Abstract
The wide use of paclitaxel and docetaxel in NSCLC clinical treatment makes it necessary to find biomarkers for identifying patients who can benefit from paclitaxel or docetaxel. In present study, NCI-H460, a NSCLC cell line with different sensitivity to paclitaxel and docetaxel, was applied to DNA microarray expression profiling analysis at different time points of lower dose treatment with paclitaxel or docetaxel. And the complex signaling pathways regulating the drug response were identified, and several novel sensitivity-realted markers were biocomputated.The dynamic changes of responding genes showed that paclitaxel effect is acute but that of docetaxel is durable at least for 48 hours in NCI-H460 cells. Functional annotation of the genes with altered expression showed that genes/pathways responding to these two drugs were dramatically different. Gene expression changes induced by paclitaxel treatment were mainly enriched in actin cytoskeleton (ACTC1, MYL2 and MYH2), tyrosine-protein kinases (ERRB4, KIT and TIE1) and focal adhesion pathway (MYL2, IGF1 and FLT1), while the expression alterations responding to docetaxel were highly co-related to cell surface receptor linked signal transduction (SHH, DRD5 and ADM2), cytokine-cytokine receptor interaction (IL1A and IL6) and cell cycleregulation (CCNB1, CCNE2 and PCNA). Moreover, we also confirmed some different expression patterns with real time PCR. Our study will provide the potential biomarkers for paclitaxel and docetaxel-selection therapy in clinical application.
Keywords: Non-small cell lung cancer, paclitaxel, docetaxel, microarray
Inroduction
Non-small cell lung cancer (NSCLC) accounts for approximately 80-85% of all cases of lung cancer [1], the leading cause of cancer-related death in both men and women in the world.Unfortunately, at the time of diagnosis, the majority of patients already have locally advanced or metastatic disease and a systemic, palliative treatment is one of few therapeutic options.
Paclitaxel alone or combination with carboplatin has been approved by FDA to treat advanced or metastatic NSCLC. Docetaxel, the second taxoid derivative, has also shown anti-tumor activity in NSCLC, and numerous clinical trials evaluating the therapeutic effect of docetaxel on NSCLC patients are ongoing (http://www.clinicaltrials.gov/). Actually, cisplatin with docetaxel is one of the most commonly used regimens to treat advanced NSCLC in the UK [2]. Several large clinical trials have shown that, when combined with cisplatin or carboplatin, paclitaxel has similar activity and efficacy with docetaxel, either one offered a significant advantage over another for advanced NSCLC [3-6]. The response rate of paclitaxel or docetaxel-based combination regimens is 16-24% [7], which means that most advanced NSCLC patients do not respond to these chemotherapies. Patients that respond to one of these regimens may also respond to another one: such crossover effects provide evidence of heterogeneity of chemosensitivity in NSCLC [2,8]. Not all NSCLC patients will benefit from the same treatment, and the involved molecular mechanisms are still largely unknown. So, it is urgent to figure out the biomarkers to define patient subsets with distinct sensitivities to paclitaxel or docetaxel.
There are different methods to investigate the potential biomarkers for chemosensitivity prediction in vitro. The mostly used is comparison of gene expression profiles between the drug-resistant strain and its parental strain. As it employs high dose drugs to treat cancer cells and needs almost half a year to construct a drug-resistant strain, it is a time-consuming method. Recently, it was suggested that information on drug dose-dependent effects at a lower, unsaturated range of concentrations would be critically important to understand drug actions in vivo [9,10]. In this method, dynamic changes of gene expression would help us identify novel predictive genes of sensitivity to these agents and the candidate genes that may be targeted to counteract drug resistance and increase therapeutic efficacy of the drugs.
In this study, NCI-H460, a NSCLC cell line showed different sensitivity to paclitaxel and docetaxel, was treated with paclitaxel or docetaxel for 24 or 48 h. And then DNA microarray expression profiling was employed to analyze the complex signaling pathways regulating the response to paclitaxel or docetaxel to identify sensitivity biomarkers of these two taxanes. The dynamic changes of responsive genes showed that the effect of paclitaxel treatment is acute but that of docetaxel is long lasting at least for 48 hours in NCI-H460, which was in line with previous reports [11]. Pathway analysis indicated that distinct pathways were active to respond to the paclitaxel and docetaxel 1548treatment. The differential expression of genes was verified by quantitative real-time PCR (qPCR) and the correlation between DNA microarray-based and qPCR-based gene expression changes was also analyzed.
Materials and methods
Cell culture
NCI-H460 cell line was purchased from ATCC (HTB-177) and maintained in DMEM medium supplemented with 10% FBS (Hyclone), penicillin (100 IU/ml) and Streptomycin (100 μg/ml) (Life Technologies).
MTS assay for NCI-H460 cell viability
NCI-H460 cells (4 × 103) were grown in 100 μl of DMEM medium containing serum per well in a 96-well plate. After 24 h, the cells were treated with paclitaxel (0, 2, 6.3, 20, 63, 200, 630, 2000 nmol/L, respectively) or docetaxel (0, 0.2, 0.63, 2, 6.3, 20, 63 and 200 nmol/L, respectively) for 72 h. Every treatment was triplicate in the same experiment. Then 20 μl of MTS (CellTiter 96 AQueous One Solution Reagent; Promega) was added to each well for 1 to 4 h at 37°C. After incubation, the absorbance was read at a wavelength of 490 nm according to the manufacturer’s protocol. The IC50 calculation was performed with GraphPad Prism 5.0 software.
