Somatic hypermutation in variable heavy chain rearranged regions is abrogated in the absence of the 3′ regulatory region enhancer, whereas transcription rate in the Ig heavy chain is only partially reduced.
Abstract
Interactions with cognate antigens recruit activated B cells into germinal centers where they undergo somatic hypermutation (SHM) in V(D)J exons for the generation of high-affinity antibodies. The contribution of IgH transcriptional enhancers in SHM is unclear. The Eμ enhancer upstream of Cμ has a marginal role, whereas the influence of the IgH 3′ regulatory region (3′RR) enhancers (hs3a, hs1,2, hs3b, and hs4) is controversial. To clarify the latter issue, we analyzed mice lacking the whole 30-kb extent of the IgH 3′RR. We show that SHM in VH rearranged regions is almost totally abrogated in 3′RR-deficient mice, whereas the simultaneous Ig heavy chain transcription rate is only partially reduced. In contrast, SHM in κ light chain genes remains unaltered, acquitting for any global SHM defect in our model. Beyond class switch recombination, the IgH 3′RR is a central element that controls heavy chain accessibility to activation-induced deaminase modifications including SHM.
During B cell ontogeny and maturation, the IgH locus undergoes successive transcription-coupled remodeling events, including V(D)J recombination, somatic hypermutation (SHM), and Ig class switch recombination (CSR; Henderson and Calame, 1998). RAG-mediated V(D)J recombination occurs during the antigen-independent step of B cell development. Interactions with cognate antigens later recruit activated B cells into germinal centers and can induce various activation-induced deaminase (AID)–mediated modifications. These include SHM in V(D)J exons for the generation of high-affinity antibodies and CSR for the synthesis of non-IgM Ig with diversified effector functions. Because all Ig gene remodeling events require transcription, IgH cis-regulatory regions and especially transcriptional enhancers are major locus regulators. Mouse models carrying targeted genomic deletions highlighted distinct roles for such regions. Deletion of regulatory elements located upstream of Ig constant gene (IgC) clusters affects mostly V(D)J recombination: the intergenic region which lies between the VH and D clusters (IGCR1) regulates ordered and tissue-specific VH to DH-JH rearrangements (Featherstone et al., 2010; Giallourakis et al., 2010; Guo et al., 2011), the 5′ DQ52 enhancer element influences D gene usage (Nitschke et al., 2001; Afshar et al., 2006), and the core Eμ element allows efficient DH to JH recombination (Sakai et al., 1999; Perlot et al., 2005). The IgH 3′ regulatory region (3′RR; encompassing the four transcriptional enhancers hs3a, hs1,2, hs3b, and hs4) located downstream of the IgC cluster is the master element controlling CSR (Vincent-Fabert et al., 2010; Pinaud et al., 2011). None of the previously studied targeted alterations of the endogenous IgH locus has affected SHM. So far the deletion of both IgH 3′ hs3b and hs4 enhancers only resulted in altered CSR (Pinaud et al., 2001; Morvan et al., 2003). Although CSR and SHM both require transcription and AID-mediated DNA lesions (Hackney et al., 2009), a role of the 3′RR on SHM only appeared in transgenes but not in the partial disruptions on the endogenous 3′RR, contrasting with their constant CSR defect; knockout mice lacking the 3′RR hs3b/hs4 elements thus robustly underwent VH gene SHM (Morvan et al., 2003). On the contrary in transgenic experiments, hs3b and hs4 (but not hs1,2) proved able to target SHM onto associated transcribed sequences (Tumas-Brundage et al., 1997; Terauchi et al., 2001; Laurencikiene et al., 2007). A study of large BAC transgenes either lacking or including the whole 3′RR found this region mandatory for V(D)J transcription and for the recruitment of SHM to the transcribed V(D)J segment (Dunnick et al., 2009). Finally, the IgH 3′RR is itself targeted by AID and undergoes SHM in activated B cells (Péron et al., 2012). To clarify the long-range role of the 3′RR on SHM in the context of the endogenous locus, we analyzed IgH 3′RR-deficient mice lacking the 30-kb extent of the 3′RR, a deletion which we previously characterized as inducing a severe CSR defect (Vincent-Fabert et al., 2010), but with normal V(D)J recombination (Rouaud et al., 2012).
