Abstract
Objective
To elucidate and categorize the murine placental hormones expressed across gestation, including the expression of hormones with previously undescribed roles.
Study design
Expression levels of all genes with known or predicted hormone activity expressed in two separate tissues, the placenta and maternal decidua, were assessed across a timecourse spanning the full lifetime of the placenta. Novel expression patterns were confirmed by in situ hybridization and protein level measurements.
Results
A combination of temporal and spatial information defines five groups that can accurately predict the patterns of uncharacterized hormones. Our analysis identified Secretin, a novel placental hormone that is expressed specifically by the trophoblast at levels many times greater than in any other tissue.
Conclusions
The characteristics of Secretin fit the paradigm of known placental hormones and suggest that it may play an important role during pregnancy.
Keywords: Placenta, Hormone expression, Secretin
1. Introduction
The placenta has evolved to mediate the fetal maternal exchange of nutrients, wastes, and gases that is required for fetal development within the mother, and to act as an endocrine organ that modulates maternal physiology during pregnancy. This is a complex task, as maternal physiology must be adapted to meet the nutritional needs of the fetus, to prevent maternal immune rejection of the fetus, and to prepare the mother to provide for the needs of the newborn following parturition [1,2]. Dozens of placental hormones have been identified, and the recent discovery of several novel placentally produced members of the prolactin gene family [3] suggests that we have yet to describe the full range of placental hormones.
In this report, we have used full genome expression profiling of two separate tissues, the placenta and maternal decidua, to define the full spectrum of murine placental hormones [4]. We assessed expression levels of all genes with known or predicted hormone activity across a timecourse spanning the full lifetime of the placenta. Using this unique dataset, we have identified novel placental hormones, including Secretin.
2. Materials and methods
2.1. Tissue preparation for microarray analysis
Placenta and decidua from pregnant Swiss Webster mice were dissected throughout the course of gestation and analyzed using Affymetrix mouse 430 2.0 Microarray. Mouse handling and dissections were approved under Stanford IACUC 13646. Placentas from e8.5, e9.0, e10.5, e12.0, e15.0, e17.0 were profiled using biological triplicates whereas e13.5, e19.0 and P0 were profiled using biological duplicates [4]. RNA was isolated from placenta and decidua using the Ambion Rnaqueous kit, treated with Dnase at 37° C for 20 min according to the manufacturers instructions, then extracted with phenol chloroform and ethanol precipitated using glycogen as a carrier. Five μg starting RNA was used for each sample and cRNA was prepared as previously described, except that double stranded cDNA was purified using a column from Affymetrix. All steps were carried out using Affymetrix reagents according to the manufacturer’s instructions. For each sample 20 μg of cRNA was fragmented using KOAc RNA fragmentation buffer, then hybridized to an Affymetrix mouse 430 2.0 microarray using standard hybridization procedures (www.affymetrix.com). The hybridization was conducted by the Stanford Protein and Nucleic Acid Biotechnology Facility.
2.2. Microarray data analysis
Arrays were normalized to the same median intensity using the dCHIP invariant set method [5] and modeling was performed using the dCHIP Perfect Match (PM) modeling algorithm. Prior to further analysis, all probe sets not expressed above background levels were eliminated. To this end, probe sets were filtered to include only those for which at least two arrays had an absolute expression value over 200 and at least two arrays were called “Present” by the dCHIP software. All expression values were then log 2 transformed, and the Statistical Analysis of Microarrays (SAM) [6] ‘multiclass’ function was used to identify probe sets for which there is a statistically significant difference between one or more timepoints using an estimated false discovery rate (FDR) of <0.1%. Of the 113 genes annotated with the molecular process term “hormone activity” by Gene Ontology (www.geneontology.org), this approach identified a total of 41 that are expressed above background levels and demonstrate a significant change in expression during the timecourse in either fetally or maternally derived placental tissues. Two of the original list of annotated hormones, Janus kinase 3 and arylsulfatase A, were removed due to strong evidence against hormone function. Hierarchical clustering was performed using dCHIP. Before clustering, dCHIP linearly scales the expression values for a gene across all samples so that they have a mean of 0 and a standard deviation of 1. These standardized values are then used to calculate the Pearson correlations between genes and to guide the merging of nodes during the clustering process. The standardized values are also used in generating the clustergrams.
