Summary
Clear evidence exists for heritability of human longevity, and much interest is focused on identifying genes associated with longer lives. To identify such longevity alleles, we performed the largest genome-wide linkage scan thus far reported. Linkage analyses included 2118 nonagenarian Caucasian sibling pairs that have been enrolled in fifteen study centers of eleven European countries as part of the Genetics of Healthy Ageing (GEHA) project. In the joint linkage analyses we observed four regions that show linkage with longevity; chromosome 14q11.2 (LOD=3.47), chromosome 17q12-q22 (LOD=2.95), chromosome 19p13.3-p13.11 (LOD=3.76) and chromosome 19q13.11-q13.32 (LOD=3.57). To fine map these regions linked to longevity, we performed association analysis using GWAS data in a subgroup of 1,228 unrelated nonagenarian and 1,907 geographically matched controls. Using a fixed effect meta-analysis approach, rs4420638 at the TOMM40/APOE/APOC1 gene locus showed significant association with longevity (p-value=9.6 × 10−8). By combined modeling of linkage and association we showed that association of longevity with APOEε4 and APOEε2 alleles explain the linkage at 19q13.11-q13.32 with p-value=0.02 and p-value=1.0 × 10−5, respectively. In the largest linkage scan thus far performed for human familial longevity, we confirm that the APOE locus is a longevity gene and that additional longevity loci may be identified at 14q11.2, 17q12-q22 and 19p13.3-p13.11. Since the latter linkage results are not explained by common variants, we suggest that rare variants play an important role in human familial longevity.
Keywords: Human familial longevity, genome-wide linkage analysis, APOE gene, association analysis, nonagenarian sibling pairs
Introduction
Nonagenarians, centenarians and their first degree family members have a life-long survival advantage (Schoenmaker et al., 2006; Perls et al., 2002) that can be attributed to a lower risk of coronary artery disease, cancer and type-2 diabetes (Terry et al., 2003; Westendorp et al., 2009). In middle age, members of long-lived families display characteristic of metabolic health such as low glucose levels and preserved insulin sensitivity (Rozing et al., 2010; Slagboom et al., 2011). The clustering of longevity in families suggests a heritable component (Perls et al., 2000; Gudmundsson et al., 2000) which has been estimated at approximately 25% in the general population (Skytthe et al., 2003) with increasing importance at the highest ages (Hjelmborg et al., 2006). However, the genetic basis of longevity, first clearly identified as a research priority by Schächter et al (Schachter et al., 1993), still remains to be elucidated.
Previously it has been demonstrated that long-lived families carry as many GWAS-identified disease susceptibility alleles as the general population (Beekman et al., 2010). It can therefore be hypothesized that long-lived families carry gene variants that promote healthy ageing and protect from disease. The genomic location of such protective variants can be identified in a genome-wide linkage scan among long-lived siblings. In the past, by genotyping 400 microsatellite markers in 137 long-lived sibships, Puca et al (Puca et al., 2001) detected linkage at 4q25 (LOD = 3.65). In a subsequent study, modest support was provided for this locus by a genome-wide linkage scan in 95 male sibling pairs concordant for healthy aging (Reed et al., 2004). However, in a targeted study of 4q25 in 164 nonagenarian sibships no linkage was observed (Beekman et al., 2006). By association of the positional candidate genes at 4q25, MTP was suggested to explain the linkage (Geesaman et al., 2003), but this association could not be replicated in other studies (Bathum et al., 2005; Nebel et al., 2005; Beekman et al., 2006). Extension of the original group of long-lived siblings as investigated by Puca et al to 279 pairs also resulted in lack of evidence for linkage at 4q25, although novel linkage regions for longevity were observed at chromosome 3p24-22 and chromosome 9q31-24 (Boyden et al., 2010).
A number of studies have performed genome-wide association (GWA) analysis for longevity to uncover common genetic variation involved in this trait. A rather large association study form the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium of 1,836 individuals aged over 90 years and 1,955 controls between 55 and 80 years did not reveal genome-wide significant loci (Newman et al., 2010) and neither did the analyses of all-cause mortality and survival free of major disease in this cohort (Walter et al., 2011). A smaller Dutch study of 403 nonagenarians and 1,670 controls younger than 65 years identified the APOE gene as a mortality locus (Deelen et al., 2011), which was confirmed in a German study of 763 long-lived individuals and 1,085 younger controls (Nebel et al., 2011) and a longitudinal study of 1,606 Danes showed that the effect size of this association increases at the highest ages (Jacobsen et al., 2010). Apparently, the influence of the common genetic variation on longevity is small which requires large meta-GWA studies for identification. Alternatively, rare genetic variants may play a more important role in longevity. Since the previous linkage studies showed contradictory results potentially due to heterogeneity in the longevity phenotype, it is expected that longevity is influenced by many private rare variants.
In this paper, we performed genome-wide linkage analysis among 2118 European nonagenarian sibships of the Genetics of Healthy Ageing (GEHA) Study (Franceschi et al., 2007; Skytthe et al., 2011). This integrated European project was initiated in 2004 with the aim of identifying genes involved in healthy ageing and longevity. The GEHA selection criterion of nonagenarian sibling pairs has been shown to result in families enriched for genetic influences on longevity by a smaller previous study of the same design (the Leiden Longevity Study) (Schoenmaker et al., 2006). This study is characterized by a survival benefit in multiple generations, low prevalence of disease and beneficial metabolic phenotypes, which is comparable to the observations reported for families of centenarian singletons such as the Longevity Study at Albert Einstein College of Medicine (Barzilai et al., 2010). Sibships over 90 years of age and controls between 55 and 75 years of age have been recruited in 10 European countries and their genomic DNA has been isolated and genotyped centrally. This logistic achievement resulted in the largest linkage study for human longevity.
Results
In the Genetics of Healthy Ageing Study (GEHA) nonagenarian sibling pairs have been recruited in eleven countries among 15 study centers (Skytthe et al., 2011). Genotypings of 5,734 SNPs (Illumina HumanLinkage12 set) were available after quality control in 4445 persons belonging to 2118 full sibships with a mean sibship size of 2.1 (Table 1).
Table 1.
Number of sibships and sibship sizes for the GEHA sample sets.