RESULTS AND DISCUSSION
SHM at the variable region of the IgH locus is abrogated in IgH 3′RR-deficient mice
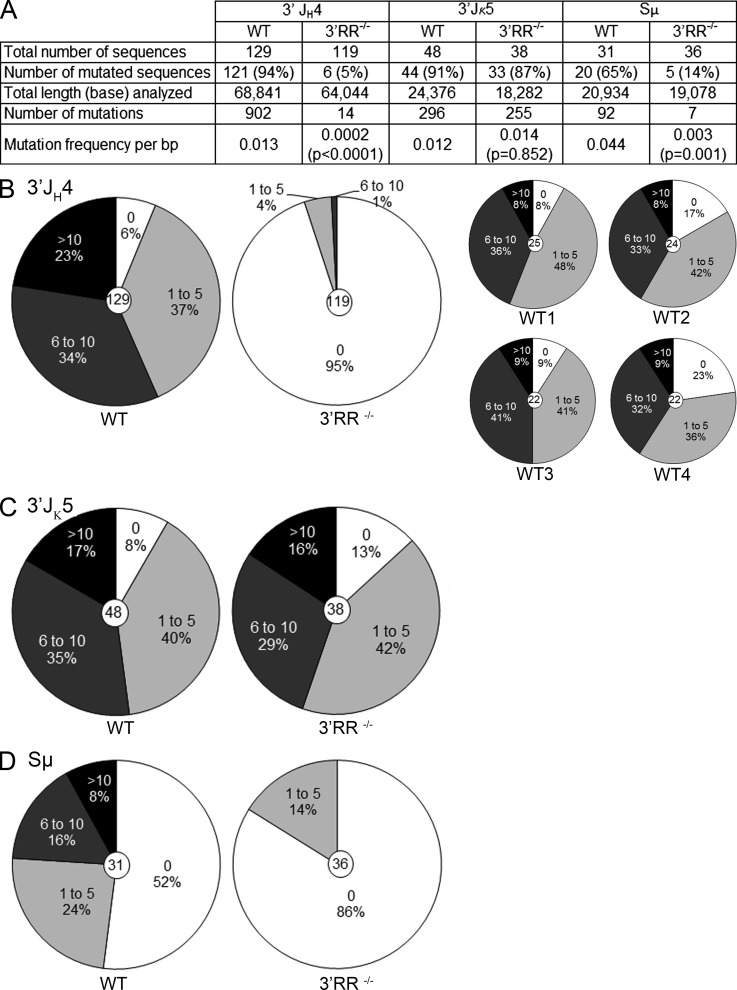
3′RR-deficient mice display normal size B cell compartments but a globally decreased Ig secretion, including a partial IgM defect and culminating in a complete blockade of classed-switched isotypes, a phenotype involving both decreased Ig secretion in plasma cells and decreased CSR frequency (Vincent-Fabert et al., 2010). Because CSR and SHM are well-known temporally and spatially associated processes, both involving AID-initiated DNA lesions (Neuberger and Rada, 2007; Schatz and Ji, 2011), we explored 3′RR-deficient mice for potential SHM defects. Mice were immunized orally with sheep red blood cells for 2 wk and intraperitoneally with 10 µg LPS for 3 d. Oral immunization with sheep red blood cells together with 3-d intraperitoneal injection of LPS corresponds to the immunization protocol that we found the most efficient to regularly obtain in vivo activated B cells in both spleen and Peyer’s patches, with high levels of somatic mutation. We are thus now routinely using this protocol as a standard for in vivo B cell stimulations. Similar occurrence (P > 0.5) of germinal center B cells (B220+PNAhighFas+) were found in Peyer’s patches of 3′RR-deficient and WT mice: 17.6 ± 3.5% vs. 18.6 ± 1.8% of PNAhighFas+ cells among B220+ cells; means obtained from groups of five mice. Numbers of Peyer’s patches were similar in 3′RR-deficient (mean of 7, range 5–10, n = 8) and WT mice (mean of 6, range 4–8, n = 8). The total number of cells in Peyer’s patches was slightly higher in 3′RR-deficient (mean of 22 × 106 cells/mouse, range 13–31, n = 8) than in WT mice (mean of 8 × 106 cells/mouse, range 5–10, n = 8). Rearranged IgH V(D)J regions from B220+PNAhighFas+ cells (10 3′RR-deficient and 8 WT mice) were cloned and sequenced to evaluate the role of the 3′RR in SHM. Ensuring that the reference SHM level in B220+PNAhighFas+ cells pooled from the eight WT mice was not biased by any “jackpot” high SHM in a few mice, four individually evaluated animals under this immunization protocol consistently and homogeneously showed the high SHM rate expected in WT mice (Fig. 1 B, right). A dramatic decrease (P < 0.0001, Mann–Whitney U test) in the relative number of mutated sequences was found in 3′RR-deficient mice (5%, 6/119) as compared with WT (94%, 121/129; Fig. 1 A). The mutation frequency in 3′RR-deficient mice (14 mutations among 64,044 bp analyzed, 0.022%) was compared with WT (902 mutations among 68,841 bp analyzed, 1.3%) and showed a >98% reduction (P < 0.0001, Mann–Whitney U test). When comparing the frequency of mutations per sequence, we found in 3′RR-deficient mice a lower frequency (1% compared with 57% in WT, P < 0.0001) of clones carrying multiple mutations (over five mutations in the analyzed region; Fig. 1 B, left). Although the number of mutations in 3′RR sequences is small, we found that the proportion of mutations at C/G did not significantly differ between WT (43.3%) and 3′RR-deficient mice (35.7%) and that A↔G transitions represented 50.8% and 41.0% for WT and 3′RR-deficient mice, respectively (not depicted). Thus, deletion of the IgH 3′RR nearly abrogates the SHM process in VH regions.
Figure 1.