2.3. RNA in situ hybridization
Placental samples for in situ hybridization were collected from pregnant Swiss Webster females at e10.0 and e17.0. Placentas were fixed in 4% paraformaldehyde, infused with sucrose, then embedded in OCT and stored at −80° C until sectioning. Digoxigenin labeled riboprobes were generated from linearized plasmids for each of the hormones (Dr. Mark Krasnow’s laboratory at Stanford University). In situ hybridization was conducted at 55° C overnight on e17.0 and e10.0 placental sections using standard procedures. Digoxigenin signal was detected using a Roche (Indianapolis, IN, USA) anti-digoxigenin alkaline phosphatase conjugated antibody. Staining was performed for 4–6 h using Roche (Indianapolis, IN, USA) NBT and BCIP; counterstaining was done with Biomeda (Foster City, CA, USA) nuclear fast red.
2.4. Western blot
Increasing amounts (25 mg, 50 mg and 75 mg) of protein from mouse placentas at different developmental stages (E7.5, E10.5, E15.5 and E18.5) were separated by SDS-PAGE, followed by immunoblot analysis with antisera against Secretin (Abbiotec, San Diego, CA #250853) and β-tubulin (loading control).
2.5. TS cell culture
Trophoblast stem (TS) cells provided by Tilo Kunath and Janet Rossant were grown at 37 °C and 5% CO2 in humidified incubators as previously described [23,24]. TS cells were grown in TS cell medium containing 20% fetal calf serum (Hyclone, Logan, UT), 1 mM sodium pyruvate (Gibco), 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 100 μM β-mercaptoethanol, 25 ng/ml human Fgf4 (R & D systems, Minneapolis, MN), and 1 μg/ml heparin (Sigma, St. Louis, MO) in DMEM/F12 with 15 mM Hepes (Invitrogen, Carlsbad, CA). To maintain undifferentiated TS cells, TS cells were either grown on irradiated mouse embryonic fibroblast feeders or grown in TS cell media supplemented with 10 ng/ml human Activin (R&D systems, Minneapolis, MN).
3. Results
3.1. Genome wide expression analysis reveals novel placental hormones
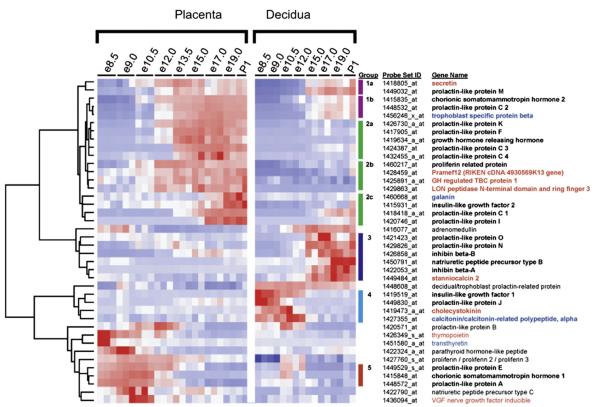
In order to define the full spectrum of murine placental hormones, we assessed a comprehensive microarray dataset of placental and decidual gene expression ([4]; GEO# GSE11224, GSE11222, GSE11220). This dataset contains nine stages throughout gestation dissected into placenta and decidua, including e8.5, e9.0, e10.5, e12.0, e13.5, e15.0, e17.0, e19.0 and P0. We examined this comprehensive dataset for all genes annotated with the molecular process term ‘hormone activity’ by Gene Ontology (www.geneontology.org), identifying 113 genes. Expression levels of these 113 predicted hormones were analyzed across a timecourse that spans the full lifetime of the placenta and decidua, from chorioallantoic fusion to term (Fig. 1). To ensure identification of hormones with significant placental roles, we first filtered the hormone list to include only genes with expression above background levels for at least one timepoint. We then used Statistical Analysis of Microarrays (SAM) to identify hormones that change significantly over our timecourse in either placental or decidual tissues, with an estimated false discovery rate (FDR) of <0.1%. Out of the 113 predicted hormones, this strategy identifies 41 hormone-encoding genes with significant placental expression. These genes are shown in Fig. 1, clustered according to their expression profiles in placental and decidual tissues. The eight genes listed in red have no previously described role in the placenta. Genes shown in black or blue have previously described roles as placental hormones. For those shown in black, placental expression has been verified at one or two timepoints by RT-PCR, northern analysis, or in situ hybridization. For those shown in blue, however, no expression pattern has been described (Fig. 1).
