Abstract
Affinity-purification mass spectrometry (AP-MS) is the preeminent technique for identification of eukaryotic protein complexes in vivo. AP-MS workflows typically express epitope-tagged bait proteins, immunopurify, and then identify associated protein complexes using mass spectrometry. However, challenges of existing strategies include the construction of expression vectors for large open reading frames and the possibility that overexpression of bait proteins may result in expression of nonphysiological levels of the bait protein with concomitant perturbation of endogenous protein complexes. To address these issues, we use human cell lines with epitope-tagged endogenous genes as AP-MS substrates to develop a platform that we call “knock-in AP-MS”, thereby avoiding the challenges of expression vector construction and ensuring that expression of tagged proteins is driven by endogenous regulatory mechanisms. Using three different bait genes (MRE11A, DNMT1 and APC), we show that cell lines expressing epitope-tagged endogenous genes make good substrates for sensitive and reproducible identification of protein interactions using AP-MS. In particular, we identify novel interactors of the important oncoprotein Adenomatous Polyposis Coli (APC), including an interaction with Flightless-1 homologue (FLII) that is enriched in nuclear fractions.
Keywords: Protein-protein interactions, protein complexes, epitope-tagging, knock-in, AP-MS, colorectal cancer, Adenomatous Polyposis Coli
INTRODUCTION
Affinity-purification mass spectrometry (AP-MS) identifies protein interactions through their participation in protein complexes, and is the method of choice for isolation and identification of protein complexes on a large scale. AP-MS work flows combine the specificity of antibody-based affinity purification with the sensitivity of mass spectrometry for protein identification.1 Although native antibodies may be used for purification of complexes of interest, epitope-tagged “bait” proteins enable standardization of affinity-purification assays, and analysis of protein complexes for which suitable antibodies are not available. AP-MS has been systematically applied to the yeast proteome, providing the first global surveys of eukaryotic protein complexes.2–4 A principal advantage of yeast as an experimental system for mapping the protein interactome is the robust and efficient means by which epitope tags can be introduced via homologous recombination. In a systematic study of the yeast protein interactome, 3′-end epitope tags were introduced into approximately 70% of known yeast open reading frames (ORFs) and protein complexes analyzed by mass spectrometry.4 In contrast, the lack of robust and efficient means of engineering epitope tags into the mammalian genome has meant that most systematic studies of mammalian protein interactomes5,6 have relied on ectopic expression of epitope-tagged bait proteins. However, bait transgenes may lack important regulatory sequences such as 3′ UTR or introns and are driven by constitutive or inducible promoters.7 Although these mammalian AP-MS studies have yielded fascinating functional insights into the mammalian protein interactome,8 an ongoing concern is the potential for artifactual interactions arising through overexpression or nonendogenous regulation of tagged bait proteins. Although there are no (to the authors’ knowledge) systematic studies of the effects of protein overexpression in mammalian cells, it has been shown in ORFeome wide studies in yeast that overexpression may lead to mislocalization of tagged proteins and morphological abnormalities.9
Workflows that specifically address the issue of bait protein expression levels have been developed, using antibodies directed against endogenous proteins,49 or epitope tags introduced into target loci within bacterial artificial chromosomes (BACs).10 These BAC clones are then transfected into mammalian cells and analyzed by AP-MS.10 Although BAC clones preserve endogenous promoters of target genes, they may not necessarily retain long distance enhancer elements, and in addition, the copy number of introduced BAC clones may vary from transfection to transfection.10 Knock-in and knock-out techniques using homologous recombination have been widely used to study gene-function in mice.50 Similar (in principle) techniques have also been applied to manipulating genes in human somatic cells,53 although applications to human cells have been hampered by the lower rates of homologous recombination than in mouse cells, and greater complexity of the human genome. Technical developments that have facilitated gene-targeting in human somatic cells include the use of recombinant adeno-associated viral vectors.56 The principal application of gene-targeting in human somatic cells has been in functional studies of genes,53 and knock-ins and knock-outs provide a powerful tool for understanding human gene function. We and others have built upon these techniques to improve the ease and efficiency of the creation of knock-in epitope-tagging in human cell lines,17,57 and showed that epitope-tagged endogenous human genes are excellent substrates for ChIP-seq applications.17 More generally, knock-in epitope-tagging of human genes will be an important technique for wider studies of protein function, including studying protein–protein interactions, as we describe here. The knock-in cell lines described here express epitope-tagged bait proteins under the control of endogenous transcriptional mechanisms in physiologically relevant cell lines (in our case, human colorectal cancer cell lines), and, as we demonstrate in this study, provide robust substrates for AP-MS. Knock-in epitope-tagging also circumvents the challenges of cloning and expression of genes with long open reading frames, and therefore provides a potentially universal platform for protein complex discovery in mammalian cells.
We selected three important colorectal-cancer associated proteins as targets for analysis using the knock-in AP-MS strategy. The target genes used are MRE11A (meiotic recombination 11), a component of the MRE11A-RAD50-NBN complex,11,12 DNMT1 (DNA methyltransferase 1), a DNA methyltransferase involved in establishment and regulation of patterns of DNA methylation13 and APC (Adenomatous Polyposis Coli), a critically important tumor suppressor gene14,15 and Wnt pathway regulator.8 Knock-in MRE11A and DNMT1 cell-lines were principally used in this study as test cases to develop the workflow, whereas the focus was to identify novel protein–protein interactions using the knock-in APC cell-line. APC is a colorectal cancer “gatekeeper” (~80% of colorectal cancers exhibit APC mutations),54 and significant efforts have been made to understand APC protein function, and the mechanisms by which APC mutations exert their effects.24 APC protein has diverse cellular functions, and localizes to multiple subcellular compartments.16,25 Although now broadly accepted, the localization of APC protein to the nucleus was initially controversial, in part because of the lack of good antibodies.30 In addition, ectopic expression of APC protein with constitutive promoters can result in accumulation of APC protein in nucleoli.55 Although nuclear APC is thought to sequester β-catenin and thereby repress Wnt signaling, the functional role of nuclear APC and nuclear APC complexes is not fully understood.30 In addition, analysis of APC protein complexes is challenging, due to the large size of the protein (2843 amino acids; 311 kDa) and length of the APC coding region (>10 kb mRNA sequence length) as well as the variability of currently available native APC antibodies.16 Knock-in AP-MS can potentially circumvent these practical challenges, by ensuring that the epitope-tagged bait proteins are expressed at endogenous levels and remain responsive to internal and external signaling cues. We show that epitope-tagged baits are expressed at similar levels to endogenous proteins; using replicate analyses, we show that knock-in cell lines coupled to AP-MS provide a robust and reproducible platform for protein complex discovery in human cells. In addition, knock-in AP-MS can be used to sensitively identify novel protein–protein interactions. In this study, we identify and characterize two APC-associated proteins, Erbb2 interacting protein (ERBB2IP) and Flightless-1 homologue (FLII), an interaction with APC that occurs more abundantly in nuclear fractions.
MATERIALS AND METHODS
Knock-In Epitope-Tagging in Human Cells
A ~1 kb genomic fragment upstream of the stop codon was PCR amplified to be used as the left homologous arm and a ~1 kb genomic fragment downstream of the stop codon was PCR amplified to be used as the right homologous arm. The left and right homologous arms were cloned into pAAV-USER-Neo-LoxP-3×Flag vector using USER system as previously described.17 The targeting AAV viruses were packaged in 293T cells (a T75 flask at 70% confluence) by transfecting equal amounts of the targeting vector, pHelper and pRC plasmids (3 μg each). Viruses were harvested 72 h post-transfection. Colon cancer cells were infected with the knock-in targeting viruses and selected with Geneticin for 20 days. The Geneticin resistant clones were then screened for homologous recombination by 35 cycles of genomic PCR with primers derived from the neomycin resistance gene and the upstream region of the left homologous arm. Confirmatory genomic PCR was also performed with positive clones identified using primers derived from the neomycin resistant gene and the downstream region of the right homologous arm. Correctly targeted clones were then infected with adenoviruses expressing the Cre-recombinase to delete the drug selection marker. While it takes around 45 days to generate 3×Flag KI clones, most of this time is required for the clones to grow up. The actual hand-on time and effort are less than 5 days (1 day for vector construction, 1/2 day for virus package, 1/2 day for virus harvest and infection, 2 h for cell plating, 1 day for targeted clone screening and ~1 day for excision of drug selection marker). The targeting rates are 3% for MRE11A, 5% for DNMT1 and 2% for APC. So far we have successfully knocked in 3×Flag sequences into ~20 loci. We have successfully targeted loci in the following colon cancer cell lines: HCT116, DLD1, RKO and LOVO and a detailed description of gene-targeting methods is provided in the Supplementary Methods in Supporting Information.
