Abstract
A popular interpretation of the major codon preference is that it reflects the operation of a regulatory device that controls the expression of individual proteins. In this popular model, rapidly translated codons are thought to promote the accumulation of the highly expressed proteins and slowly translated codons are thought to retard the expression of poorly expressed proteins. However, this widely accepted model is not supported by kinetic theory or by experimental results. A less fashionable model in which the major codon preference has nothing to do with the expression level of the individual proteins is forwarded. In this model, the major codon preference is viewed as a global strategy to support the efficient function of the translation system and thereby to maximize the growth rates of cells under favorable conditions.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson D. I., van Verseveld H. W., Stouthamer A. H., Kurland C. G. Suboptimal growth with hyper-accurate ribosomes. Arch Microbiol. 1986 Feb;144(1):96–101. doi: 10.1007/BF00454963. [DOI] [PubMed] [Google Scholar]
- Andersson S. G., Buckingham R. H., Kurland C. G. Does codon composition influence ribosome function? EMBO J. 1984 Jan;3(1):91–94. doi: 10.1002/j.1460-2075.1984.tb01766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aota S., Ikemura T. Diversity in G + C content at the third position of codons in vertebrate genes and its cause. Nucleic Acids Res. 1986 Aug 26;14(16):6345–6355. doi: 10.1093/nar/14.16.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi G. Animal mitochondrial DNA: an extreme example of genetic economy. Int Rev Cytol. 1985;93:93–145. doi: 10.1016/s0074-7696(08)61373-x. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bernardi G., Bernardi G. Codon usage and genome composition. J Mol Evol. 1985;22(4):363–365. doi: 10.1007/BF02115693. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Bernardi G. Compositional constraints and genome evolution. J Mol Evol. 1986;24(1-2):1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Ehrenberg M., Kurland C. Is translation inhibited by noncognate ternary complexes? FEBS Lett. 1988 Jun 6;233(1):95–99. doi: 10.1016/0014-5793(88)81362-0. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Kirsebom L. A., Ehrenberg M., Kurland C. G. Mutations in ribosomal proteins L7/L12 perturb EF-G and EF-Tu functions. Biochimie. 1988 May;70(5):611–618. doi: 10.1016/0300-9084(88)90244-1. [DOI] [PubMed] [Google Scholar]
- Bonekamp F., Andersen H. D., Christensen T., Jensen K. F. Codon-defined ribosomal pausing in Escherichia coli detected by using the pyrE attenuator to probe the coupling between transcription and translation. Nucleic Acids Res. 1985 Jun 11;13(11):4113–4123. doi: 10.1093/nar/13.11.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonekamp F., Dalbøge H., Christensen T., Jensen K. F. Translation rates of individual codons are not correlated with tRNA abundances or with frequencies of utilization in Escherichia coli. J Bacteriol. 1989 Nov;171(11):5812–5816. doi: 10.1128/jb.171.11.5812-5816.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonekamp F., Jensen K. F. The AGG codon is translated slowly in E. coli even at very low expression levels. Nucleic Acids Res. 1988 Apr 11;16(7):3013–3024. doi: 10.1093/nar/16.7.