Abstract
Lung cancer is the leading cause of cancer-related deaths in the US and worldwide. Better understanding of the disease is warranted for improvement in clinical management. Here we summarize the functions of small-RNA-based, posttranscriptional gene regulators, i.e. microRNAs, in the pathogenesis of lung cancers. We discuss the microRNAs that play oncogenic as well as tumor suppressive roles. We also touch on the value of microRNAs as markers for diagnosis, prognosis and the promising field of microRNA-based novel therapies for lung cancers.
Keywords: lung biology, lung cancer, microRNA
Introduction
Lung cancer has been the most fatal type of all cancers in the US for both males and females in recent decades. In the most recent statistical data from the American Cancer Society (ACS), lung and bronchus cancers are again estimated to cause the largest number of cancer-related deaths, accounting for 28% and 26% of estimated cancer deaths for US males and females respectively [1]. Globally, lung and bronchus cancers were also the most commonly diagnosed and most fatal cancers in males, while having the fourth highest incidence rate and second highest mortality rate in females [2]. Therefore, molecular studies aiming at early detection and targeted treatment of lung cancers draw intensive research interest.
MicroRNAs (miRNAs) are a group of non-coding RNA (~22nt) posttranscriptional regulators for gene expression [3]. The canonical pathway for miRNA biogenesis starts from transcription by RNA polymerase II. The resultant primary transcript pri-miRNA is cleaved by RNase III endonuclease Drosha, together with its cofactor DGCR8 [4], to generate the ~70nt long pre-miRNA. This hairpin structured transcript will be transported out of the nucleus to the cytoplasm by Exportin 5, and further processed there by another RNase III endonuclease Dicer and its cofactors to a double-stranded form. In most cases, one of the strands is selectively bound by Argonaut proteins [5] to enter the RNA induced silencing complex (RISC) while the complementary one is degraded [6]. But for some miRNAs [7], both strands can be functional. The two mature miRNAs derived from one stem loop are termed miRNA and miRNA* (or miR-5p and miR-3p), respectively. Individual mature miRNA sequences can be generated from one or more genes in the genome. The miRNA in the RISC complex recognizes target genes based on sequence complementarity primarily to the 3′ untranslated region (UTR) of target mRNAs. This binding of the RISC complex to the 3′UTR of a target RNA may result in translational inhibition or mRNA degradation [3]. Occasionally, miRNAs can bind to other regions of a mRNA in the protein-coding sequences [8] or 5′ UTR [9]. In selected cases, miRNAs were found to upregulate expression of target genes [10].
Significant progress has been made since Victor Ambros and colleagues discovered the first miRNA (lin-4) in 1993 [11]. In the latest release of the miRbase database (http://www.mirbase.org), there are 21643 mature miRNAs in 168 species. The functional importance of miRNAs is highlighted by their unique capability to simultaneously regulate multiple genes or pathways thus affecting a whole network of biological processes [12]. Since the first report of aberrant miRNA expression in cancers [13], these small RNAs are now realized to be key regulators for tumor pathogenesis and markers for novel clinical interventions. Here we review the roles that miRNAs play in the realm of lung cancers, either pro- or anti-oncogenic, and their potentials for clinical applications.
1. Oncogenic miRNAs in lung cancers
1.1 miR-17~92 cluster
This polycistronic miRNA cluster is encoded by a gene located at 13q31.3 and is comprised of 6 mature miRNA species: miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1 [14, 15]. Recently there are also some miRNA* species identified from the complementary strands of the above miRNAs [14]. The miR-17~92 cluster is also designated as oncomir-1 [16] and found to exhibit oncogenic behavior in multiple types of cancers including B-cell lymphoma, breast cancer, colon cancer, pancreatic cancer, glioblastoma, and retinoblastoma [17-22], etc. MiR-17~92 was found oncogenic first by the Hannon group [16]. However, the first piece of evidence that connected miR-17~92 to lung cancers was from the Takahashi group [19]. They showed that this cluster was markedly overexpressed in lung cancer cells and introduction of this miRNA clusterinto lung cancer cells enhanced cell growth. The Takahashi group later revealed that inhibition of miR-17-5p and miR-20a with antisense oligonucleotides induced apoptosis selectively in lung cancer cells overexpressing miR-17~92, suggesting “oncomiR addiction” [23]. The overexpression of members of this cluster in lung cancers was detected by microRNA profiling studies [24]. The expression of miR-17~92 is subject to transactivation by E2F family members [25] and MYC [26], with the latter frequently overexpressed in small cell lung cancers (SCLC) [27]. HIF-1α is a direct target of miR-17~92 identified in lung cancers [28]. This relationship in part explained the downregulation of HIF-1α by MYC. In lung cancer cellular context, this regulation affects cell proliferation under normoxia condition without influencing cellular adaptation for hypoxia. Overexpression of miR-17~92 counterbalances γ-HAX2 foci formation and reactive oxygen species (ROS) induced by knocking down the RB gene in RB wild-type lung cancer cells, suggesting a role of miR-17~92 in fine-tuning the DNA damage level in cancer cells to maintain genomic instability [29]. In order to identify other targets for miR-17~92, a proteomic approach was used in the SBC-3 SCLC cell line, which identified the RAS-related protein 14 (RAB14) as a direct target [30]. Since RAB14 silencing decreased surfactant secretion in the lung [31], downregulation of RAB14 by miR-17~92 was proposed to unguard the lung against external carcinogens, thus leading to cancer initiation and development [30]. Other targets identified for this cluster include E2F1-3, BIM, PTEN, TGFBR2, TSP1, CTGF and TNF-α [24, 25, 32–34], together with the above mentioned targets specifically studied in lung cancers rendering miR-17~92 a multifaceted oncomiR regulating cell growth and death, cell cycle progression, and angiogenesis processes in tumorigenesis.
1.2 miR-21
The miR-21 gene locates at chromosome 17 and is one of the more studied oncomiRs across cancer types. In 2006, a miRNA profiling study found that miR-21 was upregulated in all six types of solid tumors analyzed including lung cancers [24]. An antisense oligonuceotide against miR-21 inhibited growth of lung cancer cell line A549 [35]. MiRNA expression analysis of tumors derived from never-smoker lung cancer patients detected miR-21 overexpression and this overexpression was more pronounced in epidermal growth factor receptor (EGFR) gene mutant cases than in wild type cases [36]. MiR-21 level was correlated with phosphorylated EGFR (p-EGFR) level and was suppressed by EGFR tyrosine kinase inhibitors (EGFR-TKI). Antisense miR-21 enhanced EGFR-TKI induced apoptosis, with the level depending on the p-EGFR level in the host cell lines. All these data suggest miR-21 as an antiapoptotic factor regulated by the EGFR signaling pathway in lung cancers [36]. The PTEN tumor suppressor gene (TSG) was identified as a direct target of miR-21 and repression of this target by miR-21 promoted cell growth and invasion of non-small cell lung cancer (NSCLC) [37]. The mouse model-based study by Hatley et al. clearly demonstrated the oncogenicity of miR-21 in lung cancers [38]. MiR-21 overexpression in K-rasLA2 mice increased tumor proliferation, while miR-21 deletion in the same lung tumor mouse model suppressed tumor development. They also validated multiple predicted miR-21 targets including negative regulators of the RAS/MEK/ERK pathway: Spry1, Spry2, Btg2, and Pdcd4. Several pro-apoptotic targets of miR-21 were also identified: Apaf1, Faslg, RhoB, and Pdcd4. Research by Frezetti et al. [39] in thyroid and lung cancers confirmed miR-21 upregulation by RAS in vivo and showed that this upregulation requires activation of two downstream pathways: RAF/MAPK and PI3K. They found that the predicted targets of miR-21 were enriched in the regulators for cell cycle checkpoints. This finding highlights the role of miR-21 in cell cycle regulation. FOXO3a was reported to transcriptionally repress the expression of miR-21 and affect apoptosis in lung cancer cells [40]. Besides RAS and FOXO3a, miR-21 was also found to be regulated by IL-6 in multiple myeloma cells [41], IFN in prostate cancer cells [42], HER2/neu in breast cancer cell lines [43], and AP-1 in multiple types of cancer cells at the transcription level [44]. Posttranscriptionally, miR-21 is regulated by type I collagen in lung and breast cancer cells [45], PTEN in glioblastoma cells [46], and TGF-β in breast cancer cell lines [47]. Targets of miR-21 identified outside lung cancers include TPM1 and MASPRIN, which mediate tumor invasion and metastasis [48].
1.3 miR-221/222
The Apo2L/TNF-α-related apoptosis-inducing ligand (TRAIL) is a relatively new member of the TNF family known to induce apoptosis in a variety of cancers [49] and TRAIL based therapy is a promising anti-tumor agent in clinical trials [50]. In a study to identify miRNA expression signatures in TRAIL resistant NSCLC cells, upregulation of miR-221/222 was seen in NSCLC cells resistant to TRAIL [51]. MiR-221 and miR-222 are expressed from the same gene cluster on the X chromosome. They share an identical seed sequence and have overlapping targets by prediction. Introduction of inhibitors to miR-221/222 into TRAIL-resistant cells increased TRAIL sensitivity. Two genes were identified as targets of miR-221/222: Kit and p27Kip1. In NSCLC cells, a decrease in the level of p27Kip1 seems to account for the decreased sensitivity to TRAIL-induced apoptosis [51]. However, later on, additional targets of miR-221/222 – PTEN/TIMP3 [52] and PUMA [53] - were also suggested to mediate TRAIL resistance, consistent with frequent overexpression of miR-221/222 in epithelial cancers [54-56]. MiR-221/222 upregulation is common in more invasive lung cancers, and their functions in lung tumorigenesis involve promotion of invasion through the activation of AKT pathway and metallopeptidases. MiR-221/222 are subjected to activation by MET through c-JUN [52]. Interestingly, miR-130a has been linked to TRAIL-sensitivity regulation by downregulating miR-221/222 via the interaction with the 3′UTR of MET [57]. Besides MET, miR-221/222 are also positively regulated by EGFR and could decrease sensitivity of lung cancer cells to gefitinib-induced apoptosis by targeting the apoptotic peptidase activating factor 1 (APAF-1) [58].
1.4 Other miRNAs that promote lung tumorigenesis
Liu et al. [59] reported that in human bronchial epithelial cells transformed by chemical carcinogen anti-Benzo-A-Pyrene-Diol-Epoxide (anti-BPDE), miR-494 was upregulated and thus repressed tumor suppressor PTEN. The activities of caspase-3/7 were also decreased due to the alteration in miR-494 level, suggesting miR-494 as an oncomiR by repressing apoptosis of cancer cells. MiR-27a was found activated in SV40 small T antigen transformed bronchial epithelial cells, and downregulation of miR-27a in such cells decreased the ability of cells to grow in an anchorage-independent manner. MiR-27a overexpression promotes cell growth by suppression of the target gene FBXW7 which contributes to the malignant transformation of the primary human bronchial epithelial cells [60]. MiR-328 expression level in the lung tumor samples from patients with brain metastases is significantly higher than that from patients without brain metastases. Introduction of miR-328 into A549 cells promoted cell migration comparing to parental A549 cells [61]. MiR-301 is an intronic miRNA hosted in the SKA2 gene. It can positively regulate expression of its host gene through the ERK/CREB pathway. Inhibition of miR-301 or SKA2 in A549 cells increased cell proliferation and invasion as reflected by elevated mitotic index and colony formation [62]. The FUS1 gene is believed to be a TSG based on its frequent expression deficiency in lung cancers and its tumor suppressive activity in mouse model systems [63-65]. Three miRNAs - miR-93, miR-98, and miR-197 - could directly downregulate FUS1 [66]; these three miRNAs might promote lung cancer growth through repression of FUS1. MiR-20, miR-106, and miR-150 were identified as oncomiRs in lung cancer per their activities in regulating A549 cell growth and apoptosis, with miR-106 and miR-150 targeting RB and TP53 respectively [67]. Antisense oligonucleotides against miR-150 reduced volume and weight of established mouse lung tumor xenografts [68]. MiR-30b/c are often downregulated in MET or EGFR-silenced lung cancer cells [58]. Enforced expression of miR-30b/c would inhibit gefitinib-induced apoptosis by targeting BIM [58]. MiR-29a/c and miR-100 exhibit similar activities but the mechanism is not clear [58].
2. Tumor suppressive miRNAs in lung cancer
2.1 let-7 family
Let-7 was one of the early few miRNAs uncovered in C. elegans and subsequently was the first miRNA identified in humans [69, 70]. The sequences and functions of this family are well conserved across species. There are 13 members to this family in humans: let-7a-1, 7a-2, 7a-3, 7b, 7c, 7d, 7e, 7f-1, 7f-2, 7g, 7i, miR-98, and miR-202 [71]. Since there are multiple review articles dedicated to let-7s, readers are referred to them for an in-depth discussion [71-76].