The concentrations of paclitaxel and docetaxel at which NCI-H460 cell viability was suppressed by 10% or so in 24 h were determined as follow: NCI-H460 cells were treated with paclitaxel (0, 18.5, 58.5, 185, 585 and 1850 nmol/L, respectively) or docetaxel (0, 3, 9, 30, 90 and 300 nmol/L, respectively) for 24 h. Every treatment was triplicate in the same experiment. The cell viability was examined as above mentioned.
The time-course of paclitaxel or docetaxel treatment was carried out as follow: NCI-H460 cells were treated with paclitaxel (6.3 nmol/L) or docetaxel (0.63 nmol/L) for 0, 24, 48 and 72 h, respectively. Every treatment was triplicate in the same experiment. The cell viability was examined as above mentioned.
Microarray analysis
NCI-H460 cells (8 × 104) were grown in 2 ml of DMEM medium containing serum per well in a 6-well plate. After 24 h, the cells were treated with paclitaxel (6.3 nmol/L) or docetaxel (0.63 nmol/L) for 0, 24 and 48 h, respectively. Every treatment was duplicated in the same experiment. All the samples were homogenized with 1 ml Trizol (Invitrogen, Life Technologies) and total RNAs were extracted according to the manufacturer’s instruction.
500 ng total RNA was used to synthesize double-strand cDNA and in vitro transcribed to cRNA, purified 10 μg cRNA was used to synthesize 2nd-cycle cDNA and then hydrolyzed by RNase H and purified. Above steps were performed with Ambion WT Expression Kit. 5.5 μg 2nd-cycle cDNA was fragmented and the single-stranded cDNA was labeled with GeneChip2 WT Terminal Labeling Kit and Controls Kit (Affymetrix, PN 702880). About 700 ng fragmented and labeled single-stranded cDNA were hybridized to an Affymetrix GeneChip Human Gene 1.0 ST array, which was washed and stained with GeneChip2 Hybridization, Wash and Stain kit (Affymetrix).
Microarray data analysis was done using Significance Analysis of Microarrays (SAM) method, as described before [12]. Functional annotation was performed to the differential expression genes with DAVID 6.7 online software.
Quantitative real-time PCR (qPCR)
Total RNA above isolated was synthesized to cDNA using PrimeScript RT reagent kit with gDNA Eraser (Takara, RR074A) for RT-PCR with mixture of oligo-dT and Random Primer (9 mer). The primers used for qPCR validation were list in Table 1. Real-time qPCR was performed on CFX-96 (Bio-lab), with endogenous control hActb. Gene expression was calculated relative to expression of hActb endogenous control and adjusted relative to expression in untreated control cells (paclitaxel) or treated cells (docetaxel at 24 h).
Table 1.
Primers used for qPCR validation
| gene | forward | reverse |
|---|---|---|
| Actb | GCATCCCCCAAAGTTCACAA | GGACTTCCTGTAACAACGCATCT |
| ACTC1 | GTACCCTGGTATTGCTGATCG | CCTCATCGTACTCTTGCTTGCT |
| IGFBP5 | TGACCGCAAAGGATTCTACAAG | CGTCAACGTACTCCATGCCT |
| IL1A | TGTATGTGACTGCCCAAGATGAA | CCTGTGATGGTTTTGGGTATCTC |
| MMP11 | CCGCAACCGACAGAAGAGG | ATCGCTCCATACCTTTAGGGC |
| NDRG1 | CCAACAAAGACCACTCTCCTC | CCATGCCCTGCACGAAGTA |
| TIMP3 | CTACCTGCCTTGCTTTGTGACTT | GGTAACCGAAATTGGAGAGCAT |
Statistical analysis
R2 values were calculated using Pearson’s correlation coefficient. The significant difference was calculated using Student’s t-test.
Results
NCI-H460 cells showed different sensitivity to paclitaxel and docetaxel
To determine chemosensitivity of NCI-H460 to paclitaxel or docetaxel, NCI-H460 cells were treated with the two drugs at seven different concentrations for 72 h, cell viability was examined by MTS assay and IC50 to two drugs were calculated, as described in Materials and Methods. IC50 of NCI-H460 to paclitxel at 72 h is 0.183 μmol/L (R2 = 0.88), to docetaxel at 72 h is 0.030 μmol/L (R2 = 0.93) (Figure 1A). According to data reported in DTP Data Search, the mean IC50 of NCI-60 cell panel to paclitxel is 0.009-0.035 μmol/L, to docetaxel is 0.014-0.034 μmol/L. So, NCI-H460 cell line is sensitive to docetaxel, but shows resistance to paclitaxel.
Figure 1.
