Abstract
Objective
To determine whether individuals with primary ciliary dyskinesia (PCD) from unrelated Amish and Mennonite families harbor a single and unique founder mutation.
Study design
Subjects from Amish and Mennonite communities in several states were enrolled in the study. All subjects were clinically characterized, and nasal nitric oxide levels were measured. Nasal epithelial scrapings were collected from several subjects for ciliary ultrastructural analyses. DNA was isolated from patients with PCD and their unaffected first- and second-degree relatives. Genome-wide homozygosity mapping, linkage analyses, targeted mutation analyses, and exome sequencing were performed.
Results
All subjects from Old-Order Amish communities from Pennsylvania were homozygous for a nonsense mutant DNAH5 allele, c.4348C>T (p.Q1450X). Two affected siblings from an unrelated Mennonite family in Arkansas were homozygous for the same nonsense DNAH5 mutation. Children with PCD from an Amish family from Wisconsin had biallelic DNAH5 mutations, c.4348C>T (p.Q1450X) and c.10815delT (p.P3606HfsX23), and mutations in other genes associated with PCD were also identified in this community.
Conclusion
The Amish and Mennonite subjects from geographically dispersed and socially isolated communities had the same founder DNAH5 mutation, owing to the common heritage of these populations. However, disease-causing mutations in other PCD-associated genes were also found in affected individuals in these communities, illustrating the genetic heterogeneity in this consanguineous population.
The first human genetic disorder associated with ciliary dysfunction, primary ciliary dyskinesia (PCD) usually has an autosomal recessive mode of inheritance that leads to functional and, in some cases, ultrastructural abnormalities of the cilia. Impaired ciliary motility results in retention of inhaled particles and bacteria in the lung, paranasal sinuses, and middle ear, which manifest as chronic bronchitis, pansinusitis, and suppurative otitis media.1–4 Dysfunction of cilia outside of the sinopulmonary tract may cause male infertility owing to impaired movement of spermatozoa, and can play a role in laterality defects, such as situs inversus totalis, related to motility defects of nodal cilia.1–4
The variable clinical presentation of PCD is likely related to the genetic heterogeneity of this disease, which reflects the complexity of the ciliary structure. Cilia are composed of hundreds of proteins, and their chemical and physical interactions are required for motility to generate an effective and synchronous waveform.5–7 Mutations in any of these cilia proteins could potentially result in functional impairment and a spectrum of clinical manifestations. Genetic causes of PCD have been identified by sequencing human orthologs of genes discovered by screening for ciliary dysfunction in model organisms, which have provided insight into genotype–phenotype relationships in the human cilium. Using such models, mutations in several different genes, including DNAI18–10 and DNAH5,11,12 have been shown to cause PCD; these mutations account for disease in roughly half of all affected individuals.
The Old-Order Amish and Mennonite populations in North America have a common social heritage, descending from Swiss Anabaptist immigrants who emigrated seeking religious tolerance in the eighteenth century. Starting from initial settlements in Pennsylvania, these close-knit agrarian communities have spread across North America. Settlements maintain strict adherence to religious customs, and because members rarely marry outside the church, consanguinity is common. Families live in small endogamous communities that have different allele frequencies and genetic diseases due to unique founders, resulting in closed populations with a limited gene pool and an increased incidence of certain heritable conditions.13–16 In previous studies of Old Order Amish and Amish-Mennonite communities in the US, we discovered individuals with clinical manifestations consistent with PCD from several unrelated communities. Owing to consanguinity present in these geographically dispersed populations, we investigated the possibility that these communities harbor unique founder mutations.
Methods
Subjects from Amish communities in the midwest with neonatal respiratory distress, situs inversus totalis, chronic pneumonia, or rhinosinusitis were recruited to participate in the study, which has been described previously.17 Two probands from an Old-Order Amish community in Pennsylvania were enrolled and evaluated in a cross-sectional study supported by the Genetic Diseases of Mucociliary Clearance Consortium, and other family members with classic clinical features (Table I) were studied at the Clinic for Special Children. The Mennonite subjects from Arkansas were also enrolled in studies administered by the Genetic Diseases of Mucociliary Clearance Consortium and studied at the University of North Carolina at Chapel Hill. Evaluations included detailed clinical and family histories and physical examination. Nasal nitric oxide (NO) levels were measured in all subjects using established techniques with a spirometer (model CLD 88; Eco Medics, Duernten, Switzerland). Spirometry was performed on subjects aged ≥5 years. Sputum or oropharyngeal swabs were collected for microbiological studies. Nasal epithelial scrapings from inferior nasal turbinates were collected from several subjects for ultrastructural analysis of ciliary axonemes,3,18 which revealed dynein arm defects. Blood or buccal cell samples were collected for genetic analysis; DNA was extracted using commercially available kits.17 The diagnosis of PCD was based on the presence of typical clinical features, reduced nasal NO levels, and ultrastructural defects or disease-causing mutations of known genes. The relevant Institutional Review Boards approved the studies, and signed informed consent was obtained from all subjects and family members.
