Abstract
Wnt/β-catenin/TCF signaling stimulates bone formation and suppresses adipogenesis. The hallmarks of skeletal involution with age, on the other hand, are decreased bone formation and increased bone marrow adiposity. These changes are associated with increased oxidative stress and decreased growth factor production, which activate members of the FOXO family of transcription factors. FOXOs in turn attenuate Wnt/β-catenin signaling by diverting β-catenin from TCF- to FOXO-mediated transcription. We show herein that mice lacking Foxo1, -3, and -4 in bipotential progenitors of osteoblast and adipocytes (expressing Osterix1) exhibited increased osteoblast number and high bone mass that was maintained in old age as well as decreased adiposity in the aged bone marrow. The increased bone mass in the Foxo-deficient mice was accounted for by increased proliferation of osteoprogenitor cells and bone formation resulting from upregulation of Wnt/β-catenin signaling and cyclin D1 expression, but not changes in redox balance. Consistent with this mechanism, β-catenin deletion in Foxo null cells abrogated both the increased cyclin D1 expression and proliferation. The elucidation of a restraining effect of FOXOs on Wnt signaling in bipotential progenitors suggests that FOXO activation by accumulation of age-associated cellular stressors may be a seminal pathogenetic mechanism in the development of involutional osteoporosis.
Introduction
Osteoblasts, the cells responsible for the synthesis of bone matrix, are terminally differentiated postmitotic cells with a short life span (1, 2). For bone formation to continue uninterrupted throughout life, osteoblasts need to be constantly replenished with new osteoblasts originating from self-renewing mesenchymal stem cells through the replication of lineage-committed descendants of the stem cells (3). With advancing age, bone formation decreases, leading to osteoporosis. The decline of bone mass in old age is associated with a decrease in osteoblast number and an increase in bone marrow adiposity, but the molecular mechanisms behind these changes remain elusive (4).
Wnt/β-catenin signaling is indispensable for osteoblastogenesis, and loss or gain of function of this pathway is associated with a profound decrease or increase of bone mass, respectively, in humans and mice (5). In addition, Wnt/β-catenin signaling is a potent suppressor of adipogenesis (6). Wnt proteins activate the Frizzled/LRP5 or LRP6 receptor complex and thereby prevent the proteasomal degradation of the transcriptional coactivator β-catenin (7). β-catenin associates with the T cell factor (TCF) lymphoid-enhancer binding factor (LEF) family of transcription factors and regulates the expression of Wnt-target genes. Via this mechanism Wnt/β-catenin signaling promotes the progression of Osterix1-expressing (Osx1-expressing) cells to bone-producing osteoblasts (8).
In addition to its role in TCF/LEF transcription, β-catenin associates with members of the Forkhead box O (FOXO) family of transcription factors (9). FOXOs exert seminal influences on self renewal, proliferation, survival, stress resistance, tumor suppression, and longevity in model organisms (10–12). FOXO activation is normally restrained by the PI3K/Akt signaling pathway, which prevents FOXO translocation into the nucleus. On the other hand, stress conditions, such as high levels of ROS or growth factor deprivation, cause nuclear translocation and FOXO activation. Importantly, FOXOs regulate transcriptional responses independently of direct DNA binding via association with a variety of unrelated transcription factors (13). The array of FOXO transcriptional targets is large and varies widely depending on the cell type (14). Some of these genes are involved in free radical scavenging, while others control cell cycle, DNA repair, and life span (15, 16). FOXO1, -3, and -4 have broad and overlapping patterns of expression in many mammalian tissues, including bone (17, 18). Although they all recognize the same DNA target sequence (19), studies with murine models of single or combined Foxo deletion have elucidated that FOXO1, -3, and -4 exert both redundant and nonredundant functions (14, 20–23).
We have previously found that binding of β-catenin to FOXOs diverts the limited pool of β-catenin from Wnt/TCF- to FOXO-mediated transcription and decreases osteoblastogenesis in vitro (24). Growth factor depletion or high ROS levels augment the association of β-catenin with FOXOs in osteoblast progenitors as well as in other cell types (9, 24–26). Additionally, work by us and others showed that FOXOs maintain redox balance in mature osteoblasts (18, 27). However, the relevance of the diversion of β-catenin from Wnt/TCF- to FOXO-mediated transcription to skeletal homeostasis has remained unknown. We have now generated triple mutant mice lacking Foxo1, -3, and -4 in osteoblast progenitors. We show that this targeted deletion leads to an increase in bone mass that is maintained throughout life and is associated with increased β-catenin/TCF-mediated transcription.
Results
Combined deletion of Foxo1, -3, and -4 in osteoprogenitors increases bone mass.
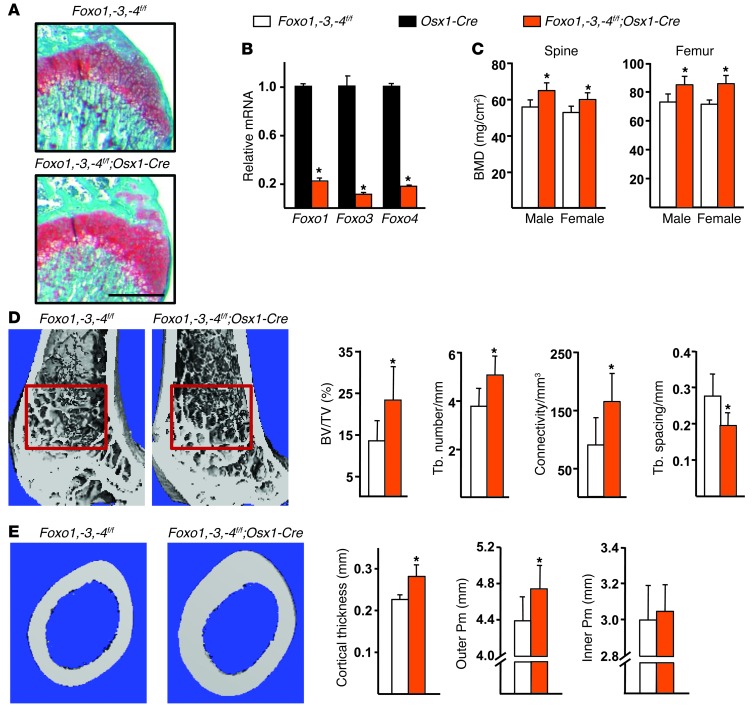
Foxo1, -3, and -4 were deleted in osteoblast precursors using transgenic mice in which a Cre-GFP fusion protein is under the control of Osx1 regulatory elements (8). The Osx1-Cre transgene is expressed in committed osteoblast progenitors present in the bone-forming regions of the perichondrium and primary spongiosa as well as in hypertrophic chondrocytes. In addition, Osx1/GFP-expressing cells are present in the thin periosteal layer overlaying the cortical bone surface (28). Mice lacking Foxo1, -3, and -4 in Osx1-Cre–expressing cells, hereafter referred to as Foxo1,-3,-4f/f;Osx1-Cre mice, were born at the expected Mendelian ratios, and their growth plates were indistinguishable from those of control Foxo1,-3,-4f/f littermates (Figure 1A). Female, but not male, mice exhibited a modest decrease in body weight (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI68049DS1). Foxo1, -3, and -4 mRNA levels were reduced by almost 90% in Osx1-GFP–positive calvaria cells from Foxo1,-3,-4f/f;Osx1-Cre mice as compared with cells from Osx1-Cre mice, isolated by FACS (Figure 1B). As expected, Foxo mRNA was unaltered in spleen and liver from Foxo1,-3,-4f/f;Osx1-Cre mice (Supplemental Figure 1B).
Figure 1. Deletion of Foxos in Osx1-Cre–expressing cells increases bone mass.
(A) Histological sections of the distal femurs of 4-week-old mice stained with Safranine-O (cartilage stains red). Scale bar: 500 μm. (B) Foxo mRNA levels in Osx1-GFP calvaria cell cultures (triplicates) determined by quantitative RT-PCR (qRT-PCR). (C) BMDs determined by DXA in 3-month-old male (n = 15–20/group) and female (n = 23–30/group) mice. (D) Micro-CT measurements of the distal femur of 3-month-old males (n = 7–10/group). Representative images of the cancellous bone region (red box). BV/TV, bone volume per tissue volume; Tb, trabecular. (E) Cortical measurements determined by micro-CT in femoral diaphysis of the samples described in D and representative images of the region analyzed. Pm, perimeter. Bars represent mean + SD. *P < 0.05 versus Osx1-Cre or Foxo1,-3,-4f/f by Student’s t test.
