double strand breaks? Cancer Lett. 229, 171–179 [DOI] [PubMed] [Google Scholar]
- 10. Dunkern T. R., Kaina B. (2002) Cell proliferation and DNA breaks are involved in ultraviolet light-induced apoptosis in nucleotide excision repair-deficient Chinese hamster cells. Mol. Biol. Cell 13, 348–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pustisek N., Situm M. (2011) UV-radiation, apoptosis and skin. Coll Antropol. 35, 339–341 [PubMed] [Google Scholar]
- 12. Ravanat J. L., Douki T., Cadet J. (2001) Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B 63, 88–102 [DOI] [PubMed] [Google Scholar]
- 13. Sinha R. P., Häder D. P. (2002) UV-induced DNA damage and repair. A review. Photochem. Photobiol. Sci. 1, 225–236 [DOI] [PubMed] [Google Scholar]
- 14. Smith M. L., Chen I. T., Zhan Q., O'Connor P. M., Fornace A. J., Jr. (1995) Involvement of the p53 tumor-suppressor in repair of UV-type DNA-damage. Oncogene 10, 1053–1059 [PubMed] [Google Scholar]
- 15. Batista L. F., Kaina B., Meneghini R., Menck C. F. (2009) How DNA lesions are turned into powerful killing structures. Insights from UV-induced apoptosis. Mutat. Res. 681, 197–208 [DOI] [PubMed] [Google Scholar]
- 16. Batty D. P., Wood R. D. (2000) Damage recognition in nucleotide excision repair of DNA. Gene 241, 193–204 [DOI] [PubMed] [Google Scholar]
- 17. Halicka H. D., Huang X., Traganos F., King M. A., Dai W., Darzynkiewicz Z. (2005) Histone H2AX phosphorylation after cell irradiation with UV-B. Relationship to cell cycle phase and induction of apoptosis. Cell Cycle 4, 339–345 [PubMed] [Google Scholar]
- 18. Zhao H., Traganos F., Darzynkiewicz Z. (2010) Kinetics of the UV-induced DNA damage response in relation to cell cycle phase. Correlation with DNA replication. Cytometry A 77, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanasoge S., Ljungman M. (2007) H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis 28, 2298–2304 [DOI] [PubMed] [Google Scholar]
- 20. Marti T. M., Hefner E., Feeney L., Natale V., Cleaver J. E. (2006) H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proceedings of the National Academy of Sciences of the United States of America 103, 9891–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oh K. S., Bustin M., Mazur S. J., Appella E., Kraemer K. H. (2011) UV-induced histone H2AX phosphorylation and DNA damage related proteins accumulate and persist in nucleotide excision repair-deficient XP-B cells. DNA Repair 10, 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allen J. D., Berns A. (1996) Complementation tagging of cooperating oncogenes in knockout mice. Seminars in Cancer Biology 7, 299–306 [DOI] [PubMed] [Google Scholar]
- 23. Amson R., Sigaux F., Przedborski S., Flandrin G., Givol D., Telerman A. (1989) The human protooncogene product P33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc. Natl. Acad. Sci. U.S.A. 86, 8857–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Claudio J. O., Masih-Khan E., Tang H., Gonçalves J., Voralia M., Li Z. H., Nadeem V., Cukerman E., Francisco-Pabalan O., Liew C. C., Woodgett J. R., Stewart A. K. (2002) A molecular compendium of genes expressed in multiple myeloma. Blood 100, 2175–2186 [DOI] [PubMed] [Google Scholar]
- 25. Cohen A. M., Grinblat B., Bessler H., Kristt D., Kremer A., Schwartz A., Halperin M., Shalom S., Merkel D., Don J. (2004) Increased expression of the hPim-2 gene in human chronic lymphocytic leukemia and non-Hodgkin lymphoma. Leuk. Lymphoma 45, 951–955 [DOI] [PubMed] [Google Scholar]