Fig. 1.
Placental hormone expression clusters. Using a microarray timecourse for placenta and decidua throughout mouse gestation 41 hormones were identified. These placental hormones are clustered according to their expression profiles in the placenta and decidua, with five co-expression groups indicated by colored bars. Novel placental hormones are shown in red, while previously described placental hormones are shown in black (expression verified in mouse placenta) or blue (no described expression pattern in the mouse placenta). Expression values for each gene across all samples were linearly scaled to have a mean of 0 and standard deviation of 1; red indicates expression greater than the mean and blue indicates expression below the mean.
3.2. Placental and decidual expression patterns define placental hormone co-expression groups
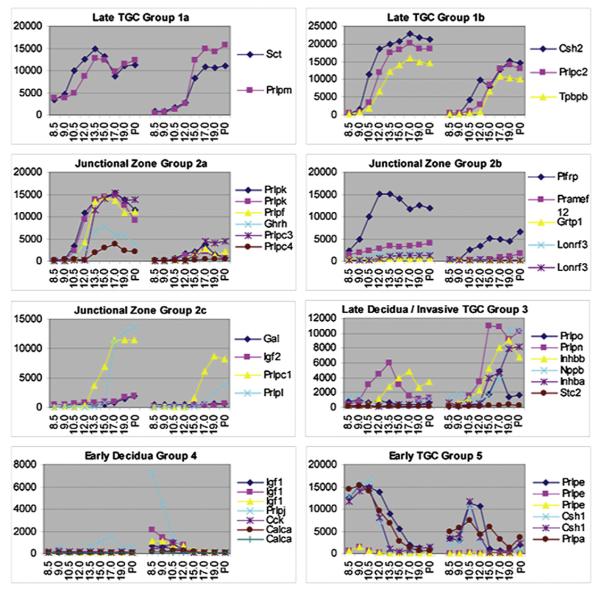
Having identified 41 placental hormones, we performed hierarchical clustering to identify co-expression groups (Fig. 2). Because these groups are based on expression data from two separate tissues, placenta and decidua, they give us information about not only the timing but also the location of hormone expression. We therefore assigned spatial expression pattern predictions to each co-expression group, as indicated by the group labels in Fig. 2. We made these assignments based on two pieces of information: first, the expression patterns we would predict based on our knowledge of specific cell types and their movements within the developing placenta, and second, the known expression patterns of characterized placental hormones within the group. For example, we predict that any hormone expressed primarily in the junctional zone or labyrinth will have expression levels that are high in placental tissues but near background in decidual tissues, whereas the reverse will be true for decidually expressed genes. These are the basic patterns we see in Group 2 (Junctional Zone), Group 3 (Late Decidua/Invasive trophoblast giant cells (TGCs)), and Group 4 (Early Decidua). Migratory TGCs, which invade the maternal decidua, are predicted to have expression in both placenta and decidua, with expression in the decidua increasing in late gestation as more cells migrate. We see two variations of this pattern in our data, as Group 1 represents early expression and Group 5 represents late expression in this cell type. Detailed expression profiles for genes in each group are shown in Fig. 2.
Fig. 2.
Placental hormone groups. Absolute expression levels across the timecourse were analyzed separately for the fetal placenta (e8.5 – P0, shown on left side of each graph) and maternal decidua (e8.5 – P0, shown on right side of each graph). Spatial expression pattern predictions for each co-expression group are indicated by the group labels: Late TGC Group 1, Junctional Zone Group 2, Late Decidua/Invasive TGC Group 3, Early Decidua Group 4, and Early TGC Group 5. Hormones that appear more than once are represented by multiple probe sets on the microarray.