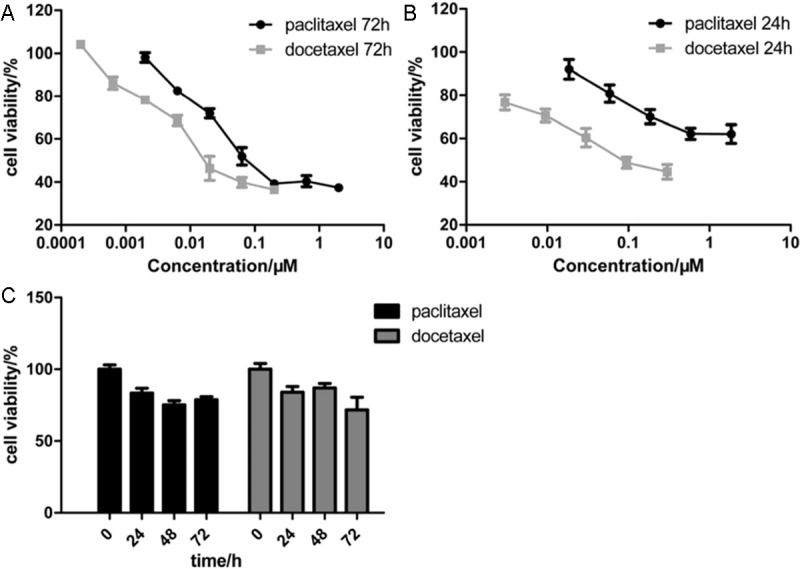
NCI-H460 cells showed different sensitivity to paclitaxel and docetaxel. A: MTS assay for NCI-H460 cells treated with paclitaxel (0, 2, 6.3, 20, 63, 200, 630, 2000 nmol/L, respectively) or docetaxel (0, 0.2, 0.63, 2, 6.3, 20, 63 and 200 nmol/L, respectively) for 72 h. B: MTS assay for NCI-H460 cells treated with paclitaxel (0, 18.5, 58.5, 185, 585 and 1850 nmol/L, respectively) or docetaxel (0, 3, 9, 30, 90 and 300 nmol/L, respectively) for 24 h. C: Time-course curve of lower dose treatment of paclitaxel or docetaxel. NCI-H460 cells were treated with paclitaxel (6.3 nmol/L) or docetaxel (0.63 nmol/L) for 0, 24, 48 and 72 h, respectively. Every treatment was triplicate in the same experiment. Error bars represent the standard deviation (SD).
Furthermore, we examined the IC50 of NCI-H460 cells to paclitaxel or docetaxel at the time point of 24 h. Five different concentrations of paclitaxel or docetaxel were administrated to NCI-H460 cells for 24 h. The IC50 of NCI-H460 to paclitaxel at 24 h is 4.496 μmol/L (R2= 0.86), to docetaxel at 24 h is 0.116 μmol/L (R2 = 0.98) (Figure 1B). The sensitivity difference of NCI-H460 cells to paclitaxel or docetaxel at 24 h is more significant than that at 72 h.
And then, a relatively lower concentration of 6.3 nmol/L of paclitaxel or 0.63 nmol/L of docetaxel was selected to treat NCI-H460 cells for 0, 24, 48 and 72 h. The concentrations were far lower than their corresponding IC50 doses. The time-course curves of paclitaxel and docetaxel treatment were present in Figure 1C. The results showed that when treated with the lower concentrations of paclitaxel or docetaxel for 24 and 48 h, NCI-H460 cell viability was suppressed by 20% or so. At these conditions, cell growth was moderately inhibited and cell apoptosis or death-associated pathways may be not activated severely and that the expression changes of chemosensitivity-involved genes may be relatively marked. Then the cell samples treated with lower concentrations of paclitaxel or docetaxel for 0, 24 and 48 h were collected for DNA microarray analysis.
Gene expression analysis
NCI-H460 cells were treated with 6.3 nmol/L paclitaxel or 0.63 nmol/L docetaxel for 0, 24 and 48 hours (Figure 1C). Following paclitaxel treatment,the expressionof 992 genes were increased (FDR ≤ 0.1%) and only that of 10 genes were decreased (FDR ≤ 0.1%) compared with the untreated cells at 24 h (Figure 2A).While there were 303 genes showed increased expression (FDR ≤ 0.1%) and 667 genes showed decreased expression (FDR ≤ 0.1%) from 24 h to 48 h. When treated with docetaxel, 153 (FDR ≤ 5%) and 1254 genes (FDR ≤ 0.01%) showed increased expression at two time stages: 0-24 h and 24-48 h respectively, yet 72 (FDR ≤ 5%) and 206 genes (FDR ≤ 0.01%) showed decreased expression at the corresponding time stages, respectively. And then we investigated the correlation of gene expression alterations between 0-24 h (stage 1) and 24-48 h (stage 2) after paclitaxel or docetaxel treatment (Figure 2B). The results showed that gene expression alterations induced by paclitaxel in the 2 stages are negatively correlated, which indicated that the cellular response to paclitaxel is fast, and the gene expression has been mostly changed in the first 24 h and then probably came back to its resting state in the second 24 h. But for docetaxel, gene expression alterations in the 2 stages are roughly correlated, which suggested that the cellular response to docetaxel is slow and could last for more than 48 h. So, it is reasonable to compare the differential expression profiles between paclitaxel from 0 to 24 h and docetaxel from 24 to 48 h to observe the distinct genes/pathways responding to these two drugs, respectively.
Figure 2.