| Sibship size |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster | Country | Center | 2 | 3 | 4 | 5 | Sibships | Mean age (yrs) |
Male sibpairs |
Female sibpairs |
| 1 | Finland | Tampere | 124 | 24 | 1 | 1 | 150 | 92.6 | 11 | 102 |
| 2 | Ukraine | Kiev | 46 | 1 | 0 | 0 | 47 | 93.2 | 1 | 38 |
| 2 | Poland | Warsaw | 131 | 4 | 0 | 0 | 135 | 93.0 | 11 | 87 |
| 2 | Germany | Kiel | 90 | 4 | 0 | 0 | 94 | 92.8 | 12 | 53 |
| 2 | Denmark | Odense | 384 | 45 | 5 | 0 | 434 | 92.3 | 50 | 215 |
| 2 | The Netherlands | Leiden | 150 | 14 | 2 | 0 | 166 | 93.0 | 20 | 82 |
| 2 | United Kingdom | Newcastle | 98 | 1 | 0 | 0 | 99 | 92.8 | 7 | 55 |
| 2 | United Kingdom | Belfast | 52 | 4 | 1 | 0 | 57 | 92.9 | 4 | 40 |
| 2 | Belgium | Louvain | 76 | 1 | 0 | 0 | 77 | 92.9 | 13 | 40 |
| 2 | France | Montpellier | 238 | 27 | 3 | 1 | 269 | 93.3 | 30 | 157 |
| 3 | Italy | Bologna | 178 | 23 | 6 | 1 | 208 | 93.4 | 14 | 126 |
| 3 | Italy | Rome | 51 | 2 | 0 | 0 | 53 | 92.5 | 6 | 32 |
| 3 | Italy | Calabria | 181 | 7 | 1 | 0 | 189 | 93.0 | 24 | 79 |
| 3 | Italy | Sassari | 43 | 2 | 0 | 0 | 45 | 93.4 | 3 | 24 |
| 3 | Greece | Athens | 92 | 3 | 0 | 0 | 95 | 92.9 | 57 | 15 |
| Total | 1934 | 162 | 19 | 3 | 2118 | 92.9 | 263 | 1145 | ||
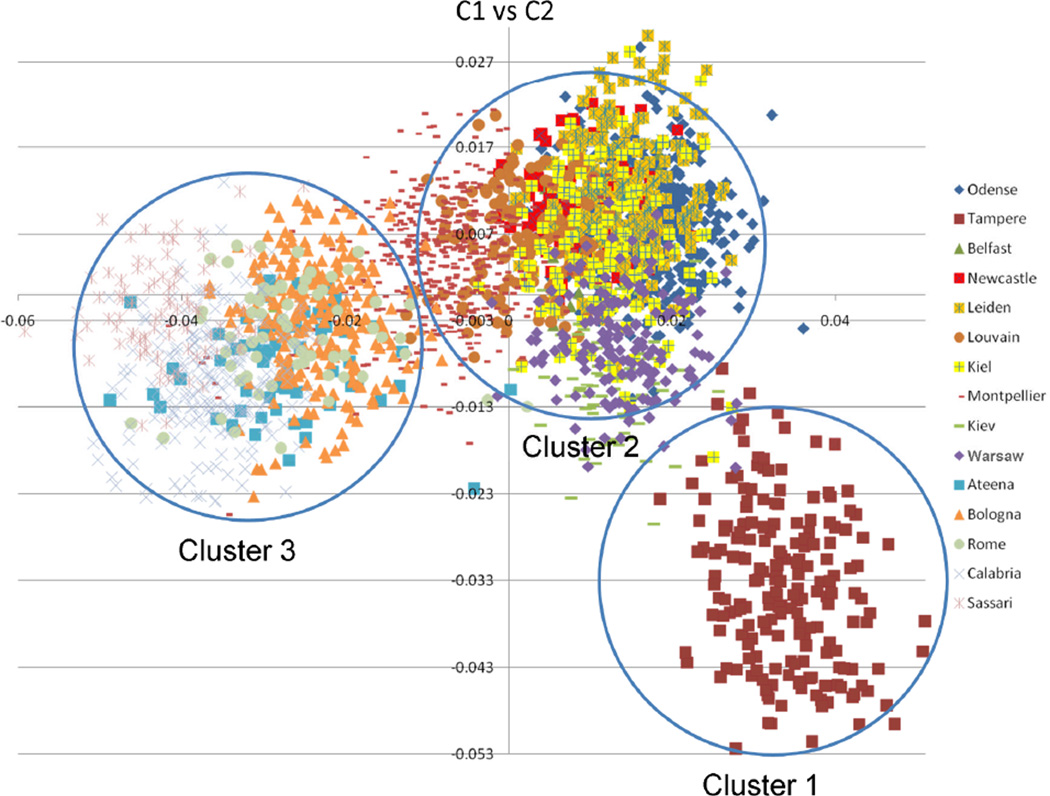
To investigate potential substructure, identical-by-state (IBS) estimates for all pairs of individuals in the dataset were computed using EIGENSTRAT (Price et al., 2006). The first two resulting principal components (C1 and C2) were plotted against each other (Fig. 1), which gives a representation of the data in two dimensions. In the resulting scatter plot each point represents an individual and each recruitment center is marked by its own symbol. After visual inspection of component 1 (C1) and component 2 (C2), C1 appeared to reflect the North-south localisation and C2 dissociated the Fins from the rest of Europe. On the basis of this genetic heterogeneity among the study samples, the linkage analyses were performed in three geographical clusters: 1) Finland with Nsibships=150; 2) Northern Europe with Nsibships=1,378, consisting of Ukraine, Poland, Germany, Denmark, The Netherlands, United Kingdom, Belgium and France; 3) Southern Europe with Nsibships=590, consisting of Italy and Greece.
Fig. 1.
Population structure among GEHA study and the grouping of countries into three clusters. Cluster 1: Finland; Cluster 2: Northern Europe consisting of Ukraine, Poland, Germany, Denmark, The Netherlands, United Kingdom, Belgium and France; Cluster 3: Southern Europe consisting of Italy and Greece.
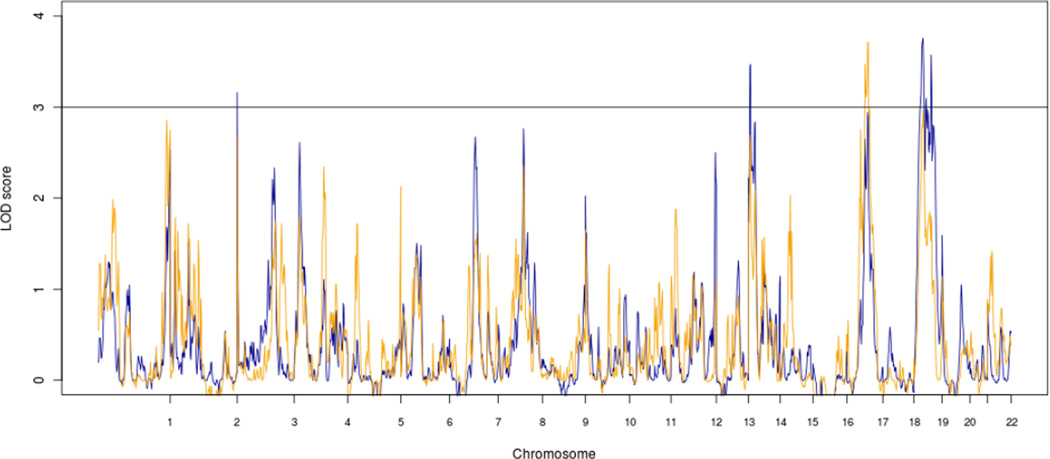
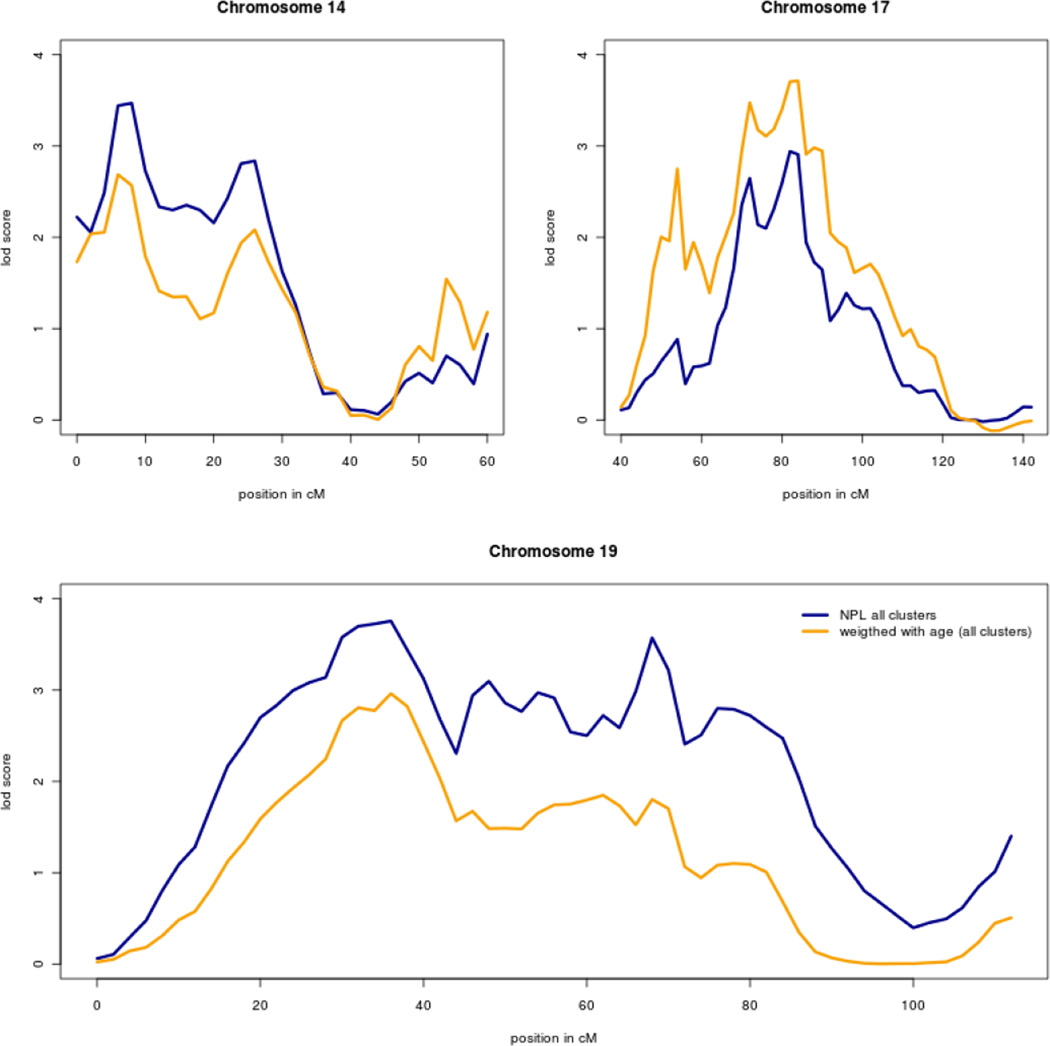
