Abstract
The aim of the present study is to determine the chemical structure and conformation of DNA adducts formed by incubation of the bioactive form of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), N-acetoxy-PhIP, with a single-stranded 11mer oligodeoxyribonucleotide. Using conditions optimized to give the C8-dG-PhIP adduct as the major product, sufficient material was synthesized for NMR solution structure determination. The NMR data indicate that in duplex DNA this adduct exists in equilibrium between two different conformational states. In the main conformer, the covalently bound PhIP molecule intercalates in the helix, whilst in the minor conformation the PhIP ligand is probably solvent exposed. In addition to the C8-dG-PhIP adduct, at least eight polar adducts are found after reaction of N-acetoxy-PhIP with the oligonucleotide. Three of these were purified for further characterization and shown to exhibit lowest energy UV absorption bands in the range 342–347 nm, confirming the presence of PhIP or PhIP derivative. Accurate mass determination of two of the polar adducts by negative ion MALDI-TOF MS revealed ions consistent with a spirobisguanidino-PhIP derivative and a ring-opened adduct. The third adduct, which has the same mass as the C8-dG-PhIP oligonucleotide adduct, may contain PhIP bound to the N2 position of guanine.
INTRODUCTION
The food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is the most mass abundant of the heterocyclic amines, a group of compounds formed during the cooking of meat and fish (1). Administration of PhIP to rodents results in DNA adduct formation and tumor development at multiple sites (2,3). Consumption of heterocyclic amines has been implicated in human cancers (4,5). Specifically, PhIP is associated with the induction of colon (6), prostate (7) and breast cancer (8). This association is supported by evidence showing DNA adduct formation in colon tissue from cancer patients undergoing surgery who received dietary relevant doses of 14C-labeled PhIP (9). In mammalian cells, PhIP induces primarily single base substitutions, predominantly G→T transversions with lesser amounts of G→A transitions (10,11). In addition, a –1 frameshift hotspot has been observed in a 5′-GGGA-3′ sequence in the Apc gene in rat colon tumors induced by PhIP and in the lacI gene of rat mammary glands (12,13).
PhIP itself is not genotoxic, it must be metabolically activated, primarily by cytochrome P450 1A2-mediated N-hydroxylation, followed by N-acetylation or sulfation, in order to bind to DNA (14,15). N-acetoxy-PhIP, which is ∼20-fold more reactive towards DNA than the N-hydroxylated metabolite (16), is thought to form DNA adducts following non-covalent groove-binding (17) and formation of a nitrenium ion, the ultimate electrophilic species (18,19). Although 32P-postlabeling indicates PhIP forms at least two major and several minor adducts in vivo (20,16), to date only the C8-dG-PhIP adduct has been identified (16,21). It has been suggested that the additional 32P-postlabeled products are adducted oligonucleotides, resistant to enzymatic hydrolysis (22). However, we have previously demonstrated, using fluorescence spectroscopy, that reaction of N-acetoxy-PhIP with calf thymus DNA results in formation of multiple adduct species (23). The structure of these adducts, which could be degradation products of the C8-dG adduct or distinct adducts containing PhIP linked to a site other than the C8 position of guanine, needs to be determined.
The synthesis of well characterized DNA adducts is often the limiting factor in efforts to study the relationship between adduct structure or conformation and biological activity. For detailed structural analysis, milligram amounts of modified oligonucleotides are usually required and, consequently, there are no experimentally determined solution structures available for any of the heterocyclic amine adducts in duplex DNA. Therefore, we have optimized the synthesis and handling of the major PhIP adduct on an 11mer oligonucleotide to enable production of sufficient quantities for NMR studies. We report the existence of two conformations of the C8-dG-PhIP adduct in duplex DNA and the spectroscopic characterization of three additional adducts formed from the in vitro reaction of N-acetoxy-PhIP with the single-stranded oligonucleotide.
MATERIALS AND METHODS
Chemicals and enzymes
Solvents, chemicals and enzymes were obtained from Sigma Chemical Co. (St Louis, MO) unless otherwise stated. N-hydroxy-PhIP was purchased from SRI International (Palo Alto, CA). The C8-dG-PhIP nucleoside adduct standard was prepared according to published methods (16). The oligonucleotide d(CCATCGCTACC) and complementary strand d(GGTAGCGATGG) were synthesized on an Applied Biosystems Model 394 DNA synthesizer using reagents acquired from PE Biosystems and purified by reverse phase HPLC (24). Oligonucleotide sequence and purity was confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) and NMR spectroscopy.
Synthesis of N-acetoxy-PhIP
N-hydroxy-PhIP (83 mg, 0.35 mmol) was dissolved in CH2Cl2 (20 ml) containing acetic acid (760 µl). While this stirring solution was maintained at 0°C, 1.89 ml acetic anhydride was added dropwise over 10 min. After a further 10 min, the reaction was quenched by addition of water (20 ml) and the organic phase was washed (3 × 20 ml H2O), dried (Na2SO4) and concentrated under vacuum. Addition of diethyl ether (10 ml) gave N-acetoxy-PhIP (60 mg, 62% yield) as an orange colored precipitate which was filtered and washed with ether. The N-acetoxy-PhIP, which was ∼83% pure as determined by HPLC, was stored in liquid nitrogen until required for further reactions.
Optimization of the C8-dG-PhIP oligonucleotide adduct synthesis
Complete structural characterization of the C8-dG-PhIP oligonucleotide adduct required large quantities of material. Initial studies were therefore undertaken to determine the reaction conditions that would give maximum yield of this adduct.
Effect of N-acetoxy-PhIP concentration and reaction pH
The general method employed for the synthesis of PhIP oligonucleotide adducts involved gradual addition of N-acetoxy-PhIP dissolved in methanol (1 mg/ml) to a solution of d(CCATCGCTACC) (∼0.078 µmol). To investigate the effect of reaction pH, the oligonucleotide was dissolved in 300 µl of 10 mM sodium citrate buffer containing 1 mM EDTA and 0.1 M NaCl, at either pH 5.0 or 7.0. Methanol was added to give a final reaction volume of 500 µl. In initial experiments, performed at both pH 5.0 and 7.0, the amount of N-acetoxy-PhIP added was varied to give molar ratios of N-acetoxy-PhIP:oligonucleotide of 1:1, 2:1, 5:1, 10:1 and 20:1. Control incubations contained N-acetoxy-PhIP or oligonucleotide only and reactions were carried out for 60 min at 37°C. Aliquots (200 µl) of each reaction were then subject to HPLC analysis using the system described below.