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouadloun F., Donner D., Kurland C. G. Codon-specific missense errors in vivo. EMBO J. 1983;2(8):1351–1356. doi: 10.1002/j.1460-2075.1983.tb01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton R., Sanfaçon H., Papayannopoulos I., Biemann K., Lapointe J. Glutamyl-tRNA synthetase of Escherichia coli. Isolation and primary structure of the gltX gene and homology with other aminoacyl-tRNA synthetases. J Biol Chem. 1986 Aug 15;261(23):10610–10617. [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. Codon usage and intragenic position. J Theor Biol. 1988 Jul 8;133(1):67–71. doi: 10.1016/s0022-5193(88)80024-9. [DOI] [PubMed] [Google Scholar]
- Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987 Feb 19;325(6106):728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- Burns D. M., Beacham I. R. Rare codons in E. coli and S. typhimurium signal sequences. FEBS Lett. 1985 Sep 23;189(2):318–324. doi: 10.1016/0014-5793(85)81048-6. [DOI] [PubMed] [Google Scholar]
- Chavancy G., Garel J. P. Does quantitative tRNA adaptation to codon content in mRNA optimize the ribosomal translation efficiency? Proposal for a translation system model. Biochimie. 1981 Mar;63(3):187–195. doi: 10.1016/s0300-9084(81)80192-7. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H., Brenner S., Klug A., Pieczenik G. A speculation on the origin of protein synthesis. Orig Life. 1976 Dec;7(4):389–397. doi: 10.1007/BF00927934. [DOI] [PubMed] [Google Scholar]
- Crick F. H. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989 Sep 5;209(1):65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J Mol Biol. 1988 Sep 5;203(1):75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- Dean A. M., Dykhuizen D. E., Hartl D. L. Fitness effects of amino acid replacements in the beta-galactosidase of Escherichia coli. Mol Biol Evol. 1988 Sep;5(5):469–485. doi: 10.1093/oxfordjournals.molbev.a040513. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M., Kurland C. G. Costs of accuracy determined by a maximal growth rate constraint. Q Rev Biophys. 1984 Feb;17(1):45–82. doi: 10.1017/s0033583500005254. [DOI] [PubMed] [Google Scholar]
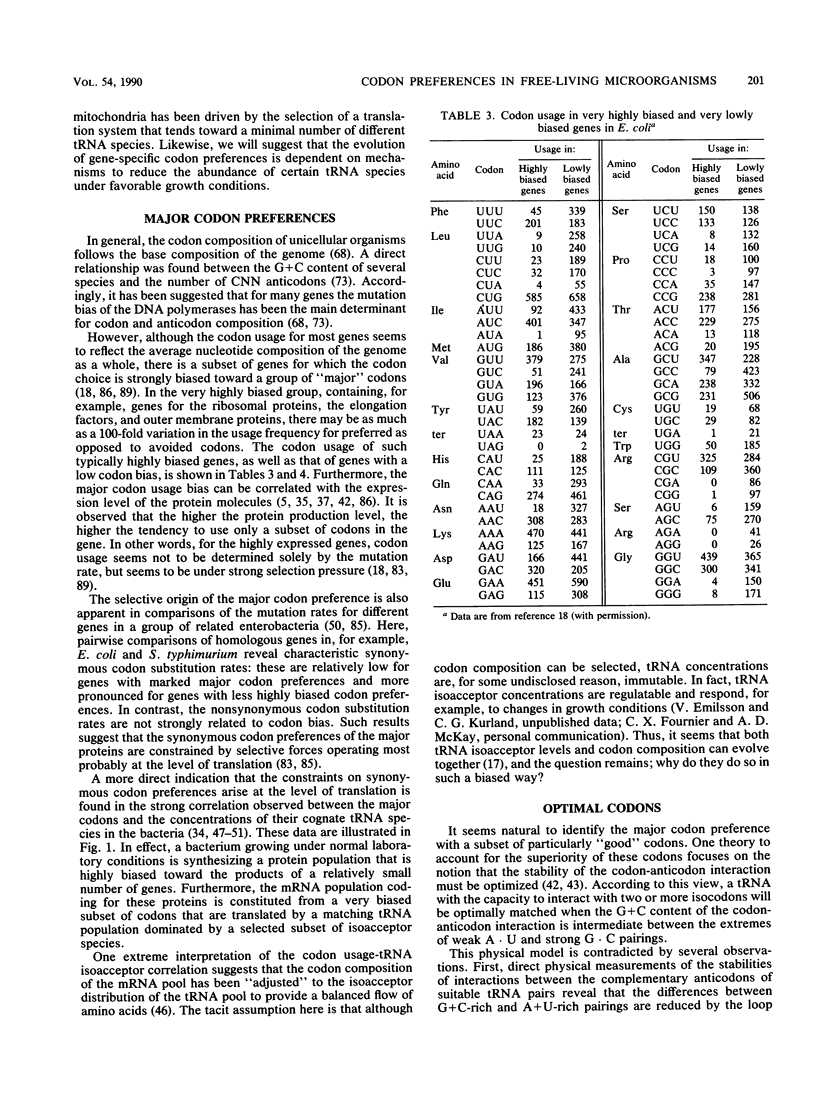
- Eigen M., Winkler-Oswatitsch R. Transfer-RNA, an early gene? Naturwissenschaften. 1981 Jun;68(6):282–292. doi: 10.1007/BF01047470. [DOI] [PubMed] [Google Scholar]
- Fast R., Eberhard T. H., Ruusala T., Kurland C. G. Does streptomycin cause an error catastrophe? Biochimie. 1987 Feb;69(2):131–136. doi: 10.1016/0300-9084(87)90245-8. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Natural variation in the genetic code. Annu Rev Genet. 1987;21:67–91. doi: 10.1146/annurev.ge.21.120187.000435. [DOI] [PubMed] [Google Scholar]
- Garel J. P. Functional adaptation of tRNA population. J Theor Biol. 1974 Jan;43(1):211–225. doi: 10.1016/s0022-5193(74)80054-8. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 1980 May 10;8(9):1893–1912. doi: 10.1093/nar/8.9.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green G. A., Jones D. S. The nucleotide sequence of a cytoplasmic tRNAPhe from Scenedesmus obliquus and comparison with a tRNATyr species. Biochem J. 1986 Jun 1;236(2):601–603. doi: 10.1042/bj2360601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell L. A. Molecular evolution. Deciphering divergent codes. Nature. 1986 Nov 13;324(6093):109–110. doi: 10.1038/324109a0. [DOI] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Sankoff D., Jou W. M., Fiers W., Cedergren R. J. Bacteriophage MS2 RNA: a correlation between the stability of the codon: anticodon interaction and the choice of code words. J Mol Evol. 1978 Dec 29;12(2):113–119. doi: 10.1007/BF01733262. [DOI] [PubMed] [Google Scholar]