3.3. Co-expression groups predict expression patterns of placental hormones
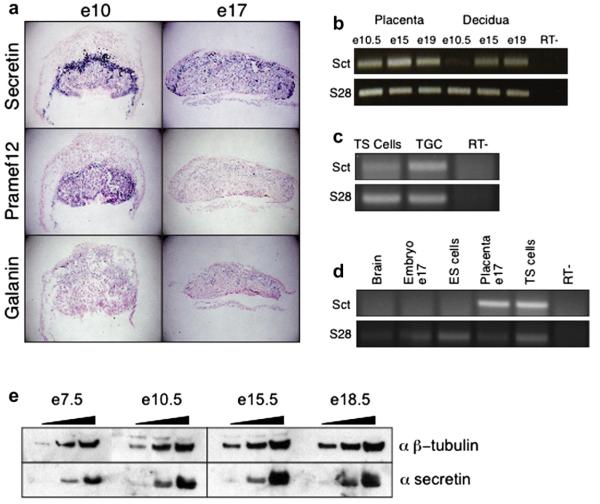
The co-expression groups allow us to predict expression patterns for several novel placental hormones, as well as for hormones that have a described role in the placenta but lack information on mouse placental expression. Among these genes, Secretin, Pramef12, and Galanin are all unknown murine placental hormones that fall within clusters containing known genes with simple expression patterns. To determine the expression patterns of these genes, we conducted in situ hybridization on e10 and e17 placental sections. As shown in Fig. 3, predictions based on co-expression group assignments are supported by the in situ hybridization results. The most striking expression pattern is that of Secretin, which demonstrated the greatest absolute expression levels on our array profiles (Figs. 1 and 2). We find that Secretin is strongly expressed in migratory TGCs and in TGCs and spongiotrophoblasts within the junctional zone at e10 and e17, with additional lighter staining within the labyrinth layer at e10 and scattered staining in the labyrinth at e17 (Fig. 3a). The expression profiles predicted Secretin’s strong expression in migratory TGCs and are consistent with its junctional zone and labyrinth expression, although they could not predict the full complexity of the pattern. Also consistent with the co-expression group predictions, Pramef12 is expressed in the junctional zone and labyrinth at e10 and in the junctional zone at e17, while Galanin has a variable pattern at e10 and is expressed in the junctional zone at e17.
Fig. 3.
Characterization of Secretin expression in placental tissues. a) In situ hybridization for Secretin, Pramef12, and Galanin was conducted using e10 and e17 placental sections. Secretin is strongly expressed in migratory TGCs and in TGCs and spongtiotrophoblasts within the junctional zone at e10 and e17, with additional lighter staining within the labyrinth layer at e10 and scattered staining in the labyrinth at e17. Pramef12 is expressed in the junctional zone and labyrinth at e10 and in the junctional zone at e17. Galanin is expressed in the junctional zone at e17. b) RT-PCR confirms the Secretin expression profile generated in the array analysis. c) RT-PCR analysis demonstrates that Secretin is preferentially expressed in differentiated trophoblast cells compared to trophoblast stem (TS) cells, in vitro. d) Secretin expression is found at much higher levels in the placenta (e17) and in TS cells than in the developing embryo (e17), adult brain, or embryonic stem cells. e) Western blot analysis confirms that Secretin protein levels mimic the RNA expression levels.