In the series of reactions described above, excess amounts of N-acetoxy-PhIP (>0.078 µmol) resulted in a reduced yield of the C8-dG-PhIP oligonucleotide adduct. This suggests that N-acetoxy-PhIP was either inhibiting C8-dG-PhIP adduct formation or causing it to decompose. To determine whether N-acetoxy-PhIP induced degradation of the C8-dG-PhIP oligonucleotide adduct, a set of five incubations, with varying amounts of N-acetoxy-PhIP were performed. To each sample containing 0.009 µmol of C8-dG-PhIP oligonucleotide adduct (in 50 µl 10 mM sodium citrate, 1 mM EDTA, 0.1 M NaCl pH 7.0 buffer) was added 20 µl of an N-acetoxy-PhIP solution (0.0, 0.028, 0.071, 0.14 or 0.21 µmol in methanol). After 1 h at 37°C, the samples were centrifuged and 20 µl removed for HPLC analysis.
Effect of reaction temperature
To investigate the effect of temperature on adduct yield, equimolar amounts of N-acetoxy-PhIP and oligonucleotide were reacted at temperatures ranging from 0–60°C in 10 mM sodium citrate buffer at pH 7.0, containing 1 mM EDTA and 0.1 M NaCl.
Stability of the single-stranded C8-dG-PhIP oligonucleotide adduct
To assess adduct stability in the buffers utilized, the C8-dG-PhIP oligonucleotide adduct (0.010 µmol) was incubated in 10 mM sodium citrate buffer containing 1 mM EDTA and 0.1 M NaCl, at both pH 7.0 and 5.0, and in 0.05 M ammonium formate pH 5.4 for 4 days. Aliquots (20 µl) were analyzed by HPLC at daily intervals and the amount of C8-dG-PhIP adduct remaining calculated by the integrated peak area.
HPLC separation of single-stranded PhIP oligonucleotide adducts
Reaction mixtures were separated by HPLC using a Shimadzu LC-10A system on either an analytical (250 × 4.6 mm) or semi-preparative (1.4 × 15 cm) Hypersil ODS column (Keystone Scientific, Bellefonte, PA) at flow rates of 1.0 or 3.0 ml/min, respectively, with UV detection at 254 and 360 nm. Elution was performed using a solvent gradient of either 0.05 M ammonium formate pH 5.4 or 20 mM sodium phosphate pH 7.0 buffer and methanol, as described in the figure legends. Adducted oligonucleotides and other PhIP derivatives could be distinguished from unmodified oligonucleotide by virtue of their absorption at 360 nm. Pure adducts required for structural characterization were obtained by repeated HPLC on an analytical column using the sodium phosphate/methanol mobile phase.
Enzymatic digestion
In order to confirm guanine as the site of PhIP binding for each of the adducts, samples of oligonucleotide adduct (20–30 µg) were digested to nucleosides using a method based on that reported by Shibutani et al. for the digestion of 8-oxo-7,8-dihydrodeoxyadenosine adducts (25). DNA samples were dissolved in 500 µl sodium acetate buffer (30 mM, pH 5.3) containing 2 mM 2-mercaptoethanol and 25 µl 20 mM zinc sulfate and incubated at 37°C with 10 U nuclease P1 and 15 U alkaline phosphatase type VII-S, for 2 h. After this time, the pH was adjusted by addition of 100 µl 0.5 M Tris–HCl pH 8.5 and the digestion continued for a further 2 h. The samples were then immersed in boiling water for 3 min then evaporated to dryness under a stream of N2. The residue was extracted with methanol (2 × 500 µl) and the organic phase concentrated and redissolved in 20 µl H2O:methanol (80:20). The digestion products were analysed by HPLC on a Hypersil ODS column using ammonium acetate (50 mM, pH 4.5) to acetonitrile gradient of 2–10% over 18 min, 10–100% over 15 min, then isocratic for 10 min.
Spectroscopic analysis
UV spectra were obtained using a Shimadzu UV-2101PC spectrophotometer. MALDI-TOF MS was performed on a PerSeptive Biosystems Voyager DE-STR equipped with a N2 laser for desorption/ionization and data was obtained in negative ion mode using the reflectron for high resolution and precise mass measurements. The oligonucleotide samples were desalted prior to analysis by drop dialysis on a 0.0025 mm Micropore filter (Millipore) for 1 h. The samples were mixed with a 3-hydroxypicolinic acid matrix and deposited on the sample stage. Accurate mass measurements of the oligonucleotide PhIP adducts were determined with a two point internal calibration using ACTH 1–17 (m/z 2091.0710) and ACTH 7–38 (m/z 3655.9132) [M – H]– peptide ions as the calibrants. For each sample, mass measurements were obtained from ten spectra and the average mass calculated. The mass measurement accuracy is consistently <5 p.p.m. error, which is adequate to determine the elemental composition of the oligonucleotide single-stranded PhIP adducts.
All NMR data sets were recorded on a Varian INOVA 600 MHz spectrometer. The duplex C8-dG-PhIP oligonucleotide adduct and duplex control 11mer DNA were prepared by annealing the modified and complementary strands at 70°C and using NMR to monitor the integrated area of the single proton resonances belonging to each strand. Knowing the concentration of the complementary DNA strand required to give a 1:1 ratio, the extinction coefficient of the C8-dG-PhIP single-stranded 11mer adduct was calculated as 105 356 L/(mol.cm). The one-dimensional proton NMR spectra of the C8-dG-PhIP 11mer duplex adduct (9.1 mg of duplex) and the corresponding control 11mer duplex (9.0 mg of duplex) were recorded in 0.1 M NaCl, 10 mM sodium phosphate and 1 mM EDTA, D2O buffer pH 7.0 at 25°C. The temperature of the sample was calibrated with an external methanol sample. Proton-carbon heteronuclear multiple quantum coherence (HMQC) spectra were recorded in D2O buffer at 25°C. The proton carrier frequency was set on the water resonance at 4.76 p.p.m. with a sweep width of 10 p.p.m., while the 13C carrier frequency was set to 75 p.p.m. with a sweep width of 150 p.p.m. The carbon spectra were referenced relative to external 3-(trimethylsilyl)-propionate using the method described by Bax and Subramanian (26).
RESULTS
Synthesis of N-acetoxy-PhIP
Published procedures for the synthesis of N-acetoxy-PhIP involve acetylation of N-hydroxy-PhIP in water miscible solvents such as methanol or dimethylformamide (21,23). The crude reaction mixture, also containing acetic anhydride and acetic acid, is then reacted directly with DNA, which could result in non-specific acetylation. We report an improved method for the synthesis of this compound, using dichloromethane as the solvent, which enables removal of excess reagents and purification of N-acetoxy-PhIP by aqueous extraction and crystallization from ether. In the solid state this derivative is stable in liquid nitrogen for at least 6 months, which facilitates large scale or repeated adduct syntheses.