V(D)J, Ig κ, and Eμ SHM in 3′RR-deficient and WT mice. (A) Expressed IgH V(D)J regions and Cκ regions from B220+PNAhighFas+ cells were cloned and sequenced. Number of mutated sequences, number of mutations, and mutation frequency are reported. Clonally related sequences were excluded. Statistical differences were investigated with the Mann–Whitney U test. Peyer’s patches from 10 3′RR-deficient and 12 WT mice were used for B220+PNAhighFas+ cell purification. Mice were 8 wk old, and immunizations were performed orally with sheep red blood cells for 2 wk and intraperitoneally with 10 µg LPS for 3 d. (B) Mutation analysis in expressed IgH V(D)J regions of 3′RR-deficient mice and WT mice. (left) Expressed IgH V(D)J region B220+PNAhighFas+ cells were cloned and sequenced (12 WT and 10 3′RR-deficient mice). (right) Mutations found in four separated WT mice. (C) Mutation analysis in expressed Cκ regions of 10 3′RR-deficient and 12 WT mice. (D) Mutation analysis in the 5′ end of Sμ. B splenocytes from five 3′RR-deficient and five WT mice were used for B220+PNAhigh cell purification. Mice were 8 wk old, and immunizations were performed orally with sheep red blood cells for 2 wk and intraperitoneally with 10 µg LPS for 3 d.
Expression of AID and SHM-related cofactors are unaffected in 3′RR-deficient mice
We investigated the transcripts encoding several proteins involved in SHM (Neuberger and Rada, 2007). Transcripts for AID, POLη, REV1, UNG, MSH2, and MSH6 were detected in similar amounts (P > 0.1) in Peyer’s patch B cells from 3′RR-deficient and WT mice, showing that at the RNA level, 3′RR-deficient B cells normally express the SHM trans-acting machinery (not depicted). Thus, lack of SHM in IgH V(D)J segments in 3′RR-deficient mice is not related to a deficiency of AID and related cofactor expression.
SHM at the variable region of the Ig κ locus is normal in 3′RR-deficient mice
To further check that the lack of SHM at the heavy chain locus is not related to a global decrease of SHM in 3′RR-deficient mouse B cells, we investigated SHM in the Ig κ locus. A similar number of mutated sequences was found in 3′RR-deficient (87%, 33/38) and WT mouse B cells (91%, 44/48; Fig. 1 A). The mutation frequency in 3′RR-deficient mice (255 mutations among 18,282 bp analyzed, 1.39%) was similar to WT (296 mutations among 24,376 bp analyzed, 1.21%). When comparing the number of mutations per sequence, we found a similar number of clones from 3′RR-deficient cells that carried multiple mutations (i.e., with over five mutations in the analyzed region: 45% for 3′RR-deficient mice compared with 52% in WT; Fig. 1 C). The proportion of transitions did not differ between WT (52.4%) and 3′RR-deficient mice (51.8%), and A↔G transitions represented 43.8% and 34.0% for WT and 3′RR-deficient mice, respectively (not depicted). Thus, deletion of the 3′RR specifically abrogates the SHM process in the heavy chain locus without significantly affecting the κ light chain locus.
IgH transcription is reduced but not abrogated in 3′RR-deficient mice
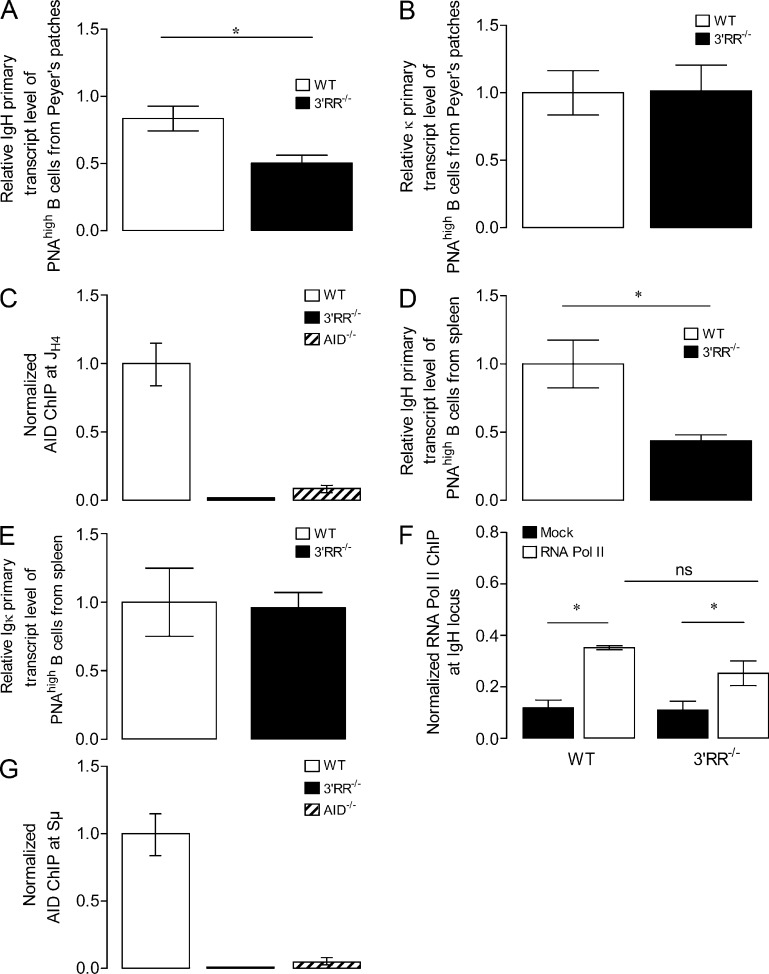
SHM correlates with transcription (Fukita et al., 1998). Alterations of the SHM rate have been reported using BAC transgenes with large deletion of the 3′RR, although transcription of these transgenes was variably affected by the deletion (Dunnick et al., 2009). In contrast, the partial IgH transcription defect observed in hs3b/4-deficient mouse resting B cells did not lead to any significant SHM decrease in germinal center B cells (Morvan et al., 2003). The situation is quite different in 3′RR-deficient mice, which showed a nearly complete V(D)J SHM blockade (98% of reduction) along with only a partial decrease (40%) of IgH primary transcripts in B220+PNAhighFas+ Peyer’s patch B cells (Fig. 2 A). In contrast, deletion of the 3′RR did not affect SHM (Fig. 1 C) and transcription at the κ light chain locus (Fig. 2 B). Our model indicates that Ig heavy chain primary transcription is maintained at a fairly high level in germinal center B cells devoid of 3′RR, although SHM is almost completely abrogated. This suggests that such a strong SHM defect cannot solely result from decreased transcription. Rather, our data suggest that the 3′RR could be directly essential for the recruitment of AID onto the IgH locus, a hypothesis which fits well with recent results showing