3.4. The placenta is the major producer of Secretin
Secretin stands out among the newly identified placental hormones. Its placental expression levels are in the range of those demonstrated by important placental hormones, and it is clearly expressed in both TGCs and spongiotrophoblasts, the expression locations most commonly associated with known placental hormones. Therefore, we used a combination of RT-PCR, western, and publicly available expression databases to further characterize the expression pattern of Secretin in the placenta and in other tissues of the body. First, we conducted RT-PCR to verify the placental expression of Secretin in the placenta and in two trophoblast cell types in culture. As shown in Fig. 3, RT-PCR confirms the Secretin expression profile generated in the array analysis and demonstrates that Secretin is preferentially expressed in differentiated trophoblast cells, rather than in trophoblast stem cells, in vitro (Fig. 3b and c). Next, we asked how the expression levels of Secretin in the placenta and in trophoblast cells in vitro compare to Secretin expression levels in other tissues. Secretin has a well characterized role in the gastrointestinal system, with expression reported mainly in duodenum and jejunum [7]; more recently expression has also been described in the brain and in many regions of the developing fetus [7,8]. Using RT-PCR, we find that placental expression of Secretin overwhelms expression in two previously reported Secretin expressers, the developing embryo (at e17) and the adult brain, as well as the expression in ES cells (Fig. 3d). Furthermore, to determine whether Secretin protein is present, we analyzed Secretin protein expression within the placenta throughout gestation (Fig. 3e). As anticipated by the RNA results, we found that Secretin protein is expressed within extraembryonic tissues as early as e7.5 and continues to be expressed within the placenta until e18.5. Overall, the Secretin expression data suggests that it may play a key role in murine placentation.
In order to expand our comparison to include additional Secretin expressers, such as the GI tract, and to include embryos of different developmental stages, we used the Novartis GeneAtlas dataset [9,10]. This dataset includes expression levels for 36,182 genes in a total of 61 different tissue types or cell populations. A comparison of Secretin expression in all 61 tissues, generated by the Novartis SymAtlas website (http://symatlas.gnf.org), demonstrates that Secretin expression in the placenta exceeds that in any other tissue, including tissues of the GI tract, developing embryo, and brain, by approximately forty fold (Fig. S1). The only other tissues with expression levels above background are the large intestine, the small intestine, embryo day 6.5, and embryo day 7.5. The Secretin expression identified in day 6.5 and 7.5 embryo samples lends additional support to the primary role of the trophectoderm in Secretin expression, since the extraembryonic ectoderm and ectoplacental cone (both of which are composed of trophectoderm cells) were included with the e6.5 and e7.5 embryo samples used in the GeneAtlas dataset, while all extraembryonic tissues were removed from later stage embryonic samples.
4. Discussion
In this report, we defined five placental hormone groups whose members demonstrate remarkably similar spatial expression patterns. These groups, constructed from expression patterns, succeeded not only in classifying hormones with previously described patterns, but also predict the in situ patterns of the three hormones tested. This success is remarkable given the complexity of placental tissues and the diversity of cell types that composes both placental and decidual tissues used in our analysis. Our ability to classify the hormones into groups that demonstrate such similar expression patterns is likely dependent on two characteristics of this gene set. First, it appears that the hormones demonstrate a preponderance of relatively simple expression patterns, minimizing pattern classification errors due to overlapping array profiles of distinct, complex patterns. Second, many hormones are expressed at very high levels and exhibit dramatic expression level changes throughout the course of pregnancy, minimizing the effects of background noise and minor sample to sample variation. This method of classification may therefore work well for other gene groups that similarly have a large representation of simple expression patterns and demonstrate dramatic expression changes.
Of the novel placental hormones identified in this analysis and investigated by in situ hybridization, one, Secretin, has a well characterized role in the gastrointestinal system and one, Pramef12, is uncharacterized. Secretin is expressed in several trophectodermal subtypes, with expression levels that dwarf the expression found in other tissues. Secretin has a well characterized role in the gastrointestinal system, and is produced by the duodenum and jejunum in response to gastric acid influx. It has been extensively studied for its role in stimulating exocrine release of water and bicarbonate from the pancreas and inhibiting gastric acid secretion and emptying [7]. In recent years, Secretin has been recognized as a neuropeptide with actions in the central nervous system [11]. There has been intense interest in the role of Secretin, particularly in the developing brain, following a report that autistic children experienced alleviation of symptoms following Secretin administration [12]. While the effect was not seen in the vast majority of subsequent clinical trials [13–15] one study found an effect among subset of autistic children with GI symptoms [16]. Both the Secretin knockout mouse [17] and the Secretin receptor knockout mouse [18] demonstrate impaired hippocampal synaptic plasticity, whereas the Secretin receptor knockout mice also demonstrated abnormal social and cognitive behaviors [18]. These studies, and the presence of Secretin receptor in the developing fetal brain [19], raise the interesting possibility that placental Secretin could act either on maternal tissues or on tissues of the developing fetus. Such action would not be unprecedented, as some placental hormones are known to be transmitted to the fetus [20]. Furthermore, peripherally administered Secretin analog can cross the blood brain barrier [21]. There is growing recognition that the placenta generates hormones critical for neural function prior to the time such hormones are produced by the fetal brain itself [22], raising the possibility that Secretin and other recently identified hormones may play significant roles in fetal neurodevelopment.