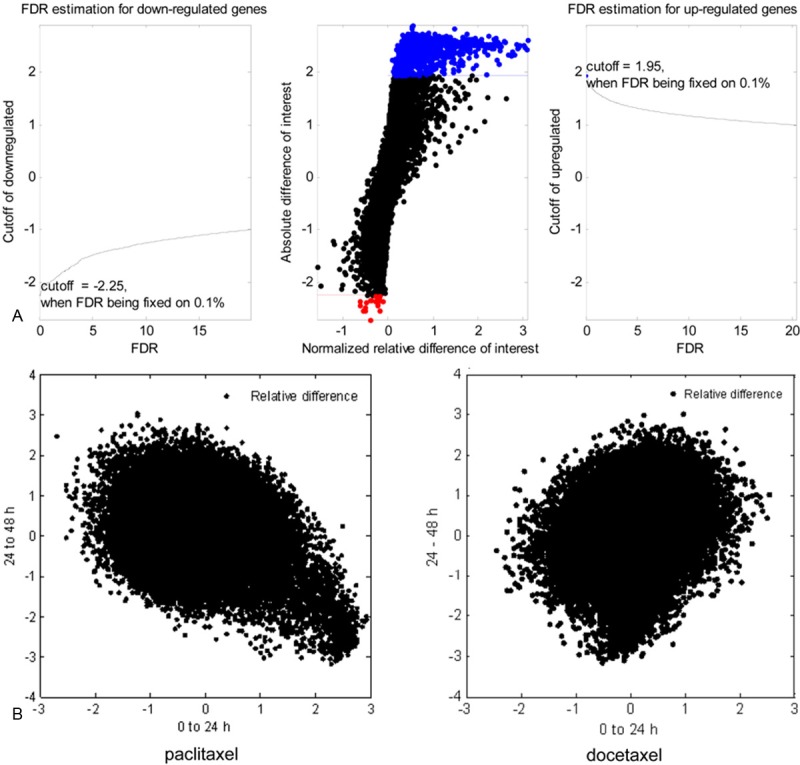
Microarray data analysis. A: Selection of significantly altered genes. The normalized relative difference of gene expression following paclitaxel treatment from 0 to 24 h was calculated by SAM method. Left: for down-regulated genes, when false discovery rate (FDR) was fixed on 0.1%, cutoff value of the normalized relative difference was -2.25. Genes with relative difference lower than -2.25 were identified as significantly down-regulated genes. Right: for up-regulated genes, when FDR was fixed on 0.1%, cutoff value of the normalized relative difference was 1.95. Genes with relative difference higher than 1.95 were identified as significantly up-regulated genes. Middle: criterions above mentioned were applied to all genes on the microarray chip, significantly up-regulated genes were labeled with blue dots and significantly down-regulated genes were dotted with red color. B: Dynamic changes of gene expression patterns between paclitaxel and docetaxel treatment at two time stages. All genes were dotted by their normalized relative difference value between two stages: 0-24 h and 24-48 h.
Functional annotation showed that genes/pathways responding to these two drugs were dramatically different. Gene expression alterations induced by paclitaxel treatment were mainly enriched in actin cytoskeleton (ACTC1, MYL2 and MYH2), cell adhesion pathway, extracellular matrix, regulation of cell migration and tyrosine-protein kinase (ERRB4, KIT and TIE1) (Table 2, Figure 3A), while gene expression changes responding to docetaxel stimulation were enriched in cell surface receptor linked signal transduction (SHH, DRD5 and ADM2), cytokine-cytokine receptor interaction (IL1A and IL6) and cell cycleregulators (CCNB1, CCNE2 and PCNA) (Table 3, Figure 3B).Following paclitaxel treatment (24 h vs 0 h), the most relevant significantly upregulated genes were involved in focal adhesion pathway (MYL2, MYLK2, IGF1, FLT1, ACTN2 and PDGFA) andtyrosine-protein kinase (FLT1, ERBB4, KIT, PDGFRA and TIE1). While after docetaxel treatment (48 h vs 24 h), the most relevant significantly upregulated genes were CCND2 (3.1-fold, p = 0.006), IL1B (2.8-fold, p = 0.0008) and those genes involved in cytokine-cytokine receptor interaction pathway (SHH, DRD5 and ADM2) and neuroactive ligand-receptor interaction pathway (CCKBR, DRD5, DRD4, HRH3, GALR3, HTR6 and GIPR), the most relevant significantly downregulated genes were CCNB1 (3.5-fold, p = 0.0007), CCNE1 (3.3-fold, p = 0.005) and those linked to RNA processing. The different genes/pathways responding to paclitaxel or docetaxel treatment may reflect to some extent the chemosensitivity of NCI-H460 cells to these two drugs.
Table 2.
GSEA for paclitaxel induced genes
| Pathways/Cellular process | P value | FDR/% | Count | Genes |
|---|---|---|---|---|
| actin cytoskeleton | 3.88E-20 | 5.35E-17 | 56 | ACTC1, MYL2, MYH2, MYBPC2, AIF1, MYBPC1, TNNC1, UTRN, PDLIM3, TTN, etc. |
| cell adhesion | 2.69E-16 | 3.89E-13 | 85 | DLC1, C1ORF38, AEBP1, CLSTN2, MYBPC2, MYBPC1, POSTN, MMRN1, CXCL12, etc. |
| myosin complex | 1.76E-13 | 2.42E-10 | 23 | CGNL1, MYBPC2, SHROOM4, MYL2, MYH1, MYL3, MYBPC1, etc. |
| extracellular matrix | 3.92E-12 | 5.40E-09 | 51 | ASPN, LTBP2, FGF9, LTBP4, POSTN, MMP2, WNT2, etc. |
| EGF-like domain | 2.80E-09 | 4.07E-06 | 33 | C7, LTBP2, MASP1, TNC, CD248, LTBP4, PAMR1, C1R, DLK1, etc. |
| Focal adhesion | 6.03E-07 | 7.14E-04 | 30 | CAV3, MYL2, TNC, LAMB2, COL6A3, PIK3CG, FLT1, MYLK2, IGF1, ACTN2, ACTN3, HGF, MAPK10, MYLK, etc. |
| regulation of cell migration | 2.85E-06 | 0.005 | 24 | DLC1, SELP, F10, ACVRL1, FLT1, ERBB4, TBX5, IGF1, KIT, etc. |
| tyrosine-protein kinase | 2.34E-04 | 0.34 | 15 | FLT1, ERBB4, BMX, KIT, DDR2, KDR, BTK, NTRK3, MUSK, DYRK1B, DYRK1A, TEK, PDGFRA, TIE1, CSF1R |
| Tight junction | 0.002 | 2.18 | 17 | MAGI2, MYL2, MYH1, MYH3, MYH2, MYLPF, MYH4, ACTN2, MYH7, MYH6, ACTN3, etc. |
Figure 3.
