Abstract
Accurate energy functions are critical to macromolecular modeling and design. We describe new tools for identifying inaccuracies in energy functions and guiding their improvement, and illustrate the application of these tools to improvement of the Rosetta energy function. The feature analysis tool identifies discrepancies between structures deposited in the PDB and low energy structures generated by Rosetta; these likely arise from inaccuracies in the energy function. The optE tool optimizes the weights on the different components of the energy function by maximizing the recapitulation of a wide range of experimental observations. We use the tools to examine three proposed modifications to the Rosetta energy function: improving the unfolded state energy model (reference energies), using bicubic spline interpolation to generate knowledge based torisonal potentials, and incorporating the recently developed Dunbrack 2010 rotamer library (Shapovalov and Dunbrack, 2011).
Keywords: Rosetta, energy function, scientific benchmarking, parameter estimation, decoy discrimination
1 Introduction
Scientific benchmarks are essential for the development and parameterization of molecular modeling energy functions. Widely used molecular mechanics energy functions such as Amber and OPLS were originally parameterized with experimental and quantum chemistry data from small molecules and benchmarked against experimental observables such as intermolecular energies in the gas phase, solution phase densities, and heats of vaporization (Weiner et al. 1984, Jorgensen et al. 1996). More recently, thermodynamic measurements and high resolution structures of macromolecules have provided a valuable testing ground for energy function development. Commonly used scientific tests include discriminating the ground state conformation of a macromolecule from higher energy conformations (Novotný et al. 1984, Park & Levitt, 1996, Simons et al., 1999), predicting amino acid sidechain conformations (Jacobson et al. 2002, Bower et al. 1997), and predicting free energy changes associated with protein mutations (Guerois et al. 2002, Gilis and Rooman 1997, Potapov et al. 2009).
Many studies have focused on optimizing an energy function for a particular problem in macromolecular modeling, for instance the FoldX energy function was empirically parameterized for predicting changes to the free energy of a protein when it is mutated (Guerois et al. 2002). Often, these types of energy functions are only well suited to the task they have been trained on. Kellog et al. showed that an energy function explicitly trained to predict energies of mutation did not produce native-like sequences when redesigning proteins (Kellogg et al. 2011). For many projects it is advantageous to have a single energy function that can be used for diverse modeling tasks. For example, protocols in the molecular modeling program Rosetta for ligand docking (Meiler and Baker, 2003), protein design (Kuhlman et al., 2003), and loop modeling (Wang et al., 2007) share a common energy function, which allowed Murphy et al. (2009) to combine them to shift an enzyme’s specificity from other substrate to another.
Sharing a single energy function between modeling applications presents both opportunities and challenges. Researchers applying the energy function to new tasks sometimes uncover deficiencies in the energy function. The opportunities are that correcting the deficiencies for the new tasks will result in improvements for the older tasks–after all, nature uses only one energy function. Sometimes, however, modifications to the energy function that improve performance at one task degrade its performance at others. The challenges are then to discriminate beneficial from deleterious modifications and reconcile task-specific objectives.
To address these challenges, we have developed three tools based on benchmarking Rosetta against macromolecular data. The first tool (Section 3), a suite we call “feature analysis”, can be used to contrast ensembles of structural details from structures in the PDB and from structures generated by Rosetta. The second tool (Section 4), a program we call “optE”, relies on fast, small-scale benchmarks to train the weights in the energy function. These two tools can help identify and fix flaws in the energy function, thus facilitating the process of integrating a proposed modification. We follow the description of these tools (Section 5) with a curated set of large-scale benchmarks meant to provide sufficient coverage of Rosetta’s applications. Use of these benchmarks will give evidence that a proposed energy function modification should be widely adopted.
To conclude in Section 6, we demonstrate our tools and benchmarks by evaluating three incremental modifications to the Rosetta energy function. First, we use the sequence profile recovery within OptE to refit the unfolded state model of Rosetta’s standard energy function, Score12. The resulting Score12′ improves sequence recovery performance. Second, we use feature analysis to identify and correct an artificial accumulation of predicted backbone torsion angles on the 10° grid lines caused by discontinuities in the derivative of the energy function. Rosetta predicted 23% of all ϕ, ψ pairs lying within .05° of a grid line. The resulting Score12Bicubic is not worse than Score12 on the prediction benchmarks. Third, we compare the 2002 version of the Dunbrack backbone-dependent sidechain rotamer library (Dunbrack, 2002), Dun02, with the 2010 version (Shapovalov and Dunbrack, 2011), Dun10. The Dun10 library improves rotamer recovery, but gives mixed results on the other prediction benchmarks.
Alongside this chapter, we have collected documentation for these two tools, and, for the benchmarks the set of input files and instructions on how to run them. They may be downloaded from URLHERE!!!!
2 Energy Function Model
The Rosetta energy function is a linear combination of terms that model interaction forces between atoms, solvation effects, and torsion energies. More specifically, Score12, (Rohl et al. 2004), the default fullatom energy function in Rosetta is composed of a Lennard-Jones term, an implicit solvation term (Lazaridis & Karplus, 1999), an orientation-dependent hydrogen bond term (Kortemme et al., 2003), sidechain and backbone torsion potentials derived from the PDB, a short-ranged knowledge-based electrostatic term, and reference energies for each of the 20 amino acids that model the unfolded state. Formally, given a molecular conformation, C the total energy of the system is given by
| (1) |
where each energy term Tj has parameters Θj and weight wj. The feature analysis tool described in Section 3, is meant to aid the refinement of the parameters Θ. The optE program described in Section 4, is meant to fit the weights, w.
3 Feature Analysis
Our aim is to facilitate the analysis of distributions of measurable properties of molecular conformations, which we call “feature analysis”. By formalizing the analysis process, we are able to create a suite of tools and benchmarks that unify the collection, visualization, and comparison of feature distributions. After motivating our work, we will describe the components (Section 3.1) and illustrate how they can be integrated into a workflow (Section 3.2) by investigating the distribution of the lengths of hydrogen bonds with hydroxyl donors.
Feature distributions, broadly construed, have long held a prominent role in structural biochemistry. The relationship between probability and energy given by the Boltzmann equation has justified the derivation of knowledge-based potentials for modeling protein energetics (Miyazawa & Jernigan, 1985, Sippl, 1990), and the implementation of knowledge-based potentials usually involves the extraction of feature distributions from protein structures. Feature distributions, for example, are central to the creation of rotamer libraries (Ponder & Richards, 1986; Dunbrack and Karplus, 1993; Lovel et al., 2000). The Boltzmann equation also motivates the need to compare the feature distributions from structures generated by an energy function against those observed in nature: for an energy function to generate geometries that are rarely observed is synonymous with that energy function wrongly assigning low energies to high-energy geometries. Many structure validation tools, such as MolProbity (Chen, et al. 2010), look for outlier features as indications of errors in the structure.
The Rosetta community, too, has a tradition of analyzing feature distributions: the extraction of features from protein structures is central to the derivation of Rosetta’s low-resolution (Simons et al., 1999) and high-resolution (Kortemme et al., 2003) knowledge-based potentials and for the analysis of packing in protein cores (Sheffler & Baker, 2009) and hydrophobic patches on protein surfaces (Jacak et al., 2012), to list only a few. However, each foray into feature analysis has been performed with ad hoc scripts that have not been preserved, preventing researchers from readily replicating the analyses of other groups. Our primary goal was to create a unified framework for feature analysis.
Feature analysis also provides a means to tune the parameters of an energy function. Recently, Song et al. (2011) observed peaks in the backbone ϕ, ψ distributions of Rosetta generated structures, absent from the ϕ, ψ distributions in crystal structures, which they attributed to double counting in knowledge-based potentials. In one case, a commonly observed loop motif has a hydrogen bond between the asparagine sidechain of residue i and the backbone of residue i+2, constraining the backbone ψ angle of residue i to 120°. The proliferation of this motif caused an artifact in Rosetta’s knowledge-based Ramachandran energy term, since E(ψ=120 | ASN) ~ −log(P(ψ=120 | ASN)), irrespective of hydrogen bond formation. To correct this, they considered the Ramachandran term as a parametric model and iteratively tuned the parameters until the asparagine ψ distribution from predicted structures matched the distribution from crystal structures. Hamelryck et al. (2010) have also observed that this iterative process is a useful way to improve an energy function. Our second major goal for the feature analysis tool is to facilitate this process of parameter tuning.
3.1 Feature Analysis Components
The feature analysis framework consists of two components. Feature reporters take a set of structures, a batch, and populate a relational database with feature data.. Next, feature analysis scripts select, estimate, visualize, and compare feature distributions from the database.
3.1.1 Features Database
To facilitate the analysis of a large number of feature distributions, we have created an relational database architecture for feature data. Typically when analyzing feature distributions, we decompose a basic feature distribution (e.g., the hydrogen–acceptor distance in hydrogen bonds) into many conditional feature distributions (e.g., hydrogen-acceptor distances in carboxylate/guanidine hydrogen bonds). By putting basic features into a relational database along with other supporting data, we can perform the expensive task of extracting features from some input batch of structures only once, while retaining the ability to examine arbitrary conditional feature distributions in the future.
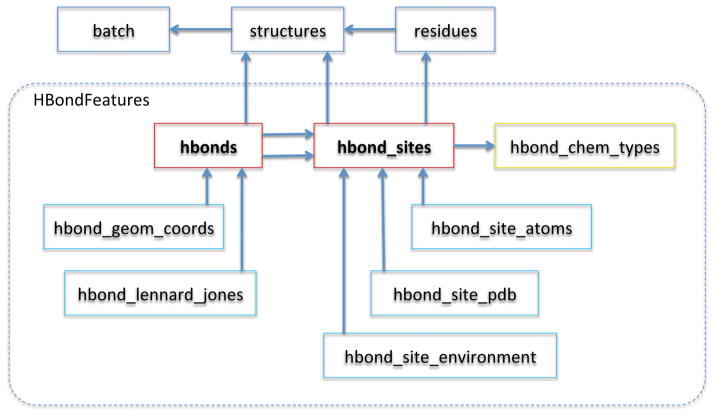
The database schema that we have built is structured as a hierarchy with the highest level tables holding very general data describing the structures the database was populated from, and with the lowest level tables describing basic features. For example, the tables describing hydrogen bond properties include foreign-key references to the more general residues and structures tables (Figure 1).
Figure 1. HBondFeatures Database Schema.