Table I.
Clinical characteristics and diagnostic testing of Amish and Mennonite subjects with PCD and DNAH5 mutant alleles
| Patient | Age | Sex | Clinical manifestations | Laterality | Ultrastructural defect | Nasal NO, nL/min* | DNAH5 mutation |
|---|---|---|---|---|---|---|---|
| Pennsylvania | |||||||
| 1 | 14 y | Female | RS, OM, HL, PC, BR | SS | ODA | 15.6 | 4348C>T/4348C>T |
| 2 | 7 y | Male | NRD, CHD, RS, OM, HL, PC, BR | SA | ODA | 5.7 | 4348C>T/4348C>T |
| 3 | 10 m | Male | OM | SS | ND | 68.0 | 4348C>T/4348C>T |
| 4 | 63 y | Female | RS, PC, BR | SS | ND | 14.3 | 4348C>T/4348C>T |
| 5 | 9 y | Female | NRD, OM, RS, PC | SS | ND | 31.5 | 4348C>T/4348C>T |
| 6 | 6 y | Male | NRD, OM, HL, RS, PC | SS | ND | 40.5 | 4348C>T/4348C>T |
| 7 | 11 y | Male | NRD, OM, HL, RS, PC | SS | ND | 26.5 | 4348C>T/4348C>T |
| 8 | 13 y | Female | NRD, OM, HL, RS, PC | SS | ND | 26.7 | 4348C>T/4348C>T |
| Arkansas | |||||||
| 1 | 13 y | Male | RDS, CHD, OM, HL, RS, PC | SA | ND | 8.9 | 4348C>T/4348C>T |
| 2 | 18 y | Male | RDS, OM, HL, RS, PC | SS | ND | 9.2 | 4348C>T/4348C>T |
| Wisconsin | |||||||
| 1 | 3 m | Male | NRD, RS, OM, PC | SS | ODA | 8.6 | 10815dT/4348C>T |
| 2 | 2 y | Female | NRD, RS, OM, HL, PC | SI | ODA | 8.1 | 10815dT/4348C>T |
BR, bronchiectasis; CHD, congenital heart disease; HL, hearing loss; ND, not done; NRD, neonatal respiratory distress or hypoxemia; ODA, outer dynein arm; OM, chronic or recurrent otitis media; PC, daily productive cough; RS, rhinosinusitis; SA, situs ambiguus; SI, situs inversus totalis; SS, situs solitus.
DNAH5 mutations: exon 27 c.4348C>T (p.Q1450X) and exon 63 c.10815delT (p.P3606HfsX23).
Normal nasal NO level, >100 nL/min.
Specimens were collected for DNA isolation from subjects with PCD and unaffected first- and second-degree relatives with known consanguinity from Plain communities from Arkansas, Illinois, Missouri, Pennsylvania, and Wisconsin. In addition, DNA from unrelated Amish and Mennonite children with other genetic conditions without symptoms consistent with PCD were analyzed, to determine the relative frequency of DNAH5 mutations in Pennsylvania settlements.
DNA was extracted from blood or buccal swab specimens, and whole genome amplification was performed when necessary to provide adequate genetic material for further analysis. Genome-wide homozygosity mapping, using high-density single nucleotide polymorphism arrays, was performed following established protocols.19 Targeted mutation analyses were conducted by sequencing 9 exons of DNAH5 and DNAI1 using a clinical genetic panel developed at the University of North Carolina at Chapel Hill, which was developed based on results from large studies that showed at least 1 mutation present in the selected exons and splice sites in DNAI1 (exons 1, 13, 16, and 17) and DNAH5 (exons 34, 50, 63, 76, and 77) in most affected individuals in whom PCD was due to the mutations in those genes.8–12 This panel reduced DNA sequencing from a total of 99 exons. When only a single mutation was detected, all of the coding exons and splice sites were interrogated for the pertinent gene to identify a possible second mutant allele. Moreover, if biallelic mutations were not detected, at least 1 affected individual from that cohort was sequenced for all coding exons and splice sites for DNAI1 and DNAH5, to detect any mutations in exons not covered initially.20
To investigate linkage with 9 candidate genes known to be involved in outer dynein arm structure (DNAI1, DNAH5, DNAH11, DNAH9, DNAI2, DNAL4, DNCL1, DNLC2A, and TCTEL1), affected individuals and family members from Midwestern Amish-Mennonite communities were genotyped for polymorphic microsatellites within and flanking these genes. Haplotypes were reconstructed such that recombinants could be visualized on pedigrees.