2.2 miR-34/449 family
In contrast to let-7 being one of the earliest studied miRNAs, the miR-34/449 family came under spotlight only in recent years. In 2007, several groups reported the connections between miRNAs and the p53 network: miR-34a and miR-34b/c were found to be under direct regulation of p53 to control apoptosis and cell cycle arrest in cancer cell lines [77-80]. MiR-34a and miR-34b/c are homologues and belong to the same family. Another miRNA family discovered later, miR-449a, 449b and 449c together abbreviated as miR-449, share identical seed sequence and secondary structures with miR-34 and therefore was assigned as part of the miR-34 family [81]. MiR-34/449 form a feedback network with p53 and E2F transcription factors [81]. P53 activates miR-34 while E2F activates miR-449. Both miRNAs directly suppress E2F but upregulate p53 by targeting dyacetylase gene SIRT1. We categorize the pleiotropic activities of these miRNAs into these classes: (a) arresting cell cycle by targeting CCND1, CCNE2, CDC25A, CDK4, CDK6, c-MYC, E2Fs, and MET; (b) conferring apoptosis by targeting BCL-2, GMNN, N-MYC, HDAC1, SIRT1 and MET; (c) activating senescence by repression of c-MYC, HDMX, and SIRT1; (d) inhibiting cancer cell migration/invasion by repression of HMGA2, AXL, SNAIL1, and SERPINE1 [81-84].
Several reports have linked miR-34 to lung cancers. In one of the early papers on miR-34 and p53 [85], researchers showed that miR-34b/c expression was significantly reduced in 6/14 (43%) NSCLC samples and restoration of miR-34 inhibited lung cancer cell growth. Greatly reduced expression of miR-34 was found in rat lungs exposed to a chemical carcinogen (nicotine-derived nitrosamine ketone (NNK)) or cigarette smoke [86, 87], in line with the tumor suppressive role of miR-34. MiR-34a and miR-15/16 synergistically induced cell cycle arrest in NSCLC cells in a RB-dependent manner [88]. AXL [83] and SNAIL1 [82] as miR-34 direct targets were identified in lung cancer cells. MiR-34 expression would inhibit lung cancer cell migration and invasion via repression of these genes. Interestingly, a very recent study of miR-34 and SNAIL in colon cancer cells found that SNAIL actually directly repressed miR-34, forming a double negative feedback loop [89]. Pulmonary tissue is a site where miR-449 is preferentially expressed (the other site is testis) [90]. But the literatures about miR-449 and lung cancer are limited. One study detected reduced expression of miR-449a/b in lung cancer tissues compared with matched normal tissues [91]. HDAC1 was realized as a target of miR-449a/b in lung cancer cells [91]. Ectopic expression o f m i R-449a/b decreased cell viability in multiple lung cancer cell lines and anchorage-independent growth of H1299 cells. Combination of either miR-449a or miR-449b with HDAC1 inhibitor elicited stronger growth inhibition than the inhibitor mono-treatment. Another group reported that introduction of miR-449a/b into H1299 cells induced apoptosis [92]. All these results confirmed that miR-34/449 family acts as tumor suppressors in lung cancers.
2.3 miR-15/16
MiR-15a and miR-16-1 are located at 13q14.3 and were the first miRNAs implicated in oncogenesis. They were found to be frequently deleted or downregulated in chronic lymphocytic leukemia (CLL) in 2002 [13], suggesting their role as tumor suppressors. Gene targets of this cluster include BCL-2, CDC2, CCND1, ETS1, JUN, MCL1, MSH2, PDCD4, PDCD6IP, RAB9B, WT1, and WNT3A, through which this miRNA cluster regulates multiple aspects of tumorigenesis, including cell survival, proliferation and invasion (reviewed in Ref. [93]). Other targets of this cluster were identified along the line of their tumor suppressor functions. MiR-15a and miR-16-1 together with miR-15b and miR-16-2 were identified as direct targets of E2F1 and controls E2F-dependent cell proliferation by targeting CCNE1 [94]. Fibroblast growth factor 2 (FGF-2) and its receptor FGFR1 were identified as novel targets which affect both stromal cells and prostate cancer cells to promote cancer cell proliferation and invasion [95]. In lung cancers, the miR-15/16 cluster was also frequently deleted or downregulated as in other types of cancers [96]. In NSCLC cell lines, the expression level of the miR-15/16 cluster was inversely correlated with expression level of Cyclin D1. Cyclins D1, D2 and E1 are directly regulated by these miRNAs in lung cancer cells and overexpression of the miR-15/16 cluster induced G1 arrest in an RB-dependent manner. When combined with miR-34a, the induction of G1 arrest was more profound than the additive effect of treating cells with the two miRNAs separately [88], indicating a cooperation between the two tumor suppressive miRNA clusters.
2.4 miR-200 family and miR-205
The miR-200 family has five members: miR-200a, miR-200b, miR-429, miR-200c, and miR-141. In humans, the first three miRNAs co-localize at chromosome 1 and the latter two co-localize at chromosome 12 [97]. This family together with miR-205 inhibited epithelial-mesenchymal transition (EMT) through targeting ZEB1 and ZEB2 [97], highlighting their significance in the regulation of cancer pathogenesis especially with regard to invasion and metastasis. In lung cancers, miR-200c overexpression reduced ZEB1 expression and derepressed the transcriptional target of ZEB1, E-cadherin, in A549 cells [98]. Murine model systems revealed that forced expression of the miR-200 family in metastasis-prone mouse lung tumor cells blocked the capability of these cells to go through EMT, thus weakening cellular invasive and metastatic potentials with concomitant alterations in the expression profiles of epithelial and mesenchymal cell marker genes [99]. Expression analysis in 39 human lung cancer cells lines detected strong correlations of miR-141 level with the expression of CDH1, CDH2, ZEB1, ZEB2, and VIM [99]. In a separate study [100], miR-200c expression was shown to be negatively correlated with invasion in a panel of 9 NSCLC cell lines and ectopic expression of miR-200c in highly invasive cells inhibited mesenchymal phenotypes and in vivo metastasis formation. MiR-200b prevented malignant transformation of human bronchial epithelial cells induced by the chemical carcinogen arsenic [101].
Further studies revealed targets beyond ZEB1 and ZEB2 that mediate miR-200s/miR-205-dependent regulation of EMT and metastasis in lung cancers. The Kurie group utilized bioinformatics tools to search for targets of miR-200s, and validated one of the 35 predicted targets [102]: Flt1 in a mouse lung cancer system. They later identified GATA3, a component of the Notch signaling pathway, as downregulated by miR-200s [103]. In a luciferase reporter assay, although GATA3 3′UTR was inhibited by miR-200s to a statistically significant level, the level of inhibition was not as profound as seen with the ZEB1 3′UTR. Thus, GATA3 may be an indirect target of miR-200s. To expand their efforts in elucidating miR-200 targets in the EMT regulation pathway, they harnessed a proteomics approach to compare expression profiles of lung cancer cells with or without miR-200s restoration [104]. Over 300 proteins showed changed expression, which can be categorized as peptidases, cell adhesion andextracellular matrix (ECM) proteins and cytoskeletal regulators. Surprisingly, Korpal et al. discovered that miR-200s promote metastatic colonization to the lung by direct targeting Sec23a [105]. The miR-200 family also regulates multidrug sensitivity in lung cancers [106]. MiR-200b/c and miR-429 were down-regulated in multidrug resistant cells and restoration of this cluster sensitized cells to anti-cancer drugs. The regulation of drug-induced apoptosis is through targeted inhibition of anti-apoptotic factors BCL-2 and XIAP.
2.5 miR-143/145
MiR-143 and miR-145 are co-transcribed from a bicistronic gene cluster on chromosome 5 [107]. They have been identified as tumor suppressors in multiple cancers such as colorectal cancer, bladder cancer, gastric cancer, cervical cancer, breast cancer, prostate cancer and leukemia [108-114]. In an analysis of miRNA expression patterns in the rodent lung exposed to carcinogenic cigarette smoke [86], miR-145 was one of the severely downregulated miRNAs, connecting this miRNA to lung carcinogenesis. Soon after that, the inhibitory effect of miR-145 on cell growth was reported in mouse lung cancer cells [115] and confirmed in humans [116]. C-MYC, EGFR, NUDT1, and OCT4 were all identified as targets and mediators of miR-145 for cell proliferation regulation in lung cancers [117-119]. Besides cell growth, miR-145 regulates cell cycle and causes G1 arrest by targeting CDK4 in lung cancers [118]. Inhibition of lung cancer metastasis by miR-145 was observed in an animal model, partly by targeting MUC1 [120]. Similarly, miR-143 was downregulated in chemical-induced mouse lung tumors [121], whereas in human lung tumor samples, expression of both miR-145 and miR-143 were reduced comparing to normal tissues [122]. In contrast to miR-145, less is known regarding the involvement of miR-143 in lung tumorigenesis.
2.6 Other miRNAs that suppress lung tumorigenesis
The miR-29 family (miR-29a, b, and c) activates p53 by targeting its negative regulators, p85α and CDC42. This family has been extensively studied in CLL [123-125]. In the pulmonary tissue, miR-29s are considered important regulatory factors for fibrosis [126]. In lung cancers, expression of miR-29s is inversely correlated to that of the DNA methyltransferases DNMT3A and -3B [127], both of which are direct targets of miR-29s. The enforced expression of miR-29s in lung cancer cell lines inhibited tumorigenicity in vitro and in vivo through demethylation and reactivation of epigenetically silenced TSGs such as FHIT and WWOX [127]. Other important targets that mediate miR-29 function as a tumor suppressor include TTP [128] (which regulates cell polarity and metastasis) and the BCL-2 family member MCL-1 [129].
MiR-1, miR-133, and miR-206 are three so called “muscle specific” miRNAs highly expressed in cardiac and smooth muscle tissues [130, 131]. They exhibit cancer suppressive activities in the lung. MiR-1 was downregulated in primary lung cancer samples and cell lines [132]. Enforced expression of miR-1 in A549 and H1299 cells inhibited cell growth, replicative potential, motility/migration, clonogenic survival and tumorigenicity in mice [132]. Targets mediating these effects include MET, PIM-1, HDAC4, and FOXP1. MiR-133a is expressed at a reduced level in lung squamous cell carcinomas (SqCC) relative to normal tissues [133]. Restoration of miR-133a in lung SqCC cell lines inhibited cell proliferation. Multiple targets were predicted and two of them (ARPC5 and GSTP1) were confirmed in lung cancer cells [133]. Analysis of miR-206 expression in lung tumors and cell lines established that miR-206 level is lower in highly metastatic tumors. Upregulation of miR-206 in cancer cell lines resulted in induction of apoptosis and inhibition of cell proliferation, migration, and invasion [131].
MiR-126 and its complementary species miR-126* are encoded by intron 7 of the Epidermal Growth Factor-Like Domain 7 (EGFL7) gene [134]. MiR-126/126* are important factors for angiogenesis and vascular integrity through repression of inhibitors of vascular epithelial growth factor (VEGF)-induced proliferation (reviewed in [134]). In terms of tumorigenesis, studies so far have assigned them as tumor suppressors. Both miR-126 and miR-126* were downregulated in lung cancers [134-137]. Through targeting VEGF, miR-126 decreased lung cancer cell growth, induced cell cycle arrest at G1 phase, and impaired tumorigenicity of mouse-hosted tumor xenografts [138]. By downregulating CRK at protein level, miR-126 retarded adhesion, migration, and invasion of NSCLC cells [139, 140]. MiR-126 also promoted irradiation-induced apoptosis in NSCLC cells [137]. In SCLC cells, miR-126 slowed down cellular proliferation via targeting SLC7A5 [141]. It is known that miR-126 negatively regulates its host gene EGFL7 and this feed-back signaling manifests itself in lung tumorigenesis both in vitro and in vivo [142].
Frequent loss of heterozygosity (LOH) for miR-128b located on chromosome 3p was discovered in NSCLC samples [143]. EGFR was realized as a direct target for this miRNA [143]. Thus, a reduction in miR-128 expression would promote carcinogenesis via derepression of an oncogene (i.e, EGFR). MiR-142-5p was suppressed in mouse and human lung cancers, and transfection of miR-142-5p significantly suppressed lung cancer growth [115]. Expression of miR-451 was reduced in human lung cancer samples and ectopic expression of this miRNA significantly inhibited NSCLC cell proliferation and colony formation as well as xenograft tumor growth in nude mice by inducing apoptosis [144]. RAB14 was a confirmed target mediating this effect [144]. A reduced level of miR-101 was seen in human lung cancers and correlated with the overexpression of either one of the two targets: EZH2 [145] or MCL-1 [146]. Introduction of miR-101 into NSCLC cells decreased cell proliferation, invasion and sensitivity to drug-induced apoptosis [145]. MiR-218 negatively impacted NSCLC cell proliferation, invasion and soft agar colony formation by repressing PXN [147]. MiR-129 repressed cell cycle progression thus cell growth of lung adenocarcinoma (AD) cells by targeting CDK6 [148]. MiR-212 induced apoptosis of NSCLC cells by targeting an antiapoptotic protein PED [149]. Since miR-519c negatively regulates the expression of HIF-1α [150] (a key factor for tumor angiogenesis [151]), this miRNA inhibits angiogenesis of lung cancers [150]. The cancer prevention activity of the green tea polyphenol epigallocatechin-3 gallate (EGCG) is believed to be partially mediated by miR-210 [152]. Two miRNAs under negative regulation of the MET oncogene, miR-103 and miR-203, were shown to promote gefitinib-induced apoptosis through inhibition of protein kinase C ε (PKC-ε) and sarcoma viral oncogene homolog (SRC), respectively [58].