Male and female Foxo1,-3,-4f/f;Osx1-Cre mice exhibited increased spinal and femoral bone mineral density (BMD) at 12 weeks of age, as measured by DXA (Figure 1C). The increased BMD was due to the loss of Foxos and not to unspecific actions of Cre recombinase, as indicated by the lack of a skeletal phenotype in Osx1-Cre mice as compared with wild-type controls obtained following a breeding strategy similar to that used to generate the Foxo1,-3,-4f/f;Osx1-Cre and Foxo1,-3,-4f/f mice (Supplemental Figure 1C). Nonetheless, Osx1-Cre mice exhibited decreased body weight (Supplemental Figure 1D), in line with previous evidence that the Osx1-Cre transgene decreases body size (29).
Micro-CT analysis of the femur (Figure 1, D and E) and vertebra (Supplemental Figure 1E) revealed that the deletion of Foxos in Osx1-expressing cells led to an increase in both cancellous and cortical bone mass. The former was detected as early as 4 weeks of age in the femur (Supplemental Table 1), and it was associated with an increase in trabecular number and connectivity and with a decrease in trabecular spacing. The higher cortical thickness of the Foxo1,-3,-4f/f;Osx1-Cre mice was detected as early as 7 weeks of age (Supplemental Table 1). This change resulted from an enlargement of the outer perimeter (Figure 1E), indicating that Foxo deletion from osteoprogenitor cells promotes periosteal apposition. No changes were detected in the inner perimeter of the femur.
We had previously shown that Foxo3 was expressed at higher levels than Foxo1 and Foxo4 in osteoblastic cells (18). To determine whether deletion of Foxo3 alone could recapitulate the skeletal effect seen with the triple Foxo deletion, we generated Foxo3f/f;Osx1-Cre mice and the respective wild-type, Foxo3f/f, and Osx1-Cre littermate controls. Foxo3 mRNA expression in cultured bone marrow–derived osteoblastic cells from Foxo3f/f;Osx1-Cre was decreased by 60%, while the expression of FOXO1 or FOXO4 was not affected (Supplemental Figure 2A). Body weight and femur BMD in Osx1-Cre or Foxo3f/f;Osx1-Cre mice were lower as compared with those in wild-type or Foxo3f/f littermate control mice (Supplemental Figure 2, B and C). Despite the smaller body size and the reduced femoral BMD in both Osx1-Cre and Foxo3f/f;Osx1-Cre mice, these 2 measures were indistinguishable between Osx1-Cre and Foxo3f/f;Osx1-Cre mice. This result indicates that, whereas Osx1-Cre expression in and of itself had an effect in decreasing body size and femoral BMD, Foxo3 deletion per se did not affect any of these measurements. Furthermore, spine BMD was indistinguishable among the 4 genotypes (Supplemental Figure 2D). These results are in agreement with the published evidence that Foxo1, -3, and -4 have redundant functions (14, 20, 21).
The high bone mass phenotype of the Foxo-deficient mice is maintained throughout life.
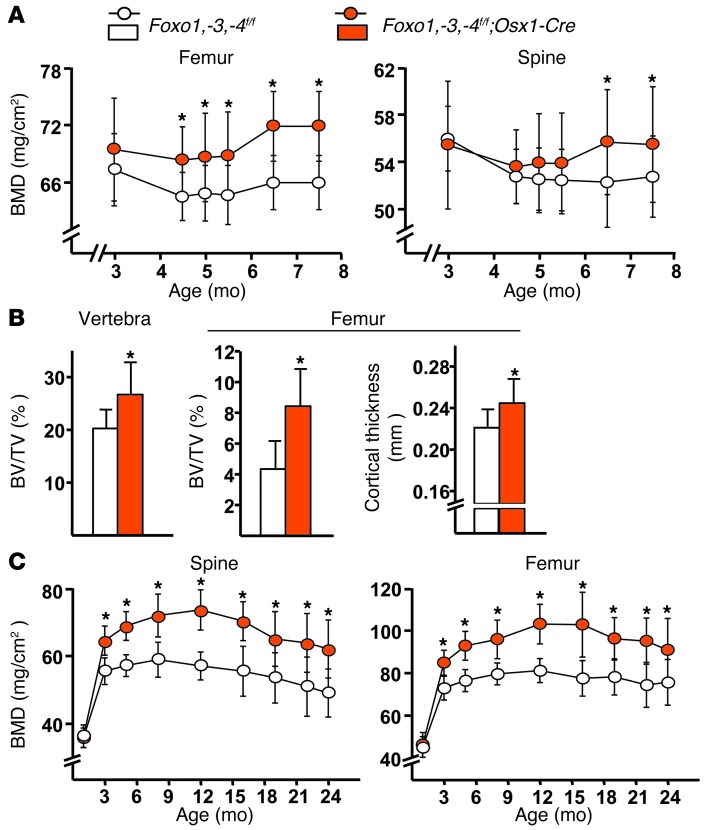
To determine whether the effect of Foxo deletion on bone mass could be elicited in an adult animal, we induced the triple Foxo deletion after most of the growth phase was completed (at 3 months of age), taking advantage of the Tet-off system incorporated into the Osx1-Cre transgene. As expected, Foxo1, -3, and -4 transcripts were significantly decreased in bone marrow–derived osteoblasts from the Foxo1,-3,-4f/f;Osx1-Cre mice (Supplemental Figure 3A). Longitudinal BMD measurements in these mice showed a significant increase in bone mass at the spine and femur 12 weeks following the deletion (Figure 2A). Moreover, micro-CT measurements revealed that deletion of Foxos at 3 months of age caused identical microarchitectural changes to those seen when Foxos were deleted from conception, including increased cancellous bone mass, trabecular number, and connectivity as well as decreased trabecular spacing at both the femur and spine (Figure 2B and Supplemental Figure 3B). Likewise, femoral cortical thickness was increased (Figure 2B). Therefore, the effect of the deletion on bone mass was not caused by changes imparted during development and growth.
Figure 2. The high bone mass of Foxo1,-3,-4f/f;Osx1-Cre mice is maintained throughout life.
(A and B) Female mice from both genotypes were maintained in a doxycycline-containing diet from conception until 3 months of age, when the diet was changed to regular chow to activate Foxo deletion (n = 10–11/group). (A) Longitudinal BMD measurements were determined by DXA. Data represent mean + SD. *P < 0.05 based on comparisons of least squares means from a mixed effects model including genotype, age, and their interaction. The interaction term was not statistically significant. (B) Micro-CT measurements in 7.5-month-old mice. Bars represent mean + SD. *P < 0.05 by Student’s t test. (C) Longitudinal DXA BMD measurements in female mice in which Foxos were deleted since conception (initial n = 24–33/group; final n = 12–18/group). Data represent mean + SD. *P < 0.05 based on comparisons of least squares means from a mixed effects model including genotype, age, age2, and the interactions of genotype with age and age2. All factors, including both interactions were statistically significant.
Age-related bone loss is associated with increased oxidative stress and decreased growth factor levels, 2 conditions that can stimulate FOXOs (24, 30). We, therefore, determined whether FOXOs in osteoprogenitors could influence bone mass during aging. To do this, we aged a cohort of female Foxo1,-3,-4f/f;Osx1-Cre mice. The high bone mass of Foxo1,-3,-4f/f;Osx1-Cre mice was maintained up to 24 months of age (Figure 2C), as determined by longitudinal DXA BMD measurements.
The high bone mass phenotype of the Foxo-deficient mice is independent of ROS.
We had previously shown that global combined Foxo1, -3, and -4 deletion increases oxidative stress in bone (18). We therefore sought to determine whether targeted deletion of Foxos in osteoprogenitors had a similar effect. The mRNA levels of known antioxidant FOXO target genes were not affected in Osx1-GFP–positive calvaria cells from Foxo1,-3,-4f/f;Osx1-Cre mice (Supplemental Table 2). In contrast, the mRNA encoding glutathione S–transferase α 4 (Gsta4) — a protein involved in the generation of the antioxidant glutathione (GSH) — was increased 4-fold. N-Acetylcysteine (NAC) administration for 12 weeks had no effect on BMD in Foxo1,-3,-4f/f control mice nor did it affect the increase in BMD at the spine and femur seen in the Foxo1,-3,-4f/f;Osx1-Cre mice (Supplemental Figure 4A). NAC, however, decreased ROS and increased GSH levels in the bone marrow (Supplemental Figure 4B) and the phosphorylation of p66shc — a marker of oxidative stress — in the vertebra (Supplemental Figure 4C). In line with the increase of Gsta4 mRNA levels, ROS and p66shc phosphorylation were decreased in the bone of Foxo1,-3,-4f/f;Osx1-Cre mice; the decrease of ROS was maintained at 24 months of age (Supplemental Figure 4D). Taken together, these results suggest that FOXOS do not attenuate ROS in osteoblast progenitors and that the increase in bone mass resulting from Foxo deletion in Osx1 cells could not be accounted for by altered redox balance.