We calculated non-parametric linkage scores over the whole genome for the three clusters combined (Fig. 2, blue line). Besides the inflated LOD score probably due to linkage disequilibrium between markers at the beginning of chromosome 3, the combined linkage score for the GEHA study exceeded the genome wide significant LOD score of 3 at chromosome 14q11.2 (8 cM), 19p13.3-p13.11 (36 cM) and 19q13.11-q13.32 (68 cM) (Fig. 3). For centers for which population based mortality rates were available, i.e. Italy, UK, Denmark, France, Netherlands, Germany and Finland, a weighted NPL statistic was computed. The weights are the products of the age and sex specific cumulative hazard for the two siblings (Houwing-Duistermaat et al., 2009) that are based on country specific mortality rates. In this weighted NPL analysis extremely old sibling pairs obtain more weight (Fig. 2, yellow line) and the LOD score at chromosome 17q12-q22 (82 cM) increased from 2.95 to 3.71 (Fig. 3; Table 2). This increase has been observed in each of the three clusters Northern Europe, Southern Europe and Finland. The northern European cluster contributes most to the linkage at 14q11.2 (8 cM), 19p13.3-p13.11 (36 cM) and 19q13.11-q13.32 (68 cM), while all three clusters contribute to linkage at 17q12-q22 (Fig. S1).
Fig. 2.
Genome wide linkage graphs of among 2118 sibships of the GEHA study. The blue line displays the NPL LOD scores and the yellow line the age-weighted LOD scores.
Fig. 3.
Chromosomal regions linked to longevity with LOD score above 3. The blue line displays the NPL LOD scores and the yellow line the age-weighted LOD scores.
Table 2.
Four linkage regions with LOD score above 3 in combined GEHA samples for NPL and age-weighted linkage analyses.
| 14q11.2 | 17q12-q22 | 19p13.3- p13.11 |
19q13.11-q13.32 | |
|---|---|---|---|---|
| 1LOD-drop interval | 0–12 cM1 | 68–95 cM1 | 20–42 cM1 | 58–85 cM1 |
| Start SNP | Rs10484218 | Rs2429990 | Rs432001 | Rs7250748 |
| End SNP | Rs977870 | Rs12947910 | Rs919333 | Rs10403760 |
| NPL | 3.47 | 2.95 | 3.76 | 3.57 |
| NPL_subsets* | 3.15 | 3.26 | 3.85 | 3.80 |
| Weighted_NPL_subsets** | 2.69 | 3.71 | 2.96 | 1.85 |
According to genome build 35
Population mortality rates were available in: Cluster 1: Finland; Cluster 2: Denmark, United Kingdom, The Netherlands, France; Cluster 3: Italy
weighted with ages of the siblings
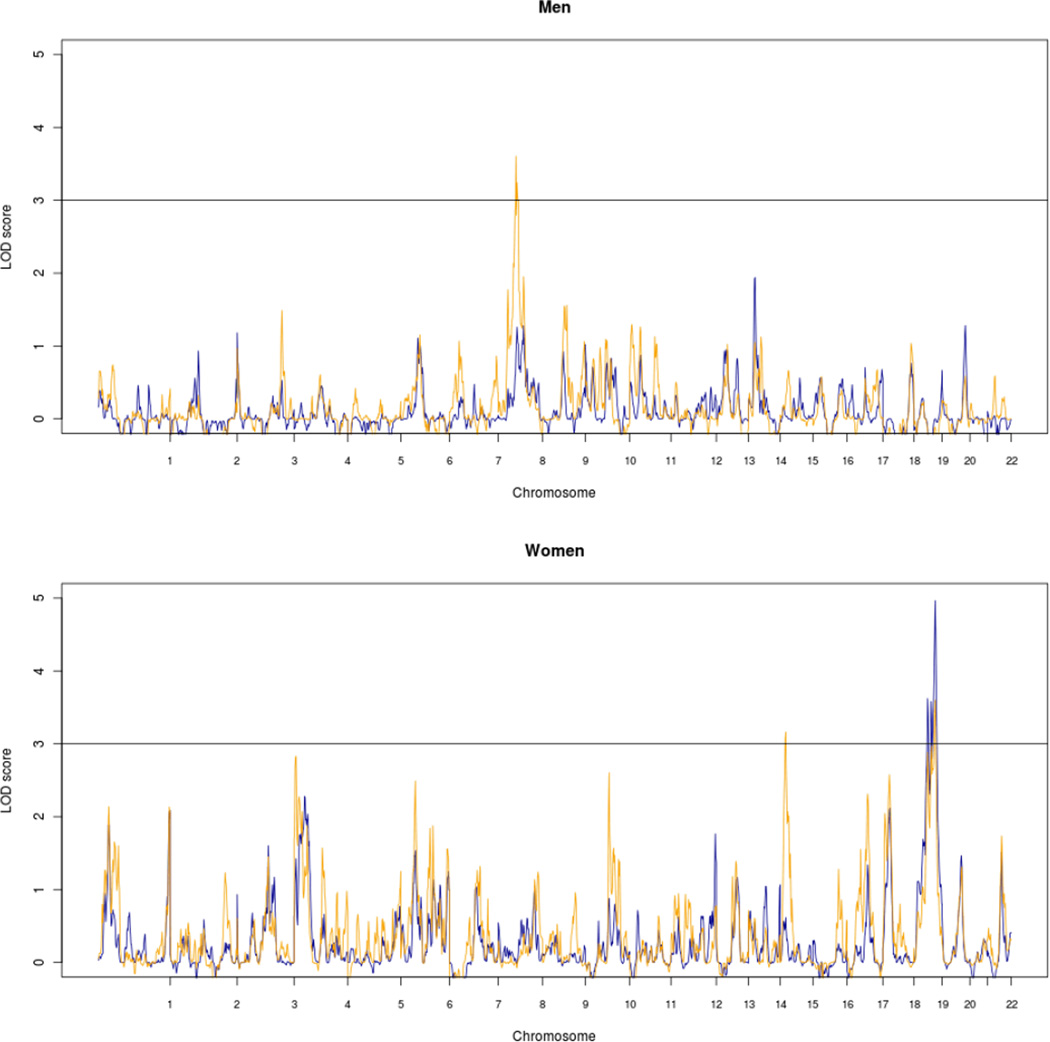
Since gender specific effects play a role in human longevity, linkage analysis was also performed in male-only (Npairs=263) and female-only (Npairs=1,145) sibling pairs. Although NPL analysis did not reveal any linkage for male-only pairs, significant linkage (Weighted-NPL 3.61) was observed at 8p11.21-q13.1 (70 cM) among the male-only pairs in the age-weighted analysis (Fig. 4, upper panel; Table 3). Among the female-only sibpairs (Fig. 4, lower panel) significant linkage was observed at chromosome 15q12-q14 (22 cM) (Weighted-NPL 3.16) and at 19q13.33-q13.41 (80 cM) (NPL=4.97) (Table 3). Further investigation of these linkage areas revealed that the Northern Europe cluster and the Finland cluster provide evidence for the 8p11.21-q13.1 and 15q12-q14 loci, and that the 19q13.33-q13.41 locus seems only to link with female longevity in the Northern Europe cluster (Fig. S2).