Plasmid Construction, Cell Targeting, and Cell Culture
Each gene used in this study was targeted by C-terminal 3×Flag tag sequence with plasmid pAAV-LoxP-Neo-3×Flag17 and knocked in target cells using Adeno-Associated Virus (rAAV) transfection system followed by Geneticin (Invitrogen) selection and Cre-mediated excision of the drug resistance marker in targeted cells. RKO, HCT116, MRE11KI-RKO, DNMT1KI-RKO and APCKI-HCT116 cells are regularly maintained in McCoy-5A media (Invitrogen, 16600-108, Carlsbad, CA) containing 10% fetal bovine serum (Invitrogen, 10438-026, Carlsbad, CA) and 1% streptomycin–penicillin (Invitrogen, 15140-148, Carlsbad, CA) at 37 °C in CO2 incubator (5% CO2, 100% H2O).
Protein Extraction and Affinity Purification
All centrifugation and incubation are performed at 4 °C and all buffers are prechilled on ice. For all cells, the media were removed from the 150 mm culture plates and the cells (3 × 107 cells were used for each affinity purification) were collected and washed three times with 25 mL of PBS. After pelleting the cells, the cells were lysed with 2 mL of cell lysis buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.1% NP-40, 1× protease inhibitor cocktail) with homogenization for 40 times. The lysate were incubated on ice for an additional 30 min before 13 000 rpm centrifugation. Next, 100 μL of packed anti-FLAG M2 affinity beads (Sigma A2220, Saint Louis, MO) was first equilibrated with 500 μL of TBS buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl) followed by 500 μL of cell lysis buffer and centrifuged at 10 000 rpm for 5 s before adding the cell lysate supernatant. The protein–bead mixtures were then incubated on a rotator at 4 °C overnight. The mixtures were centrifuged at 10 000 rpm for 5 s and the supernatant (flow though) was kept for later analyses. The beads were washed twice with cell lysis buffer, four times with TBS buffer and twice with 100 mM ammonium bicarbonate (NH4HCO3) and then eluted with elution buffer: TBS buffer containing 5 μg/μL 3×FLAG peptide (Sigma F4799, Saint Louis, MO).
In-Gel Tryptic Digestion
Standard in-gel tryptic digestion was performed according to the published method.18 The combined elution fractions were lyophilized in a SpeedVac Concentrator (Thermo Electron Corporation, Milford, MA), resuspended in 100 μL of 0.1% formic acid and further cleaned up by reverse phase chromatography using C18 column (Harvard, Southborough, MA). The final volume was reduced to 10 μL by vacuum centrifugation and addition of 0.1% formic acid.
Subcellular Fractionation
Cytoplasmic and nuclear proteins were extracted from cells using Thermo Scientific Pierce NE-PER Nuclear and Cytoplasmic Extraction Kit (78833, Pierce Technology, Rockford, IL) and used immediately in SDS-PAGE and Western blots or affinity purifications.
In-Solution Tryptic Digestion
The protein elution fractions were dried by vacuum centrifugation. Each sample was denatured with the addition of 20 μL of 8 M urea and reduced with 2 μL 100 mM DTT in 100 mM NH4CO3. The solution was mixed well and incubated at room temperature for 45 min. For each sample, 2 μL of 250 mM iodoacetamide was then added and incubated in the dark for another 45 min. Each sample was digested by adding 5 μL of the 0.2 μg/μL trypsin solution (Promega, PR-V5111) and 125 μL of 100 mM NH4CO3 solution and incubated at 37 °C overnight. The C18 column was wet with 100% methanol and balanced with 0.1% formic acid. The peptide mix from each sample was loaded on the C18 column and washed five times with 200 μL of 0.1% formic acid, and then eluted with elution buffer (50% acetonitrile and 0.1% formic acid). The elution fractions were then lyophilized in a SpeedVac Concentrator (Thermo Electron Corporation, Milford, MA) and resuspended in 10 μL of 0.1% formic acid prior to further analyses.
Mass Spectrometric Analyses and Raw Data Analysis
Tryptic peptides were separated by online reverse phase nanoscale capillary liquid chromatography (nano-LC, Dionex Ultimate 3000 series HPLC system) coupled to electoral spray injection (ESI) tandem mass spectrometer (MS–MS) with octopole collision cell (Thermo-Finnegan LTQ Orbitrap). Loaded peptides were eluted on nano-LC with 90 min gradients ranging from 6 to 73% acetonitrile in 0.5% formic acid with a flow rate of 300 nL/min. Data dependent acquisition was performed on the LTQ-Orbitrap using Xcalibur software (version2.0.6, Thermo Scientific) in the positive ion mode with a resolution of 60 000 at m/z range of 325.0–1800.0, and using 35% normalized collision energy, up to five most intensive multiple charged ions were sequentially isolated, fragmented and further analyzed. Raw LC–MS/MS data were processed by Mascot version 2.2.0 (Matrix Science, Boston, MA). Mascot was searched with a fragment ion mass tolerance of 0.8 Da and a parent ion tolerance of 15 ppm. The raw data were searched against the human International Protein Index database (released in 2009 and containing 74 017 protein sequences) with fixed modification carbamidomethyl (C) and variable modification oxidation (M). Peptides were filtered at a significance threshold of P < 0.05 (Mascot). Raw mass spectrometry chromatograms were processed and analyzed using Xcalibur Qual Browser software (Thermo Fisher Scientific, Inc., Version 2.0.7) and then manually annotated and verified.
Data Processing
Scaffold (Proteome Software, Inc., Portland, OR; version 3.00.04) was used to validate LC–MS/MS-based peptide and protein identifications.19 Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm.20 Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides.21 With these stringent parameters of Peptide Prophet and Protein Prophet, the false discovery rate was zero.21 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Selected signature peptides were verified and annotated manually.
Statistical and Interaction Network Analyses
Spectral counts for proteins identified as above were used to calculate bait/control ratios and significance. For each protein, the logarithm (base 2) of the ratio of the sum of spectral counts for replicate experiments (bait or control) was computed. For all baits, several replicate blocks of experiments were performed, where replicate experiments are biological replicates of bait and control experiments (independent affinitypurifications/cell-cultures and subsequent mass spectrometry). P-values were computed using the Wilcoxon rank sum test to identify proteins for which bait spectral counts > control spectral counts. Significant proteins were then used in subsequent interaction network analysis and visualization using the Ingenuity Pathways Analysis (IPA) tool. To generate the biological networks, both direct and indirect interactions were used and the Ingenuity knowledge base was used as the reference set. BioGRID version 3.1.78 was used in the sensitivity analysis for the APC data sets.
SDS-PAGE and Western Blot Analysis
Equal amount (20 μg) of proteins from different samples was loaded on precast 4–12% Bis-Tris gel (Invitrogen NP-0335, Carlsbad, CA) and subjected to electrophoresis. Afterward, gels were either stained with Coomassie Brilliant Blue or transferred to nitrocellulose membrane (Whatman 10402594, Dassel, Germany). Western blotting was used to detect the protein with super signal ELISA Pico chemiluminescent substrate (Pierce 37070, Rockford, IL). Primary antibodies anti-FLAG (Sigma F1804, Saint Louis, MO), anti-DNMT1 (Cell Signaling Technology 5119, Danvers, MA), anti-MRE11 (Novus Biologicals NB100-142, Littleton, CO), anti-APC (generously provided by Dr. Kristi Neufeld from University of Kansas), anti-β-catenin (Cell Signaling Technology 9581, Danvers, MA), anti-TPM1 (Cell Signaling Technology 3910, Danvers, MA), anti-FLII (Santa Cruz sc-55583), anti-ERBB2IP (Sigma 09105, Saint Louis, MO) and anti-α-tubulin (Cell Signaling Technology, Inc., 2144, Danvers, MA) as loading control were applied at 1:1000 and secondary antibodies horseradish peroxidase (HRP)-conjugated anti-mouse (Promega W4011, Madison, WI) and HRP-conjugated anti-rabbit (Novus Biologicals NB730-H, Littleton, CO) were added at 1:20 000. Chemiluminescence detection using SuperSignal* ELISA Pico Chemiluminescent Substrate (Thermo Scientific PI-37070, Rockford, IL) was applied to all westerns.