3. miRNAs that exhibit both pro- and anti-lung cancer functions
As in protein-coding genes, individual miRNAs may exhibit both pro- and anti-cancer activities in a context-dependent manner. Below, we discuss selected cases with relevance to lung cancers.
3.1 miR-7
In the human genome, there are three miR-7 host genes on chromosome 9, 15, and 19, respectively. The function of this miRNA was first studied in Drosophila [153]. In 2008, it was identified as a tumor suppressor in glioblastomas [154], directly targeting EGFR as well as downregulating the AKT pathway to decrease viability and invasiveness of cancer cells. This effect was confirmed in the A549 lung cancer cells a year later [155]. Moreover, miR-7 was also reported to inhibit A549 cell growth by targeting BCL-2 [156]. On the other hand, Chou et al. reported miR-7 as an oncomiR in lung cancers [157]. They detected an elevated miR-7 expression in human lung cancers and a significant correlation between EGFR and miR-7 expression in EGFR mutated but not in EGFR-wild-type lung cancers. EGFR can induce miR-7 expression through the RAS/ERK/MYC pathway. Enforced expression of miR-7 promoted proliferation and tumorigenicity of CL1-5 lung cancer cells by targeting Ets2 transcriptional repressor (ERF) [157].
3.2 miR-31
The host gene encoding hsa-miR-31 is at chromosome 9. The expression of miR-31 in the lung can be induced by chemical carcinogens like cigarette smoke or vinyl carbamate [121, 158]. MiR-31 is also overexpressed in both mouse and human lung cancers [158, 159]. Gain- and loss-of-function experiments established the role of miR-31 on enhancing proliferation and tumorigenicity of lung cancer cells [158, 159]. Two TSGs are targets of miR-31: LAST2 and PPP2R2A [159]. On the contrary, in established pulmonary metastasis of breast cancer, miR-31 would trigger regression of the metastasis by targeting ITGA5, RDX, and RhoA [160, 161], playing a metastasis suppressor role.
3.3 miR-125
The miR-125 family includes two members with identical seed sequence: miR-125a transcribed from a gene at chromosome 19, and miR-125b encoded by two separate genes at chromosome 11 and 21 respectively. In 2007, both microRNAs were identified as tumor suppressors in breast cancer through downregulation of ERBB2 and ERBB3 genes [162]. Their tumor suppressive functions were also confirmed in other cancers like glioblastoma [163] and gastric cancer [164]. Perplexingly, miR-125a/b also target TP53, suggesting their roles as oncomiRs [165, 166]. In lung cancers, genes encoding miR-125b were frequently deleted [167, 168]. MiR-125a-3p, a newly identified derivative of miR-125a, has an opposing function to miR-125a-5p (previously miR-125a). A reduced expression of miR-125a-3p was reported in NSCLC tissues [169] and in lung cancers cells with higher metastatic potential [170]. The role of miR-125a-3p in inhibiting lung cancer invasion and metastasis was confirmed by both loss- and gain-of-function experiments [169]. As to miR-125a-5p, findings so far have been inconclusive as miR-125a-5p has been found to be capable of inhibiting [171] or promoting [169, 172] metastasis.
3.4 miR-183-96-182
Human miR-183 family members (miR-183, miR-182, and miR-96) are co-expressed from an intergenic region on chromosome 7. They have both common and distinctive targets. All three members repress zinc transporters that regulate zinc homeostasis in cancers [173]. FOXO1 is another common target [173]. Distinctive targets for different family members include: EZRIN, ITGB1, KIF2A, PDCD4, and EGR1 for miR-183 [174-177]; KRAS for miR-96 [178]; RGS17, BRCA1, and CTTN for miR-182 [179-181]; FOXO3 for miR-96 and miR-182 [182, 183]. MiRNAs in this family regulate the development of multiple cancer types like breast cancer [184-187], colorectal cancer [188, 189], hepatocellular carcinoma [175], endometrial cancer [173], pancreatic cancer [178, 190], ovarian cancer [191], and bladder urothelial carcinoma [192], etc. All three members have been shown to have both oncogenic and tumor suppressive functions. In lung cancers, overexpression of the miR-183 family was identified as a risk factor [193]. But miR-183 was also demonstrated to inhibit lung tumor invasion and metastasis through targeting EZRIN [177]. Likewise, a similar scenario with invasion and metastasis has been reported for miR-182 and its target CTTN [181]. MiR-182 inhibited lung cancer cell proliferation and anchorage-independent growth by suppression of RGS17 [180]. Research for specific targets and functions of miR-96 has not been conducted in lung cancers. In view of the evidence that the miR-183 family keeps invasion and metastasis in check [177], the positive correlation of miR-183 overexpression with lung cancer risk may indicate an oncogenic role for miR-183 in other aspects of tumorigenesis, such as cell death control or angiogenesis.
4. miRNAs and lung developmental genes
Thyroid transcription factor 1 (TTF-1 or NKX2-1) is a member of the NKX family transcription factors characterized by the DNA binding homeodomain [194]. The expression of TTF-1 is largely restricted to lung, thyroid, and forebrain [195]. The role of TTF-1 in lung development and morphogenesis has been well established [196]. Ttf-1 null mice were born dead and lacked lung parenchyma [195]. Genetically engineered mice expressing a mutant form of TTF-1 resistant to phosphorylation at seven serine residues die immediately after birth. Although lobulation and branching morphogenesis were maintained, the mutant animals displayed pulmonary hypoplasia and abnormalities in acinar tubules [197]. These reports explicitly identified TTF-1 as a master regulator of lung development. In 2007, TTF-1 amplification in lung cancers was reported by us and others [198-201], with gain- and loss-of-function studies in cell systems suggesting TTF-1 as a lineage specific oncogene. But the subsequent reports of TTF-1 suppressing lung cancer development in a mouse model [202] and TTF-1 counteracting TGF-β-induced EMT in lung cancer cells [203] complicate this issue. Despite the complexity, the study about molecular mechanisms of TTF-1 regulation is warranted. To this end, our laboratory utilized a candidate-based approach to search for miRNAs that regulate TTF-1 [204]. We were able to validate one miRNA (miR-365) out of the ten candidates predicted by TargetScan algorithm [205]. MiR-365 is encoded by two genes located at chromosome 16 and 17. Both genes repressed the TTF-1 3′UTR reporter and reduced endogenous TTF-1 protein expression when transduced into lung cancer cells. TGF-β repression of TTF-1 seemed to be at least partially mediated by inducing miR-365. This miRNA also upregulates TGF-β2 to form a signaling loop [204]. These observations have opened up a new and miRNA-based angle to examine the biology of TTF-1. Achaete-scute homologue-1 (ASH1) is a basic helix–loop–helix transcription factor important for the development of neuroendocrine (NE) features in normal lung cells as well as lung cancer cells. Newborn mice with a disrupted ASH1 gene had no detectable pulmonary NE cells. Knockdown of ASH1 in lung cancer cells with NE features repressed expression of the NE markers [206]. Lung cancer cells with NE features are often very aggressive, and ASH1 was shown to promote airway dysplasia and tumorigenicity of SV40 large T antigen in the airway epithelium [207]. This suggests that ASH1 is more than a mediator for the induction of NE features in the lung. Recently, miR-375 was identified as a transcriptional target of ASH1. This microRNA was normally coexpressed with ASH1 in a lineage–dependent manner, and alleviated the growth inhibitory effect in lung cancers imposed by the miR-375 target gene YAP1, thus mediating tumor promoting activity of ASH1 [208].
5. Clinical implications
The intertwined network of miRNAs, oncogenes, TSGs, and lung developmental genes implicates ubiquitous involvement of miRNAs in all aspects of lung cancer pathogenesis. This raises the possibility of developing novel methods for lung cancer interventions, including diagnosis, risk evaluation, outcome prediction, or treatment using miRNAs.
5.1 miRNAs for lung cancer diagnosis
In 2006, high expression of let-7a-2 and low expression of miR-155 were used to discriminate lung cancer tissues from noncancerous lung tissue [209]. Later, circulating miRNAs in the blood plasma and serum were found to be stable [210]; and an elevated expression of miR-25 and miR-223 was identified as a biomarker for NSCLC. This finding paved the way for noninvasive diagnosis of lung cancers using blood-miRNA expression patterns. Following that, extensive studies gave rise to a burst of different “miRNA signatures” for lung cancer diagnosis. For example, simultaneous overexpression of miR-155, miR-182, and miR-197 discriminated lung cancers from cancer-free controls with 81.33% sensitivity and 86.76% specificity in a cohort of 74 patients and 68 matched controls [211]. Diagnosis of NSCLC based on elevated levels of serum miR-1254 and miR-574-5p showed over 70% sensitivity and specificity in two separate cohorts [212]. Boeri et al. [213] generated a plasma-derived signature consisting of 16 miRNA expression ratios from 13 miRNAs for lung cancer diagnosis with >80% sensitivity and specificity. Moreover, they reported a 15-miRNA signature predictive for lung cancer development in disease-free smokers, which might be even more helpful for early detection of disease.
Sputum as a unique type of body fluid in lung cancer diagnosis has been found to contain miRNAs that can also serve as a biomarker for lung cancer detection. From sputum samples, the Jiang group found that overexpression of miR-21 clearly discriminated NSCLC patients from cancer-free individuals [214]. Overexpression of miR-21, miR-200b, and miR-375 combined with a reduced level of miR-486 segregated AD patients from normal individual with 80.6% sensitivity and 91.7% specificity [214], while downregulation of three miRNAs (miR-205, miR-210, and miR-708) predicted SqCC with 73% sensitivity and 96% specificity [214]. Another miRNA signature, based on the expressions of 34 miRNAs, detected early-stage NSCLC in asymptomatic individuals with 80% accuracy [215].
Besides discriminating cancer from normal status, miRNAs can also help classify lung cancer subtypes. A miRNA expression study in 165 AD and 125 SqCC tissues led to 127 miRs with significant differential expression between AD and SqCC groups [216]. In another study with 60 SqCC patients and 62 AD patients, miR-205 was identified as a highly specific marker of SqCC, and the expression ratio of miR-205/miR-21 can differentiate SqCC against AD with over 90% accuracy and sensitivity [217]. The robustness of miR-205 and miR-21 expressions in discriminating AD and SqCC was confirmed in a separate study recently [213]. These miRNA-based diagnostic markers are of promising clinical value since SqCC and AD require different treatments. MiRNAs could also serve as a marker to differentiate primary lung tumors from lung metastases from other sites. Barshack et al. found that miR-182 was most significantly overexpressed in primary lung tumors while miR-126 was overexpressed in lung metastases in two separate sets of samples tested [218].
A factor that impedes the translation of laboratory-derived miRNA expression signatures into clinics is the lack of reproducibility [219]. This could be partly explained by discrepancies in microarray platforms, experimental operations, analysis methods, and sample properties (e.g., ethnicity of patients). Moreover, miRNA signatures in lung cancers change with disease progression [220]. But more importantly, the small number of samples analyzed in most of the profiling studies and the correlated data structure contributed to a large part of the signature instability [219]. To develop better miRNA biomarkers for clinical use, a much larger sample size and a standard operating protocol with standardized platforms and data analysis methods are essential. Markers developed in such a manner are likely to have a higher success rate for clinical adaptation.
5.2 miRNAs for lung cancer prognosis
In the first report bringing miRNAs into the lung cancer research field, a low level of let-7 expression was found to be significantly associated with poor survival in surgically-resected NSCLC patients [221], suggesting a prognostic value for miRNAs. This was corroborated by a large number of subsequent studies. The prognostic value of let-7 was initially confirmed [209] but later refuted in a set of 51 AD samples [222]. The correlation of high miR-21 level, in tumors or sera, with poor survival of lung cancer patients, was reported by several groups independently [122, 134, 223-227]. Lower expression of individual miRNAs other than let-7 predicted poor survival or high recurrence of lung cancers. These miRNAs include miR-34a [228], miR-374a [229], miR-181a [122], miR-221 [230], miR-200c [223], and miR-218 [147]. On the other hand, some miRNAs showed the opposite predictive trend (similar to miR-21). MiRNAs in this latter group include the miR-183 family (miR-182, miR-183, and miR-96) [193], miR-31 [188], miR-16 [231], miR-92a-2* [232], and miR-126 [233]. The correlation of high miR-126 level with poor prognosis is especially significant in the SqCC subtype. Also, a high level of miR-126 was correlated with increased VEGF in the tumor, and co-expression of miR-126 and VEGF had a significant diagnostic impact for poor survival. The diagnostic value for miR-155 is still ambiguous. An initial study correlated high level of miR-155 with poor prognosis [209], whereas a later report showed that miR-155 level was a negative predictor for AD, but a positive one for SqCC with lymph node metastases. If not divided by subtype, the overall patient survival in the second study was not associated with the level of miR-155 [234]. For SqCC, miR-146b alone was found to have the prediction accuracy for stratifying prognostic groups at 78% [235].