Results
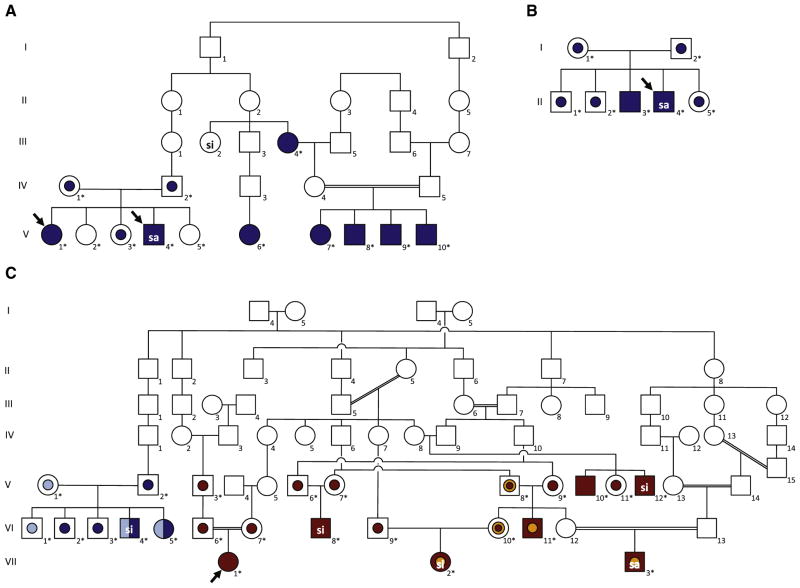
All of the subjects studied had classic features, and ultrastructural examination of the cliliary axoneme revealed dynein arm defects in several affected individuals consistent with a diagnosis of PCD (Table I). Identification of multiple members from several Amish families from central Pennsylvania facilitated genome-wide homozygosity mapping, using high-density single nucleotide polymorphism arrays. These analyses localized the disease locus to chromosome 5 in a region containing a known PCD-causing gene, DNAH5. Sequencing identified a nonsense mutation in exon 27, c.4348C>T (p.Q1450X), that was previously reported in a patient of Swiss descent.21 All of the affected individuals from this cohort were homozygous for this nonsense mutation (Figure, A; III4, V1, V4, V6, V7, V8, V9, and V10). This variant was also found in 2 affected siblings from an apparently nonconsanguineous Mennonite family in Arkansas (Figure, B; II2 and II3). Exomic analyses of 102 unrelated children without PCD from Amish and Mennonite families in Pennsylvania identified 18 missense or nonsense variants in DNAH5,11,12,21 including 1 subject heterozygous for DNAH5 c.4348C>T (p.Q1450X) (Table II).
Figure.
Family pedigrees and genetic analyses. A, Consanguineous kindred with related nuclear families in Amish communities from Pennsylvania. B, Mennonite family from Arkansas. C, Amish-Mennonite family with children with PCD from several midwestern states, in which affected individuals had 2 heteroallelic DNAH5 mutations. Solid symbols indicate affected individuals; central dots, heterozygous individuals; dark-blue symbols, c.4348C>T (p.Q1450X) DNAH5 mutation; light-blue symbols, c.10815delT (p.P3606HfsX23) DNAH5 mutation; yellow symbols, DNAI1 (g.IVS1+2_3insT) mutation; red symbols, HEATR2 (p.L795P) mutation. Arrows specify the probands. Asterisks indicate subjects who underwent genetic testing. si, situs inversus totalis; sa, situs ambiguus
Table II.