RESULTS
Overview of the Knock-In AP-MS Strategy
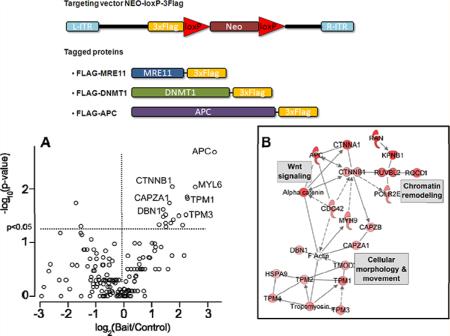
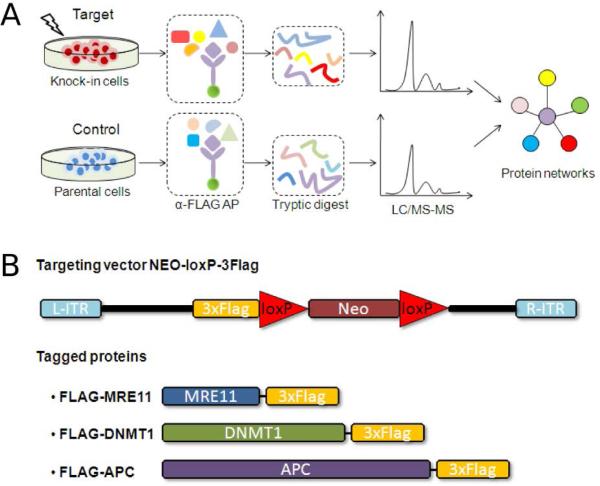
Figure 1 provides an overview of the knock-in AP-MS pipeline. AP-MS experiments are performed as shown in Figure 1A. Endogenous human target genes are epitope-tagged by introduction of triple-FLAG tags directly into the human genome in selected human cell lines as previously described.17 AP-MS experiments using these knock-in cells in parallel to control cells (the untagged parental cell-line) are performed and analyzed using label-free LC–MS/MS. The targeting vector used for generating knock-in cell lines and the resulting epitope-tagged bait proteins are shown in Figure 1B. Complete details of how knock-in epitope-tagged cell lines are created are provided in the Supplementary Methods in Supporting Information.
Figure 1.
Knock-in AP-MS workflow. (A) Experimental design of knock-in AP-MS in this study. (B) Targeting vector (previously described in Zhang et al. 2008)17 and three targeted human genes (MRE11A, DNMT1 and APC) used in this study are shown. Each gene was tagged with a 3×FLAG at the C-terminal end.
We selected 3 target human genes for development and proof of principle of the knock-in AP-MS pipeline. These 3 target genes, Meiotic Recombination 11 Homologue A (MRE11A), DNA-methyltransferase 1 (DNMT1) and Adenomatous Polyposis Coli (APC), were selected based upon their significance to human colorectal cancer, an area of interest in our laboratories, and epitope-tagged in relevant human colorectal cancer cell lines. MRE11A (meiotic recombination 11 homologue A) encodes a component of the heterotrimeric MRN complex (with Rad50 homologue, RAD50 and Nijmegen breakage syndrome 1, NBN), that plays a key role in DNA double-stranded repair and other recombination-related processes.22 The MRE11A gene was tagged in the RKO colorectal cancer cell-line (MRE11KI-RKO) and we previously showed that the subcellular localization in nuclei of epitope-tagged MRE11A in MRE11KI-RKO cells resembled that of untagged MRE11A.17 In addition, immunopurification with anti-FLAG from MRE11KI-RKO cells showed association of MRE11A with RAD50 and NBN using Western blots.17
In this study, we used the MRE11KI-RKO cell line to test knock-in AP-MS in terms of the reproducibility and significance of mass spectrometry based identification of MRN complex components. A second target gene, DNMT1, was epitope-tagged in two colorectal cancer cell lines, RKO (DNMT1KI-RKO) and HCT116 (DNMT1KI-HCT116). Subsequent AP-MS experiments were conducted using the DNMT1KI-RKO cell line. DNMT1 establishes and maintains patterns of DNA methylation during development and differentiation, and disregulation of DNMT1 is associated with many different cancers and implicated in colorectal carcinogenesis.23
The third target gene, APC, is a large (2843 amino acids; 311 kDa) multifunctional protein, best characterized as a tumor suppressor in colorectal cancer. APC is a negative regulator of the Wnt Signaling pathway and defects of the APC gene are tightly associated with the development of colorectal cancer.24 There is significant interest in identification of novel APC interacting proteins, both because of the importance of APC in colorectal cancer as well as understanding the diverse cellular functions of APC.25 Other possible AP-MS approaches to identification of APC interactors include transgenic expression of epitope-tagged APC or immunopurification of APC using native antibodies. However, neither of these approaches is straightforward. In the first case, the large size of APC makes construction and cloning of the epitope-tagged APC gene challenging, and in the second case, the variability of native APC antibodies16 hinders immunopurification of native APC. Knock-in AP-MS circumvents these issues. We knock-in tagged APC in the HCT116 cell line (APCKI-HCT116) and used this cell line to identify APC interacting proteins.
Expression of Epitope-Tagged Target Proteins
Prior to developing knock-in AP-MS, we observed considerable variability of bait protein expression levels following transient transfection of overexpression clones. For example, transient transfection of RKO cells with a FLAG-MRE11A construct showed significant variability of FLAG-MRE11A protein expression (Supplementary Figure 5A), although there is little difference between overall FLAG-MRE11A protein expression level after pooling multiple overexpression clones and FLAG-MRE11A expression in difference in MRE11KI-RKO cells (Supplementary Figure 5B in Supporting Information).
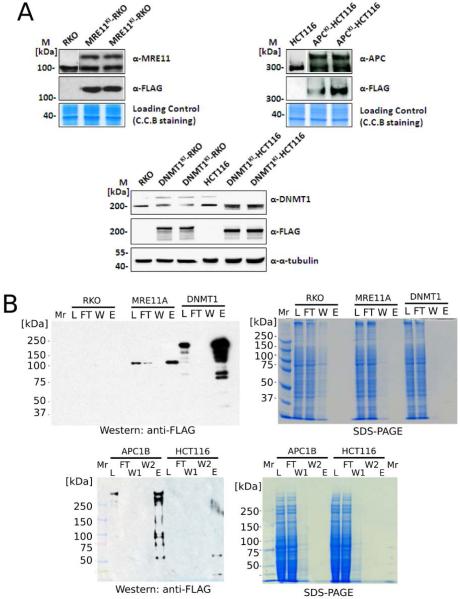
The previously described knock-in tagging approach17 was used to generate triple-FLAG epitope-tagged target proteins (Figure 1B) for the 3 loci of interest in two different human colorectal cancer cell lines (RKO and HCT116). The knock-in approach (Figure 1B) generates cell lines expressing full-length target proteins with a C-terminal triple FLAG tag (N-DYKDDDDKDYKDDDDKDYKDDDDK-C). To confirm expression of the recombinant proteins, Western blot analysis of whole cell lysates from knock-in cell lines and parental (RKO and/or HCT116) cell lines was performed (Figure 2A). For each of the 3 target genes (MRE11A, DNMT1 and APC), immunoblots of two independently tagged cell lines with either the anti-FLAG or a respective native antibody are shown. Since a key motivation for development of knock-in AP-MS is that bait protein expression should be maintained at endogenous levels, we compared expression levels of FLAG-tagged proteins with untagged equivalents and found that these are approximately similar (Figure 2A). High resolution anti-DNMT1 Western blots of the DNMT1KI-RKO and DNMT1KI-HCT116 cell-lines are provided in Supplementary Figure 7 showing untagged and tagged endogenous DNMT1 alleles.