that the IgH 3′RR is itself an AID target (Péron et al., 2012). To determine whether AID recruitment to the JH4 region is dependent on the 3′RR, we performed chromatin immunoprecipitation (ChIP) analysis using an anti-AID antibody on chromatin prepared from B splenocytes of WT, 3′RR-deficient, and AID-deficient mice (immunized orally with sheep red blood cells for 2 wk and intraperitoneally with 10 µg LPS for 3 d). We found that AID occupancy at the JH4 region was dramatically reduced in 3′RR-deficient mice as compared with WT (Fig. 2 C). Only background levels of immunoprecipitation were found both for 3′RR-deficient and AID-deficient mice, leading to the conclusion that AID enrichment to the IgH JH4 region is drastically impaired in 3′RR-deficient mice. Confirming results obtained with Peyer’s patches, deletion of the 3′RR lead only to a partial decrease (50%) of IgH primary transcripts in B220+PNAhigh splenic B cells (Fig. 2 D) without significant effect on κ primary transcripts (Fig. 2 E). The significant binding of RNA polymerase II on the IgH allele of 3′RR-deficient mice (after 3-d LPS stimulation of splenic B cells in vitro) confirms that transcription is maintained (Fig. 2 F). This work provides the first conclusive evidence that, first, AID-mediated SHM is targeted to the IgH locus by the cis-acting 3′RR and, second, that SHM within Ig genes does not imply parallel transcription. These results are reminiscent of those reported for 3′ regulatory elements located in the chicken Ig light chain (Kothapalli et al., 2008; Blagodatski et al., 2009). Similarly to the IgH 3′RR, the DIVAC (diversification activator) cis-acting element is required for SHM at the chicken Ig light chain gene (Blagodatski et al., 2009) but does not function by increasing transcription (Kohler et al., 2012). Although high levels of transcription are not sufficient for robust SHM in IgL locus (Yang et al., 2006), separated mutational and transcriptional enhancers were recently reported for IgL genes (Kothapalli et al., 2011). The mechanism of how cis-regulatory sequences activate SHM in neighboring transcription units remains speculative. As hypothesized for DIVAC (Blagodatski et al., 2009), the IgH 3′RR might promote the formation of protein complexes that first bind AID and then hand it over to the neighboring transcription initiation complex.
Figure 2.
IgH transcription and AID-ChIP in 3′RR-deficient and WT mice. (A and B) Analysis by real-time PCR of IgH (A) and κ (B) primary transcripts in B220+PNAhighFas+ cells from Peyer’s patches of immunized 3′RR-deficient and WT mice. Transcript levels were normalized to GAPDH transcripts. Results are the mean ± SEM of six WT and six 3′RR-deficient mice. (C) Accessibility of IgH locus downstream of JH4 to AID. ChIP assays were performed with splenic B cells isolated from immunized AID-deficient, 3′RR-deficient, and WT mice. For each sample, AID-Chip values were normalized to the input control, and AID-Chip signal in WT B cells was assigned an arbitrary value of 1. Data presented are from one primer pair and error bars corresponding to technical replicates. One representative experiment out of two is shown. (D and E) Analysis by real-time PCR of IgH (D) and κ (E) primary transcripts in B220+PNAhigh splenic B cells of immunized 3′RR-deficient and WT mice. Results are the mean ± SEM of four WT and five 3′RR-deficient mice. (F) Accessibility of IgH locus to RNA polymerase II (Pol II). ChIP assays were performed with 3-d LPS-stimulated splenic B cells isolated from 3′RR-deficient and WT mice. The relative enrichments (percent input) were analyzed by real-time PCR (same primers as for analysis of IgH primary transcripts) and compared with those of negative controls obtained without antibody (mock). Results are the mean ± SEM of three 3′RR-deficient and three WT mice. ns, not significant. (G) ChIP analysis for AID occupancy at the Sμ region in B splenocytes of immunized WT, 3′RR-deficient, and AID-deficient mice. For each sample, AID-Chip values (mean ± SD) were normalized to the input control, and AID-Chip signal in WT B cells was assigned an arbitrary value of 1. Data represent results obtained using two different primer pairs (error bars are indicative of the variation between the two PCRs). One representative experiment out of two is shown. *, P < 0.05; Mann–Whitney U test.