Several other signatures are based on the combined expression levels of multiple miRNAs. A high level of miR-486 and miR-30d combined with a low level of miR-1 and miR-499 in the serum could significantly predict poor survival for NSCLC [236]. And a five-miRNA signature (miR-25, miR-34a, miR-34c-5p, miR-191, and let-7e) was reported by the Wang group to predict survival for SqCC subtype specifically [216]. A plasma signature of 9 miRNAs in pre-disease samples of lung cancer patients reliably predicts the risk of aggressive development of the disease with poor prognosis [213]. Interestingly, the samples collected from the same cohort at the time of disease detection revealed a different miRNA signature for prognosis. A common feature shared by the two signatures was the downregulation of miR-486-5p [213]. This finding supports the notion that miRNA signature evolves with disease progression.
Besides miRNA level, changes in methylation status of miRNAs could independently predict lung cancer prognosis [237]. Methylation of miR-9-3 and miR-124-2/3 is individually correlated with advanced T stage of lung cancer, and a higher number of overall methylation sites is correlated with poor survival [238]. Methylation of miR-34b/c is independently correlated with high recurrence and poor overall survival (OS) for stage I NSCLC [237]. The expression levels of miRNAs could also serve as indicators for drug responses in lung cancers. For example, Ranade et al. reported that high levels of miR-92a-2*, miR-147, and miR-547-5p were correlated with chemoresistance in SCLC patients [232]. The inconsistency of reported miRNA-based biomarkers is a similar issue in prognosis as it is in diagnosis. Efforts towards solving this problem would hopefully clear up the controversies surrounding a selection of miRNAs and further increase the prognostic values of miRNAs as a group for lung cancers.
5.3 miRNAs polymorphisms, lung cancer susceptibility and prognosis
New findings in recent years have demonstrated that expression and functions of miRNAs can be altered by sequence polymorphisms in the miRNA genes, miRNA processing machinery genes or miRNA target genes [239, 240]. Emerging evidences suggest that certain miRNA polymorphisms may be indicators for cancer predispositions and outcome [240, 241]. The single nucleotide polymorphism (SNP) rs11614913 T>C in hsa-mir-196a2 (the precursor for miR-196) enhanced miRNA processing to produce more mature miRNA, resulting in an enhanced binding of miR-196a to its targets [242]. The CC genotype correlated with both heightened susceptibility to lung cancers and decreased survival in NSCLC patients in Chinese population [242, 243].
Among the miRNA processing machinery genes, three harbor SNPs associated with lung cancer risk or prognosis. In a case control study, the rs636832A>G SNP in AGO1 was associated with decreased lung cancer risk in a dose-dependent manner [244]. Moreover, a haplotype (defined as a combination of alleles) in Drosha (GTAATC), was associated with a reduced survival of lung cancer patients, and the rs640831G>T SNP included in this haplotype is correlated with expression changes of cancer-related miRNAs such as let-7s and miR-21 in AD patients [245]. In another miRNA machinery gene XPO5, AA genotype for the rs11077A>C SNP was independently associated with time to relapse (TTR) in surgically-resected stage-I NSCLC patients [246].
SNPs that affect miRNA-based regulation of the target genes and consequently lung cancer risk and prognosis were detected in four genes so far. One SNP in the let-7 binding site within KRAS 3′UTR, termed LCS6, influenced KRAS expression level and was correlated with increased lung cancer risks in moderate smokers, but not KRAS mutation or lung cancer survival [247, 248]. The CC genotype in KRT81 SNP rs3660C>G, is associated with TTR in surgically-resected NSCLC patients, especially in SqCC patients [246]. Since this SNP would alter the binding sites of these miRNAs - miR-17, miR-93, miR-20b, miR-519d, miR-520g, miR-520h, miR-519c-3p, miR-519b-3p, miR-519a, and miR-765 - the target-binding alteration in one or more of these miRNAs may be the basis of the differential prognosis. The MYCL1 oncogene contains the rs3134615G>T SNP within the miR-1827 binding site. This SNP alleviates MYCL1 repression by miR-1827 and increases susceptibility to SCLC [249]. An increased risk of lung cancers can also be associated with the CC genotype in the rs2735383G>C SNP of the NBS1 gene. This is mediated by an altered interaction of NBS1 with miR-629, resulting in a reduced level of NBS1 expression in the CC genotype carriers [250].
5.4 miRNA based therapies for lung cancer
Given the significant roles that miRNAs play in multiple pathways of lung carcinogenesis, increasing efforts are dedicated to research and development of miRNA-based therapies, including restoring functions of tumor suppressive miRNAs (“miRNA replacement therapy [251]”) or inhibiting oncogenic miRNAs.
Exogenous delivery of let-7 into mouse models of lung cancers significantly reduced tumor growth or decreased tumor burden in established tumors, implying that miRNA replacement therapy could be promising [252-254]. Also, liposome-based delivery of miR-7 into mouse xenografts of human lung tumors resulted in significant shrinking of the tumors [255]. Activation of miR-31 function diminished tumor burden in established macroscopic pulmonary metastasis of breast cancers. MiR-34a based strategy has been proved effective in mouse lung cancer models by two separate groups [256, 257]. Comparing to siRNA based therapies which are already in clinical trials, miRNAs are less toxic [258] and capable of “multi-targeting”, pointing to their potential to enter clinical practices. The critical issues for the development of this therapy are fast and effective delivery into target sites, potency of the therapy, and elimination of off-target effects. Towards solving these issues, double-stranded miRNA mimics promise an easier and cost-effective source of functional mature miRNAs with less non-specific effects comparing to miRNA-expressing viral vectors or chemically synthesized pre-miRs. Let-7 double-stranded mimics were tested in lung cancer cell lines for the inhibition of tumorigenic properties and were proven effective [259]. Lipid- and nanoparticle-based delivery systems were demonstrated to execute effective delivery to lung cancer cells or to animals locally and/or systematically [256-259]. Chemical modifications at the 3′-end of let-7 double-stranded mimics may increase the activity of the mimic [259].
To achieve therapeutic effect through targeting oncomiRs, several types of miRNA inhibitors have been developed. Chemical modifications, like 2′-O methyl and Locked Nucleic Acid (LNA), would increase oligo stability against nucleases. Antisense oligonucleotides with these modifications are termed “antagomirs” and “LNA-antimiRs”, respectively [260, 261]. A new class of miRNA inhibitors, “miRNA sponges”, can be expressed in cells from transgenes of tandem miRNA binding sites linked to a strong promoter [262]. A LNA-antimiR against miR-122 has been shown to effectively silence miR-122 in primates without evidence for toxicity, suggesting the feasibility of miRNA inhibitors for utilization in clinical settings [261]. In lung cancers, anti-miR-150 delivered intratumorally into lung tumor xenografts in mice led to tumor regression in volume and weight [68]. To inhibit families of miRNAs sharing identical seed sequences, seed-targeting 8-mer LNAs (termed “tiny LNAs”) have been developed [263], affording a tool to selectively inhibit miRNAs at the basis of individual families.
Resistance to chemo- or radio-therapy is a roadblock for the current treatment of lung cancers. Many miRNAs have been shown to alter responses of lung cancer cells to different drugs or irradiation. MiRNAs reported to alter lung cancer responses to chemotherapy drugs are summarized in Table 1. Let-7a and let-7b were shown to radiosensitize lung cancer cell A549, and probably so do other members of the let-7 family except let-7g based on the expression pattern changes after irradiation [264]. Let-7g was shown to be radioprotective for lung cancer cells in one study [264] but radiosensitizing in two others [265, 266]. More research is needed to solve this myth. Another miRNA reported to radiosensitize lung cancer cells was miR-26b [267], whereas miR-155 [268], miR-9 [265, 269], miR-7 [269], and miR-126 [136] had the opposite effect. In these situations, combining miRNA mimics or inhibitors with traditional therapies may potentiate the efficacy of the treatment. Overall, miRNA-based therapies, alone or in combination with conventional therapies, are still in its infancy; yet, its potential is promising. Inspiringly, a clinical trial in NSCLC using a let-7a-1 based therapy has been initiated [270]. Future efforts should focus on chemical modifications and delivery of the miRNAs, especially targeted delivery to tumor sites without harming non-cancer cells.
Table 1.
microRNAs experimentally validated to alter chemosensitivity of lung cancers
| Drug | MiRNAs Affecting Response to Drug |
Sensitivity Change When MiRNA Expressed |
References |
|---|---|---|---|
| TRAIL | miR-212,miR-130a | Increase | [57, 149] |
| miR-221/222 | Decrease | [52] | |
| Cisplatin | miR-181a, miR-181b | Increase | [100, 271-276] |
| miR-497, miR-451, miR-138, miR-200c miR-150 miR-134/379/495 miR-630, miR-106 |
Decrease | [67, 272] | |
| Doxorubicin | miR-1 | ||
| miR-134/379/495 | Increase | [132, 273] | |
| Docetaxel | miR-200b,miR-100 | Increase | [277, 278] |
| Cetuximab | miR-200c | Increase | [100] |
| Etoposide | miR-134/379/495 | Increase | [273] |
| MEK inhibitor | |||
| AZD6244 | miR-17 | Decrease | [279] |
| EGFR-TKI | |||
| AG1478 | miR-21 | Decrease | [36] |
| Vincristin | miR-181b, miR-497 | Increase | [275, 276] |
| DMC | miR-106, miR-150 | Decrease | [67] |
| Gefitinib | miR-221/222, miR-30b/c | Decrease | [58] |
| miR-21, miR-29a/c miR-100 miR-103, miR-203 |
Increase | [58] |
DMC, Demethylcantharidin.
6. Conclusions
Evidently, miRNAs are not only regulators for many cancer related genes, but also targets of important oncogenes or TSGs, like RAS or TP53. Moreover, each miRNA can target several cancer pathways while key factors in the cancer network are usually under heavy posttranscriptional regulation by multiple miRNAs. Through the interwoven web, miRNAs regulate a number of hallmark processes of tumorigenesis like cell growth, proliferation, invasion, and angiogenesis, acting as oncomiRs, tumor suppressors, or players of varying functions depending on the cellular context (Fig. 1). Better methods to systematically identify targets for miRNA(s) or miRNA regulators for genes of interest would produce a more complete understanding of functions of miRNAs in the signal transduction cascades of critical cancer genes. The involvement of miRNAs in cancer pathogenesis provides novel opportunities for cancer interventions. MiRNAs as biomarkers for lung cancer diagnosis, classification, diagnosis, and drug response evaluation is promising but in need of further improvement. Expression analysis in larger cohorts with standardized operational and analysis procedures are likely to generate more robust markers with enhanced sensitivity and specificity. Development of therapies based on miRNAs is still a young field but growing prosperously with the help of new delivery technologies like nanoparticles. Overall, we are optimistic that miRNA-based management of lung cancers will help tame the “Devil” in the future.
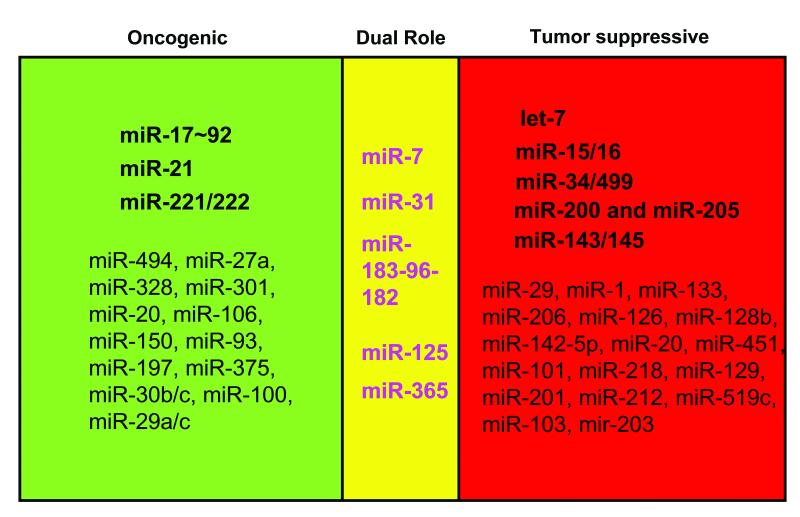
Figure 1. MiRNAs involved in the “Yin” or “Yang” side of lung cancer pathogenesis.
“Dual role” indicates those miRNAs that exhibit either pro- or anti-oncogenic activity depending on the biological context.
Acknowledgements
D.M. received research funding from NCI (CA127547). We thank Shawn Rice and Aaron Runkle for helpful discussion.