Boxes represent tables in the database and arrows represent foreign-key dependencies. The HBondFeatures class populates these tables with hydrogen bond data. For each hydrogen bond site (an acceptor atom, or a polar hydrogen), atomic coordinates, experimental data and solvent environment are reported. For each hydrogen bond that forms between two hydrogen bond sites, the geometric properties (i.e., distances and angles) and the sum of the Lennard-Jones energies for the atoms involved are also reported.
Each feature database is meant to hold the features for a single batch of conformations. Once a batch has been selected, the features of that batch can be extracted and inserted into the database using the RosettaScripts interface (Fleishman et al., 2011). The RosettaScripts interface allows components of Rosetta to be assembled from an XML file into linear protocols where each stage of the protocol is represented by a “mover.” The protocol begins when a structure is read in, and then proceeds as each mover is applied in turn to the structure. Movers can be anything, and in our case, we use a mover, the ReportToDB mover, to report data about each structure into a database. The ReportToDB mover creates the database session and initializes the database schema. When it is declared in the XML file, it is given a list of feature reporters, (See Table 1) which are responsible for defining tables and for inserting features for a given structure into those tables. This interface allows researches to populate their databases with only the features they are interested in; it also allows them to report features at any point or at multiple points in the middle of RosettaScripts protocol. Feature extraction is robust, in that it supports multiple database backends (SQLite, PostgreSQL and MySQL), database sharing and merging through universally unique identifiers, and incremental feature extraction.
Table I.
Each FeatureReporter is responsible for extracting a particular structural feature from a structure and reporting it to a relational database. The FeatureReporters that are currently implemented are in black while some FeatureReporters that would be interesting to implement in the future are in gray.
| FeatureReporter Classes |
|---|
| Meta |
| Protocol |
| Batch |
| JobData |
| PoseComments |
| Experimental Data |
| PdbData |
| PdbHeaderData |
| DDG |
| NMR |
| DensityMap |
| MultiSequenceAlignment |
| HomologyAlignment |
| Chemical |
| AtomType |
| ResidueType |
| One Body |
| Residue |
| ResidueConformation |
| ProteinResidueConformation |
| ProteinBackboneTorsionAngle |
| ResidueBurial |
| ResidueSecondaryStructure |
| GeometricSolvation |
| BetaTurn |
| RotamerBoltzmannWeight |
| ResidueStrideSecondaryStructure |
| HelixCapping |
| BondGeometry |
| ResidueLazaridisKarplusSolvation |
| ResidueGeneralizedBornSolvation |
| ResiduePoissonBoltzmannSolvation |
| Pka |
| ResidueCentroids |
| Two Body |
| Pair |
| AtomAtomPair |
| AtomInResidue- |
| AtomInResiduePair |
| ProteinBackbone- |
| AtomAtomPair |
| HBond |
| Orbital |
| SaltBridge |
| LoopAnchor |
| DFIREPair |
| ChargeCharge |
| Multi Structure |
| ProteinRMSD |
| ResidueRecovery |
| ResiduePairRecovery |
| ResidueClusterRecovery |
| Cluster |
| Multi Body |
| Structure |
| PoseConformation |
| RadiusOfGyration |
| SecondaryStructure |
| HydrophbicPatch |
| Cavity |
| GraphMotif |
| SequenceMotif |
| Rigidity |
| VoronoiPacking |
| InterfaceAnalysis |
| Energy Function |
| ScoreFunction |
| ScoreType |
| StructureScores |
| ResidueScores |
| HBondParameters |
| <EnergyTerm>Parameters |
3.1.2 Distribution Analysis
The second part of the feature-analysis suite provides tools to query feature databases, to transform features in order to correctly estimate feature distributions, and to plot those distributions. The feature analysis scripts are released with Rosetta and may be found in the rosetta/rosetta_tests/features directory.
The primary entry point when performing feature analyses is the compare_sample_sources.R script. In running the script the user provides a JSON analysis configuration file specifying a set of feature databases (extracted from separate structure batches), the analysis scripts to run, and the output formats. This wrapper script prepares the database connections and ensures that the necessary R packages have been installed on the system before it hands control-of-flow to the specified feature-analysis scripts. Feature analysis scripts usually consists of three parts: an SQL query to retrieve features from the input databases, a kernel density estimation on the extracted features (or on transformed features), and the creation of a plot using the ggplot2 grammar-of-graphics package in R.
Feature analysis scripts begin by querying the input sample sources using one or more SQL statements, ending with a SELECT statement. It is within these SQL queries that data across multiple tables can be joined to compile the data for arbitrarily complicated conditional feature distributions. The result of a typical query is a table where each row represents a feature instance, where some of the columns identify the feature (including which batch it came from along with other covariates), and where the other columns measure the feature in the feature space.
Once the features and their identifying data have been retrieved, density distributions can be computed. To do this, the feature analysis scripting framework uses the split-apply-combine strategy (Wickham, 2011): feature instances are grouped by their identifying columns, and for each group, a kernel density estimation (KDE) is computed over the measured columns. When computing density estimations over feature spaces, care must be taken that an appropriate transformation is applied to normalize the space and to handle boundary conditions. Since data normalization and boundary-reflection are common tasks in the performing feature analyses, our framework provides normalization routines for common transformations and boundary-reflection routines. It also provides strategies for selecting an appropriate kernel bandwidth, which controls the smoothness of the estimated distribution.
Once a collection of feature distributions have been estimated, they can be visualized through the grammar of graphics method (Wilkinson, 1999; Wickham, 2010). Here, a plot conceptually maps the dimensions of the data to graphical dimensions on a page, using elements such as location, color, size, and shape. This has two principal advantages: first, plot specification becomes concise and expressive, allowing rapid inspection of different visualizations. Second, it forces the separation of the data from the plotting. The latter is essential for the future reusability of a feature-analysis script.
The scripting framework also provides support for other types of feature-analysis tasks. For example, the results from prediction benchmarks such as RotamerRecovery can be encoded as feature instances and analyzed with other features as covariates using decision trees, regression, etc. Feature instances can also be aligned and exported to a PyMOL session for interactive inspection.
3.2 Feature Analysis Workflow
Feature analysis has three common stages: sample generation, feature extraction, and distribution comparison. Feature analysis can be used to optimize the energy function parameters by iteratively modifying the energy function and comparing the feature distributions from structures generated with the new energy function against those from crystal structures.
For a demonstration of this iterative process, we consider the hydrogen bond length (defined as the hydrogen-acceptor distance) for hydroxyl donors (i.e. serine and threonine). In X-ray crystal structures of proteins, the most favorable distance is 1.65 Å, while for non-hydroxyl donor H-bonds the most favorable distance is 1.85 Å. Rosetta does not correctly recapitulate this distance distribution, most likely due to the way in which the Rosetta hydrogen bond term was originally derived. Previously, to avoid the problem of inferring the hydroxyl hydrogen locations, the hydroxyl donor parameters were taken from the sidechain amide and carboxylamide donor parameters (Kortemme et al., 2003).
To start, we compared structures generated from Rosetta’s existing energy function against native structures to verify that Rosetta does not generate the correct distribution of hydroxyl hydrogen bond distances. We used as our reference source a subset of the top8000 chains dataset (Keedy et al., 2012) with a maximum sequence identity of 70%, which gave 6,563 chains. Hydrogen atom coordinates were optimized using Reduce (Word et al., 1999). We further restricted our investigation to residues with B-factors of at most 30. In the remainder of this chapter we call this the Native sample source. Then, for each candidate energy function, we relaxed each protein chain with the FastRelax protocol in Rosetta (Khatib et al., 2011).
The analysis script to look for a discrepancy for hydroxyl donor hydrogen bond lengths has three parts. First, for each sample source, the acceptor-hydrogen distance for all hydrogen bonds with non-aromatic ring hydroxyl donors (i.e., serine and theonine) with protein backbone acceptors is extracted. Second, for the instances associated with each sample source, a one-dimensional kernel density estimation is constructed normalizing for equal volume per unit length. Also, for each non-reference sample source, the Boltzmann distribution for the A-H distance term in the hydrogen bond energy term is computed. Third, the density distributions are plotted.
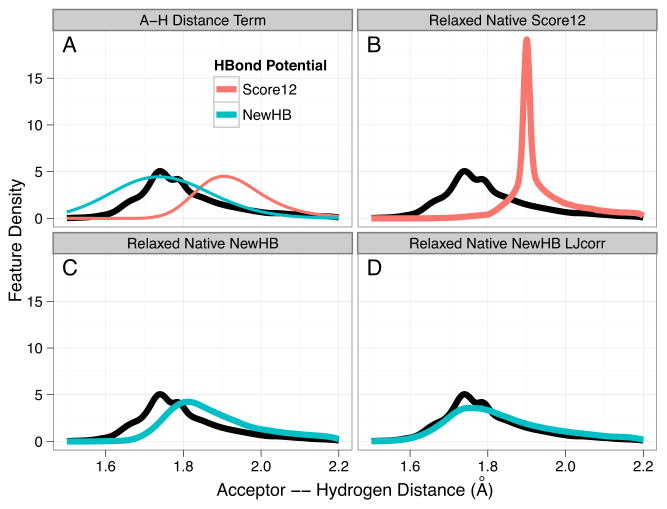
With this distribution analysis script in place, we evaluated two incremental modifications to the standard Score12 hydrogen bond term. The first, NewHB, among other modifications, adjusts parameters for the A–H Distance term for these hydrogen bond interactions so that the optimal location is consistent with the location of the peak in the Native sample source (panel A in Figure 3). As expected, this shifts the distribution of predicted hydrogen bonds towards shorter interactions. However, the predicted distribution does not move far enough to recapitulate the observed distribution (panel B to panel C in Figure 3). In Score12, the Lennard-Jones energy term between the donor and acceptor oxygen atoms has optimal energy when they are 3 Å apart. However, the peak in the O-O distance distribution is at 2.6 Å. Thus hydrogen bonds with the most favorable distance according to the NewHB parametrization experience a strong repulsion from the Lennard-Jones term. To reduce correlation between the hydrogen bond and the Lennard-Jones energy terms, we decreased the optimal distance for the Lennard-Jones term for these specific interactions to 2.6 Å. With this second modification, Rosetta recapitulates the native distribution (panel D in Figure 3).
Figure 3. Hydrogen bond A–H Distance Distributions for Hydroxyl Donors (SER/THR) to Backbone Oyxgens.