Deletion of Foxos increases bone formation.
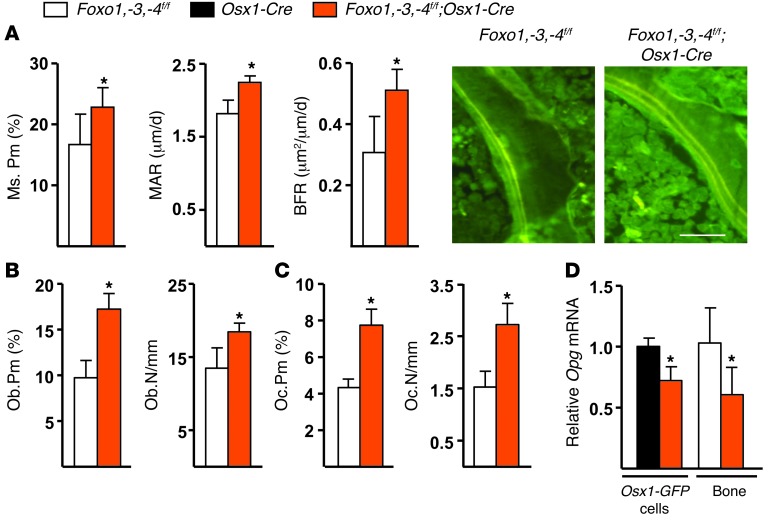
In agreement with the high–bone mass phenotype, the bone-formation rate (BFR) in cancellous bone of 7-week-old Foxo1,-3,-4f/f;Osx1-Cre mice was increased by 40% as a result of elevated mineralizing perimeter and mineral apposition rate (MAR) (Figure 3A). These changes were accompanied by increased osteoblast numbers (Figure 3B) and expression of the osteoblast specific gene Ocn in bone (Figure 3C). At 24 months of age, BFR was 10-fold lower in both littermate controls and Foxo1,-3,-4f/f;Osx1-Cre mice (Supplemental Figure 5), as compared with the 7-week-old mice, due to reduced mineralizing perimeter and MAR. Nonetheless, in spite of the persistence of higher bone mass in the aged Foxo1,-3,-4f/f;Osx1-Cre mice as compared with the littermate controls, we could no longer detect a difference in bone formation, suggesting that aging-related mechanisms overrode the positive effect of the Foxo deletion seen at 7 weeks, albeit bone mass remained higher in the Foxo1,-3,-4f/f;Osx1-Cre mice, probably because the gain made earlier in life was preserved after a steady-state remodeling rate was achieved.
Figure 3. Foxo deletion in Osx1-Cre–expressing cells increases bone formation.
(A) MAR, mineralizing surface (MS), and BFR as determined by tetracycline labels, shown in the photomicrographs, in the cancellous bone of longitudinal undecalcified vertebral (L1–L3) sections from 7-week-old mice (n = 8/9 per group). Scale bar: 20 μm. (B) Osteoblast (Ob) and (C) osteoclast (Oc) perimeter and number (N) per mm cancellous bone surface in the vertebral sections described in A (n = 4/group). (D) Opg mRNA levels in sorted Osx1-GFP calvaria cells (triplicates) and in calvaria bone from 1-month-old mice (n = 4–5/group) determined by qRT-PCR. Bars represent mean + SD; *P < 0.05 versus Foxo1,-3,-4f/f or Osx1-Cre by Student’s t test.
In line with earlier findings by us and others in mice with loss or gain of FOXO function in mature osteoblasts (18, 27), the number of osteoclasts was higher in the Foxo1,-3,-4f/f;Osx1-Cre mice (Figure 3D). However, the high bone mass phenotype of the latter indicates that the effect of the increased osteoclast number was overridden by the increase in osteoblastogenesis. FOXO1 activation in osteoblasts increases the expression of the anti-osteoclastogenic cytokine Opg (31). Opg mRNA was decreased in whole bone and in Osx1-GFP–positive calvaria cells from our Foxo1,-3,-4f/f;Osx1-Cre mice (Figure 3D); but Rankl and M-csf were unaffected (Supplemental Table 3), suggesting that a decrease in Opg may contribute to the higher osteoclast number seen in these mice.
Deletion of Foxos increases the proliferation of osteoprogenitor cells.
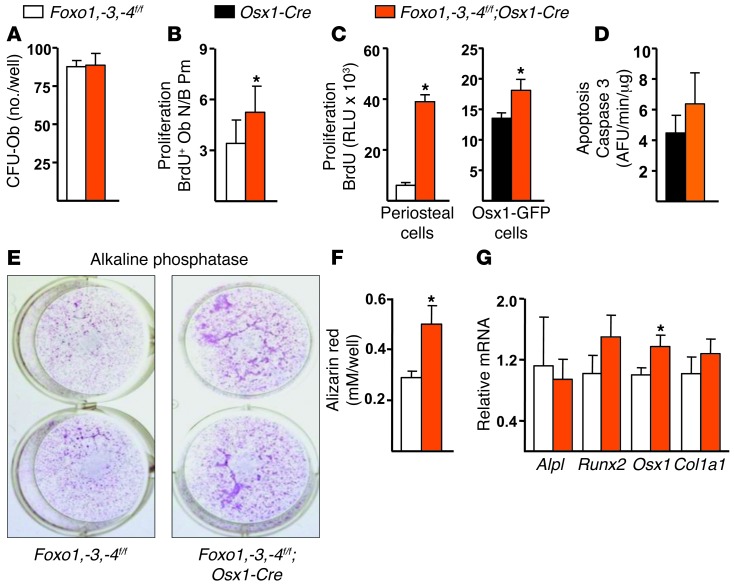
Deletion of Foxos in Osx1 cells did not affect the number of mesenchymal progenitors present in the bone marrow capable of initiating osteoblastic colonies, as indicated by the CFU osteoblast (CFU-OB) assay (Figure 4A). However, there was an increase in the number of BrdU-positive osteoblasts in cancellous bone (Figure 4B). Similarly, the rate of proliferation of cultured periosteal cells from femurs was increased (Figure 4C), consistent with an increase in periosteal apposition and the larger outer perimeter of the femur. As expected, the cultured periosteal cells displayed lower expression of Foxo1, -3, and -4 (Supplemental Figure 6A). An increased rate of proliferation was also observed in Osx1-GFP calvaria cells from the Foxo1,-3,-4f/f;Osx1-Cre mice (Figure 4C). Apoptosis was not affected in Foxo-deleted Osx1-GFP calvaria cells (Figure 4C). However, in line with our previous findings (18), calvaria-derived osteoblasts from Foxo1,-3,-4f/f;Osx1-Cre mice cultured for 10 days in ascorbic acid exhibited increased apoptosis (Supplemental Figure 6B). Nevertheless, the increased osteoprogenitor proliferation apparently overrode the increase in mature osteoblast apoptosis in Foxo1,-3,-4f/f;Osx1-Cre mice.
Figure 4. Foxo deletion in Osx1-Cre–expressing cells increases proliferation.
(A) Number of CFU-OB in the bone marrow from femurs of 3-month-old mice cultured for 21 days with 1% ascorbate (triplicates). (B) Osteoblast proliferation as determined by BrdU incorporation in decalcified paraffin-embedded vertebral (L4–L5) sections from 7-week-old mice (n = 9–11/group). (C) BrdU incorporation in cultures of periosteal cells from 7-week-old mice (left) and sorted Osx1-GFP calvaria cells (right) (triplicate wells each). (D) Caspase 3 activity in sorted Osx1-GFP calvaria cells (triplicates). (E) ALP activity and (F) Alizarin red staining (triplicate) of high-density bone marrow stromal cells cultured with 1% ascorbate for 3 and 21 days, respectively. (G) mRNA levels by qRT-PCR of osteoblast differentiation markers, in the cultures described in E. Bars represent mean + SD; *P < 0.05 versus Foxo1,-3,-4f/f or Osx1-Cre by Student’s t test.
Alkaline phosphatase (ALP) activity and mineralization were higher in bone marrow–derived osteoblastic cells from Foxo1,-3,-4f/f;Osx1-Cre mice (Figure 4, E and F). However, the expression of genes involved in osteoblast differentiation, such as Alpl, Runx2, and Col1a1, was unaffected (Figure 4G). Osx1 was modestly increased in the bone marrow–derived osteoblastic cells, but Osx1 or any of the other differentiation-associated genes was unaffected in Osx1-GFP calvaria cells (Supplemental Table 3). The lack of an effect of Foxo deletion on osteoblast differentiation in both bone marrow and calvaria cells suggests that the increases in ALP activity and mineralization observed in the bone marrow–derived osteoblastic cells from Foxo1,-3,-4f/f;Osx1-Cre mice were the result of increased osteoblast number.