Abbreviations
- ACS
American Cancer Society
- AD
adenocarcinoma
- AGO1
Argonaute-1
- AKT
v-akt murine thymoma viral oncogene homolog
- Apaf1
apoptotic peptidase activating factor 1
- ARPC5
actin related protein 2/3 complex, subunit 5
- ASH1
achaete-scute homologue-1
- AXL
AXL receptor tyrosine kinase
- BCL-2
B-cell CLL/lymphoma 2
- BIM
BCL2-like 11
- BPDE
Benzo-A-Pyrene-Diol-Epoxide
- BRCA1
breast cancer 1, early onset
- Btg2
BTG family, member 2
- CCND1
cyclin D1
- CCNE2
cyclin E2
- CDC2
cyclin-dependent kinase 1
- CDC25A
cell division cycle 25 homolog A
- CDC42
cell division cycle 42
- CDH1
E-cadherin
- CDH2
N-cadherin
- CDK4
cyclin-dependent kinase 4
- CDK6
cyclin-dependent kinase 6
- c-JUN
V-jun avian sarcoma virus 17 oncogene homolog
- CLL
chronic lymphocytic leukemia
- c-MYC
v-myc myelocytomatosis viral oncogene homolog
- CRK
v-crk sarcoma virus CT10 oncogene homolog
- CTGF
connective tissue growth factor
- CTTN
cortactin
- DGCR8
DiGeorge syndrome critical region gene 8
- Dicer
dicer 1, ribonuclease type III
- DMC
demethylcantharidin
- DNMT3A
DNA (cytosine-5-)-methyltransferase 3 alpha
- Drosha
drosha ribonuclease type III
- E2F
E2F transcription factor 1
- ECM
extracellular matrix
- EGCG
epigallocatechin-3 gallate
- EGFL7
Epidermal Growth Factor-Like Domain 7
- EGFR
epidermal growth factor receptor
- EGFR-TKI
EGFR tyrosine kinase inhibitors
- EGR1
early growth response 1
- EMT
epithelial-mesenchymal transition
- ERBB2/3
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2/3
- ERF
Ets2 transcriptional repressor
- ETS1
v-ets erythroblastosis virus E26 oncogene homolog 1
- EZH2
enhancer of zeste homolog 2
- Faslg
Fas ligand (TNF superfamily, member 6)
- FBXW7
F-box and WD repeat domain containing 7
- FGF-2
fibroblast growth factor 2
- FGFR1
FGF-2 receptor
- FHIT
fragile histidine triad gene
- Flt1
fms-related tyrosine kinase 1
- FOXO1
forkhead box O1
- FOXP1
forkhead box P1
- FUS1
fused in sarcoma
- GATA3
GATA binding protein 3
- GMNN
geminin, DNA replication inhibitor
- GSTP1
glutathione S-transferase pi 1
- HDAC1/4
histone deacetylase 1/4
- HDMX
Mdm2 p53 binding protein homolog
- HIF-1α
hypoxia inducible factor 1, alpha subunit
- HMGA2
high mobility group AT-hook 2
- IL-6
interleukin 6
- ITGA5
integrin, alpha 5
- ITGB1
integrin, beta 1
- KIF2A
kinesin heavy chain member 2A
- Kit
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
- KRT81
keratin 81
- LNA
Locked Nucleic Acid
- LOH
loss of heterozygosity
- MASPRIN
serpin peptidase inhibitor, clade B (ovalbumin), member 5
- MCL1
myeloid cell leukemia sequence 1
- MET
met proto-oncogene
- miRNA
microRNA
- MSH2
mutS homolog 2
- MUC1
mucin 1
- NBS1
nijmegen breakage syndrome 1
- NE
neuroendocrine
- NKX2-1
see TTF-1
- NNK
nicotine-derived nitrosamine ketone
- NSCLC
non-small cell lung cancer
- NUDT1
nudix (nucleoside diphosphate linked moiety X)-type motif 1
- OCT4
POU class 5 homeobox 1
- OS
overall survival
- Pdcd4
programmed cell death 4
- PDCD6IP
programmed cell death 6 interacting protein
- PED
phosphoprotein enriched in astrocytes 15
- p-EGFR
phosphorylated EGFR
- PI3K
phosphoinositide-3-kinase
- PIM-1
pim-1 oncogene (proviral integration site 1)
- PKC-ε
protein kinase C ε
- PPP2R2A
protein phosphatase 2 regulatory subunit B alpha
- PTEN
phosphatase and tensin homolog
- PUMA
p53 upregulated modulator of apoptosis
- PXN
paxillin
- RAB9B
RAB9B member RAS oncogene family
- RAB14
RAS-related protein 14
- RDX
radixin
- RGS17
regulator of G-protein signaling 17
- RhoA/B
ras homolog gene family, member A/B
- RISC
RNA induced silencing complex
- ROS
reactive oxygen species
- SCLC
small cell lung cancers
- SERPINE1
serpin peptidase inhibitor clade E
- SIRT1
sirtuin 1
- SLC7A5
solute carrier family 7
- SNAIL1
snail homolog 1
- SNP
single nucleotide polymorphism
- Spry1/2
sprouty homolog 1/2
- SqCC
squamous cell carcinomas
- SRC
sarcoma viral oncogene homolog
- TGF-β
Transforming growth factor beta
- TGFBR2
transforming growth factor, beta receptor II
- TIMP3
TIMP metallopeptidase inhibitor 3
- TNF
tumor necrosis factor
- TP53
p53 tumor suppressor
- TPM1
tropomyosin 1 (alpha)
- TRAIL
Apo2L/TNF-α-related apoptosis-inducing ligand
- TSG
tumor suppressor gene
- TSP1
tumor suppressor region 1
- TTF-1
thyroid transcription factor 1
- TTR
time to relapse
- UTR
untranslated region
- VEGF
vascular epithelial growth factor
- VIM
vimentin
- WNT3A
wingless-type
- MMTV integration site family
member 3A
- WT1
Wilms tumor 1
- WWOX
WW domain containing oxidoreductase
- XIAP
X-linked inhibitor of apoptosis
- XPO5
exportin 5
- YAP1
Yes-associated protein 1
- ZEB1/2
zinc finger E-box binding homeobox 1/2
References
- 1.Cancer Facts and Figures 2011. American Cancer Society; Available from: http://www.cancer.org. [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2010;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26(5):611–23. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110(5):563–74. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 7.Meister J, Schmidt MH. miR-126 and miR-126*: new players in cancer. ScientificWorldJournal. 2010;10:2090–100. doi: 10.1100/tsw.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 9.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Vasudevan S. Posttranscriptional Upregulation by MicroRNAs. Wiley Interdiscip Rev RNA. 2011 doi: 10.1002/wrna.121. DOI: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 11.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 12.Sozzi G, Pastorino U, Croce CM. MicroRNAs and lung cancer: from markers to targets. Cell Cycle. 2011;10(13):2045–6. doi: 10.4161/cc.10.13.15712. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42(8):1348–54. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339(2):327–35. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 16.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, MacPherson D. miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 2011;25(16):1734–45. doi: 10.1101/gad.17027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernst A, Campos B, Meier J, Devens F, Liesenberg F, Wolter M, Reifenberger G, Herold-Mende C, Lichter P, Radlwimmer B. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene. 2010;29(23):3411–22. doi: 10.1038/onc.2010.83. [DOI] [PubMed] [Google Scholar]
- 19.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 20.Kim K, Chadalapaka G, Lee SO, Yamada D, Sastre-Garau X, Defossez PA, Park YY, Lee JS, Safe S. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene. 2011 doi: 10.1038/onc.2011.296. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–57. doi: 10.4161/cbt.10.8.13083. [DOI] [PubMed] [Google Scholar]
- 22.Zhao HY, Ooyama A, Yamamoto M, Ikeda R, Haraguchi M, Tabata S, Furukawa T, Che XF, Iwashita K, Oka T, Fukushima M, Nakagawa M, Ono M, Kuwano M, Akiyama S. Down regulation of c-Myc and induction of an angiogenesis inhibitor, thrombospondin-1, by 5-FU in human colon cancer KM12C cells. Cancer Lett. 2008;270(1):156–63. doi: 10.1016/j.canlet.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 23.Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M, Nimura Y, Osada H, Takahashi T. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26(41):6099–105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 24.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282(4):2135–43. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 26.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi T, Obata Y, Sekido Y, Hida T, Ueda R, Watanabe H, Ariyoshi Y, Sugiura T, Takahashi T. Expression and amplification of myc gene family in small cell lung cancer and its relation to biological characteristics. Cancer Res. 1989;49(10):2683–8. [PubMed] [Google Scholar]
- 28.Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68(14):5540–5. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- 29.Ebi H, Sato T, Sugito N, Hosono Y, Yatabe Y, Matsuyama Y, Yamaguchi T, Osada H, Suzuki M, Takahashi T. Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene. 2009;28(38):3371–9. doi: 10.1038/onc.2009.201. [DOI] [PubMed] [Google Scholar]
- 30.Kanzaki H, Ito S, Hanafusa H, Jitsumori Y, Tamaru S, Shimizu K, Ouchida M. Identification of direct targets for the miR-17-92 cluster by proteomic analysis. Proteomics. 2011;11(17):3531–9. doi: 10.1002/pmic.201000501. [DOI] [PubMed] [Google Scholar]
- 31.Gou D, Mishra A, Weng T, Su L, Chintagari NR, Wang Z, Zhang H, Gao L, Wang P, Stricker HM, Liu L. Annexin A2 interactions with Rab14 in alveolar type II cells. J Biol Chem. 2008;283(19):13156–64. doi: 10.1074/jbc.M801532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38(9):1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu M, Wang Z, Yang S, Zhang W, He S, Hu C, Zhu H, Quan L, Bai J, Xu N. TNF-alpha is a novel target of miR-19a. Int J Oncol. 2011;38(4):1013–22. doi: 10.3892/ijo.2011.924. [DOI] [PubMed] [Google Scholar]
- 34.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fei J, Lan F, Guo M, Li Y, Liu Y. Inhibitory effects of anti-miRNA oligonucleotides (AMOs) on A549 cell growth. J Drug Target. 2008;16(9):688–93. doi: 10.1080/10611860802295946. [DOI] [PubMed] [Google Scholar]
- 36.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, Gemma A, Kudoh S, Croce CM, Harris CC. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106(29):12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411(11-12):846–52. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 38.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frezzetti D, De Menna M, Zoppoli P, Guerra C, Ferraro A, Bello AM, De Luca P, Calabrese C, Fusco A, Ceccarelli M, Zollo M, Barbacid M, Di Lauro R, De Vita G. Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene. 2011;30(3):275–86. doi: 10.1038/onc.2010.416. [DOI] [PubMed] [Google Scholar]
- 40.Wang K, Li PF. Foxo3a regulates apoptosis by negatively targeting miR-21. J Biol Chem. 2010;285(22):16958–66. doi: 10.1074/jbc.M109.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, Stadler PF, Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110(4):1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 42.Yang CH, Yue J, Fan M, Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70(20):8108–16. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang TH, Wu F, Loeb GB, Hsu R, Heidersbach A, Brincat A, Horiuchi D, Lebbink RJ, Mo YY, Goga A, McManus MT. Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J Biol Chem. 2009;284(27):18515–24. doi: 10.1074/jbc.M109.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378(3):492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Li C, Nguyen HT, Zhuang Y, Lin Y, Flemington EK, Guo W, Guenther J, Burow ME, Morris GF, Sullivan D, Shan B. Post-transcriptional up-regulation of miR-21 by type I collagen. Mol Carcinog. 2011;50(7):563–70. doi: 10.1002/mc.20742. [DOI] [PubMed] [Google Scholar]
- 46.Kim YJ, Park SJ, Choi EY, Kim S, Kwak HJ, Yoo BC, Yoo H, Lee SH, Kim D, Park JB, Kim JH. PTEN Modulates miR-21 Processing via RNA-Regulatory Protein RNH1. PLoS One. 2011;6(12):e28308. doi: 10.1371/journal.pone.0028308. DOI: 10.1371/journal.pone.0028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18(3):350–9. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 49.Schaefer U, Voloshanenko O, Willen D, Walczak H. TRAIL: a multifunctional cytokine. Front Biosci. 2007;12:3813–24. doi: 10.2741/2354. [DOI] [PubMed] [Google Scholar]
- 50.Koschny R, Walczak H, Ganten TM. The promise of TRAIL--potential and risks of a novel anticancer therapy. J Mol Med (Berl) 2007;85(9):923–35. doi: 10.1007/s00109-007-0194-1. [DOI] [PubMed] [Google Scholar]
- 51.Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27(27):3845–55. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 52.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, Pu P, Kang C. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol. 2010;37(6):1621–6. doi: 10.3892/ijo_00000816. [DOI] [PubMed] [Google Scholar]
- 54.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282(32):23716–24. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 55.Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria R, Spagnoli LG, Farace MG, Ciafre SA. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS One. 2008;3(12):e4029. doi: 10.1371/journal.pone.0004029. DOI: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14(3):791–8. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 57.Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli G, Chiariello M, Croce CM. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2011 doi: 10.1038/onc.2011.260. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, Engelman JA, Ono M, Rho JK, Cascione L, Volinia S, Nephew KP, Croce CM. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2011;18(1):74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Liu L, Jiang Y, Zhang H, Greenlee AR, Han Z. Overexpressed miR-494 down-regulates PTEN gene expression in cells transformed by anti-benzo(a)pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Life Sci. 2010;86(5-6):192–8. doi: 10.1016/j.lfs.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 60.Wang Q, Li DC, Li ZF, Liu CX, Xiao YM, Zhang B, Li XD, Zhao J, Chen LP, Xing XM, Tang SF, Lin YC, Lai YD, Yang P, Zeng JL, Xiao Q, Zeng XW, Lin ZN, Zhuang ZX, Zhuang SM, Chen W. Upregulation of miR-27a contributes to the malignant transformation of human bronchial epithelial cells induced by SV40 small T antigen. Oncogene. 2011;30(36):3875–86. doi: 10.1038/onc.2011.103. [DOI] [PubMed] [Google Scholar]