Mutation in Sμ is severely compromised in 3′RR-deficient mice
Studies reported that Sμ targets SHM; mutational frequency peaked 5′ of Sμ and decreased thereafter (Maul and Gearhart, 2010). We investigated whether deletion of the 3′RR abrogated this pattern of SHM. Because of high GC content and the repetitive nature of the Sμ sequence (Fig. S1), accurate PCR amplification across the region is known to be problematic. We thus cloned the entire Iμ-Cμ sequence (location of PCR primers in Fig. S1) and sequenced both the 5′ and the 3′ part of the PCR product encompassing Sμ. Experiments were performed with B splenocytes of WT and 3′RR-deficient mice immunized orally with sheep red blood cells and intraperitoneally with LPS. A dramatic decrease (P < 0.0001, Mann–Whitney U test) of mutated sequences 5′ to Sμ was found in 3′RR-deficient mice (14%, 5/36) as compared with WT (65%, 20/31; Fig. 1 A). The mutation frequency in 3′RR-deficient mice (7 mutations among 19,078 bp analyzed, 0.036%) was compared with WT (92 mutations among 20,934 bp analyzed, 0.44%) and showed a >90% reduction (Fig. 1, A and D). In agreement with previous results (Maul and Gearhart, 2010), no mutations were documented 3′ to Sμ in WT nor in 3′RR-deficient mice (not depicted). ChIP experiments indicated that AID occupancy at the Sμ region was markedly reduced in 3′RR-deficient splenocytes as compared with WT (Fig. 2 G). Only background levels of immunoprecipitation were found both for 3′RR-deficient mice and AID-deficient mice, leading to the conclusion that AID binding to Sμ is totally impaired in 3′RR-deficient mice.
Concluding remarks
There have been several previous genomic alterations of the IgH 3′RR, notably through individual enhancer deletions (Manis et al., 1998; Pinaud et al., 2001; Vincent-Fabert et al., 2009; Bébin et al., 2010). Partial functional redundancies of the four 3′RR enhancers have long kept their role quite elusive, especially with regard to SHM. The complete deletion now clarifies that the 3′RR is mandatory for SHM. This major SHM defect paralleled a partly decreased IgH transcription, suggesting that elements beyond primary transcription accessibility are missing for recruitment of the SHM machinery onto the 3′RR-deficient IgH locus. The very low residual SHM suggests that no additional elements cooperate with the 3′RR for SHM. Although transcription is a prerequisite for several regulatory mechanisms controlling key stages in the B cell physiology, it is clearly not sufficient to ensure Ig locus accessibility to AID. The 3′RR-deficient mice will facilitate further elucidation of how the 3′RR “enhanceosome” or the 3′RR-dependent IgH locus 3D-conformation might regulate AID accessibility beyond transcriptional regulation.
MATERIALS AND METHODS
Mice.
Our research has been approved by the ethics committee review board of our university (Limoges, France) and hospital (Comité Régional d’Ethique sur l’Expérimentation Animale du Limousin, CHU Dupuytren, Limoges, France). Generation of 3′RR-deficient mice has been previously reported (Vincent-Fabert et al., 2010) and was performed on a C57BL/6 background. Mice were bred and maintained under specific pathogen–free conditions. Age-matched littermates (8–12 wk old) were used in all experiments. C57BL/6 mice were used as WT mice.
Cell cytometry and sorting procedures for SHM experiments.
Mouse immunizations were performed orally with sheep red blood cells for 2 wk and intraperitoneally with 10 µg LPS for 3 d. Single-cell suspensions from Peyer’s patches were labeled with B220-APC–, PNA-FITC–, and Fas-PE–conjugated antibodies. Purification of B220+PNAhighFas+ cells was realized on a FACSVantage (BD). These cells were used for SHM analysis of V(D)J-rearranged fragments and Igκ light chain VJ-rearranged fragments and analysis of IgH and κ primary transcripts. Single-cell suspensions from splenocytes were labeled with B220-APC– and PNA-FITC–conjugated antibodies for purification of B220+PNAhigh B splenocytes. These cells were used for SHM analysis of Sμ.
DNA extraction and amplification for SHM experiments.