The thick curves are kernel density estimations from observed data normalized for equal volume per unit distance. The black curve in the background of each panel represents the Native sample source. (A) Boltzmann distribution for the A–H distance term in the Rosetta H-bond model with the Score12 and NewHB parameterizations. (B) Relaxed Natives with the Score12 energy function. The excessive peakiness is due to a discontinuity in the Score12 parametrization of the H-bond model. (C) Relaxed Natives with the NewHB energy function. (D) Relaxed Natives with the NewHB energy function and the Lennard-Jones minima between the acceptor and hydroxyl heavy atoms adjusted from 3.0 Å to 2.6 Å, and between the acceptor and the hydrogen atoms adjusted from 1.95 Å to 1.75 Å.
4 Maximum Likelihood Parameter Estimation with OptE
Recall that the Rosetta energy function is a weighted linear combination of energy terms that capture different aspects of molecular structure, as defined in Eq. (1). The weights, w, balance the contribution of each term to give the overall energy. Because the weights often need to be adjusted after modifying an energy term, we have developed a tool called “optE” to facilitate fitting them against scientific benchmarks. The benchmarks are small, tractable tests of the ability of Rosetta to recapitulate experimental observations given a particular assignment of weights. Although the weight sets that optE generates have not proven to be good replacements for the existing weights in Rosetta, we have found optE useful at two tasks: at identifying problems in the Rosetta energy function and at fitting the twenty amino-acid-reference-energy weights.
In the next section, we give a formula for generic likelihood-based loss functions and then describe the scientific benchmarks that are available in optE.
4.1 Loss Function Models
The full-scale scientific benchmarks described in Section 5 are sufficiently computationally demanding that they require several thousand CPU hours to run. Many of these benchmarks, however, can be scaled down to forms that are efficiently evaluated and can be used to train the energy function. Speed is an important factor for determining which benchmarks are included in optE, because optE evaluates tens of thousands of weight combinations while searching through weight space. A benchmark that takes more than a second is too expensive to include.
As mentioned in the introduction, a scientific benchmark should measure Rosetta’s ability to recapitulate experimental observations. To jointly optimize an energy function’s performance at the scientific benchmarks, we require success at each benchmark to be reported as a single number, which is called the loss. For scientific benchmarks based on recapitulating experimental observations, a common method of defining the loss is, given the weights, the negative log-probability of predicting the observed data. If the observed data are assumed to be independently sampled, the loss is the sum of the negative log-probability over all observations. Thinking of the loss as a function of the weights for a fixed set of observations, it is called the negative log-likelihood of the weights. An ideal prediction protocol will generate predictions according to the Boltzmann distribution for the energy function10. Therefore, the probability of an observation o is
| (2) |
| (3) |
where the partition function, Z(w) includes o and all possible alternatives, A. Because of the vast size of conformation space, computing Z is often intractable; this is a common problem for energy-based loss functions (LeCun & Jie, 2005). To address this problem, we rely on loss functions that do not consider all possible alternatives.
4.1.1 Recovering Native Sequences
Within the Rosetta community, we have observed that improvements to the energy function, independently conceived to fix a particular aspect of Rosetta’s behavior, have produced improvements in sequence recovery when redesigning naturally occurring proteins (Kuhlman & Baker, 2000, Morozov & Kortemme, 2005). Therefore, we attempt to increase sequence recovery to improve the energy function.
The standard sequence recovery benchmark (described in Section 5.2) looks at the fraction of the amino acids recovered after performing complete protein redesign using Rosetta’s Monte Carlo optimization technique. To turn this benchmark into a loss function like Eq. (2) would require us to compute the exact solution to the NP-Complete sidechain optimization problem (Pierce & Winfree, 2002) for the numerator, and, for an N residue protein, to repeat that computation 20N times for each possible sequence for the denominator. This is not feasible.
Instead, we approached the benchmark as a one-at-a-time optimization, maximizing the probability of the native amino acid at a single position given some fixed context. This introduces a split between the energy function whose weights are being modified to fit the native amino acid into a particular environment and the energy function which is holding that environment together. Upweighting one term may help distinguish the native amino acid from the others, but it might also cause the rest of the environment to relax into some alternate conformation in which the native amino acid is no longer optimal. To build consistency between the two energy functions, we developed an iterative protocol (described in greater detail in Section 4.2.1) that oscillates between loss-function optimization and full-protein redesign. Briefly, the iterative protocol consists of a pair of nested loops. In the outer loop, the loss function is optimized to produce a set of candidate weights. In the inner loop, the candidate weights are mixed in various proportions with the weights from the previous iteration through the outer loop, and for each set of mixed weights, complete protein redesign is performed. The redesigned structures from the last iteration through the inner loop are then used to define new loss functions for the next iteration through the outer loop.
We define the loss function for a single residue by the log-likelihood of the native amino acid defined by a Boltzmann distribution of possible amino acids at that position. We call this the PNatAA loss function.
| (4) |
| (5) |
where E(nat|w) is the energy of the best rotamer for the native amino acid and E(aa|w) is the energy of the best rotamer for amino acid aa. Rotamers are sampled from Roland Dunbrack’s backbone-dependent rotamer library from 2002 (Dunbrack, 2002), with extra samples taken at ±σ for both χ1 and χ2. The best rotamer depends on assigned weights, meaning that this function has discontinuous derivatives when two rotamers tie for the best. The energies for the rotamers at a particular position are computed in the context of a fixed surrounding; the contexts for the outer-loop iteration i are the designed structures from iteration i−1, where the first round’s context comes from the initial crystal structures.
4.1.2 Recovering Native Rotamers
As is the case for the full fledged sequence-recovery benchmark, the rotamer-recovery benchmark (described in Section 5.1) would be intractably expressed as a generic log-likelihood loss function as given in Eq. (2). Instead, we again approach the recovery test as a one-at-a-time benchmark to maximize the probability of the native rotamer at a particular position when considering all other rotamer assignments.
For residue j, the probability of the native rotamer is given by
| (6) |
where E(nat|w) is the energy of the native rotamer and the set rots contains all other rotamers built at residue j. We define a loss function, LPNatRot, for residue j as the negative log of this probability.
4.1.3 Decoy Discrimination
Benchmarking the ability of Rosetta to correctly predict protein structures from their sequences is an incredibly expensive task. In the high-resolution refinement benchmark (described in Section 5.4), non-native structures, decoys, are generated using the energy function being tested so that each decoy and each near-native structure will lie at a local minimum, but this takes ~20K CPU hours. Within optE, we instead test the ability of Rosetta to discriminate near-native structures from decoys looking only at static structures; as optE changes the weights, the property that each structure lies at a local minimum is lost.
Given a set N of relaxed, near-native structures for a protein, and a set D of relaxed decoy structures, we approximate the probability of the native structure for that protein as:
| (7) |
| (8) |
and similarly define a decoy-discrimination loss function, LPNatStruct, as the negative log of this probability. Here, nj is the “nativeness” of conformation j which is 1 if j is below 1.5 Å Cα RMSD from the crystal structure, and 0 if it is above 2 Å RMSD. The nativeness decreases linearly from 1 to 0 in the range between 1.5 and 2. Similarly, dj is the “decoyness” of conformation j which is 0 if j is below 4 Å RMSD and 1 otherwise. σ is a dynamic-range-normalization factor that prevents the widening of an energy gap between the natives and the decoys by scaling all of the weights; it is defined as σ = σ(D,w)/σ0 where σ(D,w) is the computed standard deviation for the decoy energies for a particular assignment of weights and σ0 is the standard deviation of the decoy energies measured at the start of the simulation.
In the partition function, the Rlnα− Σkdk term approximates the entropy of the decoys, an aspect that is otherwise neglected in a partition function that does not include all possible decoy conformations. This term attempts to add shadow decoys to the partition function, and the number of extra decoys added scales exponentially with the length of the chain, R. We chose 1.5 as the scale factor, α, which is relatively small given the number of degrees of freedom (DOFs) each residue adds. Counting torsions alone, there are between 3 (glycine) and 7 (arginine) extra DOFs per residue. Our choice of a small α is meant to reflect the rarity of low-energy conformations. To normalize between runs which contain differing numbers of far-from-native decoys, we added the −Σkdk term; doubling the number of decoys between two runs should not cause the partition function to double in value.
4.1.4 ΔΔG of mutation
The full benchmark for predicting ΔΔGs of mutation (described in Section 5.3) is computationally expensive, and so, similar to the decoy-discrimination loss function, we define a ΔΔG loss function which relies on static structures. This loss function is given by:
| (9) |
where muts is a set of structures for the mutant sequence, wts is a set of structures for the wild type sequence, and the experimentally observed ΔΔG is defined such that it is positive if the mutation is destabilizing and negative if it is stabilizing. Note that this loss function is convex if there is only one structure each in the muts and wts sets. This is very similar to the linear least-squares fitting, except that it is limited to a slope of one and a y-intercept of zero. The slope can be fit by introducing a scaling parameter to weight optimization, as described in Section 4.2.2 below.
4.2 Loss Function Optimization
OptE uses a combination of a particle swarm minimizer (Chen et al., 2007) and gradient-based minimization to optimize the loss function. Non-convexity of the loss function prevents perfect optimization, and independent runs sometimes result in divergent weight sets that have similar loss function values. In spite of this problem, optE tends to converge on very similar weight sets at the end of training.
4.2.1 Iterative Protocol
As described above, training with the PNatAA loss function effectively splits the energy function into two which we attempt to merge with an iterative procedure where we oscillate between loss-function optimization in an outer loop and complete protein redesign in an inner loop. Between rounds of the outer-loop, the weights fluctuate significantly, and so in each iteration of the inner loop, we create a weight set that is a linear combination of the weights resulting from round i’s loss-function optimization and the weight set selected at the end of round i−1. The weight set used for design during outer-loop iteration i, inner-loop iteration j is given by:
| (10) |
| (11) |
where wl(i) is the weight set generated by minimizing the loss function in round i, and w(i−1) is the final weight set from round i−1. In the first round, w(1) is simply assigned wl(1). This inner loop is exited if the sequence recovery rate improves over the previous round or if six iterations through this loop are completed. The weight set w(i,j) for the last iteration through the inner loop taken as the weight set w(i) for round i and is written to disk. The set of designed structures from this iteration are taken to serve as the context for the PNatAA loss function in the next round.
4.2.2 Extra Capabilities
OptE provides extra capabilities useful for exploring weight space. For instance, it is possible to weight the contributions of the various loss functions differently. Typically, we upweight the decoy-discrimination loss function by 100 when optimizing it along with the PNatAA and PNatRot loss functions. We typically train with ~100 native-set/decoy-set pairs to train on, compared to several thousand residues from which we can define PNatAA and PNatRot loss functions. Upweighting the PNatStruct loss function prevents it from being drowned out.