Exomic analysis revealing DNAH5 sequence variants on chromosome 5 in 102 unaffected individuals from Pennsylvania Amish communities
| Position | Identification no. | Reference | Alternate | Protein change | Variant | Mutation class |
|---|---|---|---|---|---|---|
| 13701536 | rs3734110 | T | C | p.I4450V | Novel | Missense |
| 13717471 | rs2277046 | T | C | p.T4220A | Novel | Missense |
| 13719089 | rs30168 | G | A | p.A4134V | Novel | Missense |
| 13719111 | rs148696723 | G | A | p.R4127C | Novel | Missense |
| 13737444 | rs17263496 | G | A | p.T3791I | Novel | Missense |
| 13754379 | - | C | A | p.L3496F | Novel | Missense |
| 13788886 | rs10513155 | C | A | p.L2862F | Novel | Missense |
| 13811775 | rs10078391 | T | C | p.Q2463R | Novel | Missense |
| 13817740 | - | A | C | p.F2302C | Novel | Missense |
| 13865784 | CM091933* | G | A | p.Q1450X | Reported | Nonsense |
| 13866375 | rs200516414 | C | T | p.S1357N | Novel | Missense |
| 13871757 | rs141168110 | G | T | p.Q1172K | Novel | Missense |
| 13871811 | - | C | T | p.E1154K | Novel | Missense |
| 13894894 | rs4701997 | T | A | p.I766L | Novel | Missense |
| 13902220 | rs1530498 | T | C | p.T558A | Novel | Missense |
| 13902245 | rs139160176 | G | C | p.N549K | Novel | Missense |
| 13931340 | rs1530496 | C | T | p.G24E | Novel | Missense |
| 13944512 | rs339445 | A | C | p.H12Q | Novel | Missense |
dbSNP, Single Nucleotide Polymorphism Database; HGMD, Human Gene Mutation Database; NCBI, National Center for Biotechnology Information.
Annotation and literature search: NCBI dbSNP, Ensembl Genome Browser (www.ensembl.org), and HGMD Public (www.hgmd.cf.ac.uk/ac/index.php).
Denotes HGMD identification number; all other identifiers are from NCBI dbSNP (build 137).
Resequencing of the DNAH5 coding region and flanking introns from subjects and family members from a large Midwestern Amish-Mennonite community revealed compound heterozygous DNAH5 mutations in 2 affected siblings: c.10815delT (p.P3606HfsX23) and c.4348C>T (p.Q1450X) (Figure, C; VI4 and VI5); however, several other family members with PCD did not have DNAH5 mutations. Genetic testing showed that 5 subjects (Figure, C; V8, VI10, VI11, VII2, and VII3) were heterozygous for the founder DNAI1 mutation, g.IVS1+2_3insT (c.48+2_3insT). Analyses of microsatellite genotype data confirmed that other affected individuals were disconcordant for microsatellite marker alleles at known PCD loci, including those associated with DNAH5 and DNAI1, indicating an unidentified PCD-causing mutation in a novel gene. Subsequently, whole-exome sequencing revealed an autosomal recessive, nonsynonymous missense mutation in HEATR2 (p.L795P), an uncharacterized gene not previously associated with ciliary assembly or function.22 Of note, 3 affected children were homozygous for the HEATR2 mutation and heterozygous for the DNAI1 mutation (VI11, VII2, and VII3), further illustrating the complex genetic basis of PCD in this community.
Discussion
PCD is a genetically heterogeneous, autosomal recessive disease resulting from impaired ciliary function. The estimated incidence is 1 in 15 000–30 000 births,23,24 with an estimated prevalence as high as 5% in children that have recurrent respiratory infections. Investigators have identified 15 different genes that when mutated produce unambiguous clinical phenotypes and in most cases ultrastructural defects of the ciliary axoneme.4,19,22,25,26
Arising from an Anabaptist movement, the Swiss Brethren, the endogamous Plain communities of North America present unique opportunities to better understand the genetic basis of PCD and identify new candidate genes. Relative isolation, detailed record keeping, and consanguineous marriages make Amish families ideal for genetic studies. We found a high prevalence of PCD in Amish settlements, based on classic clinical history, reduced nasal NO levels, and ultrastructural defects of the cilia. Many individuals harbored axonemal γ-dynein heavy chain DNAH5 mutations; affected individuals in Pennsylvania Amish communities and 2 subjects from a Mennonite family in Arkansas were homozygous for the nonsense c.4348C>T (p.Q1450X) mutation. Two Amish subjects from the midwestern US had biallelic mutations of DNAH5, specifically c.4348C>T (p.Q1450X) and frameshift c.10815dT (p.P3606HfsX23) mutations.