Genomic DNA was extracted from sorted B220+PNAhighFas+ cells. IgH V(D)J-rearranged fragments were amplified by PCR using the following primers and multistep programs: the forward primer 5′-GCGAAGCTTARGCCTGGGRCTTCAGTGAAG-3′, complementary to the VHJ558 segment, and the backward primer 5′-AGGCTCTGAGATCCCTAGACAG-3′, corresponding to a sequence 517 bp downstream of the JH4 segment using 1 cycle at 98°C for 30 s, 33 cycles (98°C for 10 s, 67°C for 30 s, and 72°C for 90 s), and 1 cycle at 72°C for 10 min. Igκ light chain VJ-rearranged fragments were amplified by PCR as previously reported (Xiang and Garrard, 2008) using the following primers and multistep programs: the forward primer 5′-GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC-3′ (consensus for Vκ) and the backward primer 5′-AGCGAATTCAACTTAGGAGACAAAAGAGAGAAC-3′ (found 557 bp downstream of the Jκ5 segment) using 1 cycle at 98°C for 30 s, 33 cycles (98°C for 10 s, 66°C for 30 s, and 72°C for 90 s), and 1 cycle at 72°C for 10 min. Sμ from B220+PNAhigh B splenocytes was amplified by PCR using Herculase II DNA polymerase (Agilent Technologies) and the following primers and multistep programs: the forward primer 5′-CTCTGGCCCTGCTTATTGTTG-3′ and the backward primer 5′-GAAGACATTTGGGAAGGACTGACT-3′ using 1 cycle at 95°C for 120 s, 40 cycles (95°C for 15 s, 61°C for 20 s, and 72°C for 180 s), and 1 cycle at 72°C for 180 s. The exact location of the PCR primers and the Sμ PCR product used for mutation analysis are shown in Fig. S1.
Cloning and sequencing.
PCR products were cloned into the Zero Blunt Topo PCR cloning vector system (Invitrogen). Plasmids were isolated using the NucleoSpin kit (Macherey-Nagel) and sequenced using an automated laser fluorescent ANA ABI-PRISM sequencer (PerkinElmer).
Transcript analysis.
B cells from spleen of IgH 3′RR-deficient or WT mice were purified using CD43-coupled beads from Miltenyi Biotec according to the manufacturer’s recommendations. Total RNA was extracted using tri-reagent (Ambion). Real-time PCR was performed in duplicate by using TaqMan assay reagents and analyzed on an ABI Prism 7000 system (Applied Biosystems). Transcripts were studied for Msh2 (reference Mn00500563-m1), Msh6 (reference Mn00487769-g1), UNG (reference Mn00449156-m1), POLη (reference Mn00453168-m1), REV1 (reference Mn00450983-m1), AID (reference Mn01184115-m1), and GAPDH (used for normalization of gene expression levels; reference Mm99999915-g1) as previously reported (Truffinet et al., 2007; Vincent-Fabert et al., 2009). B220+PNAhighFas+ cells were used for analysis of IgH and κ primary transcripts. IgH primary transcripts (probe located in the intron between the last JH and the intronic Eμ enhancer) were studied as previously reported (Tinguely et al., 2012): forward primer, 5′-TTCTGAGCATTGCAGACTAATCTTG-3′; reverse primer, 5′-CCTAGACAGTTTATTTCCCAACTTCTC-3′; and probe, 5′-CCCTGAGGGAGCCG-3′. κ primary transcripts (probe located in the intron between the last Jκ and the intronic enhancer Eiκ): κ forward primer, 5′-ACCCCCGCGGTAGCA-3′; κ reverse primer, 5′-TCCTATCACTGTGCCTCAGGAA-3′; and probe, 5′-CCCTTGCTCCGCGTGGACCA-3′. GAPDH was used for normalization of gene expression levels (reference Mm99999915-g1).
ChIP.
Mice immunizations were performed orally with sheep red blood cells for 2 wk and intraperitoneally with 10 µg LPS for 3 d. CD19 magnetic cell-sorted splenic B cells were used for AID-Chip experiments as previously described (Jeevan-Raj et al., 2011). For Sμ Q-PCR, data represent results obtained using two different primer pairs. Sμ forward 1 primer, 5′-TAAAATGCGCTAAACTGAGGTGATTACT-3′; Sμ reverse 1 primer, 5′-CATCTCAGCTCAGAACAGTCCAGTG-3′ (Kuang et al., 2009); Sμ forward 2 primer, 5′-TAGTAAGCGAGGCTCTAAAAAGCAT-3′; and Sμ reverse 2 primer, 5′-AGAACAGTCCAGTGTAGGCAGTAGA-3′ (Pavri et al., 2010). These primers were used with QuantiTect SYBR Green mix from QIAGEN. For JH4 experiments, two primers (UPL 3′JH4 forward, 5′-AGGGACTTTGGAGGCTCATT-3′; and UPL 3′JH4 reverse, 5′-CTCCAACTACAGCCCCAACT-3′) have been designed and were used in combination with UPL probe#19 and Roche LightCycler 480 Probes Master mix. Splenic B cells were purified by CD43 magnetic cell sorting from nonimmunized mice and cultured in vitro with 10 µg/ml LPS for 3 d and used for Pol II–ChIP experiments as previously described (Tinguely et al., 2012). PCR primers were those used for analysis of IgH primary transcripts (probe located in the intron between the last JH and the intronic Eμ enhancer; see sequences above).
Online supplemental material.
Fig. S1 shows Sμ sequence and location of primers for Sμ PCR experiments. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20130072/DC1.
Supplementary Material
Acknowledgments
We thank S. Desforges and B. Remerand for help with animal care.