Fig. 4.
Genome wide linkage graphs of among 1145 female-only sibships and 263 male-only sibships of the GEHA study. The blue line displays the NPL LOD scores and the yellow line the age-weighted LOD scores.
Table 3.
Sex specific linkage regions with LOD score above 3 in combined GEHA samples for NPL and age-weighted linkage analyses.
| 8p11.21- q13.1 |
15q12-q14 | 19q13.33- q13.41 |
|
|---|---|---|---|
| Sex | Men | Women | Women |
| 1LOD-drop interval | 64–82 cM1 | 14–28 cM1 | 66–90 cM1 |
| Start SNP | Rs801100 | Rs1871009 | Rs1236093 |
| End SNP | Rs4368961 | Rs580839 | Rs1661965 |
| NPL | 1.14 | 0.62 | 4.97 |
| NPL_subsets* | 1.96 | 1.44 | 3.92 |
| Weighted_NPL_subsets** | 3.61 | 3.16 | 3.60 |
According to genome build 35
Population mortality rates were available in: Cluster 1: Finland; Cluster 2: Denmark, United Kingdom, The Netherlands, France; Cluster 3: Italy
weighted with ages of the siblings
When we consider the genes in the 1-LOD drop intervals of the linkage analysis as candidate genes for longevity, we have a list of 1151 unique position candidate genes (Supplementary data GEHA_positional_candidate_genes.xls) for which we have performed gene set enrichment analysis (Table S3, Fig. S4). The 1-LOD drop intervals were investigated for association with longevity by comparison of unrelated nonagenarians to geographically matched younger controls. Of each sibpair of the centers that provided the largest contribution to the linkage results (i.e. Netherlands, Denmark, France, Italy (Bologna) and UK (Newcastle)) (Table S1), the eldest sibs were taken as cases and as controls for France and Bologna, GEHA controls were taken. As controls for UK, Netherlands and Denmark already genotyped available younger population controls were used. We performed a meta-analysis for 57,607 imputed SNPs in the seven candidate regions with longevity. We tested for differences in genotype frequencies between cases and geographically matched controls using a trend test, i.e. assuming additive genetic effects. The test statistic was adjusted for uncertainty in imputations and for the inflation factor before computing the p-values. We used a fixed effect meta-analysis approach to combine the results across centers. Finally, we used a Bonferroni correction based on the number of SNPs within the region to correct for multiple testing.
No significant association (p-value <8.7 × 10−7) was observed at the loci at chromosome 14q11.2, 17q12-q22 and 19p13.3-p13.11, and neither at the sex specific loci. At 19q13.11-q13.32 locus, rs4420638 at the TOMM40/APOE/APOC1 gene locus showed significant association with longevity (p-value=9.6 × 10−8). Since the linkage plots per center show that especially the sample from Denmark provides the statistical evidence for an increased LOD score at this position (Fig. S3), we tested in the Danish dataset with the software LAMP (Burdick et al., 2006) for residual linkage at the 19q13.11-q13.21 locus if the APOEε2ε3ε4 rs7412 and rs429358 genotypes were taken into account. It appeared that the APOEε4 allele, although with moderate significance, contributed to the linkage peak (p-value=0.02). Remarkably, with a much higher significance the APOEε2 allele also explained the linkage peak (p-value=1.0 × 10−5).
Discussion
To identify chromosomal regions involved in longevity, we performed the largest linkage analysis thus far reported. This analysis included 2118 nonagenarian sibling pairs from eleven European countries enrolled as part of the Genetics of Healthy Ageing (GEHA) project. We identified four regions linked to human longevity at 14q11.2, 17q12-q22, 19p13.3-p13.11 and 19q13.11-q13.32. In addition to these loci we identified three loci that were linked to longevity in a sex-specific manner: 8p11.21-q13.1 (men), 15q12-q14 (women) and 19q13.33-q13.41 (women). In these seven linked regions we performed association analysis using GWAS data that was available in a subset of the cohorts, and we showed that genetic variation at the APOE gene locus was significantly associated with longevity. The absence of the APOEε4 allele among the nonagenarians is obvious (Deelen et al., 2011) and contributed to the linkage, suggesting that APOE is a mortality gene. However the linkage was mainly explained by the shared presence of the APOEε2 allele, indicating that the APOE gene indeed is a longevity gene.
Affected sibpair analysis is a powerful tool to map strong dominant as well as recessive gene effects, but to map rare genetic variation with relatively small effect a group of at least 600 sibling pairs with one sibling over age 95 years should be examined (Tan et al., 2004). Since the GEHA project yielded an impressive number of 2118 genotyped nonagenarian sibling pairs to investigate for linkage (Skytthe et al., 2011), of which 899 sibling pairs contained one sib aged over 95 years of age, this linkage study has sufficient power to detect the loci with relatively small effects.
Previous studies for longevity loci detected linkage at 4q25 (Puca et al., 2001), 3p24-22, 9q31-34, 12q24 (Boyden et al., 2010), 6p12.1, 7q11.21, 14q22.1 (Edwards et al., 2011). None of these loci showed linkage in the GEHA project. The largest of the previous linkage studies examined 279 long-lived sibling pairs, which is considered a relatively small study and therefore the previous linkage results should be considered with caution, given the complexity of human longevity (Tan et al., 2004). Otherwise, the effect of previously detected loci on longevity could be private for a specific genetic background or specific environment, or even private for a small number of families.