In addition to Secretin, we found placental expression of Pramef12, an uncharacterized gene that was electronically annotated as having hormone activity. It is conserved in human, rat and chimp. We found that it is expressed at e10 in the junctional zone and labyrinth of the mouse placenta, and at e17 in the junctional zone. To our knowledge, this is the first report of a possible role for this gene as a placental hormone.
Although human and mouse placentas have many molecular and structural similarities, there are significant differences between them. This is especially true of the placental hormones, as many murine placental hormones are known to be rodent specific [4]. While Secretin is conserved between mice and humans, evidence from term human placenta in the GeneAtlas human dataset suggests that Secretin expression in human placenta at term is present but low. It will be of interest to determine whether less mature human placentas express Secretin at levels similar to those in the term mouse, since the level of development of the mouse fetus at term is quite different from that of a term infant.
5. Conclusion
As the endocrine organ responsible for mediating maternal fetal exchange and modulating maternal physiology during pregnancy, the placenta produces a wide variety of hormones. Here we have defined the full range of murine placental hormones, identified Secretin as a candidate for an important role in pregnancy.
Supplementary Material
Acknowledgments
We would like to thank Dr. Mark Krasnow and Dr. Hernan Espinoza in the Stanford University Department of Biochemistry for providing the hormone cDNA clones. Dr. Janet Rossant at the Mount Sinai Hospital in Toronto kindly provided the trophoblast stem cell line used in this study. We thank Dr. Emin Maltepe, Dr. Eric Chiao, Dr. Ichiko Nishijima and the Baker lab members for support and advice.
Funding This work was supported by the NIH R01 HD41557 and the March of Dimes (J.C.B); NIH Director’s New Innovator Award DP2OD005675 (A.A.P.); Lucile Packard Foundation for Children’s Health Pediatric Research Fund (D.L.); Stanford University Medical Scientist Training Program (K.K.).
Appendix.
Supplementary material Supplementary data related to this article can be found online at doi:10.1016/j.placenta.2011.08.013.
Footnotes
Conflict of interest The authors have no conflict of interest.
References
- [1].Linzer DI, Fisher SJ. The placenta and the prolactin family of hormones: regulation of the physiology of pregnancy. Molecular Endocrinology. 1999;13:837–40. doi: 10.1210/mend.13.6.0286. [DOI] [PubMed] [Google Scholar]
- [2].Soares MJ, Muller H, Orwig KE, Peters TJ, Dai G. The uteroplacental prolactin family and pregnancy. Biology of Reproduction. 1998;58:273–84. doi: 10.1095/biolreprod58.2.273. [DOI] [PubMed] [Google Scholar]
- [3].Wiemers DO, Shao LJ, Ain R, Dai G, Soares MJ. The mouse prolactin gene family locus. Endocrinology. 2003;144:313–25. doi: 10.1210/en.2002-220724. [DOI] [PubMed] [Google Scholar]
- [4].Knox K, Baker JC. Genomic evolution of the placenta using co-option and duplication and divergence. Genome Research. 2008;18:695–705. doi: 10.1101/gr.071407.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chey WY, Chang TM. Secretin, 100 years later. Journal of Gastroenterology. 2003;38:1025–35. doi: 10.1007/s00535-003-1235-3. [DOI] [PubMed] [Google Scholar]
- [8].Siu FK, Sham MH, Chow BK. Secretin, a known gastrointestinal peptide, is widely expressed during mouse embryonic development. Gene Expression Patterns. 2005;5:445–51. doi: 10.1016/j.modgep.2004.09.001. [DOI] [PubMed] [Google Scholar]