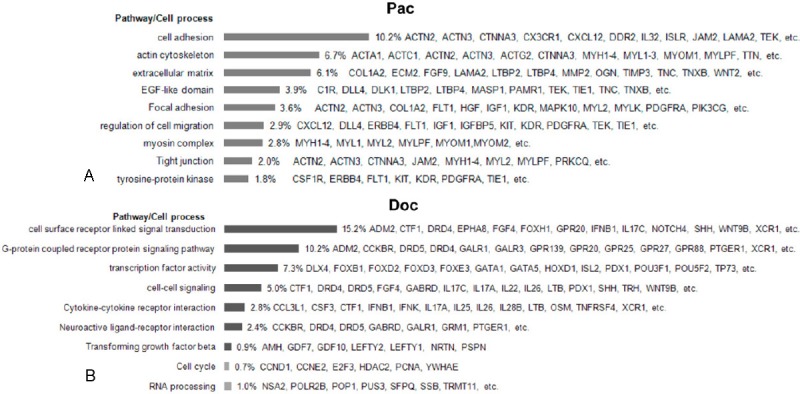
GSEA of genes responding to paclitaxel and docetaxel treatments, respectively, according to pathway/cellular process using online software DAVID 6.7. A: GSEA of genes induced by paclitaxel. B: GSEA of genes induced or repressed by docetaxel. The black column represents the induced pathway/cellular process, while the grey column represents the repressed pathway/cellular process.
Table 3.
GSEA for docetaxel induced or suppressed genes
| Pathway/cellular process | expression | P | Count | Genes |
|---|---|---|---|---|
| cell surface receptor linked signal transduction | Up | 1.21E-09 | 115 | NRTN, GDF7, ADCY8, SHH, S1PR2, GNG8, MARCO, APOA1, GALR1, etc. |
| G-protein coupled receptor protein signaling pathway | Up | 2.57E-08 | 77 | NXPH3, OR2A1, ADCY8, OR2A42, GPR88, GNG8, S1PR2, VN1R3, etc. |
| Cytokine-cytokine receptor interaction | Up | 3.18E-04 | 21 | IL1A, IL6, CSF3, IL2RB, TNFRSF25, TNFRSF13B, CTF1, IL25, IL26, etc. |
| cell-cell signaling | Up | 7.70E-04 | 38 | GAL3ST4, SYT5, OXT, DRD5, CTF1, TH, DRD4, PDX1, TRH, etc. |
| transcription factor activity | Up | 0.001 | 55 | PDX1, HOXD1, CBFA2T3, SOHLH1, GATA1, POU5F2, GATA5, etc. |
| Transforming growth factor beta | Up | 0.002 | 7 | AMH, PSPN, NRTN, GDF7, LEFTY2, GDF10, LEFTY1 |
| Neuroactive ligand-receptor interaction | Up | 0.004 | 18 | GABRD, PTGER1, CCKBR, OPRL1, DRD5, DRD4, GRM1, S1PR2, etc. |
| RNA processing | Down | 9.28E-05 | 9 | HNRNPA1L2, HNRNPM, PUS3, TRMT11, SFPQ, POP1, SSB, POLR2B, NSA2 |
| Cell cycle | Down | 0.009 | 6 | CCNB1, CCNE2, E2F3, HDAC2, PCNA, YWHAE |
FDR of all pathways list in this Table is smaller than 5%.
Validation of microarray results
Quantitative real-time PCR (qPCR) was used to verify the expression of 6 genes differentially expressed after paclitaxel or docetaxel treatment. The expression change determined by microarray and qPCR were showed in Figure 4. For ACTC1, a member of actin cytoskeleton, microarray data showed its expression was induced to 23-fold by paclitaxel but repressed by 19% following docetaxel treatment, qPCR results showed that its expression was increased to 2.4-fold after paclitaxel treatment but suppressed by 23% following docetaxel treatment. Although there was variation in change folds determined by these two methods, the expression trends were consistent. For the other five genes, IL1A, MMP11, NDRG1, TIMP3 and IGFBP5, the situation is the same. Within these genes, ACTC1 was specifically induced by paclitaxel, while IL1A, MMP11 and NDRG1 were specifically induced by docetaxel. For these 6 genes, the average fold change in gene expression levels, as determined by microarray and qPCR analysis, was log2 transformed and the correlation between both data sets examined using R2. An R2 value of 0.84 (P = 0.09) was calculated following paclitaxel treatment. While for docetaxel treatment, R2 value was 0.98 (P = 0.04). Collectively, these data suggest a strong overall concordance of expression trends between the microarray and qPCR data for both paclitaxel and docetaxel-responding genes.
Figure 4.