In the chromosomal region linked to familial longevity at 19q13.11-q13.32 we found rs4420638 to be significantly associated with familial longevity. This SNP tags the LD block harboring the TOMM40, APOE and APOC1 genes. It is known that the APOE gene exhibit three isoforms that are caused by two functional SNPs rs7412 (APOEε2) and rs429358 (APOEε4) which have been associated with longevity (Schachter et al., 1994). The APOEε4 allele frequency in GEHA nonagenarian sibling cases was 6.8%, which was significantly lower than the allele frequency of 12.7% among the geographically matched controls. APOEε4 allele carriers have about 50% lower chance to become a nonagenarian then non-APOEε4 allele carriers (OR=0.48, 95% CI 0.42–0.55). Recently, this gene region has been explored for other functional variation and multiple cis-elements are found to influence both APOE and TOMM40 promoter activity and a complex transcriptional regulatory structure has been suggested modulates regional expression (Bekris et al., 2012). Hence, future research should consider exploring these other functional variants for their role in familial longevity.
Genetic association analysis as an approach is suitable to detect the effect on a trait of common genetic variation (minor allele frequency larger than 1%). We did not observe significant associations in the families that contributed to linkage of common variants in the linked regions with longevity which can be explained by the possibility that the positive linkage signals may be based on allelic and/or locus heterogeneity, i.e. different genetic variants in the same gene or gene region contributed to the trait in different families. The less likely explanation is that the power, which was large enough for detecting linkage, may be too small to detect genetic association among 57K SNPs in the linkage regions in 1,228 cases. Thus common variants may not contribute to the longevity phenotype and by applying next generation sequencing analyses of the nonagenarians contributing to linkage rare, private genetic variation in the linkage regions may be found.
This genome-wide linkage scan did not provide substantial evidence for linkage to FOXO3A on chromosome 6, of which the T allele of rs2802295 predisposes to survive into old age (reviewed in Chung et al., 2010). This may be due to the relatively small effect sizes conferred by SNPs at that locus or it could be that the nonagenarian sibling design is underpowered to detect the association of FOXO3A with longevity, because its effect seems the strongest at ages above 95, in which age range the GEHA study has limited numbers of participants. Besides the NPL analyses we performed linkage analyses in which the contribution of the sibling pairs has been weighted for the age of the eldest sibling. Thus when a locus is involved in survival after an extremely old age, as is suggested for the FOXO3A locus, linkage signals would increase in this weighted analyses. Despite the use of these analysis tools we did not observe any LOD score above LOD score of 1 at the FOXO3A locus. We therefore conclude that genetic variation at the FOXO3A gene might not contribute to longevity in the GEHA Study.
Previously, it has been suggested that genetic variation in the FOXO1 gene is specifically contributing to human female longevity (reviewed in Chung et al., 2010). However, at chromosome 13q14.11 harboring the FOXO1 gene we found no evidence for linkage with female longevity (LOD<0.05) and at the gene position of FOXO1 we found no evidence for association in the females-only meta-analysis (p-values>0.042) in the GEHA Study. Potentially, the effect of this locus is not only influenced by gender but also by genetic background.
When we investigated the cluster specific linkage results, we observed that Northern Europe cluster contributes to all linkage loci, the Southern Europe cluster only contributes to the 17q12-q22 and 19p13.3-p13.11 loci and the Finland cluster only contributes to the 14q11.2 and 17q12-q22 loci (Fig. S3). The Southern Europe cluster and the Finland cluster did not obtain LOD scores above 3 on their own and the Northern Europe cluster obtained LOD scores above 3 only at chromosome 19 at 20 cM, 68 cM and 82 cM. This illustrates that probably, in addition to the sample size of the cluster, the environment and genetic background play a role in the complex phenotype of human familial longevity.
Besides linkage at the APOE locus, we detected linkage at 14q11.2, 17q12-q22 and 19p13.3-p13.11 considering the total GEHA study. These chromosomal regions linked to human longevity cover many genes that are potential candidate genes involved in neurodegenerative disease, auto-immune disease, cardiovascular disease and cancer. In the NCBI GWAS catalog (http://www.genome.gov/gwastudies/) genome-wide association has been reported at the 14q locus for the T-cell receptor alpha with narcolepsy. At the 17q12-q22 locus genome-wide association has been reported for MAPT and NSF with Parkinson’s disease, for CA10 with age of onset of menarche, and for STAT3 with Multiple Sclerosis and Crohn’s disease. At the chromosome 19p13.3-p13.11 locus genome-wide association has been reported for TYK2 and ICAM3 with Crohn’s disease, for LDLR with myocardial infarction, for SMARCA4 with coronary heart disease, and for ABHD8 and ANKLE1 with breast cancer.
In conclusion, we identified 4 chromosomal regions linked to human familial longevity and we are the first identifying genetic linkage with familial longevity at the APOE gene locus. Since genetic variation at the APOE gene explains the linkage, we confirm that the APOE locus is indeed a longevity gene. Identification of additional longevity genes in the linkage regions will provide biological insight into the pathways underlying human familial longevity, metabolic health and the resistance to age-related disease.
Experimental Procedures
Study subjects
The individuals investigated in the present study participate in Genetics of Healthy Ageing (GEHA) Study (Skytthe et al., 2011). Families participating in the GEHA study have at least two siblings meeting four inclusion criteria: (i) participants are at least 90 years old, (ii) participants have at least one living brother or sister who fulfils the first criterion and is willing to participate, (iii) the nonagenarian sibship has an identical mother and father, and (iv) the parents of the nonagenarian sibship are European and Caucasian. In total 2,249 sibships have been recruited.
In accordance with the Declaration of Helsinki, written informed consent was obtained from all participants prior to entering the study. Good clinical practice guidelines were maintained. The study protocol was approved by the local medical ethical committees of the eleven participating countries before the start of the study.
DNA isolation
The National Institute for Health and Welfare, (THL, Helsinki), extracted high molecular DNA from 7 ml of whole blood using automated DNA purification, Autopure LS (Qiagen), based on Puregene salting out methodology. DNA concentrations were adjusted to 50ng/µl, verified using PicoGreen dsDNA Quantitation kit (Molecular Probes, Life Technologies). The samples were then subjected to quality control by ABI 3730 DNA analyzer (Applied Biosystems, Life Technologies) using two sex specific and four autosomal microsatellite markers.
Genotyping for linkage analysis
The Centre National de Génotypage (CNG, Paris) performed genotyping of 6,090 SNPs in the nonagenarian siblings of the 2249 sibships recruited in the GEHA project with the Illumina HumanLinkage-12 Genotyping BeadChip. Genotyping call rate was larger than 95% per sample. We observed 19 duplicates that are either MZ twins or duplicate samples of which two were found in two different families. Five full sibs were identified as unrelated and 8 sample pairs that are supposed to be unrelated were identified as potential half-sibs. Of all sibships, 35 appeared to be composed of half-sibs. Finally two sample pairs that are supposed to be unrelated were found full-sibs. After removing the samples with unexpected relations, we obtained reliable genotyping for 4445 individuals from 2,118 sibships (Table 1). Since the recruitment of the GEHA study has been a major enterprise, it turned out in a later phase of the project that some cases were aged below 90 years of age at the time of interview (Table S2 and Figure S4).
We applied the EIGENSTRAT program (Price et al., 2006) with default parameters to infer axes of variation with the 6,090 SNPs per individual to identify genetic population stratification.
Affected Sibpair Analysis
Unlikely recombinants were detected using Merlin-0.10.2 (Abecasis et al., 2002) and erroneous genotypes were removed with Pedwipe. Additionally, Pearson’s r2 was estimated using the total data set. For the non-parametric (NPL) analyses, 275 SNPs were removed to obtain a genome wide SNP set of 5,734 SNPs with pairwise r2 smaller than 0.4. MERLIN-0.10.2 (Abecasis et al., 2002) was used to estimate the information content over the genome (infocontent.pdf) and to estimate IBD probabilities in the sibling pairs without parents.