Figure 2.
Western blots revealed protein expression levels in knock-in cells and monitored the efficiency of affinity purification. (A) 3×FLAG tagged proteins expressed at similar levels as endogenous proteins in knock in cells. (B) Significant enrichment of target proteins and their respective complexes by affinity purification. M refers to standard protein marker, L refers to lysate, W refers to wash, E refers to elution fraction and FT refers to flow through.
Recovery and Identification of Protein Complexes
To purify protein complexes for each target protein from total cellular extracts, we used affinity purification using anti-FLAG resin (Figure 1A). Protein complexes of protein baits were captured by affinity binding of FLAG-tagged bait protein and anti-FLAG resin and nonbinding proteins were washed off the protein–resin mixture. The protein complexes were then eluted by FLAG peptide competition to the resin. Bait proteins were identified in both whole cell lysates and elution fractions from affinity purifications (Figure 2B), indicating successful recovery of protein complexes and their respective baits. Figure 2B also shows that affinity purification using anti-FLAG significantly amplifies the bait protein signal as compared to whole cell lysates (Figure 2B). In the case of MRE11A, a trace of FLAG signal of the same protein size (~100 kDa) as FLAG-MRE11A (Figure 2B) was detected in flow through fractions (FT). This is likely due to the saturation of FLAG-tagged proteins loaded on a given amount of anti-FLAG resin. To estimate the quantitative enrichment of bait proteins that we observe in affinity purifications, we performed 3 replicate purifications from MRE11KI-RKO cells. Quantitative Western blot analysis showed an estimated 25-fold enrichment of bait proteins from affinity purification compared to whole cell lysate (Supplementary Figure 1).
Elution fractions from affinity purification were directly desalted, digested and analyzed by nano-HPLC coupled LTQ mass spectrometer. The raw data were searched against the human International Protein Index (IPI) sequence database using the Mascot search engine and analyzed with Scaffold software.20,21 Example base peak chromatograms for replicate mass spectrometry runs and annotated spectra for selected peptides corresponding to RAD50 and NBN from the MRE11A AP-MS experiments are in Supplementary Figure 3.
Knock-in Cell Lines Are Robust and Reproducible Substrates for AP-MS
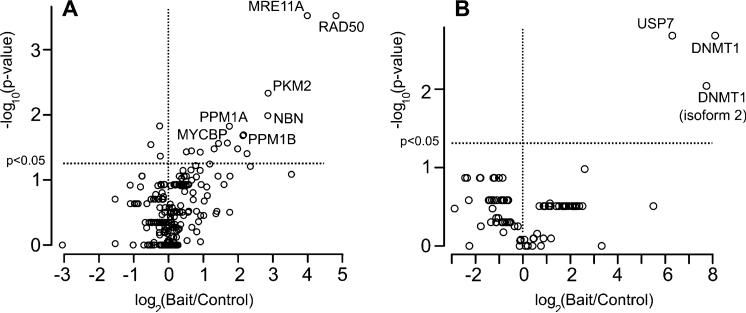
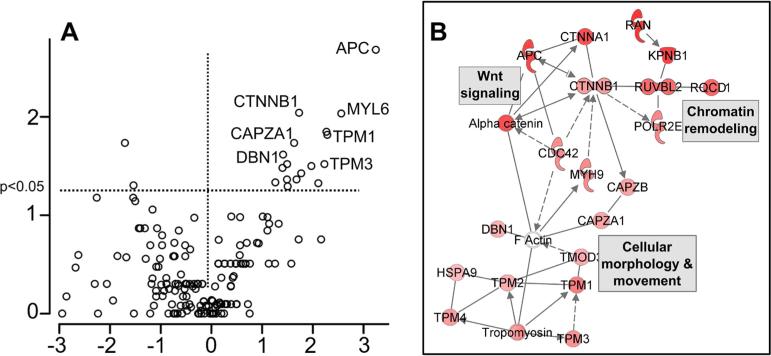
To gage the sensitivity and reproducibility of knock-in AP-MS, replicate AP-MS experiments, each consisting of a control and bait immunoprecipitation, were performed for two baits, MRE11A (n = 7) and DNMT1 (n = 5). Bait/control spectral count ratios were calculated for each identified protein (Scaffold probability ≥99%; 2 or more peptides/protein) and significance assessed for each protein based upon the replicate experiments (Wilcoxon rank sum test). The significance (y-axis) versus the log2 (bait/control) (x-axis) are plotted for each protein identified in the volcano plots in Figure 3. All proteins above the horizontal lines in each plot are significant at p < 0.05 (Wilcoxon rank sum test). Thus, putative bait-interacting proteins appear in the upper right segment of each plot, and selected proteins are labeled (protein identification and spectral count data and analysis are in Supplementary Tables 1A–D and Supplementary Tables 2A–D, respectively).
Figure 3.
Statistical analysis of knock-in AP-MS experiments. (A) MRE11A bait, (B) DNMT1 bait. Volcano plots of proteins identified from replicated AP-MS experiments are plotted with log2 (bait/control spectral counts ratio) on x-axis and –log10 P-value (Wilcoxon) on y-axis. Horizontal and vertical dashed lines indicate log2 ratio > 0 and p < 0.05, respectively. Selected significant proteins labeled with gene symbol. Complete sets of significant proteins for MRE11A and DNMT1 studies are highlighted in Supplementary Tables 2C and 2D, respectively.
Using the MRE11KI-RKO cell line and parental RKO cells as control, the MRE11A-RAD50-NBN complex was reproducibly identified. MRE11A, RAD50, and NBN peptides were uniquely detected in all replicate MRE11KI-RKO affinity purifications with p-values of p < 0.003, p < 0.003, and p < 0.01, respectively (Wilcoxon rank sum test). Thus, as a test case, knock-in AP-MS using MRE11A bait showed significant, reproducible identification of MRN complex components. We also compared the knock-in approach to overexpression of MRE11A-FLAG constructs in RKO cells. In two replicate overexpression experiments, we observed substantially similar sets of proteins as knock-in experiments, indicating that the knock-in approach performs as well as the overexpression approach, at least for the MRE11A bait (spectral count data for these experiments are provided in Supplementary Table 4).
Replicate AP-MS experiments with the DNMT1KI-RKO cell-line revealed ubiquitin-specific peptidase (USP7) as a DNMT1 associated protein (p < 0.003). We recently showed that USP7 deubiquitinates DNMT1 and functions in concert with several other proteins to regulate DNMT1 stability.26 Thus, using knock-in cell lines in conjunction with AP-MS is able to identify physiologically and functionally relevant interacting proteins. We also note the identification of both isoform 1 and 2 of DNMT1. Although DNMT1 isoform 1 is predominantly identified in the anti-DNMT1 pull-downs, we also identified DNMT1 isoform 2 with two unique peptides, suggesting that the endogenously tagged DNMT1 produces both known protein isoforms.51
Identification of APC Interacting Partners Using Knock-In AP-MS
As a key determinant of colorectal carcinogenesis, there is significant interest in identifying proteins associated with the APC protein that may further shed light on the cellular functions of this protein. We therefore used the APCKI-HCT116 cell line in AP-MS experiments and analyzed the results as shown in Figure 4A. The volcano plot in Figure 4A summarizes results from a replicated (n = 5) knock-in AP-MS experiment using APCKI-HCT116 cells. In all cases, replicate experiments refer to biological replicates, in which independent affinity purifications are performed. In a separate replicate experiment (n = 2), additional significant APC interactors were identified such as alpha-catenin (CTNNA1) (Supplementary Table 1B).
Figure 4.