OptE also offers the ability to fit two or more terms with the same weight, or to obey an arithmetic relationship as specified in an input text file. This feature was used to scale the Score12 terms by a linear factor but otherwise keep them fixed to optimize the ΔΔG of the mutation loss function (Kellogg et al., 2011). Finally, OptE allows the definition of restraints for the weights themselves to help hold them to values the user finds reasonable. This is often useful because loss functions are perfectly happy to assign negative weights to the terms in the energy function.
4.3 Energy Function Deficiencies Uncovered by OptE
OptE is particularly good at two tasks: refitting reference energies (described in Section 4.5 below) and uncovering areas where the existing Rosetta energy function falls short. Rosetta’s efficiency in searching through conformation space means it often finds decoys with lower energies than the native. Such decoys surely point to flaws in Rosetta’s energy function, however, it is not always easy to see why the natives are not at lower energy than these decoys. OptE allows efficient hypothesis driven testing: hypothesize what kind of term is absent from the Rosetta energy function, implement that term, and test that term in optE using the PNatStruct loss function. If the value of the loss function improves after the new term is added, that is strong evidence the term would improve the energy function. There are caveats, of course, because the PNatStruct loss function relies on Rosetta-relaxed native structures, some of the features present in the crystal structures might have already been erased before optE gets started.
Using optE, we found two terms that improved the decoy-discrimination loss function over Score-12. The first was a carbon-hydrogen bond potential added to Rosetta (but not included as part of Score-12) by Rhiju Das to help model RNA (Das et al., 2010). This potential was derived from a set of protein crystal structures and a set of decoy protein structures as the log of the difference in the probability of an oxygen being observed at a particular distance in a crystal structure and its probability of being observed at that distance in the decoy structures. OptE identified this term as strongly improving decoy discrimination. We followed this lead by splitting the potential into backbone/backbone, backbone/sidechain, and sidechain/sidechain contributions. Here, optE preferred to set the weight on the sidechain/sidechain and backbone/sidechain components to zero while keeping the weight on the backbone/backbone interactions high. This left exactly one interaction: the Hα hydrogen interacting with the carbonyl oxygen. This contact is observed principally in β-sheets and has been reported previously as giving evidence for a carbon hydrogen bond (Taylor & Kennard, 1982; Fabiola et al., 1997).
The original CH-bond potential proved to be a poor addition to the energy function: when used to generate new structures or to relax natives, previously observed deep minima at low RMSD (<1.5 Å) from the crystal structure were lost and were instead replaced by broad, flat, near-native minima that reached out as far as 4 Å. To improve the potential, Song et al. (2011) iteratively adjusted the parameters to minimize the difference between the observed and predicted structures. This iterative process resulted in better Hα-O distance distributions; unexpectedly, it also resulted in better distance distributions for other atom-pairs in β-sheets.
A second term identified by optE was a simple Coulombic electrostatic potential with a distance-dependent dielectric (Yanover & Bradley, 2011). After an initial signal from optE suggested the importance of this term, we again separated the term into backbone/backbone, backbone/sidechain and sidechain/sidechain components. Again, the backbone/backbone portion showed the strongest signal. From there, we separated each term into attractive and repulsive components, and optE suggested that the repulsive backbone/backbone interaction contributed the most toward improved decoy discrimination. OptE also identified which low-energy decoys in the training set were problematic in the absence of the electrostatic term. By hand, we determined that the loops in these decoys contained too-close contacts between backbone carbonyl oxygens: they were only ~3.4 Å apart, which is closer than is commonly observed in crystal structures.
4.4 Limitations
Though our original goal was to fit all the weights simultaneously, optE has not proven exceptionally useful at that task. There are a number of factors that contribute to optE’s failures here. For one, the loss function that we use is not convex, and therefore its optimization cannot be guaranteed. This sometimes leads to a divergence of the weight sets in independent trajectories, which makes interpreting the results somewhat tricky.
The biggest problem facing optE, however, is its inability to see all of the conformation space. This is true for all of the loss functions we try to optimize, but the PNatStruct loss function, in particular, can see only the decoys we give it, and typically we can afford to give optE only a few hundred decoys per protein. Thus, as optE traverses weight space, the returned weight set may be an artifact of the small sample of selected decoys. For example, optE typically tries to assign a negative weight to the Ramachandran term (described briefly in Section 6.2 below), suggesting that this term is over-optimized in our decoys compared to native structures, and, therefore, the easiest way to discriminate natives from decoys is to turn the weight negative. However, a negative weight on the Ramachandran term is clearly not good for protein structure prediction. In general, the weights that optE does produce are not good for protein modeling. However, using the protocol described in the next section, optE is fantastic at refitting reference energies.
4.5 A Sequence Profile Recovery Protocol for Fitting Reference Energies
One of the more dissatisfying results of training reference energies with the PNatAA loss function was that the profile of designed amino acids overly favored the most common amino acids (leucine, in particular) and almost never included rare amino acids (tryptophan, in particular). To address this shortcoming, we created an alternative protocol within optE for fitting only the amino acid reference energies, keeping all other weights fixed. This protocol does not use any of the loss functions described above; instead, it adjusts the reference energies directly based on the results of complete protein redesign.
The protocol iteratively performs complete protein redesign on a set of input protein structures and adjusts the reference energies upwards for amino acids it over designs and downwards for those it under designs, where the target frequencies are taken from the input set. After each round, optE computes both the sequence recovery rate and the Kullback-Leibler (KL) divergence of the designed sequence profile against the observed sequence profile, given by, , where paa is the naturally occurring frequency of amino acid aa in the test set and qaa is the frequency of amino acid aa in the redesigned structures. The final reference energies chosen are those that maximize the sum −0.1 KL-divergence + seqrec-rate.. This protocol is used to refit reference energies after each of the three energy-function changes described in Section 6.
The training set we use, which we call the “HiQ54,” is a new collection of fifty four non-redundant, monomeric proteins from the PDB through 2010 that have 60–200 residues (avg=134) and no tightly-bound or large ligands. All were required to have both resolution and MolProbity score (Chen et al., 2010) at or below 1.4, very few bond-length or angle outliers, and deposited structure-factor data. They provide reference crystal structures with a guaranteed extremely low level of incorrectly fit sidechain or backbone conformations. The HiQ54 set is available at http://kinemage.biochem.duke.edu/.
5 Benchmarks
Scientific benchmarking allows energy function comparison. The tests most pertinent to the Rosetta community often aim toward recapitulating observations from crystal structures. For example, the rotamer recovery test challenges an energy function, given the native backbone conformation, to predict the native sidechain conformations. In this section we describe a curated set of previously-published benchmarks which together provide a comprehensive view of an energy functions strengths and weaknesses. We continually test the benchmarks on the RosettaTests server to allow us to immediately detect changes to Rosetta that degrades its overall performance (Lyskov & Gray, 2012).
5.1 Rotamer Recovery
One of the most direct tests for an energy function is its ability to correctly identify the observed rotamers in a crystal structure against all other possible rotamers while keeping the backbone fixed. It is a stringent test because the majority of the torsional degrees of freedom in proteins occur in the sidechains, and the combinatorial nature of the problem produces an extremely large search space, making recovery non-trivial. Variants of the rotamer recovery test have long been used to evaluate molecular structure energy functions (Petrella et al., 1998; Liang & Grishin, 2002), including extensive use to evaluate the Rosetta energy function (Kortemme et al., 2003, Dobson et al., 2006; Dantas et al., 2007; Jacak et al., 2012), the ORBIT energy function (Sharabi et al., 2010), and the SCWRL energy function (Shapovalov & Dunbrack, 2011).
Here, we test rotamer recovery in four tests combining two bifurcated approaches to the task: discrete vs. continuous rotamer sampling and one-at-a-time vs. full-protein rotamer optimization. The discrete, one-at-a-time rotamer optimization protocol is called rotamer trials. It builds rotamers, calculates their energies in the native context, and compares the lowest-energy rotamer against the observed crystal rotamer. The continuous, one-at-a-time rotamer optimization protocol is called rtmin (Wang et al., 2005). It similarly builds rotamers in the native context but minimizes each rotamer before comparing the lowest-energy rotamer against the crystal rotamer. The discrete, full-protein optimization protocol is called pack rotamers. This builds rotamers for all positions and then seeks to find the lowest-energy assignment of rotamers to the structure using a Monte Carlo with simulated annealing protocol (Kuhlman & Baker, 2000) where at a random position, a random rotamer is substituted into the current environment, its energy is calculated, and the substitution is accepted or rejected based on the Boltzmann criterion. The rotamers in the final assignment are compared against the crystal rotamers. The continuous, full-protein rotamer optimization task is called min pack. It is very similar to the pack rotamers optimization technique except that each rotamer is minimized before the Boltzmann decision. A similar protocol has been described before (Ding & Dokholyan, 2006)).
Recovery rates are measured on a set of 152 structures each having between 50 and 200 residues and a resolution less than 1.2 Å. Rotamers are considered recovered if all their χ dihedrals are less than 20° from the crystal χ dihedrals, taking into account symmetry for the terminal dihedrals in PHE, TYR, ASP, and GLU. For the discrete optimization tests, rotamers are built at the center of the rotamer wells, with extra samples included at ±σi from χ̄i for χ1 and χ2. For the continuous optimization test, samples are only taken at χ̄i but can move away from the starting conformation through minimization.
5.2 Sequence Recovery
In the sequence recovery benchmark, we performed complete-protein fixed-backbone redesigns on a set of crystal structures, looking to recapitulate the native amino acid at each position. For this chapter, we used the test set of 38 large proteins from (Ding & Dokholyan, 2006). Sequence recovery was performed with the discrete, full-protein rotamer-and-sequence optimization protocol called PackRotamers, described above. Rotamer samples were taken from the given library (either the 2002 or 2010 library), and extra samples were chosen at ±σi for χ1, and χ2. The multi-cool annealer simulated annealing protocol (Leaver-Fay et al., 2011a) was employed instead of Rosetta’s standard simulated annealing protocol. We measured the sequence recovery rate and the KL-divergence of the designed amino-acid profile from the native amino-acid profile: the sequence recovery rate should be high, and the KL-divergence should be low.