- Hinds P. W., Blake R. D. Delineation of coding areas in DNA sequences through assignment of codon probabilities. J Biomol Struct Dyn. 1985 Dec;3(3):543–549. doi: 10.1080/07391102.1985.10508442. [DOI] [PubMed] [Google Scholar]
- Hoekema A., Kastelein R. A., Vasser M., de Boer H. A. Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol Cell Biol. 1987 Aug;7(8):2914–2924. doi: 10.1128/mcb.7.8.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. Codon usage and gene expression. Nucleic Acids Res. 1986 Apr 11;14(7):3075–3087. doi: 10.1093/nar/14.7.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Osawa S., Muto A. Divergence and directional mutation pressures. Nature. 1987 Feb 19;325(6106):668–668. doi: 10.1038/325668b0. [DOI] [PubMed] [Google Scholar]
- Kniskern P. J., Hagopian A., Montgomery D. L., Burke P., Dunn N. R., Hofmann K. J., Miller W. J., Ellis R. W. Unusually high-level expression of a foreign gene (hepatitis B virus core antigen) in Saccharomyces cerevisiae. Gene. 1986;46(1):135–141. doi: 10.1016/0378-1119(86)90177-0. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G. N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey J. C., Jr, Mullins D. W., Jr Experimental studies related to the origin of the genetic code and the process of protein synthesis--a review. Orig Life. 1983 Mar;13(1):3–42. doi: 10.1007/BF00928761. [DOI] [PubMed] [Google Scholar]
- Lehman N., Jukes T. H. Genetic code development by stop codon takeover. J Theor Biol. 1988 Nov 21;135(2):203–214. doi: 10.1016/s0022-5193(88)80074-2. [DOI] [PubMed] [Google Scholar]
- Liljenström H., von Heijne G. Translation rate modification by preferential codon usage: intragenic position effects. J Theor Biol. 1987 Jan 7;124(1):43–55. doi: 10.1016/s0022-5193(87)80251-5. [DOI] [PubMed] [Google Scholar]
- McPherson D. T. Codon preference reflects mistranslational constraints: a proposal. Nucleic Acids Res. 1988 May 11;16(9):4111–4120. doi: 10.1093/nar/16.9.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikelsaar R. A view of early cellular evolution. J Mol Evol. 1987;25(2):168–183. doi: 10.1007/BF02101759. [DOI] [PubMed] [Google Scholar]
- Miller S. L. Current status of the prebiotic synthesis of small molecules. Chem Scr. 1986;26B:5–11. [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Yamao F., Hori H., Osawa S. Gene organization of Mycoplasma capricolum. Adv Biophys. 1986;21:49–56. doi: 10.1016/0065-227x(86)90013-4. [DOI] [PubMed] [Google Scholar]
- Nagyvary J., Fendler J. H. Origin of the genetic code: a physical-chemical model of primitive codon assignments. Orig Life. 1974 Jul-Oct;5(3):357–362. [PubMed] [Google Scholar]
- Ogasawara N. Markedly unbiased codon usage in Bacillus subtilis. Gene. 1985;40(1):145–150. doi: 10.1016/0378-1119(85)90035-6. [DOI] [PubMed] [Google Scholar]
- Ohama T., Yamao F., Muto A., Osawa S. Organization and codon usage of the streptomycin operon in Micrococcus luteus, a bacterium with a high genomic G + C content. J Bacteriol. 1987 Oct;169(10):4770–4777. doi: 10.1128/jb.169.10.4770-4777.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Codon reassignment (codon capture) in evolution. J Mol Evol. 1989 Apr;28(4):271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Evolution of the genetic code as affected by anticodon content. Trends Genet. 1988 Jul;4(7):191–198. doi: 10.1016/0168-9525(88)90075-3. [DOI] [PubMed] [Google Scholar]