Analysis of knock-in AP-MS experiments with APCKI-HCT116 cell line. (A) Volcano plot of proteins identified from replicated AP-MS experiments with APCKI-HCT116 and HCT116 control cells (details as Figure 3). (B) Selected interaction network from knock-in APC AP-MS experiments. Each node in the network represents a protein identified as significant (p < 0.05) in APC AP-MS experiments. All proteins shown were found with increased abundance in APC bait experiments compared to control and node shade (red) indicates the magnitude of the bait/control ratio of spectral counts. AP-MS proteins are superimposed on known networks for corresponding genes and proteins (from Ingenuity Pathways Analysis). Edges between nodes indicate known interactions (solid edge = direct interaction, dashed edge = indirect interactions). Complete sets of proteins from APC AP-MS experiments are in Supplementary Tables 2A and 2B.
To functionally organize these data, we assembled them into interaction networks using the Ingenuity Pathways Analysis (IPA) tool. Proteins significant at p < 0.05 (Wilcoxon rank sum test) were initially organized into the network shown in Figure 4B. Several additional proteins (POLR2E, RQCD1, KPNB1 and RAN) significant at p < 0.1, were added where these extended the core functions present in the network. This network identifies the key cellular functions of APC, including interaction with chromatin,27 as a tumor suppressor in the Wnt signaling pathway,24 and as an organizer of microtubule networks in cellular mobility and migration.25
A previous proteomics study comparing colonic mucosa from patients heterozygous for an APC mutation with control patients28 identified several proteins in common with proteins identified in the knock-in AP-MS experiments. Specifically, Patel et al. identified many proteins involved in cytoskeletal and microtubule regulation that were more abundant in cells harboring the APC truncation mutation. Multiple Tropomyosin family proteins were identified by Patel et al. that were also present in the APC knock-in AP-MS data set. In addition, Patel et al. identified Ras Suppressor 1 (RSU1) as more abundant in control colonic mucosa cells than FAP colonic mucosa. In our AP-MS experiments, RSU1 was identified as more abundant in APCKI-HCT116 AP-MS experiments than in control (p < 0.1). We therefore tested several of these proteins for their presence in APC-associated protein complexes.
First, we checked the existence of β-catenin as well as APC in affinity purification elution fraction by Western blot using anti-β-catenin and anti-FLAG antisera, respectively. In two experimental replicates, FLAG-tagged APC and β-catenin were only present in elution fractions originating from APC knock-in HCT116 cells, and not in parental HCT116 cells (Figure 5), indicating that endogenously epitope-tagged APC binds to β-catenin in APCKI-HCT116 cells.
Figure 5.
Western blot verifications of selected known and novel APC interactors. Elutions from IP by α-FLAG/APC were loaded on SDS-PAGE followed by Western blot analyses with antisera against APC interactors, β-catenin, APC, and TPM1. M refers to standard protein marker and rep in sample name refers to biological replicates. Antisera α-tubulin and β-catenin were used as loading controls for equal loading of cell lysate for each IP experiment.
In addition, we used independent affinity purification and Western blot analysis using antisera against two additional proteins, Tropomyosin 1 (TPM1) and Ras Suppressor 1 (RSU1). As shown in Figure 5, TPM1 is clearly more abundant following anti-FLAG immunopurification in elution fractions from APCKI-HCT116 cells than in HCT116 cells, validating the AP-MS data and implying an association between APC and TPM1. We were unable, however, to detect RSU1 in Western blot experiments in APCKI-HCT116 cells (data not shown), and are therefore unable to conclude that APC and RSU1 occur in the same protein complexes.
Combining Subcellular Fractionation with Knock-In AP-MS Identifies Novel Nuclear APC Interactions
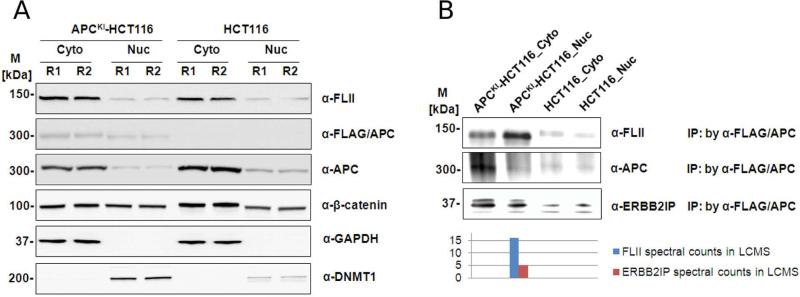
The diverse functions of APC in different subcellular locations of the cell25,29 prompted us to attempt to identify APC associated proteins in specific subcellular compartments. In particular, there is significant interest in defining the functions of APC in the nucleus and in how the nuclear functions of APC relate to colorectal cancer progression.30 We also reasoned that the sensitivity of knock-in AP-MS would be increased by an additional subcellular fractionation step. Subcellular fractionation techniques were used to generate enriched nuclear and cytosolic fractions prior to affinity purification. Western analysis using native antibodies showed that subcellular compartment-specific proteins (DNMT1 and GAPDH) were uniquely present in nuclear and cytosolic fractions, respectively (Figure 6A). Using either native anti-APC antibodies or anti-FLAG antibodies, native and epitope-tagged APC were found to be present in both cytosolic and nuclear fractions. Both antibodies show higher signal in cytosolic fractions than in nuclear, in line with other reports that wild-type APC is primarily cytoplasmic in colorectal cancer cells.29 Thus, the subcellular distribution of the knock-in epitope tagged APC allele is similar to that of the endogenous untagged APC protein, with predominant signal in the cytoplasm.
Figure 6.
Subcellular fractionation coupled to knock-in AP-MS increases sensitivity for detection of novel APC-interacting proteins. (A) Cellular fractionation of APC knock-in and parental HCT116 cells. Twenty micrograms of proteins from each fraction was loaded on SDS-PAGE followed by Western blot analyses with anti-FLII, anti-FLAG, anti-APC, anti-β-catenin, anti-α-tubulin, anti-DNMT1. M refers to standard protein marker and the numbers at end of sample names refer to biological replicates. (B) Western blot verifications of novel APC interactors Flightless-1 (FLII) and erbb2 interacting protein (ERBB2IP) in cellular fractions. Elutions from IP by α-FLAG/APC were loaded on SDS-PAGE followed by Western blot analyses with antisera against FLII. M refers to standard protein marker, R refers to biological replicates, Cyto refers to cytoplasmic fraction and Nuc refers to nuclear fraction. The Western blot was then quantified and compared to spectral counts in LCMS data (bar charts below the Western blot).
We next used AP-MS to analyze nuclear and cytosolic fractions from the APCKI-HCT116 cell line (protein identification and spectral count data from subcellular fractionation experiments are in Supplementary Tables 3A and 3B, respectively). We focused on proteins identified solely in the nuclear fractions from APCKI-HCT116 cells following AP-MS that may represent nuclear-specific interactors of APC. One such protein, Flightless-1 homologue (FLII), has previously been shown to functionally interact with β-catenin and inhibit β-catenin-mediated gene-transcription.31,32 To test whether the association between APC and FLII was exclusive to, or enriched in the nuclear fraction, we performed Western blot analysis. As shown in Figure 6A, Western analysis of cell lysates (without immunopurification) of endogenous FLII indicates significantly more FLII in the cytosolic fraction than in the nuclear fraction. However, in anti-FLAG immunoprecipitates of nuclear and cytosolic fractions, FLII is significantly enriched in the nuclear fraction (Figure 6B). As shown in Figure 6B, FLII was only detected by mass spectrometry in the nuclear fraction after anti-FLAG pull-down of APC. In addition, although there is some cytosolic signal, Western analysis (Figure 6B) indicates significant enrichment of FLII in nuclear fractions after APC pull-down. Thus, although FLII and APC proteins are both more abundant in the cytoplasm than they are in the nucleus, their coassociation is enriched in the nucleus. Given the previous studies of FLII interaction with β-catenin,31,32 it is quite probable that FLII, APC and β-catenin all co-occur in the same protein complexes in the nucleus.