5.3 ΔΔG Prediction
The ΔΔG benchmark consists of running the high-resolution protocol described in Kellogg et al. (2011), on a curated set of 1210 point mutations for which crystal structures of the wild-type protein are available. The protocol predicts a change in stability induced by the mutation by comparing the Rosetta energy for the wild type and mutant sequences after applying the same relaxation protocol to each. The Pearson correlation coefficient of the measured vs. predicted ΔΔGs is used to assess Rosetta’s performance.
5.4 High-resolution protein refinement
Rosetta’s protocol to predict protein structures from their sequence alone runs in two phases: a low-resolution phase (abinitio) relying on fragment insertion (Simons et al., 1997) to search through a relatively smooth energy landscape (Simons et al., 1999), and a high-resolution phase (relax) employing sidechain optimization and gradient-based minimization. This abrelax protocol offers the greatest ability to broadly sample conformation space in an attempt to find any non-native conformations that Rosetta prefers to near-native conformations.
Unfortunately, the abrelax protocol requires significantly more sampling than could readily be performed to benchmark a change to the energy function. The problem is that most low-resolution structures produced by the first abinitio phase do not yield low-energy structures in the second relax phase, so finding low-energy decoy conformations requires hundreds of thousands of trajectories. To reduce the required amount of sampling, Tyka et al. (2010) curated four sets of low-resolution decoys for 114 proteins by taking the low-resolution structures that generated low-energy structures after being put through high-resolution refinement. The benchmark is then to perform high-resolution refinement on these low-resolution structures with the new energy function. Each of the low-resolution sets were generated from different sets of fragments; some sets included fragments from homologues, while others included fragments from the crystal structure of the protein itself. This gave a spectrum of structures at varying distances from native structures.
These sets are used as input to the relax protocol. The decoy energies and their RMSDs from the crystal structure are used to assess the ability of Rosetta to discriminate natives from low-energy decoys. This is reported in two metrics by the benchmark: the number of proteins for which the probability of the native structure given by the Boltzmann distribution exceeds 80% (pNat > 0.8; pNat should not be confused with the PNatStruct loss function in optE), and the number of proteins for which the lowest-energy near-native conformation has a lower energy than the lowest-energy decoy conformation. For the benchmarks reported here, we relaxed 6,000 decoys randomly sampled with replacement from each of the four sets, resulting in 24,000 decoys per protein. The statistical significance of the difference between two runs can be assessed with a paired T-test, comparing pNat for each of the 114 targets.
The proteins included in this test set include many where the crystal structure of the native includes artifacts (e.g. loop rearrangements to form crystal contacts), where the protein coordinates metal ions or ligands, or where the protein forms an obligate multimer in solution. For this reason, perfect performance at this benchmark is not expected, and interpreting the results is somewhat complicated; an improvement in the benchmark is easier to interpret than a degradation.
5.5 Loop Prediction
In the loop prediction benchmark, the aim is to test Rosetta’s accuracy at de novo protein loop reconstruction. For this, we used the kinematic closure (KIC) protocol, which samples mechanically accessible conformations of a given loop by analytically determining six dihedral angles while sampling the remaining loop torsions probabilistically from Ramachandran space (Mandell et al., 2009).
We used the benchmark set of 45 12-residue loops as described in Mandell et al., (2009). For each loop, we generated 8,000 structures and calculated their Cα loop RMSD to the native.
In some cases, KIC generates multiple clusters of low-energy conformations of which one cluster is close to the native structure, while the other(s) can be several Ångstroms away. Because the Rosetta energy function does not robustly distinguish between these multiple clusters, we considered not just a single structure, but the five lowest-energy structures produced. Of these five, we used the lowest-RMSD structure when calculating benchmark performance. Overall loop reconstruction accuracy is taken as the median Cα loop RMSD of all best structures across the entire 45-loop dataset. The first and third quartile, though of lesser importance than the median, should be examined as well, as they offer a picture of the rest of the distribution.
6 Three Proposed Changes to the Rosetta Energy Function
In this final section, we describe three changes to Rosetta’s energy function. After describing each change and its rationale, we present the results of the benchmarks described above.
6.1 Score12′
We used the sequence-profile-recovery protocol in optE to fit the reference energies for Score12 to generate a new energy function we call Score12′. Refitting the Score12 reference energies in Rosetta3 (Leaver-Fay et al., 2011b) was necessary because the components of Score12 in Rosetta3 differed in several ways from those in Rosetta2. First, in Rosetta2, there was a disagreement between the energy function used during sequence optimization and the one used throughout the rest of Rosetta. In the sequence optimization module (“the packer”), the knowledge-based Ramachandran potential was disabled, and the weight on the P(aa|ϕ,ψ) was doubled from .32 to .64. In Rosetta3, there is no schism between the energy functions used in the packer and elsewhere, meaning that when Score12 is used, the packer includes the Ramachandran term and the P(aa|ϕ,ψ) term is not upweighted. Furthermore, the Lennard-Jones and implicit solvation (Lazaridis & Karplus, 1999) terms in Rosetta3 extend out to 6 Å instead of to 5.5 Å, and these terms are smoothed with splines to reach a value of 0 with a derivative of 0 at 6 Å. Previously, a linear ramp to zero starting at 5.0 Å and ending at 5.5 Å left discontinuities in the derivatives, resulting in unwanted peaks at 5.0 and 5.5 Å in atom-pair radial distribution functions (Will Sheffler & David Baker, unpublished observations). Also, the Lennard-Jones potential now starts counting the contribution of the repulsive component at the bottom of the well, instead of at the x-intercept. This change eliminates a derivative discontinuity at the x-intercept which forms if the attractive and repulsive weights differ (as they do in Score12). Rosetta3’s energy function differed from Rosetta2’s in an third way: in Rosetta2, the Cβ distance thresholds that were used to decide whether residue pairs were in range of each other were too short, causing certain interactions to be inappropriately omitted. This affected all terms in the energy function between certain pairs of residues.
Table II gives the sequence recovery rates for Rosetta2 and Rosetta3 using Score12; it also shows the sequence recovery rates obtained from training the reference energies with the PNatAA loss function (Rosetta3-PNatAA) and with the sequence-profile recovery protocol (Rosetta3-sc12′). Though the PNatAA objective function achieves satisfactory sequence-recovery rates, it over-designs leucine and lysine and never designs tryptophan (Table III). Score12′, on the other hand, does an excellent job recapitulating the sequence profile in the testing set while also outperforming the Rosetta3-PNatAA energy function at sequence recovery. The rotamer-recovery, high-resolution refinement, and loop-prediction benchmarks were not run for this proposed change to the energy function as fixed-sequence tasks are unaffected by changes to the reference energies.
Table II.
Sequence recovery rates at all, exposed (#neighbors ≤ 16), intermediately-buried (16 < #neighbors ≤ 23), and buried (23 < #neighbors) residues) and KL-Divergence of the designed sequence profile from the native sequence profile, measured on the Ding & Dokholyan-38 set. The Rosetta3-sc12 energy function represents the Score12 reference energies taken directly from Rosetta2. The Rosetta3-PNatAA weight set keeps the same weights as Score12, except the reference energies were fit by optimizing the PNatAA loss function; the Rosetta3-sc12′ (Score12′) reference energies were generated using the sequence-profile optimization protocol. Both Rosetta2 and Rosetta3-sc12′ yield high sequence recovery rates and good sequence profiles.
| Sequence Recovery and Sequence Profile Recovery Rates | |||||
|---|---|---|---|---|---|
| % Total Rec. | % Exp. Rec. | % Int. Rec. | % Bur. Rec. | KL-Divergence | |
| Rosetta2-sc12 | 38.0 | 26.9 | 45.1 | 53.9 | 0.019 |
| Rosetta3-sc12 | 32.6 | 24.2 | 38.5 | 43.4 | 0.141 |
| Rosetta3-PNatAA | 36.7 | 28.9 | 41.3 | 48.9 | 0.391 |
| Rosetta3-sc12′ | 37.0 | 27.5 | 42.5 | 51.4 | 0.008 |
Table III.
The amino acid profiles of the Ding & Dokholyan-38 test set and those generated from complete protein redesign of this set using different energy functions. The amino-acid profile that Score12′ generates matches the test-set profile quite well. In contrast, the Rosetta3-PNatAA weight set overdesigns leucine and lysine, while underdesigning phenylalanine, glutamine, and tryptophan.
| Amino Acid Profiles | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | D | E | F | G | H | I | K | L | M | N | P | Q | G | S | T | V | W | Y | |
| Test Set | 8.9 | 1.2 | 6.6 | 6.7 | 4.3 | 8.3 | 2.2 | 5.2 | 6.3 | 7.9 | 2.4 | 4.5 | 4.5 | 3.4 | 4.7 | 5.2 | 5.7 | 6.8 | 1.5 | 3.6 |
| Rosetta2-sc12 | 6.3 | 1.2 | 6.2 | 6.3 | 5.5 | 7.6 | 1.9 | 6.3 | 5.2 | 10.3 | 1.9 | 4.1 | 6.0 | 4.2 | 5.3 | 5.2 | 5.0 | 5.6 | 2.0 | 3.9 |
| Rosetta3-sc12 | 6.4 | 0.0 | 7.7 | 8.7 | 5.4 | 7.4 | 6.3 | 4.7 | 6.8 | 8.8 | 1.7 | 3.2 | 1.3 | 3.5 | 6.3 | 7.2 | 4.2 | 4.2 | 2.5 | 4.0 |
| Rosetta3-PNatAA | 9.8 | 0.0 | 6.4 | 6.2 | 0.7 | 8.3 | 0.8 | 5.7 | 11.8 | 13.5 | 0.2 | 2.7 | 7.6 | 0.2 | 3.1 | 4.7 | 6.3 | 10.6 | 0.0 | 1.5 |
| Rosetta3-sc12′ | 8.4 | 0.7 | 6.6 | 7.5 | 3.9 | 8.2 | 2.1 | 5.0 | 6.4 | 8.0 | 2.2 | 3.5 | 3.7 | 3.3 | 6.1 | 6.4 | 5.8 | 6.4 | 1.8 | 4.1 |
6.2 Interpolating Knowledge-Based Potentials with Bicubic Splines
Three knowledge-based potentials in Rosetta are defined on the φ,ψ map: the Ramachandran term which gives Erama(φ,ψ|aa) = −ln p(φ,ψ|aa), the p_aa_pp term (sometimes called the design term) which gives an energy from the log of the probability of observing a particular amino acid at a given φ,ψ, and the rotamer term (almost always called the Dunbrack term, or fa_dun), which gives Edun(χ|φ,ψ,aa). Each of these terms has in common the use of bins on the φ,ψ map to collect the data that define these potentials and the use of bilinear interpolation between the bins to define a continuous function.