- 61.Arora S, Ranade AR, Tran NL, Nasser S, Sridhar S, Korn RL, Ross JT, Dhruv H, Foss KM, Sibenaller Z, Ryken T, Gotway MB, Kim S, Weiss GJ. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int J Cancer. 2011;129(11):2621–31. doi: 10.1002/ijc.25939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia W, Li J, Li S, Chen L, Ding H, Zhao Q, Fan M, Shen B, Shao N. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun. 2010;396(4):978–82. doi: 10.1016/j.bbrc.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 63.Ji L, Nishizaki M, Gao B, Burbee D, Kondo M, Kamibayashi C, Xu K, Yen N, Atkinson EN, Fang B, Lerman MI, Roth JA, Minna JD. Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res. 2002;62(9):2715–20. [PMC free article] [PubMed] [Google Scholar]
- 64.Ivanova AV, Ivanov SV, Pascal V, Lumsden JM, Ward JM, Morris N, Tessarolo L, Anderson SK, Lerman MI. Autoimmunity, spontaneous tumourigenesis, and IL-15 insufficiency in mice with a targeted disruption of the tumour suppressor gene Fus1. J Pathol. 2007;211(5):591–601. doi: 10.1002/path.2146. [DOI] [PubMed] [Google Scholar]
- 65.Prudkin L, Behrens C, Liu DD, Zhou X, Ozburn NC, Bekele BN, Minna JD, Moran C, Roth JA, Ji L, Wistuba Loss and reduction of FUS1 protein expression is a frequent phenomenon in the pathogenesis of lung cancer. Clin Cancer Res. 2008;14(1):41–7. doi: 10.1158/1078-0432.CCR-07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, Prudkin L, Wistuba, Ji L, Roth JA, Minna JD, Pertsemlidis A. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7(8):1234–43. doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang PY, Li YJ, Zhang S, Li ZL, Yue Z, Xie N, Xie SY. Regulating A549 cells growth by ASO inhibiting miRNA expression. Mol Cell Biochem. 2010;339(1-2):163–71. doi: 10.1007/s11010-009-0380-2. [DOI] [PubMed] [Google Scholar]
- 68.Li YJ, Zhang YX, Wang PY, Chi YL, Zhang C, Ma Y, Lv CJ, Xie SY. Regression of A549 lung cancer tumors by anti-miR-150 vector. Oncol Rep. 2012;27(1):129–34. doi: 10.3892/or.2011.1466. [DOI] [PubMed] [Google Scholar]
- 69.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 70.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 71.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2009;17(1):F19–36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 72.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14(9):400–9. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 73.Jerome T, Laurie P, Louis B, Pierre C. Enjoy the Silence: The Story of let-7 MicroRNA and Cancer. Curr Genomics. 2007;8(4):229–33. doi: 10.2174/138920207781386933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osada H, Takahashi T. let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer Sci. 2010;102(1):9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- 75.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8(6):843–52. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 77.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 79.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6(13):1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 80.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lize M, Klimke A, Dobbelstein M. MicroRNA-449 in cell fate determination. Cell Cycle. 2011;10(17):2874–82. doi: 10.4161/cc.10.17.17181. [DOI] [PubMed] [Google Scholar]
- 82.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, Rowe RG, Lee S, Maher CA, Weiss SJ, Yook JI. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol. 2011;195(3):417–33. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene. 2011;30(25):2888–99. doi: 10.1038/onc.2011.13. [DOI] [PubMed] [Google Scholar]
- 84.Muth M, Hussein K, Jacobi C, Kreipe H, Bock O. Hypoxia-induced down-regulation of microRNA-449a/b impairs control over targeted SERPINE1 (PAI-1) mRNA - a mechanism involved in SERPINE1 (PAI-1) overexpression. J Transl Med. 2011;9:24. doi: 10.1186/1479-5876-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 86.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23(3):806–12. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis. 2008;29(12):2394–9. doi: 10.1093/carcin/bgn209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bandi N, Vassella E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol Cancer. 10:55. doi: 10.1186/1476-4598-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10(24) doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 90.Lize M, Herr C, Klimke A, Bals R, Dobbelstein M. MicroRNA-449a levels increase by several orders of magnitude during mucociliary differentiation of airway epithelia. Cell Cycle. 2010;9(22):4579–83. doi: 10.4161/cc.9.22.13870. [DOI] [PubMed] [Google Scholar]
- 91.Jeon HS, Lee SY, Lee EJ, Yun SC, Cha EJ, Choi E, Na MJ, Park JY, Kang J, Son JW. Combining microRNA-449a/b with a HDAC inhibitor has a synergistic effect on growth arrest in lung cancer. Lung Cancer. 2011 doi: 10.1016/j.lungcan.2011.10.012. In Press. [DOI] [PubMed] [Google Scholar]
- 92.Lize M, Pilarski S, Dobbelstein M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 2010;17(3):452–8. doi: 10.1038/cdd.2009.188. [DOI] [PubMed] [Google Scholar]
- 93.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17(2):215–20. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 94.Ofir M, Hacohen D, Ginsberg D. MiR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol Cancer Res. 2011;9(4):440–7. doi: 10.1158/1541-7786.MCR-10-0344. [DOI] [PubMed] [Google Scholar]
- 95.Musumeci M, Coppola V, Addario A, Patrizii M, Maugeri-Sacca M, Memeo L, Colarossi C, Francescangeli F, Biffoni M, Collura D, Giacobbe A, D’Urso L, Falchi M, Venneri MA, Muto G, De Maria R, Bonci D. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene. 2011;30(41):4231–42. doi: 10.1038/onc.2011.140. [DOI] [PubMed] [Google Scholar]
- 96.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69(13):5553–9. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 97.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 98.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67(17):7972–6. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 99.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, Kurie JM. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23(18):2140–51. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ceppi P, Mudduluru G, Kumarswamy R, Rapa I, Scagliotti GV, Papotti M, Allgayer H. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res. 2010;8(9):1207–16. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- 101.Wang Z, Zhao Y, Smith E, Goodall GJ, Drew PA, Brabletz T, Yang C. Reversal and prevention of arsenic-induced human bronchial epithelial cell malignant transformation by microRNA-200b. Toxicol Sci. 2011;121(1):110–22. doi: 10.1093/toxsci/kfr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbons DL, Baird BN, Alvarez C, Thilaganathan N, Liu DD, Saintigny P, Heymach JV, Creighton CJ, Kurie JM. miR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 2011;9(1):25–35. doi: 10.1158/1541-7786.MCR-10-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, Alvarez CA, Moreira DC, Creighton CJ, Gregory PA, Goodall GJ, Kurie JM. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121(4):1373–85. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schliekelman MJ, Gibbons DL, Faca VM, Creighton CJ, Rizvi ZH, Zhang Q, Wong CH, Wang H, Ungewiss C, Ahn YH, Shin DH, Kurie JM, Hanash SM. Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res. 2011;71(24):7670–82. doi: 10.1158/0008-5472.CAN-11-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17(9):1101–8. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu W, Xu H, Zhu D, Zhi H, Wang T, Wang J, Jiang B, Shu Y, Liu P. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother Pharmacol. 2011 doi: 10.1007/s00280-011-1752-3. DOI: 10.1007/s00280-011-1752-3. [DOI] [PubMed] [Google Scholar]
- 107.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23(18):2166–78. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98(12):1914–20. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125(2):345–52. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 110.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 111.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–91. [PubMed] [Google Scholar]
- 112.Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng G, Tanaka Y, Dahiya R. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis. 2011;32(5):772–8. doi: 10.1093/carcin/bgr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77(1):12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 114.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3(7):e2557. doi: 10.1371/journal.pone.0002557. DOI: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu X, Sempere LF, Galimberti F, Freemantle SJ, Black C, Dragnev KH, Ma Y, Fiering S, Memoli V, Li H, DiRenzo J, Korc M, Cole CN, Bak M, Kauppinen S, Dmitrovsky E. Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer Res. 2009;15(4):1177–83. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45(12):2197–206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 117.Cho WC, Chow AS, Au JS. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol. 2011;8(1):125–31. doi: 10.4161/rna.8.1.14259. [DOI] [PubMed] [Google Scholar]
- 118.Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng A, Hu J. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J Exp Clin Cancer Res. 2010;29:151. doi: 10.1186/1756-9966-29-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yin R, Zhang S, Wu Y, Fan X, Jiang F, Zhang Z, Feng D, Guo X, Xu L. microRNA-145 suppresses lung adenocarcinoma-initiating cell proliferation by targeting OCT4. Oncol Rep. 2011;25(6):1747–54. doi: 10.3892/or.2011.1252. [DOI] [PubMed] [Google Scholar]
- 120.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70(1):378–87. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Melkamu T, Zhang X, Tan J, Zeng Y, Kassie F. Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2010;31(2):252–8. doi: 10.1093/carcin/bgp208. [DOI] [PubMed] [Google Scholar]
- 122.Gao W, Yu Y, Cao H, Shen H, Li X, Pan S, Shu Y. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64(6):399–408. doi: 10.1016/j.biopha.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 123.Pekarsky Y, Croce CM. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget. 2010;1(3):224–7. doi: 10.18632/oncotarget.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–3. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 125.Santanam U, Zanesi N, Efanov A, Costinean S, Palamarchuk A, Hagan JP, Volinia S, Alder H, Rassenti L, Kipps T, Croce CM, Pekarsky Y. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci U S A. 2010;107(27):12210–5. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2010;45(2):287–94. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10(4):400–5. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26(42):6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang X, Ling C, Bai Y, Zhao J. MicroRNA-206 is associated with invasion and metastasis of lung cancer. Anat Rec (Hoboken) 2010;294(1):88–92. doi: 10.1002/ar.21287. [DOI] [PubMed] [Google Scholar]
- 132.Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283(48):33394–405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 133.Moriya Y, Nohata N, Kinoshita T, Mutallip M, Okamoto T, Yoshida S, Suzuki M, Yoshino I, Seki N. Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. J Hum Genet. 2012;57(1):38–45. doi: 10.1038/jhg.2011.126. [DOI] [PubMed] [Google Scholar]
- 134.Gao W, Shen H, Liu L, Xu J, Shu Y. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. 2010;137(4):557–66. doi: 10.1007/s00432-010-0918-4. [DOI] [PubMed] [Google Scholar]
- 135.Miko E, Czimmerer Z, Csanky E, Boros G, Buslig J, Dezso B, Scholtz B. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res. 2009;35(8):646–64. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- 136.Wang XC, Du LQ, Tian LL, Wu HL, Jiang XY, Zhang H, Li DG, Wang YY, Wu HY, She Y, Liu QF, Fan FY, Meng AM. Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer. 2010;72(1):92–9. doi: 10.1016/j.lungcan.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 137.Yang Y, Li X, Yang Q, Wang X, Zhou Y, Jiang T, Ma Q, Wang YJ. The role of microRNA in human lung squamous cell carcinoma. Cancer Genet Cytogenet. 2010;200(2):127–33. doi: 10.1016/j.cancergencyto.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 138.Liu B, Peng XC, Zheng XL, Wang J, Qin YW. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66(2):169–75. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 139.Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, Nuovo G, Marsh CB, Nana-Sinkam SP. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373(4):607–12. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 140.Watanabe K, Emoto N, Hamano E, Sunohara M, Kawakami M, Kage H, Kitano K, Nakajima J, Goto A, Fukayama M, Nagase T, Yatomi Y, Ohishi N, Takai D. Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int J Cancer. 2011 doi: 10.1002/ijc.26254. In Press. [DOI] [PubMed] [Google Scholar]
- 141.Miko E, Margitai Z, Czimmerer Z, Varkonyi I, Dezso B, Lanyi A, Bacso Z, Scholtz B. miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett. 2011;585(8):1191–6. doi: 10.1016/j.febslet.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 142.Sun Y, Bai Y, Zhang F, Wang Y, Guo Y, Guo L. miR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7. Biochem Biophys Res Commun. 2009;391(3):1483–9. doi: 10.1016/j.bbrc.2009.12.098. [DOI] [PubMed] [Google Scholar]