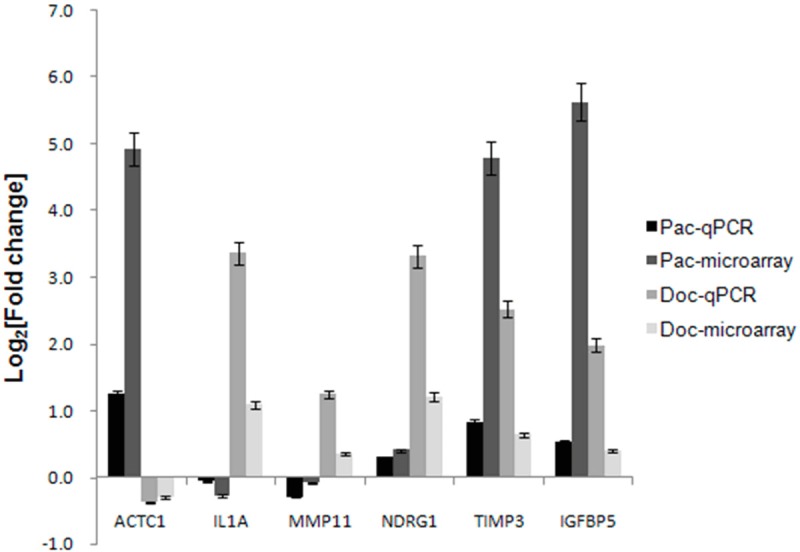
qPCR validation of microarray results. Fold change in six gene expression levels, as determined by microarray and qPCR analysis was log2 transformed in histogram. Pac-qPCR, fold change determined by qPCR after paclitaxel treatment from 0 to 24 h; Pac-microarray, fold change determined by microarray after paclitaxel treatment from 0 to 24 h; Doc-qPCR, fold change determined by qPCR after docetaxel treatment from 24 to 48 h; Doc-microarray, fold change determined by microarray after docetaxel treatment from 24 to 48 h.
Discussion
The clinical wide use of paclitaxel and docetaxel in NSCLC treatment makes it necessary to find biomarkers for identifying patients who can benefit from paclitaxel or docetaxel. In this work, we have used DNA microarray expression profiling to analyze the complex signaling pathways regulating the response to paclitaxel and docetaxel and to identify novel predictive markers for sensitivity to these agents and candidate genes that may be targeted to counteract drug resistance and increase therapeutic efficacy. The dynamic changes of gene expression between two time stages indicated that these two taxanes has different pharmacological mechanism. Subsequent functional annotation also showed that responsive genes/pathways to paclitaxel and docetaxel were totally distinct.
In this study, NCI-H460 cells were treated with different doses of paclitaxel or docetaxel for 72 or 24 h. It showed that NCI-H460 cell line was resistant to paclitaxel but sensitive to docetaxel. This different sensitivity to the two taxanes and hence are suitable for research on the mechanisms leading to this difference. Recently, it was suggested that information on drug dose-dependent effects at a lower, unsaturated range of concentrations would be critically important to understand drug actions in vivo [9]. Another reports showed that dynamic changes of gene expression at different time points following drugs treatment may help us understand deeply into the intrinsic mechanisms of drug effect [10]. Following the previous reports, a concentration of paclitaxel or docetaxel far lower than their IC50 doses at 72 h was used to treat NCI-H460 cells for 0, 24 and 48 h. It is supposed that when treated with these lower drug doses, cell growth was moderately inhibited and cell apoptosis or death-associated pathways may be not activated severely and that the expression changes of chemosensitivity-involved genes may be relatively marked. Actually, our microarray data showed that expressions of those genes linked to cell apoptosis, such as TP53, BAX and BCL2, were not significantly altered after either paclitaxel or docetaxel treatment. This result is in accord with our hypothesis in some ways. It will be more meaningful when two or more cell lines derived from the same type of cancer are used to study drug actions according to this method, i.e., lower dose and several time points treatments.
DNA microarray is a widely used tool for primary screening gene signatures differentially expressed in different tissues or cell lines, sometimes after distinct treatments [13-17]. To some extents, dynamic profiles of gene expression could be used for pharmacological analysis [18]. We found that paclitaxel and docetaxel have different pharmacological mechanisms based on the correlation analysis of differential expression patterns at different time points. The cellular response to paclitaxel is actue, but for docetaxel is slow and takes long time (more than 48 h, see Figure 2B). Our results are consistent with previous reports [11,19]. In clinic, paclitaxel is administrated to patients at 60-80 mg/m2 weekly, while docetaxel is administrated to patients at 75 or 100 mg/m2 every 3 weeks.
Our data showed that the expression alterations of genes/pathways responsive to paclitaxel (0-24 h) were mainly enriched in actin cytoskeleton, myosin complex, cell adhesion, focal adhesion and tight adhesion (Table 2, Figure 3A). It was suggested that expression of β-tubulin isotypes [20], member or regulator of the actin cytoskeleton, such as γ-actin [21] and LIMK2 [22], and the extracellular matrix protein transforming growth factor-β induced (TGFBI) [23] was correlated with paclitaxel sensitivity in various cancers. These results overlap to a considerable degree with our data. The fact that 56 members of actin cytoskeleton were significantly induced by paclitaxel indicated that actin cytoskeleton may be important to mediate paclitaxel sensitivity. Moreover, as the most relevant significantly upregulated genes were involved in focal adhesion pathway after paclitaxel treatment, these genes are also needed to be further studied. Interestingly, microarray data showed that expression of some receptor tyrosine-protein kinases (RTKs), such as ERRB4, KIT and TIE1, was significantly induced by paclitaxel, indicating that RTKs inhibition may sensitize cancer cells to paclitaxel. Report from Coley et al. showed that RTK inhibition really made ovarian cancer cells more sensitive to paclitaxel [24]. The mechanism by which RTK mediates sensitivity to paclitaxel and whether RTK has a role in paclitaxel resistance in cancer types other than ovarian cancer remain to be determined.