Reaction of N-acetoxy-PhIP with d(CCATCGCTACC)
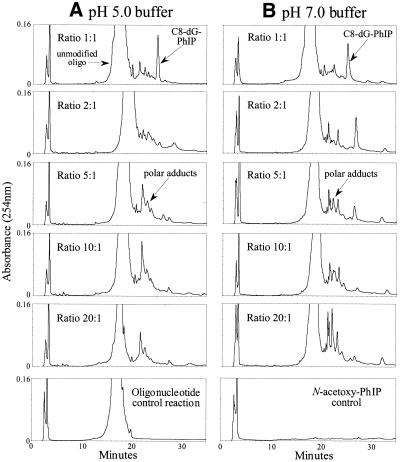
In order to investigate factors affecting adduct profile and yield, two series of reactions were performed in neutral and acidic sodium citrate buffer and the ratio of N-acetoxy-PhIP:oligonucleotide was varied. The reaction products were separated by HPLC and compared (Fig. 1). At both pH 5.0 and 7.0, when an equimolar concentration of N-acetoxy-PhIP was added to the oligonucleotide, one product predominated, which was identified as the C8-dG-PhIP adduct (see below). As the amount of N-acetoxy-PhIP added was increased, there was a reduction in this adduct, accompanied by the formation of more polar adducts. At neutral pH, at least three polar adduct peaks could be resolved following reaction with excess N-acetoxy-PhIP, whilst only one major polar peak was observed at acidic pH. Reaction pH did not appear to significantly affect the yield of C8-dG-PhIP adduct, as estimated by peak area.
Figure 1.
HPLC separation of reaction products formed from the incubation of increasing molar ratios of N-acetoxy-PhIP with d(CCATCGCTACC) at pH 5.0 (A) or pH 7.0 (B). Separations were carried out on a Hypersil C18 ODS column (250 × 4.6 mm) with an ammonium formate to methanol gradient (0–27% over 17 min, then to 31% over a further 28 min and subsequently 100% by 55 min).
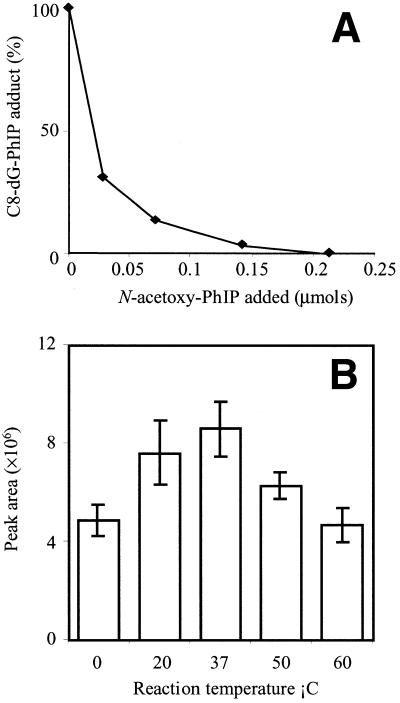
Addition of various amounts of N-acetoxy-PhIP to solutions of purified C8-dG-PhIP oligonucleotide adduct caused a concentration-dependent reduction in the C8-dG-PhIP adduct peak area and formation of a mixture of polar products (Fig. 2A). This confirmed that N-acetoxy-PhIP, or the reactive species derived from this metabolite, can actually induce adduct breakdown. The polar peaks all exhibited UV absorption at 360 nm, which indicates they contain PhIP or a related derivative. Furthermore, the majority of these peaks are more polar than the C8-dG-PhIP adduct, which suggests they are breakdown products rather than oligonucleotide adducts containing a second PhIP moiety, which would be predicted to be less polar.
Figure 2.
(A) N-acetoxy-PhIP induced decomposition of the C8-dG-PhIP oligonucleotide adduct. Various amounts of N-acetoxy-PhIP were added to adduct samples and the amount of adduct remaining determined by HPLC. (B) Effect of reaction temperature on C8-dG-PhIP oligonucleotide adduct yield. Incubations were performed in triplicate at each temperature.
Optimization of reaction conditions and stability of the single-stranded C8-dG-PhIP oligonucleotide adduct
Adduct yield was temperature-dependent, with maximum amounts of the C8-dG-PhIP oligonucleotide adduct formed at 37°C (Fig. 2B). At both higher and lower temperatures there was a reduction in the amount of C8-dG-PhIP formed, whilst the polar adducts increased. It is interesting to note that at the lower temperatures, a red precipitate was visible, possibly due to the production of bis-azo-PhIP, formed from the dimerization of two nitrenes (21). Incubation of the C8-dG-PhIP adduct in sodium citrate buffer (pH 5.0 and 7.0) and ammonium formate (pH 5.4) at 37°C for up to 4 days did not result in any significant decomposition. Peak areas varied by no more than ±1.8% over the period analyzed.
Large scale adduct synthesis requires repeated HPLC separation and concentration of adduct solutions. We find that in order to minimize adduct decomposition during handling, chromatography should be carried out using a sodium phosphate buffer rather than ammonium formate, since the latter causes adduct degradation when concentrated. In addition, adducts should be dried down under a stream of nitrogen and stored in liquid nitrogen. Using optimum reaction conditions, the yield of C8-dG-PhIP oligonucleotide adduct was consistently ∼4.0%, calculated using the extinction coefficient of 105 356 L/(mol.cm), whilst the combined yield of the polar adducts was estimated as being 3.5–4.0%.