- 143.Weiss GJ, Bemis LT, Nakajima E, Sugita M, Birks DK, Robinson WA, Varella-Garcia M, Bunn PA, Jr., Haney J, Helfrich BA, Kato H, Hirsch FR, Franklin WA. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19(6):1053–9. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- 144.Wang R, Wang ZX, Yang JS, Pan X, De W, Chen LB. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14) Oncogene. 2011;30(23):2644–58. doi: 10.1038/onc.2010.642. [DOI] [PubMed] [Google Scholar]
- 145.Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6(4):671–8. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 146.Luo L, Zhang T, Liu H, Lv T, Yuan D, Yao Y, Lv Y, Song Y. MiR-101 and Mcl-1 in non-small-cell lung cancer: expression profile and clinical significance. Med Oncol. 2011 doi: 10.1007/s12032-011-0085-8. In Press. [DOI] [PubMed] [Google Scholar]
- 147.Wu DW, Cheng YW, Wang J, Chen CY, Lee H. Paxillin predicts survival and relapse in non-small cell lung cancer by microRNA-218 targeting. Cancer Res. 2010;70(24):10392–401. doi: 10.1158/0008-5472.CAN-10-2341. [DOI] [PubMed] [Google Scholar]
- 148.Wu J, Qian J, Li C, Kwok L, Cheng F, Liu P, Perdomo C, Kotton D, Vaziri C, Anderlind C, Spira A, Cardoso WV, Lu J. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle. 2010;9(9):1809–18. doi: 10.4161/cc.9.9.11535. [DOI] [PubMed] [Google Scholar]
- 149.Incoronato M, Garofalo M, Urso L, Romano G, Quintavalle C, Zanca C, Iaboni M, Nuovo G, Croce CM, Condorelli G. miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 2010;70(9):3638–46. doi: 10.1158/0008-5472.CAN-09-3341. [DOI] [PubMed] [Google Scholar]
- 150.Cha ST, Chen PS, Johansson G, Chu CY, Wang MY, Jeng YM, Yu SL, Chen JS, Chang KJ, Jee SH, Tan CT, Lin MT, Kuo ML. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 2010;70(7):2675–85. doi: 10.1158/0008-5472.CAN-09-2448. [DOI] [PubMed] [Google Scholar]
- 151.Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci U S A. 1997;94(15):8104–9. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang H, Bian S, Yang CS. Green tea polyphenol EGCG suppresses lung cancer cell growth through upregulating miR-210 expression caused by stabilizing HIF-1alpha. Carcinogenesis. 2011;32(12):1881–9. doi: 10.1093/carcin/bgr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1(3):E60. doi: 10.1371/journal.pbio.0000060. DOI: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68(10):3566–72. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 155.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284(9):5731–41. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 156.Xiong S, Zheng Y, Jiang P, Liu R, Liu X, Chu Y. MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int J Biol Sci. 2011;7(6):805–14. doi: 10.7150/ijbs.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR, Lin SC, Chang YC, Lin SY, Chen SJ, Chen HC, Yeh SD, Wu CW. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010;70(21):8822–31. doi: 10.1158/0008-5472.CAN-10-0638. [DOI] [PubMed] [Google Scholar]
- 158.Xi S, Yang M, Tao Y, Xu H, Shan J, Inchauste S, Zhang M, Mercedes L, Hong JA, Rao M, Schrump DS. Cigarette smoke induces C/EBP-beta-mediated activation of miR-31 in normal human respiratory epithelia and lung cancer cells. PLoS One. 2010;5(10):e13764. doi: 10.1371/journal.pone.0013764. DOI: 10.1371/journal.pone.0013764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M, Dragnev KH, Li H, Direnzo J, Bak M, Freemantle SJ, Kauppinen S, Dmitrovsky E. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120(4):1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Activation of miR-31 function in already-established metastases elicits metastatic regression. Genes Dev. 2011;25(6):646–59. doi: 10.1101/gad.2004211. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 161.Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 2010;70(12):5147–54. doi: 10.1158/0008-5472.CAN-10-0410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 162.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282(2):1479–86. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 163.Cortez MA, Nicoloso MS, Shimizu M, Rossi S, Gopisetty G, Molina JR, Carlotti C, Jr., Tirapelli D, Neder L, Brassesco MS, Scrideli CA, Tone LG, Georgescu MM, Zhang W, Puduvalli V, Calin GA. miR-29b and miR-125a regulate podoplanin and suppress invasion in glioblastoma. Genes Chromosomes Cancer. 2010;49(11):981–90. doi: 10.1002/gcc.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Mori M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17(9):2725–33. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 165.Zhang Y, Gao JS, Tang X, Tucker LD, Quesenberry P, Rigoutsos I, Ramratnam B. MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 2009;583(22):3725–30. doi: 10.1016/j.febslet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23(7):862–76. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yamada H, Yanagisawa K, Tokumaru S, Taguchi A, Nimura Y, Osada H, Nagino M, Takahashi T. Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11-21 in human lung cancer. Genes Chromosomes Cancer. 2008;47(9):810–8. doi: 10.1002/gcc.20582. [DOI] [PubMed] [Google Scholar]
- 168.Nagayama K, Kohno T, Sato M, Arai Y, Minna JD, Yokota J. Homozygous deletion scanning of the lung cancer genome at a 100-kb resolution. Genes Chromosomes Cancer. 2007;46(11):1000–10. doi: 10.1002/gcc.20485. [DOI] [PubMed] [Google Scholar]
- 169.Jiang L, Huang Q, Zhang S, Zhang Q, Chang J, Qiu X, Wang E. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10(318) doi: 10.1186/1471-2407-10-318. DOI: 10.1186/1471-2407-10-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lu W, Li S, Liu B, Li Y, Luo M, Sun L, You J, Zhou Q. [Screening of Metastasis-related MicroRNAs in the Large-cell Lung Cancer Cell Lines with Different Metastastic Potentials] Zhongguo Fei Ai Za Zhi. 2011;14(11):835–40. doi: 10.3779/j.issn.1009-3419.2011.11.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang G, Mao W, Zheng S, Ye J. Epidermal growth factor receptor-regulated miR-125a-5p--a metastatic inhibitor of lung cancer. Febs J. 2009;276(19):5571–8. doi: 10.1111/j.1742-4658.2009.07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jiang L, Zhang Q, Chang J, Qiu X, Wang E. [hsa-miR-125a-5p Enhances Invasion Ability in Non-Small Lung Carcinoma Cell Lines.] Zhongguo Fei Ai Za Zhi. 2009;12(9):951–5. doi: 10.3779/j.issn.1009-3419.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 173.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2009;70(1):367–77. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Targeting of integrin beta1 and kinesin 2alpha by microRNA 183. J Biol Chem. 2009;285(8):5461–71. doi: 10.1074/jbc.M109.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li J, Fu H, Xu C, Tie Y, Xing R, Zhu J, Qin Y, Sun Z, Zheng X. miR-183 inhibits TGF-beta1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer. 2010;10:354. doi: 10.1186/1471-2407-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70(23):9570–80. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 177.Wang G, Mao W, Zheng S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008;582(25-26):3663–8. doi: 10.1016/j.febslet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 178.Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, Liu J, Yu J, Chen J. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70(14):6015–25. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 179.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, Gorospe M, Harris AL, Helleday T, Chowdhury D. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41(2):210–20. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sun Y, Fang R, Li C, Li L, Li F, Ye X, Chen H. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem Biophys Res Commun. 2010;396(2):501–7. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- 181.Zhang L, Liu T, Huang Y, Liu J. microRNA-182 inhibits the proliferation and invasion of human lung adenocarcinoma cells through its effect on human cortical actin-associated protein. Int J Mol Med. 2011;28(3):381–8. doi: 10.3892/ijmm.2011.679. [DOI] [PubMed] [Google Scholar]
- 182.Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C, Li J, Wang X, Song L. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One. 2010;5(12):e15797. doi: 10.1371/journal.pone.0015797. DOI: 10.1371/journal.pone.0015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106(6):1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284(35):23204–16. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hannafon BN, Sebastiani P, de Las Morenas A, Lu J, Rosenberg CL. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res. 2011;13(2):R24. doi: 10.1186/bcr2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, Lao K, Clarke MF. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34(4):1069–75. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 189.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, Garcia-Foncillas J. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 191.Li J, Liang SH, Lu X. [Potential role of ezrin and its related microRNA in ovarian cancer invasion and metastasis] Zhonghua Fu Chan Ke Za Zhi. 2010;45(10):787–92. [PubMed] [Google Scholar]
- 192.Wang Y, Luo H, Li Y, Chen T, Wu S, Yang L. hsa-miR-96 up-regulates MAP4K1 and IRS1 and may function as a promising. Mol Med Report. 2011;5(1):260–5. doi: 10.3892/mmr.2011.621. [DOI] [PubMed] [Google Scholar]
- 193.Zhu W, Liu X, He J, Chen D, Hunag Y, Zhang YK. Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: a case control study. BMC Cancer. 2011;11:393. doi: 10.1186/1471-2407-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Guazzi S, Price M, De Felice M, Damante G, Mattei MG, Di Lauro R. Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 1990;9(11):3631–9. doi: 10.1002/j.1460-2075.1990.tb07574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10(1):60–9. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 196.Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87(1):219–44. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 197.DeFelice M, Silberschmidt D, DiLauro R, Xu Y, Wert SE, Weaver TE, Bachurski CJ, Clark JC, Whitsett JA. TTF-1 phosphorylation is required for peripheral lung morphogenesis, perinatal survival, and tissue-specific gene expression. J Biol Chem. 2003;278(37):35574–83. doi: 10.1074/jbc.M304885200. [DOI] [PubMed] [Google Scholar]
- 198.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104(42):16663–8. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, Lee J, Choi YL, Sato M, Wang P, Hernandez-Boussard T, Gazdar AF, Petersen I, Minna JD, Pollack JR. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27(25):3635–40. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, Shimada Y, Osada H, Kosaka T, Matsubara H, Mitsudomi T, Sekido Y, Tanimoto M, Yatabe Y, Takahashi T. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67(13):6007–11. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 201.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450(7171):893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, Yoon S, Crowley D, Bronson RT, Chiang DY, Meyerson M, Jacks T. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473(7345):101–4. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Saito R-A, Watabe T, Horiguchi K, Kohyama T, Saitoh M, Nagase T, Miyazono K. Thyroid Transcription Factor-1 Inhibits Transforming Growth Factor-{beta}-Mediated Epithelial-to-Mesenchymal Transition in Lung Adenocarcinoma Cells. Cancer Res. 2009;69(7):2783–2791. doi: 10.1158/0008-5472.CAN-08-3490. [DOI] [PubMed] [Google Scholar]
- 204.Qi J, Rice SJ, Salzberg AC, Runkle EA, Liao J, Zander DS, Mu D. MiR-365 regulates lung cancer and developmental gene thyroid transcription factor 1. Cell Cycle. 2012;11(1):177–86. doi: 10.4161/cc.11.1.18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Borges M, Linnoila RI, van de Velde HJ, Chen H, Nelkin BD, Mabry M, Baylin SB, Ball DW. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature. 1997;386(6627):852–5. doi: 10.1038/386852a0. [DOI] [PubMed] [Google Scholar]
- 207.Linnoila RI, Zhao B, DeMayo JL, Nelkin BD, Baylin SB, DeMayo FJ, Ball DW. Constitutive achaete-scute homologue-1 promotes airway dysplasia and lung neuroendocrine tumors in transgenic mice. Cancer Res. 2000;60(15):4005–9. [PubMed] [Google Scholar]
- 208.Nishikawa E, Osada H, Okazaki Y, Arima C, Tomida S, Tatematsu Y, Taguchi A, Shimada Y, Yanagisawa K, Yatabe Y, Toyokuni S, Sekido Y, Takahashi T. miR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res. 2011;71(19):6165–73. doi: 10.1158/0008-5472.CAN-11-1020. [DOI] [PubMed] [Google Scholar]
- 209.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 210.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, Wang MX. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4(6):575–86. [PMC free article] [PubMed] [Google Scholar]