The higher proliferation of Foxo-deficient osteoprogenitors is due to increased β-catenin/TCF activation.
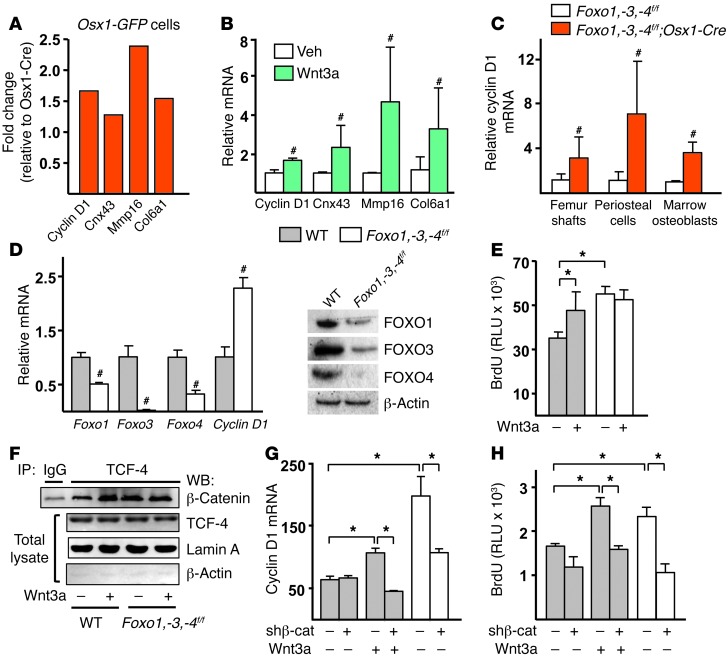
We next focused on elucidating whether the restraining effect of FOXOs on osteoblastogenesis was the result of attenuation of Wnt signaling, as our earlier work had suggested (24). Osx1-GFP calvaria cells from Foxo1,-3,-4f/f;Osx1-Cre mice exhibited increased expression of several Wnt-target genes including cyclin D1, Cnx43, Col6a1, and Mmp16 (Figure 5A). Recombinant Wnt3a increased the expression of the same genes in cells from wild-type mice, establishing that these are indeed bona fide Wnt-target genes in osteoblastic cells (Figure 5B). The increase in cyclin D1 mRNA was confirmed in bone as well as in periosteal and bone marrow cell cultures from Foxo1,-3,-4f/f;Osx1-Cre mice (Figure 5C).
Figure 5. Wnt/TCF signaling is responsible for increased osteoprogenitor proliferation.
(A) Wnt-target genes determined by microarray analysis in Osx1-GFP calvaria cells. (B) mRNA levels by qRT-PCR in calvaria cells treated with vehicle (veh) or 50 ng/ml Wnt3a overnight (triplicates). (C) mRNA levels by qRT-PCR in femoral shafts from 3-month-old mice (n = 8–9/group), and in periosteal and bone marrow osteoblastic cell cultures described in Figure 4, C and G. (D) mRNA by qRT-PCR (left, triplicates) and protein levels by Western blot (right) in calvaria-derived cells infected with Adeno-Cre to induce Foxo deletion. (E) BrdU incorporation in cells, described in D, treated with veh (–) or 50 ng/ml Wnt3a (triplicates). (F) Lysates from nuclear extracts of calvaria cells, described in D, treated with vehicle or 50 ng/ml Wnt3a, immunoprecipitated with an anti–TCF-4 or anti-IgG antibody, and probed with an anti–β-catenin antibody. (G) mRNA levels by qRT-PCR and (H) BrdU incorporation in calvaria cells infected with Adeno-Cre and with lentivirus expressing shRNA directed against β-catenin or nontargeted shRNA followed by treatment with vehicle or 50 ng/ml Wnt3a overnight (triplicates). #P < 0.05 by Student’s t test; *P < 0.05 by ANOVA with Bonferroni’s test. Bars represent mean + SD.
As was the case with cells from Foxo1,-3,-4f/f;Osx1-Cre mice, the expression of cyclin D1 was increased following Foxo deletion using Adeno-Cre (Ad-Cre) in vitro (Figure 5D). The Cre recombinase efficiently reduced Foxo1, -3, and -4 mRNA and protein levels by more than 60%. Deletion of Foxos with Ad-Cre or treatment with Wnt3a also increased the proliferation of calvaria cells (Figure 5E). Having established that Ad-Cre deletion recapitulated the effects of the in vivo triple Foxo deletion on cyclin D1 and proliferation, we determined whether the availability of β-catenin for TCF binding increased in the absence of FOXOs. To do this, we immunoprecipitated β-catenin from control or Foxo-deleted calvaria cells and assessed the amount of TCF-4 bound to β-catenin in nuclear extracts (Figure 5F) or in total cell lysates (Supplemental Figure 7). The abundance of total β-catenin and TCF-4 did not differ between the 2 genotypes. However, the amount of β-catenin associated with TCF-4 increased in the absence of Foxos. As expected, Wnt3a promoted the association of β-catenin with TCF-4 in cells from wild-type mice. We then examined the contribution of β-catenin activity to the increased expression of cyclin D1 and proliferation by silencing β-catenin (Supplemental Figure 8A). shRNA-mediated knockdown of β-catenin prevented the Wnt3a-induced increase in cyclin D1 (Figure 5G). Silencing of β-catenin also abrogated the increased cyclin D1 expression in Foxo-deleted cells. In agreement with these findings, silencing of β-catenin prevented Wnt3A-induced proliferation as well as the increased proliferation seen in Foxo-deleted osteoblastic cells (Figure 5H). These results, nonetheless, could not exclude the possibility that FOXOs attenuated Wnt signaling by upregulating the expression of Wnt-signaling inhibitors, such as DKK1, SOST, SFRP1, and SFRP2 (21, 32). To address this alternative mechanism, we searched for and found that the expression of all these Wnt-signaling inhibitors was unaffected in bone or calvaria cells from Foxo1,-3,-4f/f;Osx1-Cre mice (Supplemental Figure 8B and Supplemental Table 3). Similarly, deletion of Foxos did not alter the expression of several members of the Wnt protein family, except for a decrease in Wnt5a (Supplemental Table 3). Taken together, these results support the idea that FOXOs inhibit osteoprogenitor cell proliferation by attenuating the expression of the β-catenin/TCF-target cyclin D1.
Deletion of Foxos increases marrow adiposity in old age.
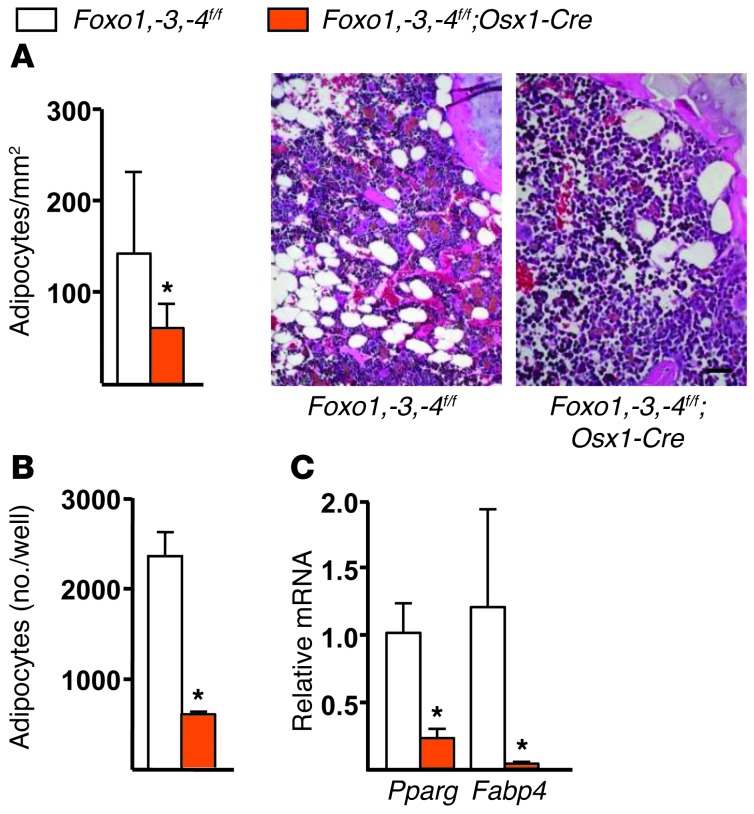
Finally, consistent with the evidence that Wnt/β-catenin signaling is a potent suppressor of adipogenesis (6) and that the Osx1-Cre transgene is active in cells capable of becoming either osteoblasts or adipocytes (33), the number of adipocytes present in the bone marrow was decreased in 24-month-old Foxo1,-3,-4f/f;Osx1-Cre mice as compared with the littermate controls (Figure 6A). In addition, the number of adipocytes formed in response to rosiglitazone in cultured bone marrow stromal cells from Foxo1,-3,-4f/f;Osx1-Cre mice was reduced (Figure 6B) as was the expression of the adipocyte markers Pparγ and Fabp4 (Figure 6C).