Next, proportion of alleles shared identical by descent were estimated in Merlin (Abecasis et al., 2002) allowing for center specific allele frequencies. Nonparametric linkage analysis (NPL) (Kruglyak et al., 1996) was performed using a score test statistic for affected sibling pairs (Callegaro et al., 2009) that was computed at a grid of 2 cM. NPL statistics per cluster were obtained by summing the NPL statistics per involved centre. This statistic compares the actual proportion of alleles shared identical by descent by a sibling pair with its expectation of 0.5 under Mendelian segregation. The variance of the statistic was obtained via simulation in Merlin. By simulation the variance is adjusted for incomplete marker informativeness and correlation of sibling pairs within families. The statistic assumes that there is no linkage disequilibrium between the markers. When this assumption is violated, the peaks will be inflated.
For centers for which population based mortality rates were available, i.e. Italy, United Kingdom, Denmark, France, Netherlands, Germany and Finland, a weighted NPL statistic was computed. The weights are the products of the age and sex specific cumulative hazard for the two siblings (Houwing-Duistermaat et al., 2009). These cumulative hazards are based on country specific mortality rates. In this analysis extremely old sibling pairs obtain more weight in the analysis.
Subjects for association analysis within linked regions
To fine map the 1-LOD drop linkage regions, association analysis was performed in the largest centres and those most contributing to the linkage results. From each pair the eldest siblings from the United Kingdom, Netherlands, Denmark, France and Bologna, and GEHA controls from France and Bologna were genotyped. As controls for United Kingdom, Netherlands and Denmark available geographically matched control datasets were used: TwinUK for UK nonagenarians (Perola et al., 2007), controls from the Leiden Longevity Study (LLS) for Dutch nonagenarians (Beekman et al., 2010), Danish GOYA controls (Paternoster et al., 2011) for Danish nonagenarians. The characteristics of the samples are displayed in table S1.
Genotyping for association analysis within linked regions
The GEHA nonagenarians from the UK, Netherlands, Denmark and France (CNG, Paris, France) and the LLS controls (genotyping facility Erasmus MC, Rotterdam, The Netherlands) have been genotyped using Illumina Infinium HD Human660W-Quad BeadChips, the GEHA nonagenarians from Bologna, their controls and the France controls have been genotyped using Illumina HumanOmniExpress BeadChips (genotyping facility Erasmus MC, Rotterdam, The Netherlands) and the Danish controls (GOYA) and UK controls (TwinsUK) have been genotyped using Illumina Infinium HD Human610-Quad BeadChips. Autosomal SNPs were included in analysis if they had less than 5% missing data, Hardy-Weinberg P-values in cases and controls greater than 1 × 10−4 and minor allele frequency (MAF) was larger than 1% (if ncases and ncontrols ≥ 200) or 5%. SNPs that passed this quality control in both cases and controls were used as input for imputation to HAPMAP2 release 22 using IMPUTE2. SNPs with a R2T value lower than 40 and a MAF lower than 1% (if ncases and ncontrols ≥ 200) or 5% were excluded from analyses (Uh et al., 2011).
Association analysis within linked regions
Association analysis has been performed using CC-assoc applying score test (Uh et al., 2011). For meta-analysis, a fixed-effect approach was used and adjustment for population stratification per study was performed by multiplying the variances with the genomic inflation factor (λ), which measures over-dispersion of test statistics from association tests. Scores and adjusted variances of the five countries were combined to obtain a single meta-statistic. We tested 57,607 SNPs within the 1-LOD drop intervals and according to Bonferoni adjustment for multiple testing, P-values below 8.7 × 10−7 were considered genome-wide significant.
Combined linkage and association at APOEε2ε3ε4 locus
To investigate whether the linkage identified can be explained by the APOEε2ε3ε4 polymorphisms, we performed linkage and association modelling in pedigrees (LAMP) (Li et al., 2005) in the Danish sample in which rs7412 and rs429358 have been genotyped by TaqMan using standard protocol of the supplier.
Gene set enrichment analyses
For each linkage region the 1-LOD drop intervals have been determined. Next, the genes located in these 1-LOD drop intervals have been listed and tested for pathway enrichment using default settings in DAVID (http://david.abcc.ncifcrf.gov/).
Supplementary Material
Acknowledgments
Funding
The work described in this paper was funded mainly by the EU GEHA Project contract no. LSHM-CT-2004-503-270. The work has additionally been supported by the following programs and agencies:
The Competitive Research Funding of the Tampere University Hospital and Academy of Finland (Tampere); United States National Institute of Aging (PO1-AG08761) (Odense); The Innovation Oriented Research Program on Genomics (Senter-Novem IGE05007), the Centre for Medical Systems Biology (CMSB) and the National Institute for Healthy Ageing (NCHA 05060810), all in the framework of the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) (Leiden); The Institute for Ageing and Health and the UK NIHR Biomedical Research Centre for Ageing and Age-related disease award to the Newcastle-upon-Tyne Foundation Hospitals NHS Trust (Newcastle); Fondation Caisse d'Epargne Rhône-Alpes Lyon CERAL (2004–2007) (Montpellier); Regione Autonoma della Sardegna (Sassari), European Union's Seventh Framework Programme (FP7/2007–2011) IDEAL-ageing under grant agreement n° 259679.
Access to genotype data from the TwinsUK cohort was kindly provided by the Department of Twin Research (DTR) and Genetic Epidemiology at King’s College London. The TwinsUK cohort acknowledges funding from the Wellcome Trust, European Community’s FP7 Programme (HEALTH-F2-2008-201865 GEFOS and HEALTH-F4-2007-201413 ENGAGE projects) and the FP5 Programme (GenomEUtwin Project QLG2-CT-2002-01254), BBSRC project grant G20234, and the National Eye Institute via an NIH/CIDR genotyping project (PI: Terri Young). We thank the staff of the Twin Research Unit for their help and support in undertaking this project. We would also like to thank all patients, their families, and control subjects for their voluntary contribution to this research project.
Footnotes
Supporting Information Listing
Fig. S1 Linkage regions in the three geographically clusters at chromosome 14 (upper left panel), chromosome 17 (upper right panel) and chromosome 19 (lower panel).
Fig. S2 Sex-specific linkage regions in the three geographically clusters. Light blue line represents Northern Europe cluster, orange line represents Southern Europe cluster, purple line represents Finland.
Fig. S3 Linkage at chromosome 19 for the three largest centers from the Northern European cluster.
Fig. S4 Histogram of ages in the GEHA study.
Table S1 Nonagenarian cases and controls for association analyses in candidate longevity regions.
Table S2 Mean age and age range of GEHA nonagenarian sibling cases.
Table S3 Gene Ontology term enrichment among the genes resided in regions linked to longevity.