The p_aa_pp term in Score12 is given by
| (12) |
For any particular φ and φ, the energy is given as the negative log of the bilinearly interpolated p(aa|φ,ψ) divided by p(aa). The Ramachandran term similarly defines the energy for the off-grid-point φ and ψ values as the negative log of the bilinearly interpolated p(φ,ψ|aa). Both the p_aa_pp and the Ramachandran term place their bin centers every ten degrees starting from 5°.
The Dunbrack term in Score12 is given by
| (13) |
where the rotamer bin, rot, which gives the probability of the rotamer, p(rot|φ,ψ,aa), is computed from the assigned χ dihedrals, and both χ̄i (φ,ψ|aa,rot;) and σi (φ,ψ|aa,rot) are the measured mean χ values and standard deviations for the rotamer. This effectively models the probability for the sidechain conformation as the product of the rotamer probability and several (height-unnormalized) Gaussians. The 2002 library gives the p(rot|φ,ψ,aa), χ̄i (φ,ψ|aa,rot), and σi (φ,ψ|aa,rot) every ten degrees. Given a particular assignment of φ and ψ, the values for p(rot|φ,ψ,aa), χ̄i (φ,ψ|aa,rot), and σi (φ,ψ|aa,rot) and are bilinearly interpolated from the four surrounding bin centers. The Dunbrack term also divides the φ,ψ plane into ten-degree bins, starting from 0°.
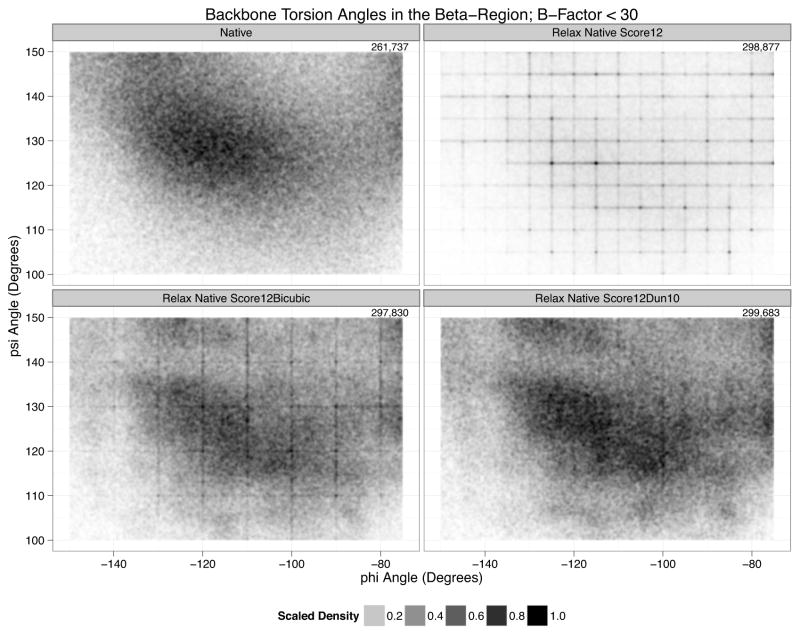
Bilinear interpolation leaves derivative discontinuities every 5° in the plane. These discontinuities frustrate the minimizer causing pile-ups at the bin boundaries. Looking at the distribution for non-helical residues, the grid boundaries are unmistakable (Figure 4B).
Figure 4. Backbone Torsion Angles in the Beta-Region with B-Factors less than 30.
A. The distribution for the top8000. B. In Score12. density accumulates on the 5° bins due to derivative discontinuities caused by bilinear interpolation. C. Score12Bicubic has only a few remaining artifacts on the 10° bin boundaries due to the continued use of bilinear interpolation for parts of the Dunbrack energy. D. Score12Dun10 has very few remaining artifacts.
Our proposed fix for this problem is to use bicubic splines to interpolate between the grid points. We fit bicubic splines with periodic boundary conditions for both the Ramachandran and p_aa_pp terms on the energies, interpolating in energy space. For the Dunbrack energy, we fit bicubic splines for the −ln(p(rot|φ,ψ,aa) portion, but, to avoid increasing our memory footprint too much, continued to use bilinear interpolation for the χ-mean and χ-standard-deviations. We refit the reference energies using the sequence-profile recovery protocol to create a new energy function, that for this chapter, we refer to as Score12Bicubic, Figure 4C shows that bicubic spline interpolation dramatically reduces the pileups. Accumulation on the 10° grid boundaries starting at 5° produced by the Ramachandran and p_aa_pp terms is completely gone. Modest accumulation on the 10° grid boundaries starting at 0° persists because bicubic splines were not used to interpolate the χ-mean and χ-standard deviations.
6.3 Replacing the 2002 Rotamer Library with the Extended 2010 Rotamer Library
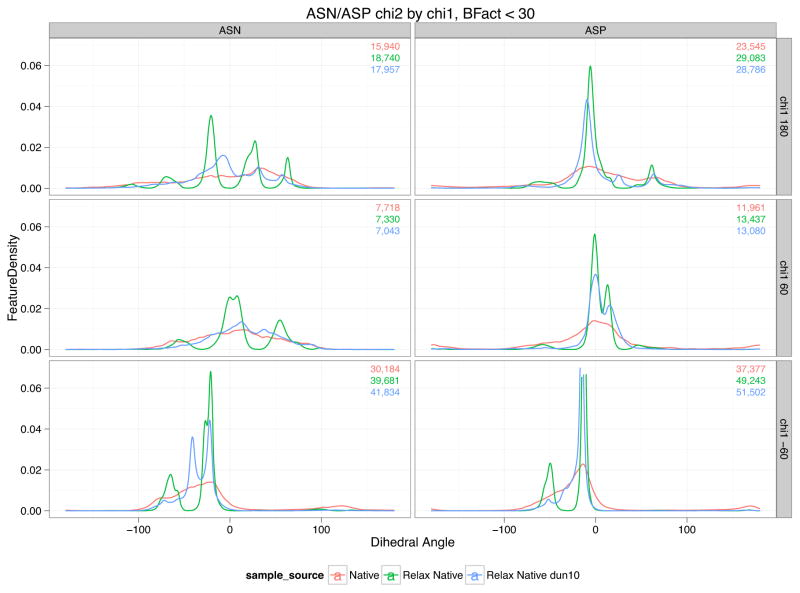
In 2010, Shapovalov and Dunbrack (Shapovalov & Dunbrack, 2011) defined a new rotamer library that differs significantly from the 2002 library in the way the terminal χ are handled for eight amino acids: ASP, ASN, GLU, GLN, HIS, PHE, TYR, and TRP. Because these terminal χ dihedrals are about bonds between sp3-hybridized atoms and sp2-hybridized atoms, there are no well-defined staggered conformations. Instead of modeling the probability landscape for the terminal χ within a particular bin as a Guassian, the new library instead provides a continuous probability distribution over all the bins. This last χ is in effect non-rotameric, though the rest of the χ in the sidechain are still rotameric; these eight amino acids can be called semirotameric. In the 2002 library, there were discontinuities in both the energy function and in the derivatives when crossing over these grid boundaries. Once a rotamer boundary is crossed, an entirely different set of χ̄ and σi are used to evaluate the rotamer energy. Such crossing would in fact have been likely given high standard deviations for the terminal χ of the eight amino acids listed above. For example, an asparagine with φ,ψ = (−120,130) in rotamer (g−,3)11, χ̄2 is −34.1, and σ2 is 11.2°, so that the mean is less than one standard deviation away from the lower grid boundary at −45°. In contrast, χ̄1 for the same rotamer is 8.6 standard deviations from the closest grid boundary. Rotamer-boundary crossings should be common for the terminal χ for these semirotameric amino acids, but our model makes such crossings difficult. Furthermore, Rosetta’s treatment of the terminal χ probability distributions as Gaussians means that Rosetta structures display Gaussian distributions (the green lines in Figure 5) that do not resemble the native distributions (the red lines in Figure 5).
Figure 5. ASN/ASP χ2 distribution by χ1 bin.
Comparison of χ2 distributions for the semirotameric amino acids ASN and ASP, broken down by χ1 rotamer. Rosetta’s implementation of the 2002 library produces Gaussian-like distributions for χ2 in relaxed natives (green), though the native distributions do not resemble Gaussians (red). Using the 2010 library (blue), the distributions improve considerably though they remain too peaky in places.
With the 2010 library, the energy for a rotameric residue is computed in the same way as for the 2002 library, and the energy for one of the semirotameric amino acids is computed as:
| (14) |
where T denotes the terminal χ, and where χ̄i (φ,ψ|aa,rot), and σi (φ,ψ|aa,rot) from Eq. (13) have been abbreviated as χ̄i and σi, though they retain their dependence on the rotamer and amino acid and are a function of φ and ψ. The 2010 library provides data every 10 degrees in φ,ψ plane, and provides data for pχT(rot|φ,ψ,aa) every 10° for φ and ψ, and every 5 or every 10 degrees for χT depending on whether aa is symmetric about χT. We fit tricubic splines to interpolate in −ln(p(rot|φ,ψ,aa)pχT(rot|φ,ψ,aa)) in φ,ψ, and χT . As in the 2002 library, χ̄i and σi are interpolated bilinearly from the four surrounding grid points. Interpolating with tricubic splines avoids the density-accumulation-at-derivative-discontinuities artifacts for χT that are apparent using trilinear interpolation. We refit the reference energies using the sequence-profile recovery protocol to create a new energy function that for this chapter we refer to as Score12Dun10. Score12Dun10 builds on top of Score12Bicubic.
6.4 Benchmark Results
The results of the benchmarks show that Score12′ is a clear improvement over Score12, substantially improving sequence recovery (Table VI), and that Score12Bicubic is a clear improvement over Score-12′, behaving as well as Score12′ on most benchmarks (Tables VI, VII, and IX) and giving a slight, but statistically insignificant improvement at the high-resolution refinement benchmark (p = 0.07, Table VIII). Score12Dun10 shows mixed results: at the rotamer recovery benchmark (Table IV), it shows a clear improvement over Score12Bicubic, and the improvement can be most clearly seen in the rotamer recovery rates for the semirotameric amino acids (Table V).
Table VI.