Identification of the C8-dG-PhIP oligonucleotide adduct
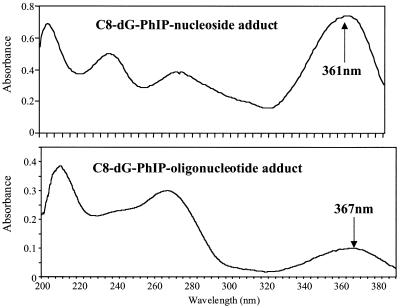
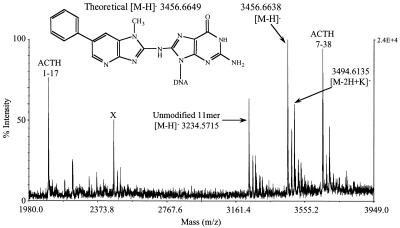
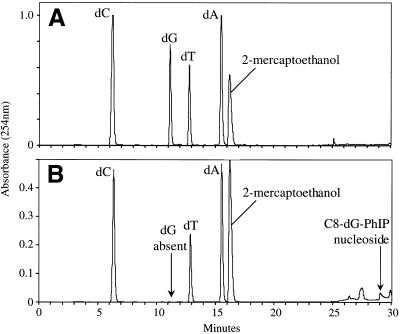
UV analysis of the major adduct peak revealed an absorption band at 367 nm, slightly red shifted relative to the C8-dG-PhIP nucleoside adduct standard (361 nm) (Fig. 3). This may be attributed to interaction of the PhIP ligand with aromatic rings of the DNA bases. The theoretical monoisotopic mass of the single-stranded C8-dG-PhIP oligonucleotide adduct (C117 N41 O64 P10 H145) is 3457.6727 with the [M – H]– ion having a theoretical monoisotopic mass of 3456.6649. The experimental mass of the [M – H]– ion determined by MALDI-TOF MS is 3456.6638 with an absolute error of –0.0011 Da or 0.33 p.p.m. (Fig. 4). Upon enzymatic digestion of the adducted oligonucleotide, unmodified deoxyguanosine was not detected, but several peaks eluting between 26 and 30 min, including one with a retention time identical to that of the C8-dG-PhIP nucleoside standard, were observed (Fig. 5). The additional peaks are probably degradation products of the C8-dG-PhIP adduct formed during enzymatic hydrolysis, since extra peaks were also observed when the nucleoside standard was subjected to the same incubation conditions (data not shown). All these data taken together confirm the structure of the major oligonucleotide adduct as the C8-dG-PhIP adduct.
Figure 3.
Comparison of the UV spectra of C8-dG-PhIP nucleoside adduct with the major oligonucleotide adduct peak. Spectra were recorded in 70% 0.05 M ammonium formate pH 5.4, 30% methanol.
Figure 4.
Accurate mass determination of the single-stranded C8-dG-PhIP oligonucleotide adduct by negative ion MALDI MS. Internal calibration was performed using the two ACTH peptide ions. Unmodified 11mer oligonucleotide was added to the sample for comparison and a third peptide (marked X), was also included but not used for calibration.
Figure 5.
HPLC separation of digested oligonucleotides. (A) Unmodified oligonucleotide. (B) C8-dG-PhIP oligonucleotide adduct. The mercaptoethanol is contained in the sodium acetate reaction buffer.
NMR spectral characteristics of the C8-dG-PhIP 11mer duplex adduct.
While detailed accounts of the NMR spectra and solution conformation of the C8-dG-PhIP 11mer duplex adduct will be published elsewhere (K.Brown, B.E.Hingerty, E.A.Guenther, V.V.Krishnan, S.Broyde, K.W.Turteltaub and M.Cosman, manuscript submitted), we report here some relevant aspects of the NMR characteristics that will not be emphasized in that manuscript. In particular, we present a comparison of the proton and carbon chemical shifts of the PhIP ligand in the nucleoside adduct recorded in DMSO (21), with those in the 11mer duplex adduct in aqueous buffer. The numbering schemes of the PhIP protons and nucleotide residues in the duplex are shown in Figure 6.
Figure 6.
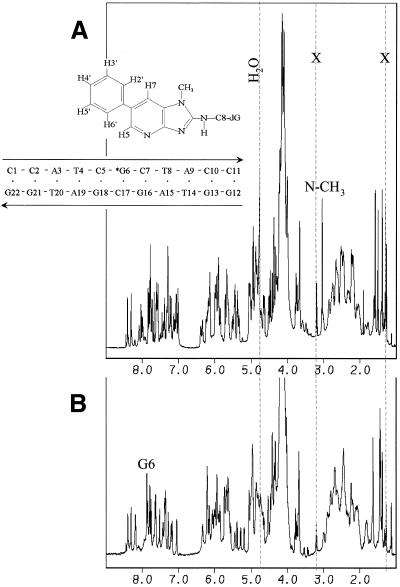
Proton one-dimensional NMR spectra of the C8-dG-PhIP-11mer duplex adduct (A) and control 11mer duplex (B). The residual H2O signal is shown and resonances due to the presence of acetate from DNA purification are indicated with an X. The positions of the G6(H8) proton in the spectrum of the control 11mer duplex (A) and the PhIP(N-CH3) group in the spectrum of the 11mer duplex adduct are also indicated.
The full, one-dimensional proton NMR spectra in D2O buffer of the C8-dG-PhIP 11mer duplex adduct and the corresponding 11mer unmodified duplex (Fig. 6) exhibit narrow and well resolved resonances. The dG6(H8) resonance at 7.86 p.p.m. in the unmodified control duplex (Fig. 6B), is not present, as expected, in the PhIP-modified duplex as determined by analyses of two-dimensional NMR experiments (data not shown). The PhIP ligand and the central d(C5-G6PhIP-C7).d(G16-C17-G18) segment undergo an equilibrium between two different conformations, as evidenced by the presence of exchange crosspeaks in the NMR data. The ratio of major to minor conformer present in solution was approximately 90:10. The few DNA resonances that could be observed for the minor conformer exhibit 1H DNA chemical shift values similar to those found for the unmodified control DNA. This result suggests that the modified DNA conformation remains predominantly B-DNA and relatively unperturbed despite the presence of the covalently linked PhIP ligand. In addition, the chemical shift perturbations for the PhIP proton resonances in the minor conformation are closer to their values in the nucleoside adduct, suggesting that the ligand is solvent exposed (Table 1).
Table 1. Comparison of the 1H-13C-chemical shifts pairs in C8-dG-PhIP in the nucleoside adduct in DMSO versus the 11mer duplex adduct in aqueous buffer.
| |
Nucleosidea |
11mer duplex (major) |
11mer duplex (minor) |
| N-CH3 | 3.65–28.11 | 3.04–29.47 | 3.74–31.38 |
| H7 | 8.01–113.42 | 7.09–114.73 | 7.99–ndb |
| H5 | 8.41–139.48 | 7.04–138.07 | 8.40–138.31 |
| H6′/H2′ | 7.76–126.79 | 7.29–130.40 | 7.84–ndb |
| H5′/H3′ | 7.49–128.97 | 7.06–128.21 | 7.81–129.81 |
| H4’ | 7.38–127.34 | 7.29–131.28 | 7.29–131.28 |
aValues reported by Frandsen et al. (21).
bNot detected.