Previous studies have demonstrated that p-glycoprotein, CYP3A4, genes linked to cell cycle (HER2, BRCA1 and AURKA), apoptosis (p53, Bcl-2, thioredoxin) and cell proliferation (MIB-1, nuclear grade) were associated to docetaxel sensitivity in breast cancer [25]. Recently, it was reported that genes associated with drug resistance and stemness were correlated to docetaxel resistance in NSCLC cell lines [26]. Our microarray data showed that the expression level changes of gene responding to docetaxel were enriched in cell surface receptor linked signal transduction, cytokine-cytokine receptor interaction, cell cycle regulation and RNA processing (Table 3, Figure 3B). Many genes associated to cytokine-cytokine receptor interaction were induced by docetaxel, such as IL1B, IL26 and IL28B. Recently, Yang reported that TR4-Oct4-IL1Ra axis may play a critical role in the development of chemo-resistance in the prostate cancerstem/progenitor cells [27]. Sakai reported that IL6 scilencing senstisized prostate cancer cell PC3 to docetaxel [28]. So, it is possible that IL1B, IL26 and IL28B can play roles in mediating sensitivity of NSCLC cells to docetaxel. Additional, our data also showed that docetaxel could suppress the expression of some cell cycle genes, such as CCNB1 and CCNE2. As NCI-H460 cells were sensitive to docetaxel, the decreased expression of CCNB1 and CCNE2 may indicate that NCI-H460 cells are on the verge of being arrested in cell cycle.
Collectively, our data suggested that NCI-H460 cells were responded to paclitaxel and docetaxel through totally different pathways, respectively, and these different pathways may be potential biomarkers for paclitaxel and docetaxel. The roles of many interesting genes, such as RTKs and focal adhesion-associated genes for paclitaxel as well as interlukins and cyclins for docetaxel, warrant functional validation in more NSCLC cell lines.
Acknowledgments
We gratefully acknowledge Michael Li, Simon Xiong, and Yi-hui Lin for helpful discussion, William Niu for the technical supports of microarray data analysis.
References
- 1.Gridelli C, Aapro M, Ardizzoni A, Balducci L, De Marinis F, Kelly K, Le Chevalier T, Manegold C, Perrone F, Rosell R, Shepherd F, De Petris L, Di Maio M, Langer C. Treatment of advanced non-small-cell lung cancer in the elderly: results of an international expert panel. J. Clin. Oncol. 2005;23:3125–3137. doi: 10.1200/JCO.2005.00.224. [DOI] [PubMed] [Google Scholar]
- 2.Glaysher S, Yiannakis D, Gabriel FG, Johnson P, Polak ME, Knight LA, Goldthorpe Z, Peregrin K, Gyi M, Modi P, Rahamim J, Smith ME, Amer K, Addis B, Poole M, Narayanan A, Gulliford TJ, Andreotti PE, Cree IA. Resistance gene expression determines the in vitro chemosensitivity of non-small cell lung cancer (NSCLC) BMC Cancer. 2009;9:300. doi: 10.1186/1471-2407-9-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH Eastern Cooperative Oncology G. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 4.Kelly K, Crowley J, Bunn PA Jr, Presant CA, Grevstad PK, Moinpour CM, Ramsey SD, Wozniak AJ, Weiss GR, Moore DF, Israel VK, Livingston RB, Gandara DR. Randomized phase III trial of paclitaxel plus carboplatin versus vinorelbine plus cisplatin in the treatment of patients with advanced non--small-cell lung cancer: a Southwest Oncology Group trial. J. Clin. Oncol. 2001;19:3210–3218. doi: 10.1200/JCO.2001.19.13.3210. [DOI] [PubMed] [Google Scholar]
- 5.Fossella F, Pereira JR, von Pawel J, Pluzanska A, Gorbounova V, Kaukel E, Mattson KV, Ramlau R, Szczesna A, Fidias P, Millward M, Belani CP. Randomized, multinational, phase III study of docetaxel plus platinum combinations versus vinorelbine plus cisplatin for advanced non-small-cell lung cancer: the TAX 326 study group. J. Clin. Oncol. 2003;21:3016–3024. doi: 10.1200/JCO.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 6.Socinski MA, Schell MJ, Peterman A, Bakri K, Yates S, Gitten R, Unger P, Lee J, Lee JH, Tynan M, Moore M, Kies MS. Phase III trial comparing a defined duration of therapy versus continuous therapy followed by second-line therapy in advanced-stage IIIB/IV non-small-cell lung cancer. J. Clin. Oncol. 2002;20:1335–1343. doi: 10.1200/JCO.2002.20.5.1335. [DOI] [PubMed] [Google Scholar]
- 7.Ramalingam S, Belani C. Systemic chemotherapy for advanced non-small cell lung cancer: recent advances and future directions. Oncologist. 2008;13(Suppl 1):5–13. doi: 10.1634/theoncologist.13-S1-5. [DOI] [PubMed] [Google Scholar]
- 8.Pusztai L. Markers predicting clinical benefit in breast cancer from microtubule-targeting agents. Ann Oncol. 2007;18(Suppl 12):xii15–20. doi: 10.1093/annonc/mdm534. [DOI] [PubMed] [Google Scholar]