GEHA_positional_candidate_genes.xls
Supplementary_data_DAVID.xls
genomewidelinkageresults_geha_all_phys_gen_map.xls
Beekman_infocontent.pdf
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Gabriely I, Atzmon G, Suh Y, Rothenberg D, Bergman A. Genetic studies reveal the role of the endocrine and metabolic systems in aging. J Clin Endocrinol Metab. 2010;95:4493–4500. doi: 10.1210/jc.2010-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathum L, Christiansen L, Tan Q, Vaupel J, Jeune B, Christensen K. No evidence for an association between extreme longevity and Microsomal Transfer Protein polymorphisms in a longitudinal study of 1651 nonagenarians. Eur J Hum Genet. 2005;13:1154–1158. doi: 10.1038/sj.ejhg.5201468. [DOI] [PubMed] [Google Scholar]
- Beekman M, Blauw GJ, Houwing-Duistermaat JJ, Brandt BW, Westendorp RG, Slagboom PE. Chromosome 4q25, microsomal transfer protein gene, and human longevity: novel data and a meta-analysis of association studies. J Gerontol A Biol Sci Med Sci. 2006;61:355–362. doi: 10.1093/gerona/61.4.355. [DOI] [PubMed] [Google Scholar]
- Beekman M, Nederstigt C, Suchiman HE, Kremer D, van der BR, Lakenberg N, Alemayehu WG, De Craen AJ, Westendorp RG, Boomsma DI, de Geus EJ, Houwing-Duistermaat JJ, Heijmans BT, Slagboom PE. Genome-wide association study (GWAS)-identified disease risk alleles do not compromise human longevity. Proc Natl Acad Sci U S A. 2010;107:18046–18049. doi: 10.1073/pnas.1003540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekris LM, Lutz F, Yu CE. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J Hum Genet. 2012;57:18–25. doi: 10.1038/jhg.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden SE, Kunkel LM. High-density genomewide linkage analysis of exceptional human longevity identifies multiple novel loci. PLoS ONE. 2010;5:e12432. doi: 10.1371/journal.pone.0012432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nat Genet. 2006;38:1002–1004. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegaro A, van Houwelingen HC, Houwing-Duistermaat JJ. Score test for age at onset genetic linkage analysis in selected sibling pairs. Stat Med. 2009;28:1913–1926. doi: 10.1002/sim.3596. [DOI] [PubMed] [Google Scholar]
- Chung WH, Dao RL, Chen LK, Hung SI. The role of genetic variants in human longevity. Ageing Res Rev. 2010;9(Suppl 1):S67–S78. doi: 10.1016/j.arr.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, Kremer D, van der BR, Suchiman HE, Lakenberg N, van den Akker EB, Passtoors WM, Tiemeier H, van HD, De Craen AJ, Rivadeneira F, de Geus EJ, Perola M, van der Ouderaa FJ, Gunn DA, Boomsma DI, Uitterlinden AG, Christensen K, van Duijn CM, Heijmans BT, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10:686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DR, Gilbert JR, Jiang L, Gallins PJ, Caywood L, Creason M, Fuzzell D, Knebusch C, Jackson CE, Pericak-Vance MA, Haines JL, Scott WK. Successful aging shows linkage to chromosomes 6, 7, and 14 in the Amish. Ann Hum Genet. 2011;75:516–528. doi: 10.1111/j.1469-1809.2011.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Bezrukov V, Blanche H, Bolund L, Christensen K, De BG, Deiana L, Gonos E, Hervonen A, Yang H, Jeune B, Kirkwood TB, Kristensen P, Leon A, Pelicci PG, Peltonen L, Poulain M, Rea IM, Remacle J, Robine JM, Schreiber S, Sikora E, Slagboom PE, Spazzafumo L, Stazi MA, Toussaint O, Vaupel JW. Genetics of healthy aging in Europe: the EU-integrated project GEHA (GEnetics of Healthy Aging) Ann N Y Acad Sci. 2007;1100:21–45. doi: 10.1196/annals.1395.003. [DOI] [PubMed] [Google Scholar]
- Geesaman BJ, Benson E, Brewster SJ, Kunkel LM, Blanche H, Thomas G, Perls TT, Daly MJ, Puca AA. Haplotype-based identification of a microsomal transfer protein marker associated with the human lifespan. Proc Natl Acad Sci U S A. 2003;100:14115–14120. doi: 10.1073/pnas.1936249100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson H, Gudbjartsson DF, Frigge M, Gulcher JR, Stefansson K. Inheritance of human longevity in Iceland. Eur J Hum Genet. 2000;8:743–749. doi: 10.1038/sj.ejhg.5200527. [DOI] [PubMed] [Google Scholar]
- Hjelmborg JV, Iachine I, Skytthe A, Vaupel JW, McGue M, Koskenvuo M, Kaprio J, Pedersen NL, Christensen K. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119:312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- Houwing-Duistermaat JJ, Callegaro A, Beekman M, Westendorp RG, Slagboom PE, van Houwelingen JC. Weighted statistics for aggregation and linkage analysis of human longevity in selected families: the Leiden Longevity Study. Stat Med. 2009;28:140–151. doi: 10.1002/sim.3421. [DOI] [PubMed] [Google Scholar]
- Huang dW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen R, Martinussen T, Christiansen L, Jeune B, ndersen-Ranberg K, Vaupel JW, Christensen K. Increased effect of the ApoE gene on survival at advanced age in healthy and long-lived Danes: two nationwide cohort studies. Aging Cell. 2010;9:1004–1009. doi: 10.1111/j.1474-9726.2010.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES. Limits on fine mapping of complex traits. Am J Hum Genet. 1996;58:1092–1093. [PMC free article] [PubMed] [Google Scholar]
- Kuningas M, Magi R, Westendorp RG, Slagboom PE, Remm M, van HD. Haplotypes in the human Foxo1a and Foxo3a genes; impact on disease and mortality at old age. Eur J Hum Genet. 2007;15:294–301. doi: 10.1038/sj.ejhg.5201766. [DOI] [PubMed] [Google Scholar]
- Li M, Boehnke M, Abecasis GR. Joint modeling of linkage and association: identifying SNPs responsible for a linkage signal. Am J Hum Genet. 2005;76:934–949. doi: 10.1086/430277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebel A, Croucher PJ, Stiegeler R, Nikolaus S, Krawczak M, Schreiber S. No association between microsomal triglyceride transfer protein (MTP) haplotype and longevity in humans. Proc Natl Acad Sci U S A. 2005;102:7906–7909. doi: 10.1073/pnas.0408670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanche H, Junge O, Wittig M, Ellinghaus D, Flachsbart F, Wichmann HE, Meitinger T, Nikolaus S, Franke A, Krawczak M, Lathrop M, Schreiber S. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech Ageing Dev. 2011;132:324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Newman AB, Walter S, Lunetta KL, Garcia ME, Slagboom PE, Christensen K, Arnold AM, Aspelund T, Aulchenko YS, Benjamin EJ, Christiansen L, D'Agostino RB, Sr, Fitzpatrick AL, Franceschini N, Glazer NL, Gudnason V, Hofman A, Kaplan R, Karasik D, Kelly-Hayes M, Kiel DP, Launer LJ, Marciante KD, Massaro JM, Miljkovic I, Nalls MA, Hernandez D, Psaty BM, Rivadeneira F, Rotter J, Seshadri S, Smith AV, Taylor KD, Tiemeier H, Uh HW, Uitterlinden AG, Vaupel JW, Walston J, Westendorp RG, Harris TB, Lumley T, van Duijn CM, Murabito JM. A meta-analysis of four genome-wide association studies of survival to age 90 years or older: the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. J Gerontol A Biol Sci Med Sci. 2010;65:478–487. doi: 10.1093/gerona/glq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternoster L, Evans DM, Nohr EA, Holst C, Gaborieau V, Brennan P, Gjesing AP, Grarup N, Witte DR, Jorgensen T, Linneberg A, Lauritzen T, Sandbaek A, Hansen T, Pedersen O, Elliott KS, Kemp JP, St PB, McMahon G, Zelenika D, Hager J, Lathrop M, Timpson NJ, Smith GD, Sorensen TI. Genome-wide population-based association study of extremely overweight young adults--the GOYA study. PLoS ONE. 2011;6:e24303. doi: 10.1371/journal.pone.0024303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perls T, Shea-Drinkwater M, Bowen-Flynn J, Ridge SB, Kang S, Joyce E, Daly M, Brewster SJ, Kunkel L, Puca AA. Exceptional familial clustering for extreme longevity in humans. J Am Geriatr Soc. 2000;48:1483–1485. [PubMed] [Google Scholar]