Figure 6. Foxo deletion decreases bone marrow adipogenesis.
(A) Adipocytes (arrow) in distal femurs of 24 month-old mice (n = 6/group) were quantified by histomorphometry of decalcified sections stained with H&E. Scale bar: 20 μm. (B) Number of adipocytes, stained with oil red O, in bone marrow stromal cells, cultured for 6 days with 1 μM rosiglitazone (triplicates). (C) mRNA levels of the indicated genes by qRT-PCR in cells cultured as described in B. Bars represent mean + SD. *P < 0.05 by Student’s t test.
Discussion
FOXOs divert β-catenin from TCF- to FOXO-mediated transcription in diverse cell types including osteoblastic and colon cancer cell models (24, 25). Based on this evidence, we and others had hypothesized earlier that this molecular switch may represent a pathogenetic mechanism for osteoporosis and perhaps several other degenerative diseases (13, 34–37). In support of this hypothesis, we now present genetic evidence that FOXOs do indeed attenuate β-catenin/TCF transcription in osteoblast progenitors in mice (Figure 7). Through this mechanism, FOXOs restrain the proproliferative effects of Wnt signaling and thereby attenuate bone formation. Restraining the proproliferative β-catenin/TCF transcriptional signal by FOXOs is probably a defense mechanism against diverse stress stimuli in many tissues, as evidenced by the increased association of β-catenin with FOXOs in bone and colon cancer cells exposed to H2O2 as well as in the liver of starved mice (24–26). Indeed, besides its effect on bone homeostasis described herein, diversion of β-catenin from TCF- to FOXO-mediated transcription stimulates gluconeogenesis in the liver in response to starvation (26). The same mechanism has been proposed as a potential culprit of the formation of β-amyloid plaques in Alzheimer disease (38) as well as in age-related heart disease (39), insulin resistance, and obesity (40).
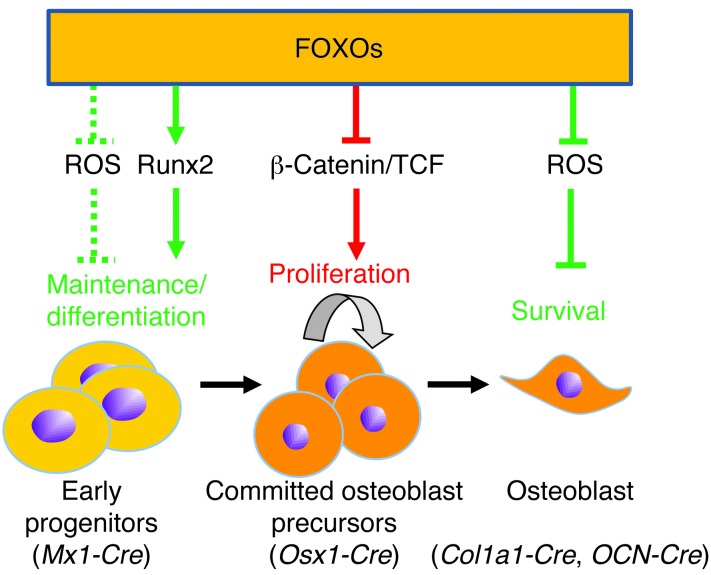
Figure 7. Model of the role of FOXOs in cells of the osteoblast lineage.
The model depicts the 3 distinct stages of the osteoblast differentiation lineage in which the role of FOXOs has been addressed by genetic manipulation in mice (26, 27), along with the respective Cre transgenes used to target them, and the mechanism of FOXO action. Green and red lines indicate a positive and negative influence on bone mass, respectively. The dashed line indicates an effect deduced from other cell lineages (15, 16), but not directly demonstrated in bone.
An increase in the proliferation of osteoblast progenitors in our Foxo-deficient mice is consistent with extensive evidence that FOXOs regulate several genes involved in cell division, including cyclin D1 (41). Nonetheless, the mechanism by which FOXOs attenuate cyclin D1 transcription remains unclear, as repression of cyclin D1 transcription does not involve direct binding of FOXOs to the cyclin D1 promoter (42). In the present work, increased cyclin D1 expression and proliferation rate in Foxo-deficient osteoprogenitors was abrogated by silencing β-catenin. This result, along with evidence that cyclin D1 is a primary target of Wnt signaling and closely correlates with TCF gene activation (43, 44), suggests that suppression of cyclin D1 by FOXOs results from the attenuation of β-catenin/TCF activity. Foxo deletion in Osx1-expressing osteoblast progenitors increased periosteal cell proliferation. This effect most likely resulted from unleashing β-catenin/TCF activity and is in agreement with evidence that β-catenin promotes the proliferation of Osx1-expressing cells that reside within the periosteal envelope by stimulating cyclin D1 (8, 45). Similar to our findings in the Foxo1,-3,-4-deficient mice, deletion of Foxo1 in enteroendocrine progenitor cells increases proliferation and the expression of several Wnt targets, including cyclin D1 (46).
Bone marrow adiposity increases with advancing age in humans and rodents and so does the expression of PPARγ2 — a transcription factor that is essential for adipogenesis — in murine bone (47, 48). The triple Foxo mutant mice of the present report exhibited decreased bone marrow adiposity at 24 months of age as compared with the control littermates. Likewise, bone marrow–derived osteoblasts from the mutant mice showed decreased expression of PPARγ2 in response to rosiglitazone. Osteoblasts and bone marrow adipocytes arise from a common progenitor, and lineage allocation into adipocytes or osteoblasts is reciprocally exclusive and dictated by β-catenin/TCF-mediated transcription (6, 49). Indeed, Wnt/β-catenin signaling is a strong inhibitor of adipogenesis (6). Conversely, deletion of β-catenin in osteoprogenitors after growth increases PPARγ2 expression and bone marrow adiposity and decreases bone mass in mice (33). The findings reported here strongly suggest that, by inhibiting β-catenin/TCF-mediated transcription, FOXOs may not only contribute to the decrease in bone mass but also to the increased marrow adiposity of the aging skeleton. The same mechanism may also contribute to the bone loss caused by glucocorticoids and the inflammatory cytokine TNF-α, 2 agents that inhibit Wnt signaling in a FOXO-dependent manner (50). Be that as it may, the murine model we used here was not designed to, and cannot, address whether FOXO activation changes with age in osteoblast progenitors or whether the stimulus responsible for FOXO activation in this cell type is oxidative stress, growth factor deprivation, hormonal changes (4), some other heretofore unknown pathologic stimulus, or all of the above. Future work with alternative models will be required to answer these questions.
The high bone-mass phenotype of the mice with the targeted Foxo deletion of the present report was maintained up to 24 months of age, suggesting that the increase in β-catenin/TCF-mediated transcription was maintained late in life. Consistent with this contention, the number of adipocytes present in the bone marrow of Foxo-deleted mice at this late age was lower as compared with the controls. Nonetheless, the BFR in the 24-month-old mice was not different between experimental and control animals, indicating that the high bone formation observed in young mice lacking Foxos in osteoblast progenitors had declined by that age. The most likely explanation for this finding is that mechanisms of aging other than the diversion of β-catenin by FOXOs overrode the advantage provided by the deletion of Foxos. In support of this scenario, we have previously shown that an increase in endogenous glucocorticoids contributes to the decrease in bone formation in old mice (51). Foxo deletion could alternatively affect a critical and unique stage in bone accrual of very young mice and have no effect in older animals. The latter scenario, however, is unlikely in view of the fact that induction of Foxo deletion in adult mice also led to an increase in bone mass.