- 9.Coser KR, Chesnes J, Hur J, Ray S, Isselbacher KJ, Shioda T. Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc Natl Acad Sci USA. 2003;100:13994–13999. doi: 10.1073/pnas.2235866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer J, Allen WL, McLean EG, Wilson PM, McCulla A, Moore S, Longley DB, Caldas C, Johnston PG. Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer. Cancer Res. 2006;66:2765–2777. doi: 10.1158/0008-5472.CAN-05-2693. [DOI] [PubMed] [Google Scholar]
- 11.Crown J, O’Leary M. The taxanes: an update. Lancet. 2000;355:1176–1178. doi: 10.1016/S0140-6736(00)02074-2. [DOI] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arribas AJ, Campos-Martin Y, Gomez-Abad C, Algara P, Sanchez-Beato M, Rodriguez-Pinilla MS, Montes-Moreno S, Martinez N, Alves-Ferreira J, Piris MA, Mollejo M. Nodal marginal zone lymphoma: gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood. 2012;119:e9–e21. doi: 10.1182/blood-2011-02-339556. [DOI] [PubMed] [Google Scholar]
- 14.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW 2nd. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 16.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, Salvatierra O Jr. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Zhang L, Yi Y, Kang HK, Datta SK. Human lupus T cells resist inactivation and escape death by upregulating COX-2. Nat Med. 2004;10:411–415. doi: 10.1038/nm1005. [DOI] [PubMed] [Google Scholar]
- 18.Marcotte ER, Srivastava LK, Quirion R. DNA microarrays in neuropsychopharmacology. Trends Pharmacol Sci. 2001;22:426–436. doi: 10.1016/s0165-6147(00)01741-7. [DOI] [PubMed] [Google Scholar]
- 19.Clarke SJ, Rivory LP. Clinical pharmacokinetics of docetaxel. Clin Pharmacokinet. 1999;36:99–114. doi: 10.2165/00003088-199936020-00002. [DOI] [PubMed] [Google Scholar]
- 20.Rosell R, Scagliotti G, Danenberg KD, Lord RV, Bepler G, Novello S, Cooc J, Crino L, Sanchez JJ, Taron M, Boni C, De Marinis F, Tonato M, Marangolo M, Gozzelino F, Di Costanzo F, Rinaldi M, Salonga D, Stephens C. Transcripts in pretreatment biopsies from a three-arm randomized trial in metastatic non-small-cell lung cancer. Oncogene. 2003;22:3548–3553. doi: 10.1038/sj.onc.1206419. [DOI] [PubMed] [Google Scholar]
- 21.Verrills NM, Po’uha ST, Liu ML, Liaw TY, Larsen MR, Ivery MT, Marshall GM, Gunning PW, Kavallaris M. Alterations in gamma-actin and tubulin-targeted drug resistance in childhood leukemia. J Natl Cancer Inst. 2006;98:1363–1374. doi: 10.1093/jnci/djj372. [DOI] [PubMed] [Google Scholar]
- 22.Dan S, Tsunoda T, Kitahara O, Yanagawa R, Zembutsu H, Katagiri T, Yamazaki K, Nakamura Y, Yamori T. An integrated database of chemosensitivity to 55 anticancer drugs and gene expression profiles of 39 human cancer cell lines. Cancer Res. 2002;62:1139–1147. [PubMed] [Google Scholar]
- 23.Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, Massie CE, Iyer NG, McGeoch A, Crawford R, Nicke B, Downward J, Swanton C, Bell SD, Earl HM, Laskey RA, Caldas C, Brenton JD. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coley HM, Shotton CF, Ajose-Adeogun A, Modjtahedi H, Thomas H. Receptor tyrosine kinase (RTK) inhibition is effective in chemosensitising EGFR-expressing drug resistant human ovarian cancer cell lines when used in combination with cytotoxic agents. Biochem Pharmacol. 2006;72:941–948. doi: 10.1016/j.bcp.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 25.Noguchi S. Predictive factors for response to docetaxel in human breast cancers. Cancer Sci. 2006;97:813–820. doi: 10.1111/j.1349-7006.2006.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasini A, Paganelli G, Tesei A, Zoli W, Giordano E, Calistri D. Specific Biomarkers Are Associated with Docetaxeland Gemcitabine-Resistant NSCLC Cell Lines. Transl Oncol. 2012;5:461–468. doi: 10.1593/tlo.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang DR, Ding XF, Luo J, Shan YX, Wang R, Lin SJ, Li G, Huang CK, Zhu J, Chen Y, Lee SO, Chang C. Increased chemo-sensitivity via targeting testicular nuclear receptor 4 (TR4)-Oct4-interleukin 1 receptor antagonist (IL1Ra) axis in prostate cancer CD133+ stem/progenitor cells to battle prostate cancer. J Biol Chem. 2013;288:16476–83. doi: 10.1074/jbc.M112.448142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai I, Miyake H, Terakawa T, Fujisawa M. Inhibition of tumor growth and sensitization to chemotherapy by RNA interference targeting interleukin-6 in the androgen-independent human prostate cancer PC3 model. Cancer Sci. 2011;102:769–775. doi: 10.1111/j.1349-7006.2011.01854.x. [DOI] [PubMed] [Google Scholar]