All three energy functions perform equally well at this test; the column meanings are given, along with a comparison of Score12′ against Score12, in Table II.
| Sequence Recovery Benchmark Results | |||||
|---|---|---|---|---|---|
| Energy Function | % Total Rec. | % Exp. Rec. | % Int. Rec. | % Bur. Rec. | KL-Divergence |
| Score12′ | 37.0 | 27.5 | 42.5 | 51.4 | 0.008 |
| Score12Bicubic | 37.6 | 28.3 | 42.8 | 52.4 | 0.010 |
| Score12Dun10 | 37.6 | 28.0 | 43.0 | 52.5 | 0.009 |
Table VII.
Each change to the energy function has only the mildest of impacts on the measured correlation coefficient. Though Score12 outperforms Score12′ by a small margin, Score12′ s poor performance at the sequence recovery benchmark in comparison is much worse. Kellogg et al. (2011) demonstrated that training the reference energies for ΔΔG prediction yielded modest improvements in correlation, but degrades sequence recovery considerably (to ~31%). In contrast, training reference energies for sequence recovery yields decent ΔΔG predictions.
| Correlation coefficient R-values from the ΔΔG’s benchmark | ||||
|---|---|---|---|---|
| Score12 | Score12′ | Score12Bicubic | Score12Dun10 | |
| ΔΔG Prediction: R-Value | 0.69 | 0.67 | 0.68 | 0.67 |
Table IX.
Performance at the loop modeling benchmark is measured by taking lowest-RMSD of the 5 lowest-energy structures for each of the loops in the benchmark and computing the median. The first, second (median), and third quartiles are reported (Å). The median values were not significantly different between the three energy functions, but Score12Bicubic and Score12Dun10 improved the third quartile RMSD.
| Loop-Modeling Benchmark Results | |||
|---|---|---|---|
| Energy Function | First Quartile | Median | Third Quartile |
| Score12 | 0.468 | 0.637 | 1.839 |
| Score12Bicubic | 0.499 | 0.644 | 1.636 |
| Score12Dun10 | 0.461 | 0.677 | 1.463 |
Table VIII.
pNat is given by exp(E(nat))/(E(nat) + Σ exp(E(d))) with E(nat) representing the best energy of any structure under 2Å RMSD from the crystal structure, and d representing any structure greater than 2Å RMSD. Score12Bicubic shows a small but statistically insignificant improvement over Score12 (p = 0.07). However, Score12Dun10, which builds on top of Score12Bicubic, performs worse than Score12Bicubic (p=0.01).
| High Resolution Refinement Benchmark Results | |||
|---|---|---|---|
| Energy Function | #(pNat > 0.8) | Σ pNat | #(eNat < eDec) |
| Score12 | 67 | 74.62 | 104 |
| Score12Bicubic | 68 | 77.88 | 105 |
| Score12Dun10 | 60 | 72.01 | 104 |
Table IV.
Percent of rotamers recovered by prediction algorithm for Score12 and for the two modifications proposed in this chapter: the use of bicubic interpolation and the 2010 rotamer library. Column names: pr, pack rotamers; mp, min pack; rt, rotamer trials; rtm, rt-min. A rotamer is considered recovered if no χ differs from the native by more than 20°.
| Rotamer Recovery Rates | ||||
|---|---|---|---|---|
| pr | mp | rt | rtm | |
| Score12 | 66.19 | 69.07 | 71.49 | 73.12 |
| Score12bicubic | 66.24 | 67.51 | 71.52 | 73.15 |
| Score12Dun10 | 67.82 | 70.50 | 72.60 | 74.23 |
Table V.
Percentage rotamer recovery by amino acid. Rotameric amino acids are listed on the left; semirotameric amino acids on the right. The 2010 rotamer library shows greatest improvements for the semirotameric amino acids.
| Rotamer Recovery By Amino Acid Type | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | K | M | I | L | S | T | V | N | D | Q | E | H | W | F | Y | |
| Score12 | 24.7 | 31.1 | 51.3 | 84.0 | 86.7 | 71.8 | 92.9 | 94.4 | 55.2 | 59.2 | 21.8 | 28.5 | 51.8 | 78.7 | 84.5 | 79.9 |
| Score12Bicubic | 25.7 | 31.8 | 51.1 | 85.2 | 86.8 | 71.5 | 92.9 | 94.4 | 54.8 | 58.7 | 20.5 | 28.7 | 52.0 | 80.1 | 83.3 | 79.9 |
| Score12Dun10 | 26.7 | 31.7 | 49.6 | 85.4 | 87.5 | 72.5 | 92.6 | 94.3 | 56.8 | 60.4 | 30.7 | 33.6 | 55.0 | 85.0 | 85.4 | 82.9 |
Score12Dun10 performed worse at the high-resolution refinement benchmark than Score12Bicubic (Table VIII). The two principle metrics for this benchmark are slightly worse: the number of proteins where the lowest-energy near-native structure has a lower energy than the lowest-energy decoy (#(eNat < eDec); 104 vs. 105), and the number of proteins where the probability of the native structure calculated by the Boltzmann distribution is greater than 80% (#(pNat > 0.8); 67 vs. 60). To estimate the significance of these results, we compared the distribution of (pNatsc12dun10 - pNatsc12bicubic) for each of the 114 targets against the null hypothesis that this distribution had a mean of 0, using a two-tailed t-test. This gave a p-value for the difference of 0.01. Because this benchmark includes proteins whose accurate prediction is unlikely given protocol limitations (disulfides are not predicted), and crystal artifacts (loops which adopt conformations supported only by crystal contacts), its results are more difficult to interpret. We therefore restricted our focus to 29 proteins in the set which are absent of these issues (Supplemental Table VI) and repeated the comparison of (pNatsc12dun10 - pNatsc12bicubic). Here too, Score12Dun10 showed a statistically significant degradation relative to Score12Bicubic, with a p-value of 0.04. The mean difference of the pNat statistic for this subset (0.06) was similar to the mean difference over the entire set (0.05).
Table VII shows the results of the ΔΔG benchmark. For each energy function tested, the starting crystal structures were pre-minimized and then run through the protocol described in row 16 of Table I in Kellogg et al. (2011). The differences between the four tested energy functions are very slight. The performance of Score12′ is somewhat degraded relative to Score12, though this should be weighed against the dramatic improvement Score12′ shows at sequence recovery (Table II). The other two energy functions perform in the same range as Score12′.
At the loop modeling benchmark, the differences between the three methods were slight. To estimate the significance of the differences, we took 100 bootstrap samples and measured the three RMSD quartiles. The differences in median RMSDs between Score12 and Score12Bicubic (p < .74), and Score12Bicubic and Score12Dun10 (p < .11) were not statistically significant. However, the third quartile improved for both Score12Bicubic and Score12Dun10; In two cases, (e.g., 1cyo and 1exm, Supplemental Figures 5 and 7), KIC simulations using Score12Dun10 correctly identified near-native structures by their lowest energy, where Score12 simulations did not.
It is not immediately clear why the 2010 rotamer library causes a degradation in Rosetta’s ability to discriminate native structures from decoys. A possible reason for this might be pointed to in the distribution of off-center χ angles. Structures refined with the new library have χ-angles more tightly distributed around the reported χ̄i. The 2010 library, which was generated using more stringent data filters than the 2002 library, reports smaller standard deviations on average: for example, the mean σ1 for leucine for rotamers in all φ,ψ bins with probability >5% is 10.8° (with a median of 13.9°) in the 2002 library, but is down to 7.9° (with a median of 7.2°) in the 2010 library. (For all leucine rotamers, the 2002 library reports a 13.1° mean, and a 9.9° median; the 2010 library reports a 9.2° mean, and a 9.9° median). By decreasing the weight on the fa_dun term or by merely weakening the “off-rotamer penalty” (the component of Eq. (14)), that the distributions may broaden and performance at the high-resolution refinement benchmark might improve. Encouragingly, decreasing the fa_dun weight down to one-half of its Score12 weight does not substantially worsen rotamer recovery for the 2010 library. There is still significant work, however, before we are ready to conclude that the new library should be adopted for general use in Rosetta.
7 Conclusion
Here we have shown three tools that can be used to evaluate and improve the Rosetta energy function. Comparing features from crystal structures and Rosetta-generated structures can be used to identify inaccuracies in the Rosetta energy function. New or re-parameterized energy terms can be rapidly tested with optE to determine if the change improves structure prediction and sequence design. When a new term is ready to be rigorously tested, we can test for unintended changes to feature distributions by relying upon the existing set of feature-analysis scripts, refit reference energies for protein design using the sequence-profile recovery protocol in optE, and measure the impact of the new term on a wide array of Rosetta protocols by running the benchmarks curated here.
Of the three changes we benchmarked in this paper, we confidently recommend that the first two should be adopted: Score12′ should be used in place of Score12, and Score12Bicubic should be used in place of Score12′.
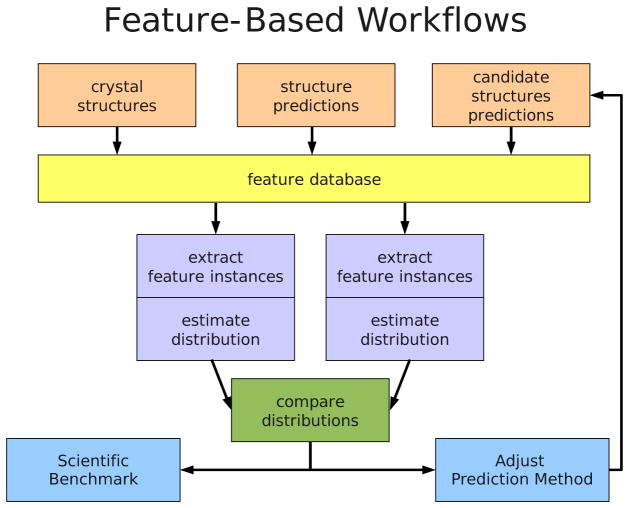
Figure 2. Workflow for feature analysis.
Example usage workflow for the feature analysis tool. Layer 1: Each sample source consists of a batch of molecular conformations, e.g., experimentally determined or predicted conformations. Layer 2: Features from each sample source are extracted into a relational database. Layer 3: Conditional feature distributions are estimated from feature instances queried from the database. Layer 4: The distributions are compared graphically. Layer 5: The results of the comparisons are used as scientific benchmarks or to inform modifications to the structure prediction protocol and energy function, where the cycle can begin again.