- 26. Dai H., Li R., Wheeler T., Diaz de Vivar A., Frolov A., Tahir S., Agoulnik I., Thompson T., Rowley D., Ayala G. (2005) Pim-2 upregulation. Biological implications associated with disease progression and perinueral invasion in prostate cancer. Prostate 65, 276–286 [DOI] [PubMed] [Google Scholar]
- 27. Dhanasekaran S. M., Barrette T. R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2001) Delineation of prognostic biomarkers in prostate cancer. Nature 412, 822–826 [DOI] [PubMed] [Google Scholar]
- 28. Nawijn M. C., Alendar A., Berns A. (2011) For better or for worse. The role of Pim oncogenes in tumorigenesis. Nat. Rev. Cancer 11, 23–34 [DOI] [PubMed] [Google Scholar]
- 29. Wang Z., Bhattacharya N., Weaver M., Petersen K., Meyer M., Gapter L., Magnuson N. S. (2001) Pim-1. A serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J. Vet. Sci. 2, 167–179 [PubMed] [Google Scholar]
- 30. Yoshida S., Kaneita Y., Aoki Y., Seto M., Mori S., Moriyama M. (1999) Identification of heterologous translocation partner genes fused to the BCL6 gene in diffuse large B-cell lymphomas. 5′-RACE and LA-PCR analyses of biopsy samples. Oncogene 18, 7994–7999 [DOI] [PubMed] [Google Scholar]
- 31. Allen J. D., Verhoeven E., Domen J., van der Valk M., Berns A. (1997) Pim-2 transgene induces lymphoid tumors, exhibiting potent synergy with c-myc. Oncogene 15, 1133–1141 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y., Wang Z., Li X., Magnuson N. S. (2008) Pim kinase-dependent inhibition of c-Myc degradation. Oncogene 27, 4809–4819 [DOI] [PubMed] [Google Scholar]
- 33. Möröy T., Verbeek S., Ma A., Achacoso P., Berns A., Alt F. (1991) E mu N- and E mu L-myc cooperate with E mu pim-1 to generate lymphoid tumors at high frequency in double-transgenic mice. Oncogene 6, 1941–1948 [PubMed] [Google Scholar]
- 34. Yan B., Zemskova M., Holder S., Chin V., Kraft A., Koskinen P. J., Lilly M. (2003) The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J. Biol. Chem. 278, 45358–45367 [DOI] [PubMed] [Google Scholar]
- 35. Fox C. J., Hammerman P. S., Cinalli R. M., Master S. R., Chodosh L. A., Thompson C. B. (2003) The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 17, 1841–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hammerman P. S., Fox C. J., Cinalli R. M., Xu A., Wagner J. D., Lindsten T., Thompson C. B. (2004) Lymphocyte transformation by Pim-2 is dependent on nuclear factor-kappaB activation. Cancer Res. 64, 8341–8348 [DOI] [PubMed] [Google Scholar]
- 37. Ren K., Zhang W., Shi Y., Gong J. (2010) Pim-2 activates API-5 to inhibit the apoptosis of hepatocellular carcinoma cells through NF-κB pathway. Pathol. Oncol. Res. 16, 229–237 [DOI] [PubMed] [Google Scholar]
- 38. Wang Z., Zhang Y., Gu J. J., Davitt C., Reeves R., Magnuson N. S. (2010) Pim-2 phosphorylation of p21(Cip1/WAF1) enhances its stability and inhibits cell proliferation in HCT116 cells. Int. J. Biochem. Cell Biol. 42, 1030–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levy D., Davidovich A., Zirkin S., Frug Y., Cohen A. M., Shalom S., Don J. (2012) Activation of cell cycle arrest and apoptosis by the proto-oncogene Pim-2. PLoS One 7, e34736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morishita D., Katayama R., Sekimizu K., Tsuruo T., Fujita N. (2008) Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 68, 5076–5085 [DOI] [PubMed] [Google Scholar]
- 41. Potter A. J., Gollahon K. A., Palanca B. J., Harbert M. J., Choi Y. M., Moskovitz A. H., Potter J. D., Rabinovitch P. S. (2002) Flow cytometric analysis of the cell cycle phase specificity of DNA damage induced by radiation, hydrogen peroxide and doxorubicin. Carcinogenesis 23, 389–401 [DOI] [PubMed] [Google Scholar]