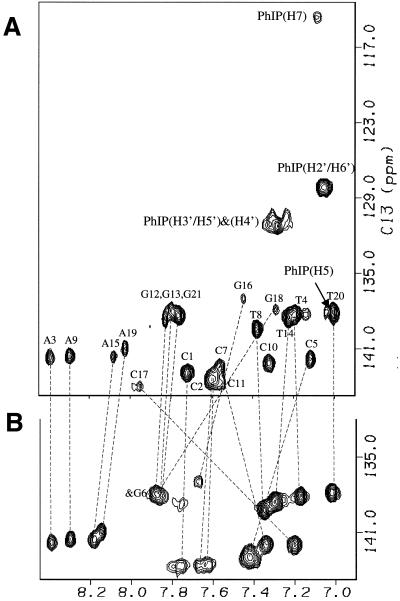
In the major conformation, several significant 1H chemical shift differences between the control and modified DNA major groove base protons located in the central 5 bp segment are evident (e.g. the H6/H8 base protons shown in Fig. 7). In particular, dC5(H6), dG18(H8) and dG16(H8) protons exhibit upfield chemical shifts, while the dC7(H6) and dC17(H6) protons display downfield shifts (Fig. 7) relative to their values in the unmodified 11mer duplex. In addition, the PhIP proton resonances are shifted upfield relative to their values in the nucleoside adduct (Table 1). These unusually shifted proton resonances can be readily accounted for by the ring current contributions of the aromatic guanine purine and PhIP rings. Based on these chemical shift differences between the control and modified DNA, the covalently attached PhIP appears to intercalate between the flanking d(C5-G18) and d(C7-G16) base pairs by displacing the modified G6 base into the major groove for the major adduct conformation (27).
Figure 7.
Expanded contour plots of the aromatic regions of the 1H-13C-HMQC spectra of the C8-dG-PhIP-11mer duplex adduct (A) and control 11mer duplex (B). The relative positions of the base H6/H8 proton in the control and modified 11mer duplex are shown by dashed lines. The position for the PhIP 1H-13C resonances are also shown. Note that the phenyl (H3′ and H5′) and (H2′ and H6′) proton pairs resonate at equivalent positions due to symmetry.
Characterization of polar adducts
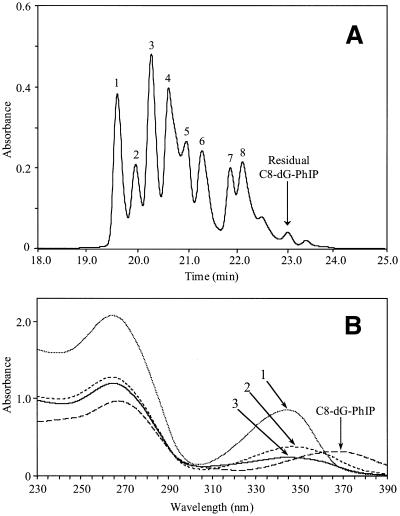
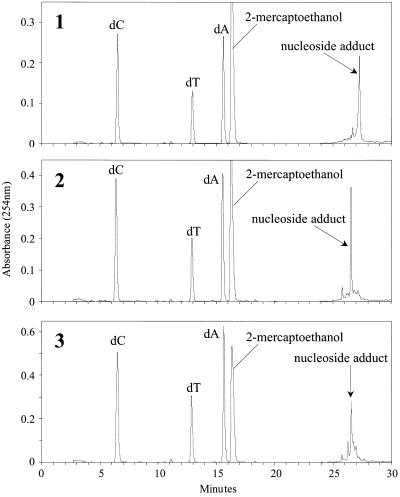
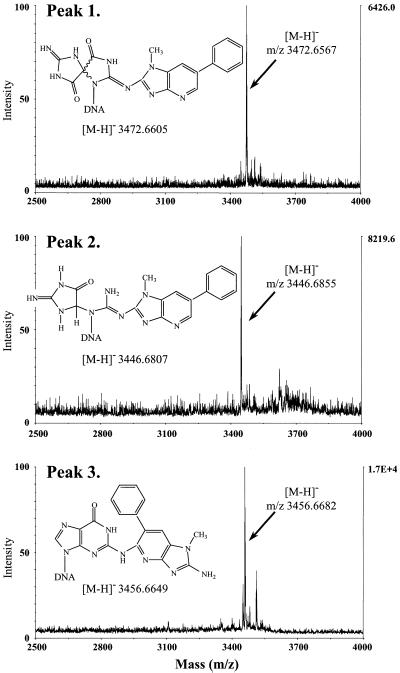
Following large scale synthesis of the C8-dG-PhIP oligonucleotide adduct, the polar adduct peaks collected from these reactions were pooled and separated by HPLC into at least eight products (Fig. 8). The three most polar adducts, numbered 1, 2 and 3, were purified by repeated chromatography on an analytical column for further analysis. All three adducts exhibited lowest energy absorption bands in the range 342–347 nm, confirming the presence of PhIP or PhIP derivative. However, these peak maxima correspond to a substantial shift to lower wavelengths compared to the C8-dG-PhIP adduct. Hydrolysis to nucleosides confirmed guanine as the adducted base for each peak, since unmodified deoxyguanosine was absent upon HPLC analysis of the digests (Fig. 9). Furthermore, for each adduct, a major peak was observed eluting at a retention time similar to the C8-dG-PhIP nucleoside adduct (Fig. 5). These peaks, which all absorb at 360 nm (data not shown) are presumably nucleoside forms of the oligonucleotide adducts. Accurate mass data obtained by MALDI-TOF MS determines the elemental composition of oligonucleotide adducts 1, 2 and 3 and potential structures consistent with this experimental data are shown in Figure 10. Analysis of peak 1 revealed the major [M – H]– ion at m/z 3472.6567 (1.10 p.p.m. error), 16 Da higher than the C8-dG-PhIP adduct and equivalent to the addition of an oxygen atom to this adduct. The proposed structure is that of a spirobisguanidino-PhIP derivative, analogous to the spirodiasteroisomers formed from oxidative degradation of the C8-dG-aminofluorene adduct reported by Shibutani et al. (28). The predominant [M – H]– ion in the spectrum of peak 2 has a m/z of 3446.6855 (1.4 p.p.m. error), ∼10 Da less than the C8-dG-PhIP adduct. Similarly, the proposed structure of this adduct is based on a ring-opened deoxyguanosine-aminofluorene adduct previously described by Shibutani et al. (28). Peak 3 has the same molecular mass as the C8-dG-PhIP oligonucleotide adduct, exhibiting a [M – H]– ion at m/z 3456.6682 (0.97 p.p.m. error). This suggests that adduct 3 contains PhIP linked to an intact guanine via a site other than the C8 carbon, possibly the exocyclic amino group. A potential structure is shown in Figure 10. This adduct was the most difficult to purify since it was not completely resolved from the adjacent peaks by HPLC. Consequently, also present in the mass spectrum are two additional ions of low intensity, due to trace amounts of peak 2 and a product of higher mass, probably derived from adduct peak 4.