We observed that many of our knock-in AP-MS interactions were also found in a recent yeast-two-hybrid study34 using APC as a bait (see next section). A feature of yeast-two-hybrid is the sensitivity with which direct interactions between potentially low-abundance proteins can be assayed. Erbb2 interacting protein (ERBB2IP) was found in both the yeast-two-hybrid study and in our knock-in AP-MS subcellular fractionation study. The PDZ domain of ERBB2IP was previously shown to mediate its interaction with both APC and β-catenin.36 ERBB2IP was also shown to negatively regulate β-catenin dependent gene expression.37 ERBB2IP is a paralog of scribble (SCRIB), both members of the LAP (LRR and PDZ domain containing family), and SCRIB functions in the Wnt signaling pathway and also interacts with APC via a PDZ domain.38 Peptides corresponding to ERBB2IP were identified only in the nuclear fraction from APCKI-HCT116 cells. To further validate the APC–ERBB2IP association, Western blot analysis using anti-ERBB2IP antibodies was performed as shown in Figure 6B. This analysis revealed significant enrichment of ERBB2IP protein in anti-FLAG immunoprecipitates from APCKI-HCT116 cells. Although ERBB2IP signal was found only in the nuclear fraction in AP-MS experiments, the antibody shows cytosolic signal as well, possibly indicating that the sensitivity of our AP-MS experiments could be further increased. Interestingly, while in the process of preparing this manuscript, another study identified ERBB2IP as an APC interactor, and showed that the APC-ERBB2IP was increased in the presence of Wnt stimulation.39
Comparing Knock-In AP-MS Interactions versus Known
To benchmark knock-in AP-MS, we compared the APC data sets to physical interactions in BioGRID, a resource that collates interactions acquired using diverse techniques.33 BioGRID lists 98 distinct APC interactions from 21 different studies where APC was used as a bait. The majority of these 98 interactions have been identified from single studies, with only 3 (CTNNB1, CTNNA1 and JUP) identified as APC interactors in more than one study. We used the BioGRID data set to benchmark knock-in AP-MS data sets as follows. First, we computed the overlap between our data sets and the BioGRID data set at varying statistical thresholds. Figure 7A plots the proportion of proteins identified that are present in the BioGRID data set at different p-value thresholds (Wilcoxon rank sum test, as in Figure 3), and shows how this proportion increases with increasing statistical stringency. Thus, at p-value <0.05, ~1/3 of the proteins identified are already in the APC BioGRID data set. Similarly, we plotted the proportion of identified proteins that are in BioGRID versus the bait/control ratio threshold (Supplementary Figure 7). Overall, ~20% (22 proteins) of the existing BioGRID set is found in the knock-in AP-MS data sets, and of these 22 proteins, 19 have log2 (bait/control ratio) greater than 0). We observed that a recent yeast-two-hybrid study34 showed a particularly high overlap with the set of knock-in AP-MS APC proteins. Indeed, 17 of 22 proteins present both in the knock-in AP-MS data set and BioGRID are also represented in the yeast-two-hybrid of Bandyopadhyay et al.34 This is a significant overlap, given that previous comparisons of AP-MS and yeast two-hybrid studies from different laboratories have tended to show minimal overlap.35
Figure 7.
Benchmarking knock-in AP-MS against known interactions. (A) The proportion of interactions identified that are already known (BioGRID) vs the bait/control p-value for the APC (unfractionated) replicated data sets. At p < 0.05, approximately 1/3 of proteins identified are known APC interactions according to BioGRID. (B) Overlap between APC BioGRID interactions and proteins identified using knock-in AP-MS (all proteins with bait/control ratio ≥2), with or without prior subcellular fractionation. Proteins in bold were also identified as APC interactors in the Bandyopadhyay et al.34 study using yeast two-hybrid.
We next ascertained whether subcellular fractionation enhances the sensitivity of knock-in AP-MS experiments. All proteins in fractionated or unfractionated data sets with 2-fold or greater bait/control ratios were compared to the BioGRID data set (198 proteins from fractionated experiments and 44 proteins from unfractionated experiments). The subcellular fractionation experiments identified 15 BioGRID proteins, whereas the unfractionated experiments identified only 5 BioGRID interactions, 4 of which were also identified in the fractionated experiments (Figure 7B shows a Venn diagram of this analysis). Therefore, subcellular fractionation substantially increased the sensitivity of knock-in AP-MS to detect protein interactions, as judged by the number of known interactions detected. In addition, although we cannot directly compute the rates of false positives in these data sets, the proportion of true positives is approximately similar between fractionated and unfractionated experiments (8% and 11%, respectively), suggesting that the larger numbers of proteins identified in the fractionated experiments does not necessarily equate to larger numbers of false positives.
Since one of our goals in these experiments is to identify nuclear APC-associated proteins, we also established whether subcellular fractionation measurably increases the proportion of known nuclear proteins. Of the 198 proteins in the subcellular fractionation experiments with a bait/control ratio >2-fold (either in cytosolic or nuclear fractions), 57% were classed as cytosolic-specific, 34% as nuclear-specific, and 9% as neither (based upon spectral count signal in the nuclear or cytosolic fraction exceeding 50% of the total spectral counts from all other fractionated samples. Within the “nuclear-specific” proteins, we observed that 41% of proteins are annotated with the Gene Ontology term “nucleus”. In contrast, considering all proteins identified, only 31% are annotated with “nucleus”, suggesting that subcellular fractionation when combined with knock-in AP-MS does improve identification of nuclear proteins. In summary, this analysis shows that knock-in AP-MS is able to identify a significant fraction of known interactions, and that subcellular fractionation substantially increases the sensitivity of the approach.
DISCUSSION
Here we describe knock-in AP-MS, a technique combining knock-in epitope-tagged cell lines and affinity-purification mass spectrometry (AP-MS) for identification of protein complexes and networks. Knock-in techniques were used to endogenously tag genes in human cancer cells by expressing C-terminal FLAG-tagged proteins at endogenous levels. Label-free mass spectrometry was used to identify interacting proteins for three target proteins, MRE11A, DNMT1 and APC. Our results show that knock-in AP-MS with endogenous target proteins expressed at physiological levels provides a robust tool for mapping protein interactions.
Knock-in AP-MS reproducibly identifies all components of the MRN complex, of which the bait MRE11A is a component. Utilizing knock-in AP-MS to identify DNMT1 associated proteins, we identified USP7 as a reproducibly occurring protein in AP-MS experiments. We previously showed that USP7 forms a complex with DNMT1 regulators UHRF1, Tip60, HDAC1 and PCNA and that this complex regulates DNMT1 stability through deubiquitination and acetylation.26 Knock-in AP-MS enabled identification of multiple APC interacting proteins. Several of these were previously reported, including multiple proteins only previously observed in APC yeast two-hybrid experiments. We show that knock-in AP-MS is able to identify APC interacting proteins such as Flightless-1 homologue (FLII), and that the association between APC and FLII occurs more abundantly in nuclear fractions. AP-MS, in common with other protein–protein interaction techniques, is susceptible to false-negatives (failure to identify already known protein interactions). These may be biological (interactions only occurring in specific cells or tissues) or technical, such as lack of sensitivity (AP-MS may fail to detect transient interactions). For example, we did not identify ARHGEF4 protein in anti-FLAG pull-downs from APCKI-HCT116 cells, and ARHGEF4 (Asef) was previously shown to link APC to G-protein signaling.52
A primary motivation for developing knock-in AP-MS as a platform for protein interaction network mapping is to exploit the efficiency of epitope-tagging for immunopurification while expressing tagged proteins at physiological levels. The most commonly used approaches for identification of protein complexes using AP-MS use either ectopic expression of epitope-tagged cDNAs (driven by generic promoters) or native antibodies for endogenous proteins. These methods have been widely applied in multiple model organisms such as Oryza sativa,40Caenorhabditis elegans,41Saccharomyces cerevisiae3,42 and Homo sapiens5,6 and have provided significant insights into eukaryotic interaction network biology. However, both the cDNA approach and native antibody approach have important drawbacks. In the cDNA approach, inappropriate expression of epitope-tagged proteins may alter natively occurring protein interactions and networks. A previous systematic study of protein overexpression in yeast showed that approximately 15% of yeast ORFs when overexpressed resulted in reduced growth rate.43 Using native antibodies circumvents the issue of protein overexpression, but may suffer from the variability or lack of antibodies for all proteins, thus not being scalable to the complete proteome. Methods that combine the efficiency and scalability of epitope-tagging while ensuring that tagged proteins are expressed at physiologically relevant levels will therefore be powerful, scalable tools for accurately mapping eukaryotic interaction networks. A promising approach that makes use of recombineering to engineer large bacterial artificial chromosomes (BACs) constructs containing the tagged target gene and its associated regulatory sequences was previously described.10 The engineered BACs integrate into the host genome and express the epitope-tagged transgene at endogenous levels.10 The QUBIC platform combines expression of tagged genes from BAC constructs with quantitative proteomics and has been shown to provide a sensitive platform for identification of in vivo protein interactions.44 As noted by the authors, BAC TransgeneOmics does not necessarily guarantee integration and expression of a single BAC transgene per cell.10 In addition, untagged endogenous proteins may compete with the epitope-tagged protein for incorporation into multiprotein complexes decreasing the sensitivity of the assay.45 It was previously shown in Drosophila AP-MS experiments that RNAi depletion of endogenous, untagged target protein increased the sensitivity of AP-MS and enabled detection of novel interactors.45 Knock-in AP-MS, as described here, results in epitope-tagging of an endogenous allele of the target gene, therefore, potentially reducing the overall expression of untagged target gene.17 In addition, if desired, the knock-in technique may be used to sequentially target both alleles of the target gene (data not shown), further increasing the sensitivity of AP-MS. Though knock-in cell lines can be fairly rapidly produced (3 months from start to finish in the Wang laboratory) and would be amenable to being scaled up, the throughput may not ultimately match that of BAC transgene engineering.