This work was supported by grants from the Association pour la Recherche sur le Cancer (ARC; SL 220100601332), Conseil Régional du Limousin, and Agence Nationale de la Recherche (Projets Blanc 2011). C. Vincent-Fabert was supported by a grant from the ARC.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 3′RR
- 3′ regulatory region
- AID
- activation-induced deaminase
- ChIP
- chromatin immunoprecipitation
- CSR
- class switch recombination
- SHM
- somatic hypermutation
References
- Afshar R., Pierce S., Bolland D.J., Corcoran A., Oltz E.M. 2006. Regulation of IgH gene assembly: role of the intronic enhancer and 5’DQ52 region in targeting DHJH recombination. J. Immunol. 176:2439–2447 [DOI] [PubMed] [Google Scholar]
- Bébin A.G., Carrion C., Marquet M., Cogné N., Lecardeur S., Cogné M., Pinaud E. 2010. In vivo redundant function of the 3′ IgH regulatory element HS3b in the mouse. J. Immunol. 184:3710–3717 10.4049/jimmunol.0901978 [DOI] [PubMed] [Google Scholar]
- Blagodatski A., Batrak V., Schmidl S., Schoetz U., Caldwell R.B., Arakawa H., Buerstedde J.M. 2009. A cis-acting diversification activator both necessary and sufficient for AID-mediated hypermutation. PLoS Genet. 5:e1000332 10.1371/journal.pgen.1000332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick W.A., Collins J.T., Shi J., Westfield G., Fontaine C., Hakimpour P., Papavasiliou F.N. 2009. Switch recombination and somatic hypermutation are controlled by the heavy chain 3′ enhancer region. J. Exp. Med. 206:2613–2623 10.1084/jem.20091280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone K., Wood A.L., Bowen A.J., Corcoran A.E. 2010. The mouse immunoglobulin heavy chain V-D intergenic sequence contains insulators that may regulate ordered V(D)J recombination. J. Biol. Chem. 285:9327–9338 10.1074/jbc.M109.098251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukita Y., Jacobs H., Rajewsky K. 1998. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 9:105–114 10.1016/S1074-7613(00)80592-0 [DOI] [PubMed] [Google Scholar]
- Giallourakis C.C., Franklin A., Guo C., Cheng H.L., Yoon H.S., Gallagher M., Perlot T., Andzelm M., Murphy A.J., Macdonald L.E., et al. 2010. Elements between the IgH variable (V) and diversity (D) clusters influence antisense transcription and lineage-specific V(D)J recombination. Proc. Natl. Acad. Sci. USA. 107:22207–22212 10.1073/pnas.1015954107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Yoon H.S., Franklin A., Jain S., Ebert A., Cheng H.L., Hansen E., Despo O., Bossen C., Vettermann C., et al. 2011. CTCF-binding elements mediate control of V(D)J recombination. Nature. 477:424–430 10.1038/nature10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney J.A., Misaghi S., Senger K., Garris C., Sun Y., Lorenzo M.N., Zarrin A.A. 2009. DNA targets of AID evolutionary link between antibody somatic hypermutation and class switch recombination. Adv. Immunol. 101:163–189 10.1016/S0065-2776(08)01005-5 [DOI] [PubMed] [Google Scholar]
- Henderson A., Calame K. 1998. Transcriptional regulation during B cell development. Annu. Rev. Immunol. 16:163–200 10.1146/annurev.immunol.16.1.163 [DOI] [PubMed] [Google Scholar]
- Jeevan-Raj B.P., Robert I., Heyer V., Page A., Wang J.H., Cammas F., Alt F.W., Losson R., Reina-San-Martin B. 2011. Epigenetic tethering of AID to the donor switch region during immunoglobulin class switch recombination. J. Exp. Med. 208:1649–1660 10.1084/jem.20110118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler K.M., McDonald J.J., Duke J.L., Arakawa H., Tan S., Kleinstein S.H., Buerstedde J.M., Schatz D.G. 2012. Identification of core DNA elements that target somatic hypermutation. J. Immunol. 189:5314–5326 10.4049/jimmunol.1202082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli N., Norton D.D., Fugmann S.D. 2008. Cutting edge: a cis-acting DNA element targets AID-mediated sequence diversification to the chicken Ig light chain gene locus. J. Immunol. 180:2019–2023 [DOI] [PubMed] [Google Scholar]
- Kothapalli N.R., Collura K.M., Norton D.D., Fugmann S.D. 2011. Separation of mutational and transcriptional enhancers in Ig genes. J. Immunol. 187:3247–3255 10.4049/jimmunol.1101568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang F.L., Luo Z., Scharff M.D. 2009. H3 trimethyl K9 and H3 acetyl K9 chromatin modifications are associated with class switch recombination. Proc. Natl. Acad. Sci. USA. 106:5288–5293 10.1073/pnas.0901368106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencikiene J., Tamosiunas V., Severinson E. 2007. Regulation of ε germline transcription and switch region mutations by IgH locus 3′ enhancers in transgenic mice. Blood. 109:159–167 10.1182/blood-2006-02-005355 [DOI] [PubMed] [Google Scholar]