We and others had shown earlier that broad deletion of Foxo1, -3, and -4 (using Mx1-Cre) or specific deletion of Foxo1 in mature osteoblasts (using Col1a1-Cre) causes an osteoporotic phenotype (18, 27). Consistent with evidence that Foxos in some cell types attenuate ROS, those mice exhibited increased oxidative stress in bone and increased osteoblast apoptosis. Conversely, overexpression of a Foxo3 transgene in mature osteoblasts decreased oxidative stress and osteoblast apoptosis and increased bone mass (18). In contrast to these earlier findings, targeted deletion of Foxos in osteoblast progenitors (and inexorably their progeny) in the present work did not alter the expression of known Foxo antioxidant genes or apoptosis nor did increase oxidative stress in bone, indicating that the effects of FOXOs in osteoblast progenitors are independent of ROS. Nonetheless, in agreement with our previous findings, mature osteoblast cultured from the Foxo1,-3,-4f/f;Osx1-Cre mice did exhibit increased apoptosis, supporting the conclusion that in mature osteoblasts, FOXOs attenuate ROS. These results, along with the evidence that the high bone mass phenotype seen with the targeted Foxo1,-3,-4 deletion in Osx1-Cre–expressing cells is opposite to the low bone mass phenotype of the mice with triple Foxo deletion in Mx1-Cre–expressing cells, indicate that FOXOs have different functions at distinct stages of the osteoblast lineage (Figure 7). In support of this notion, FOXO1 ablation in pancreatic progenitor cells, but not in committed endocrine progenitors or terminally differentiated β cells, increased the number of juxtaductal β cells (52). In addition, Tothova et al., using the triple Foxo deletion in Mx1-Cre–expressing cells, have shown that in hematopoietic stem cells, FOXOs maintain redox homeostasis by decreasing ROS (20). In contrast, FOXOs have no impact on ROS in myeloid progenitor cells, indicating that FOXOs do indeed act to maintain redox homeostasis in some cells, but not others, irrespective of whether the activation stimulus could still be oxidative stress.
Within the mesenchymal lineage, the Mx1-Cre transgene targets an early progenitor upstream of the committed osteoblasts that express Osx1 (3). We had shown before that Foxo deletion using Mx1-Cre decreases the expression of markers of osteoblast differentiation, including Runx2 and Osx1 (ref. 18 and Figure 7). Consistent with our findings in that model, FOXOs stimulate Runx2 expression and activity, which in turn promote the transcription of genes responsible for osteoblast differentiation (53, 54). In mice with targeted deletion of Foxos in committed osteoblast progenitors of the present report, however, the markers of osteoblast differentiation were unaffected. This indicates that Foxo deletion in Osx1 cells spared the upstream progenitors that were affected by our earlier Foxo deletion model in which we had used the Mx1-Cre transgene. More important, the difference between the 2 models suggests that while FOXOs restrain the replication of osteoblast progenitors expressing Osx1, they help the maintenance and/or differentiation of earlier osteoprogenitors or perhaps mesenchymal stem cells. In support of this suggestion, FOXOs promote the maintenance of hematopoietic, neuronal, spermatogonial, and embryonic stem cells (20, 21, 55, 56). Furthermore, in both the hematopoietic and neuronal lineages FOXOs constrain the proliferation of lineage-committed progenitors while promoting stem cell maintenance (20, 21). Deletion of Foxos using Mx1-Cre in our earlier work inexorably decreased FOXOs in osteoprogenitors expressing Osx1 and mature osteoblasts. Therefore, the increased osteoprogenitor replication, elucidated in the present report with the deletion of Foxos in Osx1 cells, must have been overridden in that previous model by the adverse effects of the deletion in earlier progenitors. In other words, increased replication of Osx1 cells could not compensate for the decreased supply of earlier progenitors.
In closing, our findings suggest that in lineage-committed osteoblast progenitors expressing Osx1, FOXOs attenuate Wnt signaling, thereby decreasing the number of matrix-synthesizing osteoblasts and amount of bone mass. The elucidation of bone gain in mice in which Foxos were deleted in Osx1-expressing cells as opposed to the osteoporotic phenotype of mice with loss of FOXO function in Msx1 or Col1a1 cells (18, 27) provides strong genetic support for the notion that the overriding function of FOXOs in bone and other tissues is to provide an optimal balance among the maintenance of self-renewing stem cells, the replication of lineage-committed intermediates, and the survival of the terminally differentiated progeny, most likely for the purpose of compensatory adaptations to stresses that accumulate in bone with advancing age. Nonetheless, as is the case with several other defense responses against aging, FoxO activation can eventually aggravate the effects of aging on bone and become a culprit of involutional osteoporosis (57).
Methods
Animal experimentation.
The experimental mice were generated by a 2-step breeding strategy. Hemizygous Osx1-Cre transgenic mice in which a Cre-GFP fusion protein is under the control of Osx1 regulatory elements (8) (mixed background backcrossed into C57BL/6 for 5 generations) were crossed with Foxo1,-3,-4f/f mice (mixture of FVBn and 129Sv), a gift from Ronald DePinho (University of Texas, MD Anderson Cancer Center, Houston, Texas, USA) (14), to generate mice heterozygous for Foxo1, -3, and -4 conditional alleles with and without the Cre allele. The Foxo1,-3,-4f/+;Osx1-Cre were backcrossed with the Foxo1,-3,-4f/f mice for 2 successive generations to obtain the experimental Foxo1,-3,-4f/f;Osx1-Cre mice and the Foxo1,-3,-4f/f control littermates. For subsequent studies, we intercrossed the Foxo1,-3,-4f/f mice with and without the Cre alleles. To generate the wild-type and Osx-Cre control mice in a mixed background similar to that of the Foxo1,-3,-4f/f;Osx1-Cre mice, Foxo1,-3,-4f/+ mice were bred with Osx1-Cre mice and the resulting progeny carrying Foxo wild-type alleles were intercrossed for 2 generations. Genotypes of the offspring were determined by PCR using primers specific for Cre 5′-GCGGTCTGGCAGTAAAAACTATC-3′ and 5′-GTGAAACAGCATTGCTGTCACTT-3′, product size 102 bp, and that detected the wild-type and floxed Foxo1, -3, and 4 alleles (14). To induce Foxo1, -3, and -4 deletion after birth, adult Foxo1,-3,-4f/f;Osx1-Cre and Foxo1,-3,-4f/f mice were fed a diet containing doxycycline (Bio-Serv) beginning 1 week before breeding to suppress expression of the Cre transgene in the resulting offspring. After birth, the offspring were maintained on the doxycycline-containing diet until 3 months of age, when they were switched to regular chow or a diet containing 100 mg kg–1 NAC for 4.5 months. To quantify bone formation, mice were injected with tetracycline (15 mg/kg body weight) 8 and 4 days before harvesting. ROS were quantified as previously described (30) using bone marrow cells flushed from femurs and washed with PBS. The marrow content of reduced GSH was determined using a kit from Cayman Chemical.
Bone imaging and histology.
DXA BMD measurements were performed using a PIXImus densitometer (GE Lunar) as previously described (58). Femur and vertebra microarchitecture were analyzed with μ-CT (model μCT40; Scanco Medical). The lumbar vertebrae (L1–L3) were fixed in 10% Millonig’s formalin, transferred to 100% ethanol, and embedded undecalcified in methyl methacrylate. Histomorphometric examination was performed in longitudinal sections using the OsteoMeasure Analysis System (OsteoMetrics Inc.) as previously described (2, 59). Static and dynamic histomorphometry measurements of the cancellous bone were restricted to the secondary spongiosa. To visualize cartilage and enumerate adipocyte, femurs were decalcified with 14% EDTA and embedded in paraffin;longitudinal sections were stained with Safranine-O or H&E, respectively. Approximately 200 adipocytes were counted in 2.17 ± 0.47 mm2 tissue area of the secondary spongiosa. To quantify proliferating osteoblasts, mice were implanted with BrdU pellets (25 mg/pellet; Innovative Research of America) subcutaneously 4 days before harvest. BrdU incorporation was detected on paraffin-embedded L4–L5 vertebral sections using a BrdU-staining kit (Invitrogen).
Cell culture.
Periosteal cells scraped from femora of 7-week-old mice and osteoblastic cells derived from neonatal calvaria, as described previously (60), were cultured for 8 days in α-MEM supplemented with 10% FBS and 1% PSG. Proliferation was assayed by BrdU incorporation with a kit from Roche Diagnostics. CFU-OB number was determined in the bone marrow pooled from 3–5 mice/genotype as previously described (61), using guinea pig feeder cells (62), 15% FBS, and 1 mM ascorbate-2-phosphate. Half of the medium was replaced every 5 days. CFU-OBs were enumerated at 25 days of culture after von Kossa staining. For Alizarin red assay, bone marrow cells were seeded in 12-well tissue culture plates at 5 × 106 cells per well and cultured for 3 weeks; the mineralized matrix was stained with 40 mM Alizarin red solution. To visualize alkaline phosphatase activity, stromal bone marrow cells were cultured for 3 days in the presence of ascorbate and stained with a kit from Sigma-Aldrich (86R-1). For adipogenesis, stromal bone marrow cells were cultured for 6 days in the presence of 1 μM rosiglitazone and adipocytes enumerated after oil red O staining. For all assays cells, were plated in triplicate.