Whether epitope-tagging of proteins significantly alters protein function or interaction is best evaluated on a case-by-case basis. A previous global study of protein localization of budding yeast46 found excellent (~80%) overlap with previously published localization data arguing that the C-terminal tag does not widely alter appropriate subcellular localization. There are, however, individual examples of epitope tags interfering with endogenous protein function.4 The epitope-tagged proteins used in our study appear to function similarly to untagged, endogenous equivalents. FLAG-MRE11A was nuclear localized17 and associated with known protein partners (this paper). FLAG-STAT3 retained the ability to activate the expression of a target gene47 and FLAG-DNMT1 maintains its regulatory pathway through coordinated deubiquitination and acetylation-driven ubiquitination.26 In addition, Western analysis (Figure 6A) indicated that both native APC and FLAG-APC are present in nuclear fractions, but are more abundant in cytosolic than nuclear fractions. These examples suggest that epitope-tagging, at least for the genes investigated in this study does not radically disrupt protein function or interactions.
In the current study, we use knock-in AP-MS to identify novel proteins associated with the important oncoprotein APC. These include FLII, Flightless-1 homologue, previously discovered to inhibit β-catenin dependent gene transcription. The identification of novel nuclear APC interacting proteins is significant because the nuclear functions of APC are not well understood.30 Studying the APC protein interactome using AP-MS is challenging because of the size of the full-length APC protein (~312 kDa) and variability of available antibodies.29 Endogenous tagging of APC through knock-in techniques circumvents these issues and provides a platform for discovery of APC interacting proteins in an environment in which the endogenous APC expression mechanisms are maintained.
The potential of knock-in AP-MS is underscored by our identification of multiple APC interacting proteins only previously identified using yeast two-hybrid techniques.34 Although two-hybrid methods are the preferred technique for identification of direct physical interactions between low-abundance proteins, the downside is that experiments are typically performed in a heterologous system (yeast). Our experiments confirm that multiple APC interacting proteins, previously only identified through yeast-two-hybrid, do indeed occur in human cells. In addition, we show that by performing subcellular fractionation in conjunction with knock-in AP-MS, we are able to identify interactions enriched in different subcellular compartments, which is of significant interest because APC exists in multiple different cellular pools with diverse functions.16,30
We also anticipate that knock-in AP-MS will be a useful tool for identifying proteins associated with mutated oncoproteins. In the case of APC, mutations in the Mutation Cluster Region encode truncated APC proteins and occur frequently in colorectal cancer tissues. The truncated APC protein product is hypothesized to be selected for by cancer cells, potentially to maintain an optimal level of Wnt signaling.48 Knock-in tagging of endogenous mutant APC protein could be a powerful means of uncovering mutant APC specific functions and protein interactions, although we recognize that the C-terminal tagging approach described here may not be appropriate for mutations that truncate the protein. Finally, using knock-in AP-MS in conjunction with signaling pathway stimuli or drugs will enable the dynamics of protein complexes and networks to be unraveled. As a key component of the Wnt signaling pathway, understanding how APC interactions and/or functions may be controlled is of significant biomedical interest.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Kristi Neufeld from Department of Molecular Biosciences in University of Kansas for kindly providing the anti-APC M2 antisera. This work was supported in part by the Cleveland Foundation and NIH/NCI 1R21CA160060-01A1 grant (to R.M.E.) and by NIH RO1 CA127590, R01-HG004722 and 1R21CA160060-01A1 grants (to Z.W.).
ABBREVIATIONS
- AP-MS
Affinity-purification mass spectrometry
Footnotes
Supporting Information
Additional informations as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Collins MO, Choudhary JS. Mapping multiprotein complexes by affinity purification and mass spectrometry. Curr. Opin. Biotechnol. 2008;19(4):324–330. doi: 10.1016/j.copbio.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 3.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang LY, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CWV, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 4.Krogan NJ, Cagney G, Yu HY, Zhong GQ, Guo XH, Ignatchenko A, Li J, Pu SY, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MHY, Butland G, Altaf-Ui AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 5.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark DP, Pazdemik NJ. Biotechnology: Applying the Genetic Revolution. Academic Press; Burlington, MA: 2009. ISBN 978-0-12-17552-2. [Google Scholar]
- 8.Major MB, Camp ND, Berndt JD, Yi XH, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, MacCoss MJ, Angers S, Moon RT. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316(5827):1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- 9.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, Horinouchi S, Yoshida M. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24(7):841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 10.Poser I, Sarov M, Hutchins JRA, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, Pelletier L, Kittler R, Hua S, Naumann R, Augsburg M, Sykora MM, Hofemeister H, Zhang YM, Nasmyth K, White KP, Dietzel S, Mechtler K, Durbin R, Stewart AF, Peters JM, Buchholz F, Hyman AA. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee AYL, Liu E, Wu X. The Mre11/Rad50/Nbs1 complex plays an important role in the prevention of DNA rereplication in mammalian cells. J. Biol. Chem. 2007;282(44):32243. doi: 10.1074/jbc.M705486200. [DOI] [PubMed] [Google Scholar]
- 12.Olson E, Nievera CJ, Lee AYL, Chen L, Wu X. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J. Biol. Chem. 2007;282(31):22939. doi: 10.1074/jbc.M702162200. [DOI] [PubMed] [Google Scholar]
- 13.Damelin M, Bestor TH. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol. Cell. Biol. 2007;27(11):3891–3899. doi: 10.1128/MCB.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceol CJ, Pellman D, Zon LI. APC and colon cancer: two hits for one. Nat. Med. 2007;13(11):1286–1287. doi: 10.1038/nm1107-1286. [DOI] [PubMed] [Google Scholar]
- 15.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 2001;1(1):55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 16.Brocardo M, Nathke IS, Henderson BR. Redefining the subcellular location and transport of APC: new insights using a panel of antibodies. EMBO Rep. 2005;6(2):184–190. doi: 10.1038/sj.embor.7400329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XD, Guo CG, Chen YT, Shulha HP, Schnetz MP, LaFramboise T, Bartels CF, Markowitz S, Weng ZP, Scacheri PC, Wang ZH. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat. Methods. 2008;5(2):163–165. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez CR, Huang L, Qiu Y, Burlingame AL. Current Protocols in Protein Science. John Wiley and Sons Inc.; New York: 1998. [Google Scholar]
- 19.Searle BC. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 20.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 21.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 22.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, Hays L, Morgan WF, Petrini JHJ. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93(3):477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 23.De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855–3860. [PubMed] [Google Scholar]