- Manis J.P., van der Stoep N., Tian M., Ferrini R., Davidson L., Bottaro A., Alt F.W. 1998. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 188:1421–1431 10.1084/jem.188.8.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul R.W., Gearhart P.J. 2010. Controlling somatic hypermutation in immunoglobulin variable and switch regions. Immunol. Res. 47:113–122 10.1007/s12026-009-8142-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan C.L., Pinaud E., Decourt C., Cuvillier A., Cogné M. 2003. The immunoglobulin heavy-chain locus hs3b and hs4 3′ enhancers are dispensable for VDJ assembly and somatic hypermutation. Blood. 102:1421–1427 10.1182/blood-2002-12-3827 [DOI] [PubMed] [Google Scholar]
- Neuberger M.S., Rada C. 2007. Somatic hypermutation: activation-induced deaminase for C/G followed by polymerase eta for A/T. J. Exp. Med. 204:7–10 10.1084/jem.20062409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke L., Kestler J., Tallone T., Pelkonen S., Pelkonen J. 2001. Deletion of the DQ52 element within the Ig heavy chain locus leads to a selective reduction in VDJ recombination and altered D gene usage. J. Immunol. 166:2540–2552 [DOI] [PubMed] [Google Scholar]
- Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V., et al. 2010. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 143:122–133 10.1016/j.cell.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlot T., Alt F.W., Bassing C.H., Suh H., Pinaud E. 2005. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl. Acad. Sci. USA. 102:14362–14367 10.1073/pnas.0507090102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péron S., Laffleur B., Denis-Lagache N., Cook-Moreau J., Tinguely A., Delpy L., Denizot Y., Pinaud E., Cogné M. 2012. AID-driven deletion causes immunoglobulin heavy chain locus suicide recombination in B cells. Science. 336:931–934 10.1126/science.1218692 [DOI] [PubMed] [Google Scholar]
- Pinaud E., Khamlichi A.A., Le Morvan C., Drouet M., Nalesso V., Le Bert M., Cogné M. 2001. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 15:187–199 10.1016/S1074-7613(01)00181-9 [DOI] [PubMed] [Google Scholar]
- Pinaud E., Marquet M., Fiancette R., Péron S., Vincent-Fabert C., Denizot Y., Cogné M. 2011. The IgH locus 3′ regulatory region: pulling the strings from behind. Adv. Immunol. 110:27–70 10.1016/B978-0-12-387663-8.00002-8 [DOI] [PubMed] [Google Scholar]
- Rouaud P., Vincent-Fabert C., Fiancette R., Cogné M., Pinaud E., Denizot Y. 2012. Enhancers located in heavy chain regulatory region (hs3a, hs1,2, hs3b, and hs4) are dispensable for diversity of VDJ recombination. J. Biol. Chem. 287:8356–8360 10.1074/jbc.M112.341024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai E., Bottaro A., Davidson L., Sleckman B.P., Alt F.W. 1999. Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc. Natl. Acad. Sci. USA. 96:1526–1531 10.1073/pnas.96.4.1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., Ji Y. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11:251–263 10.1038/nri2941 [DOI] [PubMed] [Google Scholar]
- Terauchi A., Hayashi K., Kitamura D., Kozono Y., Motoyama N., Azuma T. 2001. A pivotal role for DNase I-sensitive regions 3b and/or 4 in the induction of somatic hypermutation of IgH genes. J. Immunol. 167:811–820 [DOI] [PubMed] [Google Scholar]
- Tinguely A., Chemin G., Péron S., Sirac C., Reynaud S., Cogné M., Delpy L. 2012. Cross talk between immunoglobulin heavy-chain transcription and RNA surveillance during B cell development. Mol. Cell. Biol. 32:107–117 10.1128/MCB.06138-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truffinet V., Pinaud E., Cogné N., Petit B., Guglielmi L., Cogné M., Denizot Y. 2007. The 3′ IgH locus control region is sufficient to deregulate a c-myc transgene and promote mature B cell malignancies with a predominant Burkitt-like phenotype. J. Immunol. 179:6033–6042 [DOI] [PubMed] [Google Scholar]
- Tumas-Brundage K.M., Vora K.A., Manser T. 1997. Evaluation of the role of the 3’alpha heavy chain enhancer [3’alpha E(hs1,2)] in Vh gene somatic hypermutation. Mol. Immunol. 34:367–378 10.1016/S0161-5890(97)00065-5 [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C., Truffinet V., Fiancette R., Cogné N., Cogné M., Denizot Y. 2009. Ig synthesis and class switching do not require the presence of the hs4 enhancer in the 3′ IgH regulatory region. J. Immunol. 182:6926–6932 10.4049/jimmunol.0900214 [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C., Fiancette R., Pinaud E., Truffinet V., Cogné N., Cogné M., Denizot Y. 2010. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 116:1895–1898 10.1182/blood-2010-01-264689 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Garrard W.T. 2008. The Downstream Transcriptional Enhancer, Ed, positively regulates mouse Ig kappa gene expression and somatic hypermutation. J. Immunol. 180:6725–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.Y., Fugmann S.D., Schatz D.G. 2006. Control of gene conversion and somatic hypermutation by immunoglobulin promoter and enhancer sequences. J. Exp. Med. 203:2919–2928 10.1084/jem.20061835 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.