Homologous to HCγ in the biflagellated alga Chlamydomonas reinhardtii, DNAH5 is one of the first genes to have been implicated with disease.11,12 Since the discovery of DNAH5 and its involvement in PCD, more than 60 disease-causing mutations in the gene have been identified. Previously, investigators from Europe and the North America reported that 53% of patients with PCD and known outer dynein arm defects had mutations clustered in 6 exons of DNAH5,11 and DNAH5 is the most common gene linked to PCD with outer dynein arm defects,11 although the actual frequency of these mutations may be lower than reported.20
In addition, we previously shown that some affected individuals from the midwestern Amish families were heterozygous for a founder mutation, g.IVS1+2_3insT (c.48+2_3insT) in DNAI1 (Figure, C), the human ortholog of Chlamydomonas reinhardtii IC78 and the first gene associated with PCD identified using the candidate gene approach.17 This variant was not disease-causing in these families, because microsatellite marker analysis excluded DNAI1. Moreover, a second mutation was not identified after whole-exome sequencing.17
Even though we found PCD-causing mutations in DNAI1 and DNAH5 in these Anabaptist communities, most subjects in the midwestern Amish families had an unidentified mutation in a novel gene. Whole-exome sequencing revealed a single autosomal recessive missense mutation in HEATR2 (p.L795P). Airway epithelial cells isolated from individuals with PCD had markedly reduced cytoplasmic HEATR2 levels, absent dynein arms, and virtually immotile cilia. Recent analyses indicated a role in dynein arm transport and assembly.22
In conclusion, we have found that DNAH5 mutations are common to individuals with PCD from socially and geographically isolated Amish and Mennonite communities in the US. In addition, we have identified a known DNAI1 splice mutation and novel missense mutation in HEATR2 in these families. Taken together, a panel of these mutations could be a useful diagnostic tool for such patients who present with clinical features consistent with PCD.
Acknowledgments
Supported by the Children’s Discovery Institute (to T.F.) and the National Institutes of Health (NIH; R21 HL082657 [to T.F.], R01 HL071798 [to M.K. and M.Z.], and U54 HL096458 [to T.F., J.C., M.L., M.K., and M.Z.]). The last grant supports Genetic Disorders of Mucociliary Clearance Consortium (part of NIH Rare Diseases Clinical Research Network), which receives programmatic support from the National Heart, Lung, and Blood Institute and the NIH Office of Rare Diseases Research. The views expressed do not necessarily reflect the official policies of the Department of Health and Human Services; mention of trade names, commercial practices, or organizations does not imply endorsement by the US government.
We thank Carlos Perez, MD, for his insights into lung diseases that affect children from Pennsylvania Amish communities, as well as Susan Minnix, Jane Quante, Marianna Schmajuk, Adriana Lori, Michael Armstrong, Kimberly Burns, and Hilda Metjian for their clinical assistance and expert technical support.
Glossary
- NO
Nitric oxide
- PCD
Primary ciliary dyskinesia
Footnotes
The authors declare no conflicts of interest.
References
- 1.Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193:317–9. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 2.Leigh MW, Pittman JE, Carson JL, Ferkol TW, Dell SD, Davis SD, et al. Clinical and genetic aspects of primary ciliary dyskinesia and Kartagener syndrome. Genet Med. 2009;11:473–87. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noone PG, Leigh MW, Sannuti A, Minnix SL, Carson JL, Hazucha M, et al. Primary ciliary dyskinesia: diagnostic and phenotypic features. Am J Respir Crit Care Med. 2004;169:459–67. doi: 10.1164/rccm.200303-365OC. [DOI] [PubMed] [Google Scholar]
- 4.Ferkol T, Leigh M. Ciliopathies: the central role of cilia in pediatric disorders. J Pediatr. 2012;160:366–71. doi: 10.1016/j.jpeds.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowles MR, Boucher RC. Mucus clearance as a primary innate defense mechanism for mammalian airways. J Clin Invest. 2002;109:571–7. doi: 10.1172/JCI15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsui H, Grubb BR, Tarran R, Randell SH, Gatzy JT, Davis CW, et al. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell. 1998;95:1005–15. doi: 10.1016/s0092-8674(00)81724-9. [DOI] [PubMed] [Google Scholar]