Figure 8.
(A) HPLC separation of the polar oligonucleotide adducts. Following large scale synthesis and isolation of the C8-dG-PhIP adduct, fractions containing the polar adducts were combined and separated on a semi-preparative column using a sodium phosphate to methanol gradient (0–27% over 17 min, isocratic for 13 min then to 100% over 5 min). Peaks 1, 2 and 3 were collected and purified by repeated HPLC. (B) Comparison of the UV spectra of oligonucleotide adduct peaks 1, 2 and 3 with the C8-dG-PhIP oligonucleotide adduct. Spectra were recorded in sodium phosphate (20mM, pH 7.0).
Figure 9.
HPLC separation of digested oligonucleotide polar adduct peaks 1, 2 and 3.
Figure 10.
Accurate mass determination of the single-stranded oligonucleotide adduct peaks 1, 2 and 3 by negative ion MALDI MS. Potential structures and values for the theoretical [M – H]– ions are shown.
DISCUSSION
In this study, we investigated factors influencing formation of PhIP DNA adducts and optimized our methods to enable production of sufficient quantities of the C8-dG-PhIP oligonucleotide adduct (∼5 mg) for determination of the NMR solution structure. Initial studies demonstrated the major influence that the N-acetoxy-PhIP:oligonucleotide ratio has on adduct profile and yield. Addition of excess N-acetoxy-PhIP to the oligonucleotide induces decomposition of the C8-dG-PhIP adduct at both neutral and acidic pH. This reduction in C8-dG-PhIP adduct levels was associated with the appearance of increasing amounts of polar products. These early eluting peaks appear to be pH sensitive, with fewer resolvable peaks detectable at pH 5.0 compared to pH 7.0. The maximum amount of adduct was generated by gradual addition of an equimolar concentration of N-acetoxy-PhIP to the oligonucleotide at 37°C. Even under these optimal conditions, the yield of C8-dG-PhIP oligonucleotide was still relatively low, requiring adduct synthesis to be repeated many times to obtain the amounts necessary for NMR analysis. This was made possible by the development of an improved method for the synthesis of N-acetoxy-PhIP, which affords the reactive intermediate as a stable solid.
The single-stranded C8-dG-PhIP oligonucleotide adduct was stable for at least 4 days in neutral and weakly acidic buffer, but decomposed when concentrated under vacuum in the presence of air. Fortunately, concentration of adduct solutions under a stream of nitrogen minimized degradation during the large scale synthesis of the single-stranded C8-dG-PhIP 11mer adduct. Under our digestion conditions, the C8-dG-PhIP adduct partially decomposed to yield several products in addition to the expected nucleoside adduct, which is consistent with reports that the C8-dG-PhIP nucleoside adduct breaks down to various polar derivatives during digestion or hydrolysis of adducted DNA (16). This instability suggests that some of the unidentified adducts detected by 32P-postlabeling of in vitro or in vivo PhIP modified DNA may be derived from degradation of the C8-dG adduct during DNA isolation and processing. Studies are continuing to fully understand the mechanisms involved in decomposition of the C8-dG-PhIP adduct and to elucidate the structures of the resulting products.
Determination of the conformation of the C8-dG-PhIP adduct in duplex DNA by NMR spectroscopy has previously been precluded by the requirement for milligram quantities of highly pure material. In this study, the C8-dG-PhIP oligonucleotide adduct positioned opposite dC in a DNA duplex provides NMR data of sufficient quality to enable us to pursue full structural characterization. However, comparison of the one-dimensional proton spectrum of the adduct duplex with that of the control unmodified duplex shows the presence of a number of minor peaks in addition to the expected number of sharp resonances. Further characterization of the C8-dG-PhIP adduct duplex by two-dimensional homonuclear and heteronuclear NMR spectroscopy revealed the minor peaks were not due to the presence of impurities or decomposition products, but rather to an equilibrium between two conformations (K.Brown, B.E.Hingerty, E.A.Guenther, V.V.Krishnan, S.Broyde, K.W.Turteltaub and M.Cosman, manuscript submitted). Indeed, the adduct remained stable as evidenced by the integrity and high quality of the NMR spectra collected over a 3 week period. The full structural characterization of the major conformer will be published as a separate manuscript. Conformational exchange has previously been observed for several bulky DNA adducts derived from benzo[a]pyrene, acetylaminofluorene and aminofluorene (reviewed in 27,29). Since a site-specific C8-dG-PhIP adduct in a CGC sequence has been shown to generate a variety of point mutations, predominantly G→T transversions with minor amounts of G→A transitions and G→C transversions (30), the presence of two conformers may, in part, explain this variability. The ratio of minor to major conformer for the aminofluorene adducts has also been found to be highly dependent on DNA sequence context (29). Similarly, the identity of neighboring bases near the PhIP modified guanine may also influence the proportion of each conformation that is present, and thus affect the mutational spectra for this adduct.
The reaction of N-acetoxy-PhIP with DNA is known to result in formation of a number of other products in addition to the C8-dG-PhIP adduct, which have not yet been fully characterized (23,31). At the nucleoside level, N-acetoxy-PhIP readily binds to deoxyguanosine but not other bases (16,21), although very low levels of adducts have been detected following incubation with either adenine or cytosine containing polynucleotides (32). Under our optimized reaction conditions, at least eight additional products, more polar than the C8-dG-PhIP adduct were detected. Of these, the first three peaks were sufficiently well resolved to enable purification and further analyses. Whilst enzymatic hydrolysis of peaks 1, 2 and 3 revealed guanine as the modified base for all three adducts, it is feasible that some of the remaining unidentified peaks contain PhIP bound to an alternative nucleoside. The increased polarity of peaks 1, 2 and 3 along with the blue shift of the UV spectra relative to the C8-dG-PhIP oligonucleotide adduct suggests structures with a less conjugated π-electron system, as would result from a decrease in aromaticity of the guanine, such as ring opening. Other aromatic amines including 2-amino-fluorene, 2-amino-3-methylimidiazo[4,5-f]quinoline fluorene (IQ) and 2-amino-3,8-dimethylimidiazo[4,5-f]quinoxaline (MeIQx) bind to the exocyclic amino group of guanine in addition to the C8 carbon (33,34). The N2-dG adducts of these compounds are formed in vitro at lower levels and are typically more polar than C8-dG adducts (33,35). Furthermore, Rindgen et al. (31) have previously detected two isomers of the C8-dG adduct formed from the in vitro reaction of N-acetoxy-PhIP with calf thymus DNA, one of which was described as a potential N2-guanine adduct (31). It is therefore likely that peak 3, which has the same mass as the C8-dG-PhIP oligonucleotide adduct, contains PhIP bound to the N2 position of guanine.