Acknowledgments
Support for ALF, MJO, and BK came from GM073151 and GM073960. Support for JSR came from NIH R01 GM073930. Thanks to Steven Combs for bringing the bicubic-spline implementation to Rosetta.
Footnotes
This assumption needs to be checked, e.g. by comparing distribution of structural features against the Boltzmann distributions defined by the energy function.
The 3 in this rotamer designation simply refers to the indexing in the 2002 library, which for χ1 in g− is the bin [−45°,15°].
Contributor Information
Andrew Leaver-Fay, Email: leaverfa@email.unc.edu.
Matthew J. O’Meara, Email: momeara@cs.unc.edu.
Mike Tyka, Email: mtyka@u.washington.edu.
Ron Jacak, Email: rjacak@iavi.org.
Yifan Song, Email: yfsong@u.washington.edu.
Elizabeth H. Kellogg, Email: ekellogg@u.washington.edu.
James Thompson, Email: tex@u.washington.edu.
Ian W. Davis, Email: ian.w.davis@gmail.com.
Roland A. Pache, Email: roland.pache@ucsf.edu.
Sergey Lyskov, Email: sergey.lyskov@jhu.edu.
Jeffrey J. Gray, Email: jgray@jhu.edu.
Tanja Kortemme, Email: kortemme@cgl.ucsf.edu.
Jane S. Richardson, Email: jsr@kinemage.biochem.duke.edu.
James J. Havranek, Email: havranek@genetics.wustl.edu.
Jack Snoeyink, Email: snoeyink@cs.unc.edu.
David Baker, Email: dabaker@u.washington.edu.
Brian Kuhlman, Email: bkuhlman@email.unc.edu.
References
- Bower MJ, Cohen FE, Dunbrack RL., Jr Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol. 1997;267:1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- Chen HM, Liu BF, Huang HL, Hwang SF, Ho SY. Sodock: Swarm optimization for highly flexible protein-ligand docking. J Comp Chem. 2007;28:612–623. doi: 10.1002/jcc.20542. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy Da, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas G, Corrent C, Reichow SL, Havranek JJ, Eletr ZM, Isern NG, Kuhlman B, Varani G, Merritt Ea, Baker D. High-resolution structural and thermodynamic analysis of extreme stabilization of human procarboxypeptidase by computational protein design. J Mol Biol. 2007;366:1209–21. doi: 10.1016/j.jmb.2006.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–82. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson N, Dantas G, Baker D, Varani G. High-resolution structural validation of the computational redesign of human U1A protein. Structure. 2006;14:847–56. doi: 10.1016/j.str.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Dunbrack RL, Jr, Karplus M. Backbone dependent rotamer library for proteins: application to side chain prediction. J Mol Biol. 1993;230:543–74. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]
- Dunbrack RL., Jr Rotamer libraries in the 21st century. Curr Opin Struc Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- Fabiola GF, Krishnaswarmy S, Nagarajan V, Pattabhi V. C-h···o hydrogen bonds in β-sheets. Acta Crystallogr Sect D: Biol Crystallogr. 1997;53:316–320. doi: 10.1107/S0907444997000383. [DOI] [PubMed] [Google Scholar]
- Fersht A, Shi J, Knill-Jones J, Lowe D, Wilkinson A, Blow D, Brick P, Carter P, Waye M, Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J, Baker D. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PloS One. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilis D, Rooman M. Predicting protein stability changes upon mutation using database-derived potentials: solvent accessibility determines the importance of local versus non-local interactions along the sequence. J Mol Biol. 1997;272:276–90. doi: 10.1006/jmbi.1997.1237. [DOI] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–87. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Hamelryck T, Borg M, Paluszewski M, Paulsen J, Frellsen J, Andreetta C, Boomsma W, Bottaro S, Ferkinghoff-Borg J. Potentials of mean force for protein structure prediction vindicated, formalized and generalized. PloS One. 2010;5:e13714. doi: 10.1371/journal.pone.0013714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacak R, Leaver-Fay A, Kuhlman B. Computational protein design with explicit consideration of surface hydrophobic patches. Proteins. 2012;80:825–38. doi: 10.1002/prot.23241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LC, Tawfik DS. Conformational diversity and protein evolution--a 60-year-old hypothesis revisited. Trends Biochem Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- Jacobson MP, Kaminski GA, Friesner RA, Rapp CS. Force field validation using protein side chain prediction. J Phys Chem B. 2002;106:11673–11680. [Google Scholar]
- Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- Kaufmann KW, Lemmon GH, DeLuca SL, Sheehan JH, Meiler J. Practically useful: What the rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keedy DA, Arendall WB, III, Chen VB, Williams CJ, Headd JJ, Echols N, Richardson JS, Richardson DC. Torsional bioinformatics: 1.5 million quality-filtered residues for better Ramachandran validation. 2012. In Preparation. [Google Scholar]
- Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins: Struct, Funct, Bioinf. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib F, Cooper S, Tyka M, Xu K, Makedon I, Popovic Z, Baker D, Foldit Players. Algorithm discovery by protein folding game players. Proc Natl Acad Sci U S A. 2011;109:5277–5282. doi: 10.1073/pnas.1115898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T, Morozov AV, Baker D. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J Mol Biol. 2003;326:1239–1259. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
- Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc Natl Acad Sci U S A. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton G, Varani G, Stoddard B, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins: Struct, Funct, Genet. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Leaver-Fay A, Jacak R, Stranges PB, Kuhlman B. A generic program for multistate protein design. PloS One. 2011a;6:e20937. doi: 10.1371/journal.pone.0020937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith Ca, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YEA, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popović Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011b;487:545–74. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Jie F. Loss functions for discriminative training of energy-based models. Proc. 10th Inter. Works. A. I. Stats.; 2005. (AIStats’05) [Google Scholar]
- Liang S, Grishin N. Side-chain modeling with an optimized scoring function. Protein Sci. 2002;11:322– 331. doi: 10.1110/ps.24902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins: Struct, Funct, Genet. 2000;40:389–408. [PubMed] [Google Scholar]
- Lyskov S, Gray JJ. RosettaTests. 2012 URL http://rosettatests.graylab.jhu.edu.
- Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler J, Baker D. Rapid protein fold determination using unassigned nmr data. Proc Natl Acad Sci U S A. 2003;100:15404–15409. doi: 10.1073/pnas.2434121100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa S, Jernigan RL. Estimation of effective interresidue contact energies from protein crystal structures: quasi-chemical approximation. Macromolecules. 1985;18:534–552. [Google Scholar]
- Morozov AV, Kortemme T. Potential functions for hydrogen bonds in protein structure prediction and design. Adv Protein Chem. 2005;72:1–38. doi: 10.1016/S0065-3233(05)72001-5. [DOI] [PubMed] [Google Scholar]
- Murphy PM, Bolduc JM, Gallaher JL, Stoddard BL, Baker D. Alteration of enzyme specificity by computational loop remodeling and design. Proc Natl Acad Sci U S A. 2009;106:9215–20. doi: 10.1073/pnas.0811070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotný J, Bruccoleri R, Karplus M. An analysis of incorrectly folded protein models. Implications for structure predictions. J Mol Biol. 1984;177:787– 818. doi: 10.1016/0022-2836(84)90049-4. [DOI] [PubMed] [Google Scholar]
- Park B, Levitt M. Energy functions that discriminate X-ray and near native folds from well-constructed decoys. J Mol Biol. 1996;258:367–392. doi: 10.1006/jmbi.1996.0256. [DOI] [PubMed] [Google Scholar]
- Petrella RJ, Lazaridis T, Karplus M. Protein sidechain conformer prediction: a test of the energy function. Folding Des. 1998;3:353–377. doi: 10.1016/S1359-0278(98)00050-9. [DOI] [PubMed] [Google Scholar]
- Pierce N, Winfree E. Protein design is NP-hard. Protein Engineering. 2002;15:779–82. doi: 10.1093/protein/15.10.779. [DOI] [PubMed] [Google Scholar]
- Ponder JW, Richards FM. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987;193:775–91. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Potapov V, Cohen M, Schreiber G. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Eng Des Sel. 2009;22:553–60. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- Sellers BD, Zhu K, Zhao S, Friesner RA, Jacobson MP. Toward better refinement of comparative models: predicting loops in inexact environments. Proteins. 2008;72:959–971. doi: 10.1002/prot.21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapovalov MV, Dunbrack RL. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi OZ, Yanover C, Dekel A, Shifman JM. Optimizing energy functions for protein-protein interface design. J Comp Chem. 2010;32:23–32. doi: 10.1002/jcc.21594. [DOI] [PubMed] [Google Scholar]
- Sheffler W, Baker D. RosettaHoles: rapid assessment of protein core packing for structure prediction, refinement, design and validation. Protein Sci. 2009;18:229–239. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and bayesian scoring functions. J Mol Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Sippl MJ. Calculation of conformational ensembles from potentials of mean force: an approach to the knowledge-based prediction of local structures in globular proteins. J Mol Biol. 1990;213:859–83. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- Song Y, Tyka M, Leaver-Fay A, Thompson J, Baker D. Structure guided forcefield optimization. Proteins: Struct, Funct, Bioinf. 2011;79:1898–1909. doi: 10.1002/prot.23013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R, Kennard O. Crystallographic evidence for the existence of the c-h···o, c-h···n and c-h···cl hydrogen bonds. J Am Chem Soc. 1982;104:5063–5070. [Google Scholar]
- Tyka MD, Keedy DA, André I, Dimaio F, Song Y, Richardson DC, Richardson JS, Baker D. Alternate States of Proteins Revealed by Detailed Energy Landscape Mapping. J Mol Biol. 2010;405:607–618. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373:503–19. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- Wang C, Schueler-Furman O, Baker D. Improved side-chain modeling for protein-protein docking. Protein Sci. 2005;14:1328–1339. doi: 10.1110/ps.041222905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner P. A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc. 1984;106:765–784. [Google Scholar]
- Wickham H. A layered grammar of graphics. J Comput Graph Stat. 2010;19:3–28. [Google Scholar]
- Wickham H. The split-apply-combine strategy for data analysis. J Stat Softw. 2011;40:1–29. [Google Scholar]
- Wilkinson L. The grammar of graphics. Springer-Verlag New York, Inc; New York, NY, USA: 1999. [Google Scholar]
- Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: Using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- Yanover C, Bradley P. Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2 zinc fingers. Nucleic Acids Res. 2011;39:4564–76. doi: 10.1093/nar/gkr048. [DOI] [PMC free article] [PubMed] [Google Scholar]