- Perls TT, Wilmoth J, Levenson R, Drinkwater M, Cohen M, Bogan H, Joyce E, Brewster S, Kunkel L, Puca A. Life-long sustained mortality advantage of siblings of centenarians. Proc Natl Acad Sci U S A. 2002;99:8442–8447. doi: 10.1073/pnas.122587599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perola M, Sammalisto S, Hiekkalinna T, Martin NG, Visscher PM, Montgomery GW, Benyamin B, Harris JR, Boomsma D, Willemsen G, Hottenga JJ, Christensen K, Kyvik KO, Sorensen TI, Pedersen NL, Magnusson PK, Spector TD, Widen E, Silventoinen K, Kaprio J, Palotie A, Peltonen L. Combined genome scans for body stature in 6,602 European twins: evidence for common Caucasian loci. PLoS Genet. 2007;3:e97. doi: 10.1371/journal.pgen.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Puca AA, Daly MJ, Brewster SJ, Matise TC, Barrett J, Shea-Drinkwater M, Kang S, Joyce E, Nicoli J, Benson E, Kunkel LM, Perls T. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc Natl Acad Sci U S A. 2001;98:10505–10508. doi: 10.1073/pnas.181337598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed T, Dick DM, Uniacke SK, Foroud T, Nichols WC. Genome-wide scan for a healthy aging phenotype provides support for a locus near D4S1564 promoting healthy aging. J Gerontol A Biol Sci Med Sci. 2004;59:227–232. doi: 10.1093/gerona/59.3.b227. [DOI] [PubMed] [Google Scholar]
- Rozing MP, Westendorp RG, De Craen AJ, Frolich M, de Goeij MC, Heijmans BT, Beekman M, Wijsman CA, Mooijaart SP, Blauw GJ, Slagboom PE, van HD. Favorable glucose tolerance and lower prevalence of metabolic syndrome in offspring without diabetes mellitus of nonagenarian siblings: the Leiden longevity study. J Am Geriatr Soc. 2010;58:564–569. doi: 10.1111/j.1532-5415.2010.02725.x. [DOI] [PubMed] [Google Scholar]
- Schachter F, Cohen D, Kirkwood T. Prospects for the genetics of human longevity. Hum Genet. 1993;91:519–526. doi: 10.1007/BF00205074. [DOI] [PubMed] [Google Scholar]
- Schachter F, Faure-Delanef L, Guenot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- Schoenmaker M, De Craen AJ, de Meijer PH, Beekman M, Blauw GJ, Slagboom PE, Westendorp RG. Evidence of genetic enrichment for exceptional survival using a family approach: the Leiden Longevity Study. Eur J Hum Genet. 2006;14:79–84. doi: 10.1038/sj.ejhg.5201508. [DOI] [PubMed] [Google Scholar]
- Skytthe A, Pedersen NL, Kaprio J, Stazi MA, Hjelmborg JV, Iachine I, Vaupel JW, Christensen K. Longevity studies in GenomEUtwin. Twin Res. 2003;6:448–454. doi: 10.1375/136905203770326457. [DOI] [PubMed] [Google Scholar]
- Skytthe A, Valensin S, Jeune B, Cevenini E, Balard F, Beekman M, Bezrukov V, Blanche H, Bolund L, Broczek K, Carru C, Christensen K, Christiansen L, Collerton JC, Cotichini R, De Craen AJ, Dato S, Davies K, De BG, Deiana L, Flachsbart F, Gampe J, Gilbault C, Gonos ES, Haimes E, Hervonen A, Hurme MA, Janiszewska D, Jylha M, Kirkwood TB, Kristensen P, Laiho P, Leon A, Marchisio A, Masciulli R, Nebel A, Passarino G, Pelicci G, Peltonen L, Perola M, Poulain M, Rea IM, Remacle J, Robine JM, Schreiber S, Scurti M, Sevini F, Sikora E, Skouteri A, Slagboom PE, Spazzafumo L, Stazi MA, Toccaceli V, Toussaint O, Tornwall O, Vaupel JW, Voutetakis K, Franceschi C. Design, recruitment, logistics, and data management of the GEHA (Genetics of Healthy Ageing) project. Exp Gerontol. 2011;46:934–945. doi: 10.1016/j.exger.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagboom PE, Beekman M, Passtoors WM, Deelen J, Vaarhorst AA, Boer JM, van den Akker EB, van HD, De Craen AJ, Maier AB, Rozing M, Mooijaart SP, Heijmans BT, Westendorp RG. Genomics of human longevity. Philos Trans R Soc Lond B Biol Sci. 2011;366:35–42. doi: 10.1098/rstb.2010.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Zhao JH, Iachine I, Hjelmborg J, Vach W, Vaupel JW, Christensen K, Kruse TA. Power of non-parametric linkage analysis in mapping genes contributing to human longevity in long-lived sib-pairs. Genet Epidemiol. 2004;26:245–253. doi: 10.1002/gepi.10304. [DOI] [PubMed] [Google Scholar]
- Terry DF, Wilcox M, McCormick MA, Lawler E, Perls TT. Cardiovascular advantages among the offspring of centenarians. J Gerontol A Biol Sci Med Sci. 2003;58:M425–M431. doi: 10.1093/gerona/58.5.m425. [DOI] [PubMed] [Google Scholar]
- Uh HW, Deelen J, Beekman M, Helmer Q, Rivadeneira F, Hottenga JJ, Boomsma DI, Hofman A, Uitterlinden AG, Slagboom PE, Bohringer S, Houwing-Duistermaat JJ. How to deal with the early GWAS data when imputing and combining different arrays is necessary. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2011.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S, Atzmon G, Demerath EW, Garcia ME, Kaplan RC, Kumari M, Lunetta KL, Milaneschi Y, Tanaka T, Tranah GJ, Volker U, Yu L, Arnold A, Benjamin EJ, Biffar R, Buchman AS, Boerwinkle E, Couper D, De Jager PL, Evans DA, Harris TB, Hoffmann W, Hofman A, Karasik D, Kiel DP, Kocher T, Kuningas M, Launer LJ, Lohman KK, Lutsey PL, Mackenbach J, Marciante K, Psaty BM, Reiman EM, Rotter JI, Seshadri S, Shardell MD, Smith AV, van DC, Walston J, Zillikens MC, Bandinelli S, Baumeister SE, Bennett DA, Ferrucci L, Gudnason V, Kivimaki M, Liu Y, Murabito JM, Newman AB, Tiemeier H, Franceschini N. A genome-wide association study of aging. Neurobiol Aging. 2011;32:2109–2128. doi: 10.1016/j.neurobiolaging.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorp RG, van HD, Rozing MP, Frolich M, Mooijaart SP, Blauw GJ, Beekman M, Heijmans BT, De Craen AJ, Slagboom PE. Nonagenarian Siblings and Their Offspring Display Lower Risk of Mortality and Morbidity than Sporadic Nonagenarians: The Leiden Longevity Study. J Am Geriatr Soc. 2009;57:1634–1637. doi: 10.1111/j.1532-5415.2009.02381.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.