The C8-dG adduct arising from 2-aminofluorene is known to undergo aerial oxidation under alkaline conditions resulting in two spirodiasteroisomers and a ring-opened product (28,36). It is probable that these degradation pathways also occur with other C8-dG substituted carcinogenic amines including PhIP. Accordingly, the accurate mass of polar adduct 1 is consistent with a spiro structure whilst the mass of peak 2 is consistent with the ring-opened structure. In support of these assignments is the identification by Rindgen et al. (31) of a breakdown product of the C8-dG-PhIP adduct, which is believed to arise via decomposition of a ring-opened adduct similar to the structure we have proposed for peak 2. This study represents the first identification of these ring-opened PhIP adducts and provides further evidence for the formation of an N2 adduct. Work is ongoing to complete the characterization of the polar PhIP adducts and to elucidate the pathways involved in their formation.
Acknowledgments
ACKNOWLEDGEMENTS
This work was performed under the auspices of the US DOE by LLNL (W-7405-ENG-48) and supported by NIH grants CA55861 and RR13461.
References
- 1.Layton D.W., Bogen,K.T., Knize,M.G., Hatch,F.T., Johnson,V.M. and Felton,J.S. (1995) Cancer risk of heterocyclic amines in cooked foods: an analysis and implications for research. Carcinogenesis, 16, 39–52. [DOI] [PubMed] [Google Scholar]
- 2.Ito N., Hasegawa,R., Tamano,S., Esumi,H., Takayama,S. and Sugimura,T. (1991) A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Carcinogenesis, 12, 1503–1506. [DOI] [PubMed] [Google Scholar]
- 3.Takayama K., Yamashita,K., Wakabayashi,K., Sugimura,T. and Nagao,M. (1989) DNA modification by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in rats. Jpn. J. Cancer Res., 80, 1145–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wakabayashi K., Nagao,M., Esumi,H., and Sugimura,T. (1992) Food-derived mutagens and carcinogens. Cancer Res., 52 (suppl.), 2091s-2098s. [PubMed] [Google Scholar]
- 5.Felton J.S., Malfatti,M.A., Knize,M.G., Salmon,C.P., Hopmans,E.C. and Wu,R.W. (1997) Health risks of heterocyclic amines. Mutat. Res., 376, 37–41. [DOI] [PubMed] [Google Scholar]
- 6.Ochiai M., Watanabe,M., Kushida,H., Wakabayashi,K., Sugimura,T. and Nagao,M. (1996) DNA adduct formation, cell proliferation and aberrant crypt focus formation induced by PhIP in male and female rat colon with relevance to carcinogenesis. Carcinogenesis, 17, 95–98. [DOI] [PubMed] [Google Scholar]
- 7.Shirai T., Sano,M., Tamano,S., Takahashi,S., Hirose,M., Futakuchi,M., Hasegawa,R., Imaida,K., Matsumoto,K., Wakabayashi,K., Sugimura,T. and Ito,N. (1997) The prostate: a target for carcinogenicity of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) derived from cooked foods. Cancer Res., 57, 195–198. [PubMed] [Google Scholar]
- 8.Sinha R., Gustafson,D.R., Kulldorff,M., Wen,W., Cerham,J.R. and Zheng,W. (2000) 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, a carcinogen in high-temperature-cooked meat, and breast cancer risk. J. Natl Cancer Inst., 92, 1352–1354. [DOI] [PubMed] [Google Scholar]
- 9.Dingley K.H., Curtis,K.D., Nowell,S., Felton,J.S., Lang,N.P. and Turteltaub,K.W. (1999) DNA and protein adduct formation in the colon and blood of humans after exposure to a dietary-relevant dose of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Cancer Epidemiol. Biomarkers Prev., 8, 507–512. [PubMed] [Google Scholar]
- 10.Carothers A.M., Yuan,W., Hingerty,B.E., Broyde,S., Grunberger,D. and Snyderwine,E.G. (1994) Mutation and repair induced by the carcinogen 2-(hydroxyamino)-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-OH-PhIP) in the dihydrofolate reductase gene of Chinese hamster ovary cells and conformational modeling of the dG-C8-PhIP adduct in DNA. Chem. Res. Toxicol., 7, 209–218. [DOI] [PubMed] [Google Scholar]
- 11.Yadollahi-Farsani M., Gooderham,N.J., Davies,D.S. and Boobis,A.R. (1996) Mutational spectra of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) at the Chinese hamster hprt locus. Carcinogenesis, 17, 617–624. [DOI] [PubMed] [Google Scholar]
- 12.Kakiuchi H., Watanabe,M., Ushijima,T., Toyota,M., Imai,K., Weisburger,J.H., Sugimura,T. and Nagao,M. (1995) Specific 5′-GGGA-3′→ 5′-GGA-3′ mutation of the Apc gene in rat colon tumours induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Proc. Natl Acad. Sci. USA, 92, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okochi E., Watanabe,N., Shimada,Y., Takahashi,S., Wakazono,K., Shirai,T., Sugimura,T., Nagao,M. and Ushijima,T. (1999) Preferential induction of guanine deletion at 5′-GGGA-3′ in rat mammary glands by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Carcinogenesis, 20, 1933–1938. [DOI] [PubMed] [Google Scholar]
- 14.Boobis A.R., Lynch,A.M., Murray,S., de la Torre,R., Solans,A., Farré,M., Segura,J., Gooderham,N.J. and Davies,D.S. (1994) CYP1A2-catalyzed conversion of dietary heterocyclic amines to their proximate carcinogens is their major route of metabolism in humans. Cancer Res., 54, 89–94. [PubMed] [Google Scholar]
- 15.Buonarati M.H., Turteltaub,K.W., Shen,N.H. and Felton,J.S. (1990) Role of sulfation and acetylation in the activation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine to reactive intermediates which bind to DNA. Mutat. Res., 140, 61–65. [DOI] [PubMed] [Google Scholar]
- 16.Lin D., Kaderlik,K.R., Turesky,R.J., Miller,D.W., Lay,J.O. and Kadlubar,F.F. (1992) Identification of N-(deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine as the major adduct formed by the food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, with DNA. Chem. Res. Toxicol., 5, 691–697. [DOI] [PubMed] [Google Scholar]
- 17.Marsch G.A., Ward,R.L., Colvin,M. and Turteltaub,K.W. (1994) Non-covalent DNA groove-binding by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Nucleic Acids Res., 22, 5408–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford G.P. and Griffin,G.R. (1992) Relative stabilities of nitrenium ions derived from heterocyclic amine food carcinogens: relationship to mutagenicity. Chem. Biol. Interact., 81, 19–33. [DOI] [PubMed] [Google Scholar]
- 19.Kadlubar F.F. and Beland,F.A. (1985) Chemical properties of ultimate carcinogenic metabolites of arylamines and arylamides, In Harvey,R.G. (ed.), Polycyclic Hydrocarbons and Carcinogenesis, A.C.S Monograph 283. American Chemical Society, Washington, DC, pp. 341–370.