RNA isolation and quantitative RT-PCR analysis.
Soft tissues from mice were frozen immediately upon harvest. Osteocyte-enriched bone was prepared as described previously (63). Total RNA was extracted from tissues and cultured cells using Ultraspec (Biotecx Laboratories), and the mRNA was reverse-transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems) according to the manufacturer’s instructions. Primers and probes for the different genes were manufactured by the TaqMan Gene Expression Assays service (Applied Biosystems). The mRNA levels were calculated by normalizing to the housekeeping gene ribosomal protein S2 using the ΔCt method (64). Details can be found in Supplemental Methods.
Immunoprecipitation and Western blot.
The nuclear fraction was isolated from the total cell lysates using the Nuclear Extraction Kit (Active Motif). To analyze β-catenin–TCF-4 interactions, cell lysates were immunoprecipitated with an anti–TCF-4 antibody (#2569S; Cell Signaling) or an IgG nonspecific control antibody (sc-2003; Santa Cruz Biotechnology Inc.). Immunoprecipitates were resolved by SDS-PAGE, and coimmunoprecipitating β-catenin was analyzed by Western blotting with anti–β-catenin antibody (#610154; BD Transduction Laboratories). Antibodies against FOXO1, FOXO3, FOXO4 ( #2880, #9476 and #9472, respectively; Cell Signaling), lamin A (sc-20680; Santa Cruz Biotechnology Inc.), and β-actin (sc-4778; Santa Cruz Biotechnology Inc.) were used to detect their corresponding protein levels in cell lysates. Phosphorylated p66Shc was quantified in vertebral bone extracts by Western blotting using a mouse monoclonal antibody that recognizes Ser36 phosphorylated p66Shc (EMD Biosciences; Calbiochem).
Isolation of Osx1-GFP cells and microarray analysis.
Calvaria cells isolated from 3–4 day old Foxo1,-3,-4f/f;Osx1-Cre or Osx1-Cre control mice, generated as described above, were cultured in T175 flasks in the presence of 1% ascorbic acid. At approximately 80% confluence, GFP-positive cells were sorted using a BD FACS Aria Cell Sorter (BD Biosciences). Calvaria cells from wild-type mice were used as controls to set the gates for sorting. The sorted cells were cultured in triplicate until 90% confluence, and caspase-3 activity was quantified by determining the degradation of the fluorometric substrate DEVD (Biomol Research Labs). Protein concentration was measured using a Bio-Rad detergent–compatible kit, as described previously (65).
Cultured sorted cells were also harvested for RNA isolation as described above, and 1 μg total RNA per sample was hybridized to MouseRef-8 v1 Expression Beadchips (Illumina) following protocols listed on the Gene Expression and Genomics Unit of the National Institute on Aging ( http://www.grc.nia.nih.gov/branches/rrb/dna/index/protocols.htm). Microarray florescent signals were extracted using an Illumina BeadArray 500GX reader. The signals on each sample are normalized by log z transformation to obtain z-scores and tests for distributions as previously described (66). Correlation analysis, sample clustering analysis, and principal component analysis including all probes were performed to identify/exclude any possible outliners. The resulting data set was next analyzed with DIANE 6.0, a spreadsheet based microarray analysis program, value statistic for z-score reliability below 0.05, and mean background-corrected signal intensity greater than zero.
Adenovirus infection and β-catenin silencing.
Calvaria cells from wild-type or Foxo1,-3,-4f/f mice were infected with adenovirus encoding Cre recombinase (Ad-Cre) (Vector Biolabs) at an MOI of 30 for 6 hours and allowed to recover for 72 hours. The cells were trypsinized and replated in 6-well plates for RNA analysis. For protein analysis, cells were seeded in 10-cm dishes and at approximately 60% confluence were serum starved and treated with vehicle or recombinant 50 ng/ml Wnt3a (R&D Systems) for 6 hours. For proliferation assays, cells were seeded in 96-well plates and treated with vehicle or Wnt3a for 3 days.
For silencing β-catenin, lentivirus vectors expressing shRNAs for β-catenin (Mission RNAi; Sigma-Aldrich) and control nontargeted shRNA transduction particles were coinfected with Ad-Cre for 6 hours, allowed to recover, and placed under puromycin selection for 10 days. At approximately 80% confluence, cells were trypsinized, replated in 12-well or 96-well plates, and treated with vehicle or Wnt3a overnight for RNA and proliferation assay.
Statistics.
Group mean values were compared as appropriate by Student’s 2-tailed t test or 1-way ANOVA with Bonferroni’s multiple comparison test after determining that the data were normally distributed and exhibited equivalent variances. Longitudinal BMD measurements were analyzed using the procedure for mixed effects models in the SAS software package. Comparisons of BMD values among experimental groups at individual time points in the longitudinal studies were based on differences in least squares means unadjusted for multiple comparisons. For the experiment in which FOXOs were deleted since conception (Figure 2C), some animals did not have measurements taken at baseline. To facilitate the statistical modeling, baseline measurements for these animals were imputed based on the slope between baseline and 3-month measurements of animals in the same experimental group for which measurements at both time points were available. P ≤ 0.05 was considered significant for all statistical comparisons.
Study approval.
The protocols involving genetically modified mice and their wild-type littermates were approved by the Institutional Animal Care and Use Committees of the University of Arkansas for Medical Sciences and the Central Arkansas Veterans Healthcare System.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01 AR56679, P01 AG13918, and R01 AR49794, F32 AR061956); Department of Veterans Affairs grants from the Biomedical Laboratory Research and Development Service of the VA Office of Research and Development to R.L. Jilka. (I01 BX000514), R.S. Weinstein (I01 BX000436), C.A. O’Brien (I01 BX000294), and S.C. Manolagas (I01 BX001405); the University of Arkansas for Medical Sciences Translational Research Institute and Tobacco Settlement funds. E. Ambrogini was supported by a fellowship from the University of Pisa, Italy. R. de Cabo is funded by the Intramural Research Program of the National Institute on Aging. We are grateful to R. DePinho for providing the Foxo1,-3,-4f/f and Foxo3f/f mice. We thank A. Warren, A. DeLoose, S. Berryhill, and J. Crawford for technical assistance and Leah Elrod for help with the preparation of the manuscript.
Footnotes
Conflict of interest: Stavros C. Manolagas serves on the scientific advisory board (SAB) of Radius Health Inc. He has ownership of equity in this company and is compensated for his SAB service.
Citation for this article: J Clin Invest. 2013;123(8):3409–3419. doi:10.1172/JCI68049.