- 42. Thyagarajan B., Anderson K. E., Lessard C. J., Veltri G., Jacobs D. R., Folsom A. R., Gross M. D. (2007) Alkaline unwinding flow cytometry assay to measure nucleotide excision repair. Mutagenesis 22, 147–153 [DOI] [PubMed] [Google Scholar]
- 43. Berton T. R., Mitchell D. L., Guo R., Johnson D. G. (2005) Regulation of epidermal apoptosis and DNA repair by E2F1 in response to ultraviolet B radiation. Oncogene 24, 2449–2460 [DOI] [PubMed] [Google Scholar]
- 44. Guo R., Chen J., Zhu F., Biswas A. K., Berton T. R., Mitchell D. L., Johnson D. G. (2010) E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair. J. Biol. Chem. 285, 19308–19315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stiff T., Walker S. A., Cerosaletti K., Goodarzi A. A., Petermann E., Concannon P., O'Driscoll M., Jeggo P. A. (2006) ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 25, 5775–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yajima H., Lee K. J., Zhang S., Kobayashi J., Chen B. P. (2009) DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J. Mol. Biol. 385, 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bakkenist C. J., Kastan M. B. (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 48. Banin S., Moyal L., Shieh S., Taya Y., Anderson C. W., Chessa L., Smorodinsky N. I., Prives C., Reiss Y., Shiloh Y., Ziv Y. (1998) Enhanced phosphorylation of p53 by ATN in response to DNA damage. Science 281, 1674–1677 [DOI] [PubMed] [Google Scholar]
- 49. Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D. C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM and KAP-1 dependent pathway. Nat. Cell Biol. 8, 870–876 [DOI] [PubMed] [Google Scholar]
- 50. Luo L. Z., Gopalakrishna-Pillai S., Nay S. L., Park S. W., Bates S. E., Zeng X., Iverson L. E., O'Connor T. R. (2012) DNA repair in human pluripotent stem cells is distinct from that in non-pluripotent human cells. PLoS One 7, e30541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Devary Y., Rosette C., DiDonato J. A., Karin M. (1993) NF-κB activation by ultraviolet light not dependent on a nuclear signal. Science 261, 1442–1445 [DOI] [PubMed] [Google Scholar]
- 52. Lin W. C., Lin F. T., Nevins J. R. (2001) Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15, 1833–1844 [PMC free article] [PubMed] [Google Scholar]
- 53. Wang B., Liu K., Lin F. T., Lin W. C. (2004) A role for 14–3-3 tau in E2F1 stabilization and DNA damage-induced apoptosis. J. Biol. Chem. 279, 54140–54152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Alarcon-Vargas D., Tansey W. P., Ronai Z. (2002) Regulation of c-myc stability by selective stress conditions and by MEKK1 requires aa 127–189 of c-myc. Oncogene 21, 4384–4391 [DOI] [PubMed] [Google Scholar]
- 55. Powers J. T., Hong S., Mayhew C. N., Rogers P. M., Knudsen E. S., Johnson D. G. (2004) E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis. Mol Cancer Res. 2, 203–214 [PubMed] [Google Scholar]
- 56. Xiaofei E., Pickering M. T., Debatis M., Castillo J., Lagadinos A., Wang S., Lu S., Kowalik T. F. (2011) An E2F1-mediated DNA damage response contributes to the replication of human cytomegalovirus. PLoS Pathog. 7, e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hsu J. L., Leong P. K., Ho Y. F., Hsu L. C., Lu P. H., Chen C. S., Guh J. H. (2012) Pim-1 knockdown potentiates paclitaxel-induced apoptosis in human hormone-refractory prostate cancers through inhibition of NHEJ DNA repair. Cancer Lett. 319, 214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.