- 20.Mauthe R.J., Marsch,G.A. and Turteltaub,K.W. (1996) Improved high-performance liquid chromatography analysis of 32P-postlabeled 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-DNA adducts using in-line precolumn purification. J. Chromatogr. B., 679, 91–101. [DOI] [PubMed] [Google Scholar]
- 21.Frandsen H., Grivas,S., Andersson,R., Dragsted,L. and Larsen,J.C. (1992) Reaction of the N2-acetoxy derivative of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) with 2′-deoxyguanosine and DNA. Synthesis and identification of N2-(2′-deoxyguanosin-8-yl)-PhIP. Carcinogenesis, 13, 629–635. [DOI] [PubMed] [Google Scholar]
- 22.Pfau W., Brockstedt,U., Söhren,K.D. and Marquardt,H. (1994) 32P-post-labelling analysis of DNA adducts formed by food-derived heterocyclic amines: evidence for incomplete hydrolysis and a procedure for adduct pattern simplification. Carcinogenesis, 15, 877–882. [DOI] [PubMed] [Google Scholar]
- 23.Marsch G.A., Goldman,E.N., Fultz,E., Shen,N.H. and Turteltaub,K.W. (1995) Heterogeneous DNA adduct formation in vitro by the acetylated food mutagen 2-(acetoxyamino)-1-methyl-6-phenylimidazo[4,5-b]pyridine: a fluorescence spectroscopic study. Chem. Res. Toxicol., 8, 659–670. [DOI] [PubMed] [Google Scholar]
- 24.Norman D., Abuaf,P., Hingerty,B.E., Live,D., Grunberger,D., Broyde,S. and Patel,D.J. (1989) NMR and computational characterization of the N-(deoxyguanosin-8-yl)aminofluorene [(AF)G] opposite adenosine in DNA: (AF)G[syn]:A[anti] pair formation and its pH dependence. Biochemistry, 28, 7462–7476. [DOI] [PubMed] [Google Scholar]
- 25.Shibutani S., Bodepudi,V., Johnson,F. and Grollman,A.P. (1993) Translesional synthesis on DNA templates containing 8-oxo-7,8-dihydrodeoxyadenosine. Biochemistry, 32, 4615–4621. [DOI] [PubMed] [Google Scholar]
- 26.Bax A. and Subramanian,S. (1986) Sensitivity-enhanced two-dimensional heteronuclear shift correlation NMR spectroscopy. J. Magn. Reson., 67, 565–570. [Google Scholar]
- 27.Geacintov N.E., Cosman,M., Hingerty,B., Amin,S., Broyde,S. and Patel,D.J. (1997) NMR solution structures of stereoisomeric covalent polycyclic aromatic carcinogen-DNA adducts: principles, patterns and diversity. Chem. Res. Toxicol., 10, 111–146. [DOI] [PubMed] [Google Scholar]
- 28.Shibutani S., Gentles,R.G., Iden,C.R. and Johnson,F. (1990) Facile aerial oxidation of the DNA-base adduct N-(2′-deoxyguanosin-8-yl)-2-aminofluorene [dG(C8)AF]. J. Am. Chem. Soc., 112, 5667–5668. [Google Scholar]
- 29.Patel D.J., Mao,B., Gu,Z., Hingerty,B.E., Gorin,A., Basu,A.K. and Broyde,S. (1998) Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem. Res. Toxicol., 11, 391–407. [DOI] [PubMed] [Google Scholar]
- 30.Shibutani S., Fernandes,A., Suzuki,N., Zhou,L., Johnson,F. and Grollman,A.P. (1999) Mutagenesis of the N-(deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine DNA adduct in mammalian cells. J. Biol. Chem., 274, 27433–27438. [DOI] [PubMed] [Google Scholar]
- 31.Rindgen D., Turesky,R.J. and Vouros,P. (1995) Determination of in vitro formed DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine using capillary liquid chromatography/electrospray ionization/tandem mass spectrometry. Chem. Res. Toxicol., 8, 1005–1013. [DOI] [PubMed] [Google Scholar]
- 32.Snyderwine E.G., Davis,C.D., Nouso,K., Roller,P.P. and Schut,H.A.J. (1993) 32P-postlabeling analysis of IQ, MeIQx and PhIP adducts formed in vitro in DNA and polynucelotides and found in vivo in hepatic DNA from IQ-, MeIQx- and PhIP-treated monkeys. Carcinogenesis, 14, 1389–1395. [DOI] [PubMed] [Google Scholar]
- 33.Turesky R.J., Rossi,S.C., Welti,D.H., Lay,J.O.,Jr and Kadlubar,F.F. (1992) Characterization of DNA adducts formed in vitro by reaction of N-hydroxy-2-amino-3- methylimidiazo[4,5-f]quinoline and N-hydroxy-2-amino-3,8-dimethylimidiazo[4,5-f]quinoxaline at the C-8 and N2 atoms of guanine. Chem. Res. Toxicol., 5, 479–490. [DOI] [PubMed] [Google Scholar]
- 34.Westra J.G., Kriek,E. and Hittenhausen,H. (1976) Identification of the persistently bound form of the carcinogen N-acetyl-2-aminofluorene to rat liver in vivo. Chem. Biol. Interact., 15, 149–164. [DOI] [PubMed] [Google Scholar]
- 35.Shibutani S., Gentles,R., Johnson,F. and Grollman,A.P. (1991) Isolation and characterization of oligodeoxynucleotides containing dG-N2-AAF and oxidation products of dG-C8-AF. Carcinogenesis, 12, 813–818. [DOI] [PubMed] [Google Scholar]
- 36.Johnson F., Huang,C.-Y. and Yu,P.-L. (1994) Synthetic and oxidative studies on 8-(arylamino)-2′-deoxyguanosine and -guanosine derivatives. Environ. Health Perspect., 102 (suppl.), 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]