References
- 1.Parfitt AM. The Osteoblast and Osteocyte. 1990. Bone-forming cells in clinical conditions. In: Hall BK, ed. Bone. Volume 1. ; pp. 351–429. Boca Raton, Florida, USA: Telford Press and CRC Press; [Google Scholar]
- 2.Weinstein RS, Jilka RL, Parfitt AM, Manolagas SC. Inhibition of osteoblastogenesis and promotion of apoptosis of osteoblasts and osteocytes by glucocorticoids: potential mechanisms of their deleterious effects on bone. J Clin Invest. 1998;102(2):274–282. doi: 10.1172/JCI2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park D, et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10(3):259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolagas SC. From estrogen-centric to aging and oxidative stress: a revised perspective of the pathogenesis of osteoporosis. Endocr Rev. 2010;31(3):266–300. doi: 10.1210/er.2009-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monroe DG, McGee-Lawrence ME, Oursler MJ, Westendorf JJ. Update on Wnt signaling in bone cell biology and bone disease. Gene. 2012;492(1):1–18. doi: 10.1016/j.gene.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prestwich TC, MacDougald OA. Wnt/β-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol. 2007;19(6):612–617. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133(16):3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 9.Essers MA, Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308(5725):1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 10.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366(6454):461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 11.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278(5341):1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 12.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389(6654):994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 13.van der Vos KE, Coffer PJ. FOXO-binding partners: it takes two to tango. Oncogene. 2008;27(16):2289–2299. doi: 10.1038/onc.2008.22. [DOI] [PubMed] [Google Scholar]
- 14.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128(2):309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8(6):440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 16.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20(2):126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 18.Ambrogini E, et al. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab. 2010;11(2):136–146. doi: 10.1016/j.cmet.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obsil T, Obsilova V. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene. 2008;27(16):2263–2275. doi: 10.1038/onc.2008.20. [DOI] [PubMed] [Google Scholar]
- 20.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Paik JH, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5(5):540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renault VM, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5(5):527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuchiya K, et al. Expanded granulocyte/monocyte compartment in myeloid-specific triple FoxO knockout increases oxidative stress and accelerates atherosclerosis in mice. Circ Res. 2013;112(7):992–1003. doi: 10.1161/CIRCRESAHA.112.300749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almeida M, Han L, Martin-Millan M, O’Brien CA, Manolagas SC. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting beta-catenin from T cell factor- to forkhead box O-mediated transcription. J Biol Chem. 2007;282(37):27298–27305. doi: 10.1074/jbc.M702811200. [DOI] [PubMed] [Google Scholar]
- 25.Hoogeboom D, Essers MA, Polderman PE, Voets E, Smits LM, Burgering BM. Interaction of FOXO with beta-catenin inhibits beta-catenin/T cell factor activity. J Biol Chem. 2008;283(14):9224–9230. doi: 10.1074/jbc.M706638200. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, et al. Wnt signaling regulates hepatic metabolism. Sci Signal. 2011;4(158):ra6. doi: 10.1126/scisignal.2001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rached MT, et al. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab. 2010;11(2):147–160. doi: 10.1016/j.cmet.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maes C, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19(2):329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davey RA, et al. Decreased body weight in young Osterix-Cre transgenic mice results in delayed cortical bone expansion and accrual. Transgenic Res. 2011;21(4):885–893. doi: 10.1007/s11248-011-9581-z. [DOI] [PubMed] [Google Scholar]
- 30.Almeida M, et al. Skeletal involution by age-associated oxidative stress and its acceleration by loss of sex steroids. J Biol Chem. 2007;282(37):27285–27297. doi: 10.1074/jbc.M702810200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferron M, et al. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell. 2010;142(2):296–308. doi: 10.1016/j.cell.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potente M, et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115(9):2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song L, Liu M, Ono N, Bringhurst FR, Kronenberg HM, Guo J. Loss of wnt/beta-catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J Bone Miner Res. 2012;27(11):2344–2358. doi: 10.1002/jbmr.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manolagas SC, Almeida M. Gone with the Wnts: beta-catenin, T-cell factor, forkhead box O, and oxidative stress in age-dependent diseases of bone, lipid, and glucose metabolism. Mol Endocrinol. 2007;21(11):2605–2614. doi: 10.1210/me.2007-0259. [DOI] [PubMed] [Google Scholar]
- 35.Hoogeboom D, Burgering BM. Should I stay or should I go: β-catenin decides under stress. Biochim Biophys Acta. 2009;1796(2):63–74. doi: 10.1016/j.bbcan.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Decarolis NA, Wharton KA, Jr, Eisch AJ. Which way does the Wnt blow? Exploring the duality of canonical Wnt signaling on cellular aging. BioEssays. 2008;30(2):102–106. doi: 10.1002/bies.20709. [DOI] [PubMed] [Google Scholar]
- 37.Jin T. The WNT signalling pathway and diabetes mellitus. Diabetologia. 2008;51(10):1771–1780. doi: 10.1007/s00125-008-1084-y. [DOI] [PubMed] [Google Scholar]
- 38.Manolopoulos KN, Klotz LO, Korsten P, Bornstein SR, Barthel A. Linking Alzheimer’s disease to insulin resistance: the FoxO response to oxidative stress. Mol Psychiatry. 2010;15:1046–1052. doi: 10.1038/mp.2010.17. [DOI] [PubMed] [Google Scholar]
- 39.Naito AT, Shiojima I, Komuro I. Wnt signaling and aging-related heart disorders. Circ Res. 2010;107(11):1295–1303. doi: 10.1161/CIRCRESAHA.110.223776. [DOI] [PubMed] [Google Scholar]
- 40.Palsgaard J, Emanuelli B, Winnay JN, Sumara G, Karsenty G, Kahn CR. Cross-talk between insulin and Wnt signaling in preadipocytes: role of Wnt co-receptor low density lipoprotein receptor-related protein-5 (LRP5). J Biol Chem. 2012;287(15):12016–12026. doi: 10.1074/jbc.M111.337048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho KK, Myatt SS, Lam EW. Many forks in the path: cycling with FoxO. Oncogene. 2008;27(16):2300–2311. doi: 10.1038/onc.2008.23. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt M, et al. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol. 2002;22(22):7842–7852. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shtutman M, et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96(10):5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398(6726):422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 45.Liu B, Yu HM, Hsu W. Craniosynostosis caused by Axin2 deficiency is mediated through distinct functions of beta-catenin in proliferation and differentiation. Dev Biol. 2007;301(1):298–308. doi: 10.1016/j.ydbio.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talchai C, Xuan S, Kitamura T, DePinho RA, Accili D. Generation of functional insulin-producing cells in the gut by Foxo1 ablation. Nat Genet. 2012;44(4):406–412. doi: 10.1038/ng.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almeida M, Ambrogini E, Han L, Manolagas SC, Jilka RL. Increased lipid oxidation causes oxidative stress, increased PPAR{gamma} expression and diminished pro-osteogenic Wnt signaling in the skeleton. J Biol Chem. 2009;284(40):27438–27448. doi: 10.1074/jbc.M109.023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lecka-Czernik B, Rosen CJ, Kawai M. Skeletal aging and the adipocyte program: New insights from an “old” molecule. Cell Cycle. 2010;9:3648–3654. doi: 10.4161/cc.9.18.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawai M, Rosen CJ. PPARgamma: a circadian transcription factor in adipogenesis and osteogenesis. Nat Rev Endocrinol. 2010;6(11):629–636. doi: 10.1038/nrendo.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Almeida M, Han L, Ambrogini E, Weinstein RS, Manolagas SC. Glucocorticoids and tumor necrosis factor (TNF) alpha increase oxidative stress and suppress WNT signaling in osteoblasts. J Biol Chem. 2011;286:44326–44335. doi: 10.1074/jbc.M111.283481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinstein RS, et al. Endogenous glucocorticoids decrease skeletal angiogenesis, vascularity, hydration, and strength in aged mice. Aging Cell. 2010;9(2):147–161. doi: 10.1111/j.1474-9726.2009.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitamura T, et al. Regulation of pancreatic juxtaductal endocrine cell formation by FoxO1. Mol Cell Biol. 2009;29(16):4417–4430. doi: 10.1128/MCB.01622-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teixeira CC, Liu Y, Thant LM, Pang J, Palmer G, Alikhani M. Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J Biol Chem. 2010;285(40):31055–31065. doi: 10.1074/jbc.M109.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tseng PC, et al. Resveratrol promotes osteogenesis of human mesenchymal stem cells by upregulating RUNX2 gene expression via the SIRT1/FOXO3A axis. J Bone Miner Res. 2011;26(10):2552–2563. doi: 10.1002/jbmr.460. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, et al. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat Cell Biol. 2011;13(9):1092–1099. doi: 10.1038/ncb2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goertz MJ, Wu Z, Gallardo TD, Hamra FK, Castrillon DH. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J Clin Invest. 2011;121(9):3456–3466. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez-Otin C, Biasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Brien CA, Jilka RL, Fu Q, Stewart S, Weinstein RS, Manolagas SC. IL-6 is not required for parathyroid hormone stimulation of RANKL expression, osteoclast formation, and bone loss in mice. Am J Physiol Endocrinol Metab. 2005;289(5):E784–E793. doi: 10.1152/ajpendo.00029.2005. [DOI] [PubMed] [Google Scholar]
- 59.Weinstein RS, et al. Promotion of osteoclast survival and antagonism of bisphosphonate-induced osteoclast apoptosis by glucocorticoids. J Clin Invest. 2002;109(8):1041–1048. doi: 10.1172/JCI14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jilka RL, Weinstein RS, Bellido T, Parfitt AM, Manolagas SC. Osteoblast programmed cell death (apoptosis): modulation by growth factors and cytokines. J Bone Miner Res. 1998;13(apoptosis):793–802. doi: 10.1359/jbmr.1998.13.5.793. [DOI] [PubMed] [Google Scholar]
- 61.DiGregorio G, et al. Attenuation of the self-renewal of transit amplifying osteoblast progenitors in the murine bone marrow by 17β-estradiol. J Clin Invest. 2001;107:803–812. doi: 10.1172/JCI11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuznetsov S, Robey PG. Species differences in growth requirements for bone marrow stromal fibroblast colony formation in vitro. . Calcif Tissue Int. 1996;59:265–270. doi: 10.1007/s002239900121. [DOI] [PubMed] [Google Scholar]
- 63.Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O’Brien CA. Matrix-embedded cells control osteoclast formation. Nat Med. 2011;17(10):1235–1241. doi: 10.1038/nm.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 65.Plotkin LI, Weinstein RS, Parfitt AM, Roberson PK, Manolagas SC, Bellido T. Prevention of osteocyte and osteoblast apoptosis by bisphosphonates and calcitonin. J Clin Invest. 1999;104(10):1363–1374. doi: 10.1172/JCI6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5(2):73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.