- 7.Ostrowski LE, Dutcher SK, Lo CW. Cilia and models for studying structure and function. Proc Am Thorac Soc. 2011;8:423–9. doi: 10.1513/pats.201103-027SD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennarun G, Escudier E, Chapelin C, Bridoux AM, Cacheux V, Roger G, et al. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am J Hum Genet. 1999;65:1508–19. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guichard C, Harricane MC, Lafitte JJ, Godard P, Zaegel M, Tack V, et al. Axonemal dynein intermediate-chain gene (DNAI1) mutations result in situs inversus and primary ciliary dyskinesia (Kartagener syndrome) Am J Hum Genet. 2001;68:1030–5. doi: 10.1086/319511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zariwala MA, Leigh MW, Ceppa F, Kennedy MP, Noone PG, Carson JL, et al. Mutations of DNAI1 in primary ciliary dyskinesia: evidence of founder effect in a common mutation. Am J Respir Crit Care Med. 2006;174:858–66. doi: 10.1164/rccm.200603-370OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornef N, Olbrich H, Horvath J, Zariwala MA, Fliegauf M, Loges NT, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med. 2006;174:120–6. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olbrich H, H€affner K, Kispert A, Volkel A, Volz A, Sasmaz G, et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat Genet. 2002;30:143–4. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 13.Zariwala MA, Omran H, Ferkol TW. The emerging genetics of primary ciliary dyskinesia. Proc Am Thorac Soc. 2011;8:430–3. doi: 10.1513/pats.201103-023SD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strauss KA, Puffenberger EG. Genetics, medicine, and the Plain people. Ann Rev Genomics Hum Genet. 2009;10:513–36. doi: 10.1146/annurev-genom-082908-150040. [DOI] [PubMed] [Google Scholar]
- 15.Puffenberger EG. Genetic heritage of the Old Order Mennonites of southeastern Pennsylvania. Am J Med Genet. 2003;121C:18–31. doi: 10.1002/ajmg.c.20003. [DOI] [PubMed] [Google Scholar]
- 16.Morton DH, Morton CS, Strauss KA, Robinson DL, Puffenberger EG, Hendrickson C, et al. Pediatric medicine and the genetic disorders of the Amish and Mennonite people of Pennsylvania. Am J Med Genet. 2003;121C:5–17. doi: 10.1002/ajmg.c.20002. [DOI] [PubMed] [Google Scholar]
- 17.Lie H, Zariwala MA, Helms C, Bowcock AM, Carson JL, Brown DE, et al. Primary ciliary dyskinesia in Amish communities. J Pediatr. 2010;156:1023–5. doi: 10.1016/j.jpeds.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olin JT, Burns K, Metjian H, Carson JL, Davis SD, Dell SD, et al. Diagnostic yield of nasal scrape biopsies in primary ciliary dyskinesia: a multicenter experience. Pediatr Pulmonol. 2011;46:483–8. doi: 10.1002/ppul.21402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strauss KA, Puffenberger EG, Craig DW, Panganiban CB, Lee AM, Hu-Lince D, et al. Genome-wide SNP arrays as a diagnostic tool: clinical description, genetic mapping, and molecular characterization of Salla disease in an Old Order Mennonite population. Am J Med Genet. 2005;138A:262–7. doi: 10.1002/ajmg.a.30961. [DOI] [PubMed] [Google Scholar]
- 20.Zariwala M, Knowles M, Leigh MW. Primary ciliary dyskinesia. [Accessed October 29, 2012];Gene-Reviews at GeneTests. Available at: http://www.genetests.org.
- 21.Failly M, Bartoloni L, Letourneau A, Munoz A, Falconnet E, Rossier C, et al. Mutations in DNAH5 account for only 15% of a non-preselected cohort of patients with primary ciliary dyskinesia. J Med Genet. 2009;46:281–6. doi: 10.1136/jmg.2008.061176. [DOI] [PubMed] [Google Scholar]
- 22.Horani A, Druley TE, Zariwala MA, Levinson BT, Van Arendonk LG, Thornton KC, et al. Whole exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am J Hum Genet. 2012;91:685–93. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torgersen J. Situs inversus, asymmetry, and twinning. Am J Hum Genet. 1950;2:361–70. [PMC free article] [PubMed] [Google Scholar]
- 24.Katsuhara K, Kawamoto S, Wakabayashi T, Belsky JL. Situs inversus totalis and Kartagener’s syndrome in a Japanese population. Chest. 1972;61:56–61. doi: 10.1378/chest.61.1.56. [DOI] [PubMed] [Google Scholar]
- 25.Mitchison HM, Schmidts M, Loges NT, Freshour J, Dritsoula A, Hirst RA, et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat Genet. 2012;44:381–9. doi: 10.1038/ng.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kott E, Duquesnoy P, Copin B, Legendre M, Dastot-Le Moal F, Montantin G, et al. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am J Hum Genet. 2012;91:958–64. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]