- 24.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 25.Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J. Cell Sci. 2007;120(19):3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- 26.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao H-Y, Xu Y, Willis J, Markowitz SD, Sedwick D, Ewing RM, Wang Z. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci. Signaling. 2010;3(146):ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouzmenko AP, Takeyama K, Kawasaki Y, Akiyama T, Kato S. Truncation mutations abolish chromatin-associated activities of adenomatous polyposis coli. Oncogene. 2008;27(36):4888–4899. doi: 10.1038/onc.2008.127. [DOI] [PubMed] [Google Scholar]
- 28.Patel BB, Li X-M, Dixon MP, Blagoi EL, Nicolas E, Seeholzer SH, Cheng D, He YA, Coudry RA, Howard SD, Riddle DM, Cooper HC, Boman BM, Conrad P, Crowell JA, Bellacosa A, Knudson A, Yeung AT, Kopelovich L. APC ± alters colonic fibroblast proteome in FAP. Oncotarget. 2011;2(3):197–208. doi: 10.18632/oncotarget.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brocardo M, Hendersonj BR. Detection of cytoplasmic and nuclear localization of adenomatous polyposis coli (APC) protein in cells. Methods Mol. Biol. 2008;468:77–89. doi: 10.1007/978-1-59745-249-6_6. [DOI] [PubMed] [Google Scholar]
- 30.Neufeld K. Nuclear APC. Adv. Exp. Med. Biol. 2009;656:13–29. doi: 10.1007/978-1-4419-1145-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y-H, Stallcup MR. Interplay of bac-I and FLAP1 for regulation of beta-catenin dependent transcription. Nucleic Acids Res. 2006;34(18):5052–5059. doi: 10.1093/nar/gkl652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Bang AG, Kintner C, Orth AP, Chanda SK, Ding S, Schultz PG. Identification of the Wnt signaling activator leucine-rich repeat in Flightless interaction protein 2 by a genome-wide functional analysis. Proc. Natl. Acad. Sci. U.S.A. 2005;102(6):1927–1932. doi: 10.1073/pnas.0409472102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark C, Breitkreutz B-J, Chatr-aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, Reguly T, Rust JM, Winter A, Dolinski K, Tyers M. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39(Suppl. 1):D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandyopadhyay S, Chiang C.-y., Srivastava J, Gersten M, White S, Bell R, Kurschner C, Martin CH, Smoot M, Sahasrabudhe S, Barber DL, Chanda SK, Ideker T. A human MAP kinase interactome. Nat. Methods. 2010;7(10):801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen LJ, Bork P. Not Comparable, But Complementary. Science. 2008;322(5898):56–57. doi: 10.1126/science.1164801. [DOI] [PubMed] [Google Scholar]
- 36.Ress A, Moelling K. Interaction partners of the PDZ domain of erbin. Protein Pept. Lett. 2006;13(9):877–881. doi: 10.2174/092986606778256126. [DOI] [PubMed] [Google Scholar]
- 37.Ress A, Moelling K. The PDZ protein erbin modulates beta-catenin-dependent transcription. Eur. Surg. Res. 2008;41(3):284–289. doi: 10.1159/000148241. [DOI] [PubMed] [Google Scholar]
- 38.Takizawa S, Nagasaka K, Nakagawa S, Yano T, Nakagawa K, Yasugi T, Takeuchi T, Kanda T, Huibregtse JM, Akiyama T, Taketani Y. Human scribble, a novel tumor suppressor identified as a target of high-risk HPV E6 for ubiquitin-mediated degradation, interacts with adenomatous polyposis coli. Genes Cells. 2006;11(4):453–464. doi: 10.1111/j.1365-2443.2006.00954.x. [DOI] [PubMed] [Google Scholar]
- 39.Hilger M, Mann M. Triple SILAC to determine stimulus specific interactions in the Wnt pathway. J. Proteome Res. 2012;11(2):982–994. doi: 10.1021/pr200740a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohila JS, Chen M, Chen S, Chen J, Cerny R, Dardick C, Canlas P, Xu X, Gribskov M, Kanrar S, Zhu J-K, Ronald P, Fromm ME. Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J. 2006;46(1):1–13. doi: 10.1111/j.1365-313X.2006.02671.x. [DOI] [PubMed] [Google Scholar]
- 41.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain P-O, Han J-DJ, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual J-F, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, van den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A Map of the Interactome Network of the Metazoan C. elegans. Science. 2004;303(5657):540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suter B, Fetchko MJ, Imhof R, Graham CI, Stoffel-Studer I, Zbinden C, Raghavan M, Lopez L, Beneti L, Hort J, Fillingham J, Greenblatt JF, Giaever G, Nislow C, Stagljar I. Examining protein-protein interactions using endogenously tagged yeast arrays: The Cross-and-Capture system. Genome Res. 2007;17(12):1774–1782. doi: 10.1101/gr.6667007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, Boone C, Andrews B. Mapping Pathways and Phenotypes by Systematic Gene Overexpression. Mol. Cell. 2006;21(3):319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 44.Hubner NC, Ren S, Mann M. Peptide separation with immobilized pI strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics. 2008;8(23–24):4862–4872. doi: 10.1002/pmic.200800351. [DOI] [PubMed] [Google Scholar]
- 45.Forler D, Kocher T, Rode M, Gentzel M, Izaurralde E, Wilm M. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 2003;21(1):89–92. doi: 10.1038/nbt773. [DOI] [PubMed] [Google Scholar]
- 46.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 47.Zhang XD, Guo AL, Yu JS, Possemato A, Chen YT, Zheng WP, Polakiewicz RD, Kinzler KW, Vogelstein B, Velculescu VE, Wang ZHJ. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc. Natl. Acad. Sci. U.S.A. 2007;104(10):4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segditsas S, Sieber O, Deheragoda M, East P, Rowan A, Jeffery R, Nye E, Clark S, Spencer-Dene B, Stamp G, Poulsom R, Suraweera N, Silver A, Ilyas M, Tomlinson I. Putative direct and indirect Wnt targets identified through consistent gene expression changes in APC-mutant intestinal adenomas from humans and mice. Hum. Mol. Genet. 2008;17(24):3864–3875. doi: 10.1093/hmg/ddn286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, O'Malley BW, Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc. Natl. Acad. Sci. U.S.A. 2010;107(6):2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griep AE, John MC, Ikeda S, Ikeda A. Gene targeting in the mouse. Methods Mol. Biol. 2011;770:293–312. doi: 10.1007/978-1-61779-210-6_11. [DOI] [PubMed] [Google Scholar]
- 51.Bonfils C, Beaulieu N, Chan E, Cotton-Montpetit J, MacLeod AR. Characterization of the human DNA methyltransferase splice variant Dnmt1b. J. Biol. Chem. 2000;275:10754–10760. doi: 10.1074/jbc.275.15.10754. [DOI] [PubMed] [Google Scholar]
- 52.Kawasaki Y, Senda T, Ishidate T, Koyama R, Morishita T, Iwayama Y, Higuchi O, Akiyama T. Asef, a link between the tumor suppressor APC and G-protein signaling. Science. 2000;289:1194–1197. doi: 10.1126/science.289.5482.1194. [DOI] [PubMed] [Google Scholar]
- 53.Rago C, Vogelstein B, Bunz F. Genetic knockouts and knockins in human somatic cells. Nat. Protoc. 2007;2:2734–2746. doi: 10.1038/nprot.2007.408. [DOI] [PubMed] [Google Scholar]
- 54.Kinzler KW, Vogelstein B. Lessons from Hereditary Colorectal Cancer. Cell. 1996;87(2):159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 55.Henderson BR. Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat. Cell Biol. 2000;2(9):653–660. doi: 10.1038/35023605. [DOI] [PubMed] [Google Scholar]
- 56.Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 2004;32(1):PMC373311. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J-S, Bonifant C, Bunz F, Lane WS, Waldman T. Epitope tagging of endogenous genes in diverse human cell lines. Nucleic Acids Res. 2008;36(19):e127. doi: 10.1093/nar/gkn566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.