- [9].Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, et al. Large-scale analysis of the human and mouse transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chu JY, Yung WH, Chow BK. Secretin: a pleiotrophic hormone. Annals of the New York Academy of Sciences. 2006;1070:27–50. doi: 10.1196/annals.1317.013. [DOI] [PubMed] [Google Scholar]
- [12].Horvath K, Stefanatos G, Sokolski KN, Wachtel R, Nabors L, Tildon JT. Improved social and language skills after Secretin administration in patients with autistic spectrum disorders. Journal of the Association for Academic Minority Physicians: the official publication of the Association for Academic Minority Physicians. 1998;9:9–15. [PubMed] [Google Scholar]
- [13].Esch BE, Carr JE. Secretin as a treatment for autism: a review of the evidence. Journal of Autism and Developmental Disorders. 2004;34:543–56. doi: 10.1007/s10803-004-2549-6. [DOI] [PubMed] [Google Scholar]
- [14].Sturmey P. Secretin is an ineffective treatment for pervasive developmental disabilities: a review of 15 double-blind randomized controlled trials. Research in Developmental Disabilities. 2005;26:87–97. doi: 10.1016/j.ridd.2004.09.002. [DOI] [PubMed] [Google Scholar]
- [15].Williams KW, Wray JJ, Wheeler DM. Intravenous Secretin for autism spectrum disorder. Cochrane Database of Systematic Reviews. 2005:CD003495. doi: 10.1002/14651858.CD003495.pub2. [DOI] [PubMed] [Google Scholar]
- [16].Kern JK, Van Miller S, Evans PA, Trivedi MH. Efficacy of porcine Secretin in children with autism and pervasive developmental disorder. Journal of Autism and Developmental Disorders. 2002;32:153–60. doi: 10.1023/a:1015441428154. [DOI] [PubMed] [Google Scholar]
- [17].Yamagata T, Urano H, Weeber EJ, Nelson DL, Nishijima I. Impaired hippocampal synaptic function in Secretin deficient mice. Neuroscience. 2008;154:1417–22. doi: 10.1016/j.neuroscience.2008.04.037. [DOI] [PubMed] [Google Scholar]
- [18].Nishijima I, Yamagata T, Spencer CM, Weeber EJ, Alekseyenko O, Sweatt JD, et al. Secretin receptor-deficient mice exhibit impaired synaptic plasticity and social behavior. Human Molecular Genetics. 2006;15:3241–50. doi: 10.1093/hmg/ddl402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Siu FK, Sham MH, Chow BK. The prenatal expression of Secretin receptor. Annals of the New York Academy of Sciences. 2006;1070:561–5. doi: 10.1196/annals.1317.081. [DOI] [PubMed] [Google Scholar]
- [20].Freemark M, Kirk K, Pihoker C, Robertson MC, Shiu RP, Driscoll P. Pregnancy lactogens in the rat conceptus and fetus: circulating levels, distribution of binding, and expression of receptor messenger ribonucleic acid. Endocrinology. 1993;133:1830–42. doi: 10.1210/endo.133.4.8404626. [DOI] [PubMed] [Google Scholar]
- [21].Banks WA, Goulet M, Rusche JR, Niehoff ML, Boismenu R. Differential transport of a Secretin analog across the blood-brain and blood-cerebrospinal fluid barriers of the mouse. The Journal of Pharmacology and Experimental Therapeutics. 2002;302:1062–9. doi: 10.1124/jpet.102.036129. [DOI] [PubMed] [Google Scholar]
- [22].Bonnin A, Goeden N, Chen K, Wilson ML, King J, Shih JC, et al. A transient placental source of serotonin for the fetal forebrain. Nature. 2011;472:347–50. doi: 10.1038/nature09972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Erlebacher A, Price KA, Glimcher LH. Maintenance of mouse trophoblast stem cell proliferation by TGF-beta/activin. Dev Biol. 2004;275(1):158–69. doi: 10.1016/j.ydbio.2004.07.032. [DOI] [PubMed] [Google Scholar]
- [24].Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282(5396):2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.