- 212.Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6(3):482–8. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 213.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabro E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108(9):3713–8. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67(2):170–6. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Bianchi F, Nicassio F, Marzi M, Belloni E, Dall’olio V, Bernard L, Pelosi G, Maisonneuve P, Veronesi G, Di Fiore PP. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. 2011;3(8):495–503. doi: 10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola FM, Tucker MA, Bertazzi PA, Pesatori AC, Caporaso NE, McShane LM, Wang E. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16(2):430–41. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, Nonaka D, Li J, Spector Y, Rosenfeld N, Chajut A, Cohen D, Aharonov R, Mansukhani M. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27(12):2030–7. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 218.Barshack I, Lithwick-Yanai G, Afek A, Rosenblatt K, Tabibian-Keissar H, Zepeniuk M, Cohen L, Dan H, Zion O, Strenov Y, Polak-Charcon S, Perelman M. MicroRNA expression differentiates between primary lung tumors and metastases to the lung. Pathol Res Pract. 2010;206(8):578–84. doi: 10.1016/j.prp.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 219.Xu JZ, Wong CW. Hunting for robust gene signature from cancer profiling data: sources of variability, different interpretations, and recent methodological developments. Cancer Lett. 2010;296(1):9–16. doi: 10.1016/j.canlet.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 220.Keller A, Leidinger P, Gislefoss R, Haugen A, Langseth H, Staehler P, Lenhof HP, Meese E. Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 2011;8(3):506–16. doi: 10.4161/rna.8.3.14994. [DOI] [PubMed] [Google Scholar]
- 221.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 222.Inamura K, Togashi Y, Nomura K, Ninomiya H, Hiramatsu M, Satoh Y, Okumura S, Nakagawa K, Ishikawa Y. let-7 microRNA expression is reduced in bronchioloalveolar carcinoma, a non-invasive carcinoma, and is not correlated with prognosis. Lung Cancer. 2007;58(3):392–6. doi: 10.1016/j.lungcan.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 223.Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, Zeng F, Zhou JH, Zhang YK. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2011 doi: 10.1007/s12032-011-9923-y. DOI: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 224.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54(10):1696–704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 225.Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, Bowman ED, Mathe EA, Takenoshita S, Yokota J, Haugen A, Harris CC. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17(7):1875–82. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, Saito M, Dunant A, Pham TC, Petrini I, Lee A, Khan MA, Hainaut P, Pignon JP, Brambilla E, Popper HH, Filipits M, Harris CC, Giaccone G. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res. 2010;70(21):8288–98. doi: 10.1158/0008-5472.CAN-10-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang ZX, Bian HB, Wang JR, Cheng ZX, Wang KM, De W. Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. J Surg Oncol. 2011;104(7):847–51. doi: 10.1002/jso.22008. [DOI] [PubMed] [Google Scholar]
- 228.Gallardo E, Navarro A, Vinolas N, Marrades RM, Diaz T, Gel B, Quera A, Bandres E, Garcia-Foncillas J, Ramirez J, Monzo M. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30(11):1903–9. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 229.Vosa U, Vooder T, Kolde R, Fischer K, Valk K, Tonisson N, Roosipuu R, Vilo J, Metspalu A, Annilo T. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer. 2011;50(10):812–22. doi: 10.1002/gcc.20902. [DOI] [PubMed] [Google Scholar]
- 230.Duncavage E, Goodgame B, Sezhiyan A, Govindan R, Pfeifer J. Use of microRNA expression levels to predict outcomes in resected stage I non-small cell lung cancer. J Thorac Oncol. 2010;5(11):1755–63. doi: 10.1097/JTO.0b013e3181f3909d. [DOI] [PubMed] [Google Scholar]
- 231.Navarro A, Diaz T, Gallardo E, Vinolas N, Marrades RM, Gel B, Campayo M, Quera A, Bandres E, Garcia-Foncillas J, Ramirez J, Monzo M. Prognostic implications of miR-16 expression levels in resected non-small-cell lung cancer. J Surg Oncol. 2011;103(5):411–5. doi: 10.1002/jso.21847. [DOI] [PubMed] [Google Scholar]
- 232.Ranade AR, Cherba D, Sridhar S, Richardson P, Webb C, Paripati A, Bowles B, Weiss GJ. MicroRNA 92a-2*: a biomarker predictive for chemoresistance and prognostic for survival in patients with small cell lung cancer. J Thorac Oncol. 2010;5(8):1273–8. doi: 10.1097/JTO.0b013e3181dea6be. [DOI] [PubMed] [Google Scholar]
- 233.Donnem T, Lonvik K, Eklo K, Berg T, Sorbye SW, Al-Shibli K, Al-Saad S, Andersen S, Stenvold H, Bremnes RM, Busund LT. Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer: coexpression with vascular endothelial growth factor-A predicts poor survival. Cancer. 2011;117(14):3193–200. doi: 10.1002/cncr.25907. [DOI] [PubMed] [Google Scholar]
- 234.Donnem T, Eklo K, Berg T, Sorbye SW, Lonvik K, Al-Saad S, Al-Shibli K, Andersen S, Stenvold H, Bremnes RM, Busund LT. Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. J Transl Med. 2011;9:6. doi: 10.1186/1479-5876-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69(14):5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 236.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28(10):1721–6. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 237.Wang Z, Chen Z, Gao Y, Li N, Li B, Tan F, Tan X, Lu N, Sun Y, Sun J, Sun N, He J. DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol Ther. 2011;11(5):490–6. doi: 10.4161/cbt.11.5.14550. [DOI] [PubMed] [Google Scholar]
- 238.Kitano K, Watanabe K, Emoto N, Kage H, Hamano E, Nagase T, Sano A, Murakawa T, Nakajima J, Goto A, Fukayama M, Yatomi Y, Ohishi N, Takai D. CpG island methylation of microRNAs is associated with tumor size and recurrence of non-small-cell lung cancer. Cancer Sci. 2011;102(12):2126–31. doi: 10.1111/j.1349-7006.2011.02101.x. [DOI] [PubMed] [Google Scholar]
- 239.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16(9):1124–31. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 240.Megiorni F, Pizzuti A, Frati L. Clinical Significance of MicroRNA Expression Profiles and Polymorphisms in Lung Cancer Development and Management. Patholog Res Int. 2011;2011:780652. doi: 10.4061/2011/780652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, Wojcik SE, Ferdin J, Kunej T, Xiao L, Manoukian S, Secreto G, Ravagnani F, Wang X, Radice P, Croce CM, Davuluri RV, Calin GA. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70(7):2789–98. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tian T, Shu Y, Chen J, Hu Z, Xu L, Jin G, Liang J, Liu P, Zhou X, Miao R, Ma H, Chen Y, Shen H. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1183–7. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 243.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, Chen Y, Shen H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118(7):2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kim JS, Choi YY, Jin G, Kang HG, Choi JE, Jeon HS, Lee WK, Kim DS, Kim CH, Kim YJ, Son JW, Jung TH, Park JY. Association of a common AGO1 variant with lung cancer risk: a two-stage case-control study. Mol Carcinog. 2010;49(10):913–21. doi: 10.1002/mc.20672. [DOI] [PubMed] [Google Scholar]
- 245.Rotunno M, Zhao Y, Bergen AW, Koshiol J, Burdette L, Rubagotti M, Linnoila RI, Marincola FM, Bertazzi PA, Pesatori AC, Caporaso NE, McShane LM, Wang E, Landi MT. Inherited polymorphisms in the RNA-mediated interference machinery affect microRNA expression and lung cancer survival. Br J Cancer. 2010;103(12):1870–4. doi: 10.1038/sj.bjc.6605976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Campayo M, Navarro A, Vinolas N, Tejero R, Munoz C, Diaz T, Marrades R, Cabanas ML, Gimferrer JM, Gascon P, Ramirez J, Monzo M. A dual role for KRT81: a miR-SNP associated with recurrence in non-small-cell lung cancer and a novel marker of squamous cell lung carcinoma. PLoS One. 2011;6(7):e22509. doi: 10.1371/journal.pone.0022509. DOI: 10.1371/journal.pone.0022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68(20):8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Nelson HH, Christensen BC, Plaza SL, Wiencke JK, Marsit CJ, Kelsey KT. KRAS mutation, KRAS-LCS6 polymorphism, and non-small cell lung cancer. Lung Cancer. 2010;69(1):51–3. doi: 10.1016/j.lungcan.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Xiong F, Wu C, Chang J, Yu D, Xu B, Yuan P, Zhai K, Xu J, Tan W, Lin D. Genetic variation in an miRNA-1827 binding site in MYCL1 alters susceptibility to small-cell lung cancer. Cancer Res. 2011;71(15):5175–81. doi: 10.1158/0008-5472.CAN-10-4407. [DOI] [PubMed] [Google Scholar]
- 250.Yang L, Li Y, Cheng M, Huang D, Zheng J, Liu B, Ling X, Li Q, Zhang X, Ji W, Zhou Y, Lu J. A functional polymorphism at microRNA-629-binding site in the 3′-untranslated region of NBS1 gene confers an increased risk of lung cancer in Southern and Eastern Chinese population. Carcinogenesis. 2011 doi: 10.1093/carcin/bgr272. In Press. [DOI] [PubMed] [Google Scholar]
- 251.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70(18):7027–30. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.He XY, Chen JX, Zhang Z, Li CL, Peng QL, Peng HM. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J Cancer Res Clin Oncol. 2010;136(7):1023–8. doi: 10.1007/s00432-009-0747-5. [DOI] [PubMed] [Google Scholar]
- 253.Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, Homer R, Brown D, Bader AG, Weidhaas JB, Slack FJ. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29(11):1580–7. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 255.Rai K, Takigawa N, Ito S, Kashihara H, Ichihara E, Yasuda T, Shimizu K, Tanimoto M, Kiura K. Liposomal delivery of MicroRNA-7-expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol Cancer Ther. 2011;10(9):1720–7. doi: 10.1158/1535-7163.MCT-11-0220. [DOI] [PubMed] [Google Scholar]
- 256.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18(9):1650–6. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–30. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009;17(1):169–75. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wang QZ, Lv YH, Gong YH, Li ZF, Xu W, Diao Y, Xu R. Double-stranded Let-7 mimics, potential candidates for cancer gene therapy. J Physiol Biochem. 2011 doi: 10.1007/s13105-011-0124-0. In Press. [DOI] [PubMed] [Google Scholar]
- 260.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 261.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 262.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ, Kauppinen S. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43(4):371–8. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67(23):11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Arora H, Qureshi R, Jin S, Park AK, Park WY. miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFkappaB1. Exp Mol Med. 2011;43(5):298–304. doi: 10.3858/emm.2011.43.5.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Jeong SH, Wu HG, Park WY. LIN28B confers radio-resistance through the posttranscriptional control of KRAS. Exp Mol Med. 2009;41(12):912–8. doi: 10.3858/emm.2009.41.12.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Arora H, Qureshi R, Park AK, Park WY. Coordinated regulation of ATF2 by miR-26b in gamma-irradiated lung cancer cells. PLoS One. 2011;6(8):e23802. doi: 10.1371/journal.pone.0023802. DOI: 10.1371/journal.pone.0023802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Babar IA, Czochor J, Steinmetz A, Weidhaas JB, Glazer PM, Slack FJ. Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer Biol Ther. 2011;12(10) doi: 10.4161/cbt.12.10.17681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lee KM, Choi EJ, Kim IA. microRNA-7 increases radiosensitivity of human cancer cells with activated EGFR-associated signaling. Radiother Oncol. 2011;101(1):171–6. doi: 10.1016/j.radonc.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 270.Amir BJaH. Therapy analysis—microRNA; update analysis. Pharmaprojects. 2008;29(4):1–4. [Google Scholar]
- 271.Bian HB, Pan X, Yang JS, Wang ZX, De W. Upregulation of microRNA-451 increases cisplatin sensitivity of non-small cell lung cancer cell line (A549) J Exp Clin Cancer Res. 2011;30:20. doi: 10.1186/1756-9966-30-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupe P, Robert T, Ripoche H, Lazar V, Harel-Bellan A, Dessen P, Barillot E, Kroemer G. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70(5):1793–803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 273.Guo L, Liu Y, Bai Y, Sun Y, Xiao F, Guo Y. Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur J Cancer. 2010;46(9):1692–702. doi: 10.1016/j.ejca.2010.02.043. [DOI] [PubMed] [Google Scholar]
- 274.Wang Q, Zhong M, Liu W, Li J, Huang J, Zheng L. Alterations of microRNAs in cisplatin-resistant human non-small cell lung cancer cells (A549/DDP) Exp Lung Res. 2011;37(7):427–34. doi: 10.3109/01902148.2011.584263. [DOI] [PubMed] [Google Scholar]
- 275.Zhu W, Shan X, Wang T, Shu Y, Liu P. miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int J Cancer. 2010;127(11):2520–9. doi: 10.1002/ijc.25260. [DOI] [PubMed] [Google Scholar]
- 276.Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang B, Shu Y, Liu P. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2011 doi: 10.1007/s12032-010-9797-4. DOI: 10.1007/s12032-010-9797-4. [DOI] [PubMed] [Google Scholar]
- 277.Feng B, Wang R, Chen LB. MiR-100 resensitizes docetaxel-resistant human lung adenocarcinoma cells (SPC-A1) to docetaxel by targeting Plk1. Cancer Lett. 2011 doi: 10.1016/j.canlet.2011.11.024. DOI; 10.1016/j.canlet.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 278.Feng B, Wang R, Song HZ, Chen LB. MicroRNA-200b reverses chemoresistance of docetaxel-resistant human lung adenocarcinoma cells by targeting E2F3. Cancer. 2011 doi: 10.1002/cncr.26560. DOI: 10.1002/cncr.26560. [DOI] [PubMed] [Google Scholar]
- 279.Dai B, Meng J, Peyton M, Girard L, Bornmann WG, Ji L, Minna JD, Fang B, Roth JA. STAT3 mediates resistance to MEK inhibitor through microRNA miR-17. Cancer Res. 2011;71(10):3658–68. doi: 10.1158/0008-5472.CAN-10-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]