- Osawa S., Ohama T., Jukes T. H., Watanabe K. Evolution of the mitochondrial genetic code. I. Origin of AGR serine and stop codons in metazoan mitochondria. J Mol Evol. 1989 Sep;29(3):202–207. doi: 10.1007/BF02100203. [DOI] [PubMed] [Google Scholar]
- Osawa S., Ohama T., Jukes T. H., Watanabe K., Yokoyama S. Evolution of the mitochondrial genetic code. II. Reassignment of codon AUA from isoleucine to methionine. J Mol Evol. 1989 Nov;29(5):373–380. doi: 10.1007/BF02602907. [DOI] [PubMed] [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989 Sep;53(3):273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis I. J., Bettany A. J., Santiago T. C., Coggins J. R., Duncan K., Eason R., Brown A. J. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J Mol Biol. 1987 Jan 20;193(2):413–417. doi: 10.1016/0022-2836(87)90230-0. [DOI] [PubMed] [Google Scholar]
- Randall L. L., Josefsson L. G., Hardy S. J. Novel intermediates in the synthesis of maltose-binding protein in Escherichia coli. Eur J Biochem. 1980 Jun;107(2):375–379. doi: 10.1111/j.1432-1033.1980.tb06039.x. [DOI] [PubMed] [Google Scholar]
- Robinson M., Lilley R., Little S., Emtage J. S., Yarranton G., Stephens P., Millican A., Eaton M., Humphreys G. Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res. 1984 Sep 11;12(17):6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986 Oct 10;14(19):7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987 May;4(3):222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 1987 Oct 12;15(19):8023–8040. doi: 10.1093/nar/15.19.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanjaard R. A., van Duin J. Translation of the sequence AGG-AGG yields 50% ribosomal frameshift. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7967–7971. doi: 10.1073/pnas.85.21.7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M. A., Kurland C. G., Pedersen S. Codon usage determines translation rate in Escherichia coli. J Mol Biol. 1989 May 20;207(2):365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Translation framing code and frame-monitoring mechanism as suggested by the analysis of mRNA and 16 S rRNA nucleotide sequences. J Mol Biol. 1987 Apr 20;194(4):643–652. doi: 10.1016/0022-2836(87)90241-5. [DOI] [PubMed] [Google Scholar]
- Varenne S., Buc J., Lloubes R., Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984 Dec 15;180(3):549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Varenne S., Lazdunski C. Effect of distribution of unfavourable codons on the maximum rate of gene expression by an heterologous organism. J Theor Biol. 1986 May 7;120(1):99–110. doi: 10.1016/s0022-5193(86)80020-0. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. The unc operon. Nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984 Sep 6;768(2):164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Hauser C. A., Hatfield G. W. The nucleotide sequence of the ilvBN operon of Escherichia coli: sequence homologies of the acetohydroxy acid synthase isozymes. Nucleic Acids Res. 1985 Jun 11;13(11):3995–4010. doi: 10.1093/nar/13.11.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. On the evolution of the genetic code. Proc Natl Acad Sci U S A. 1965 Dec;54(6):1546–1552. doi: 10.1073/pnas.54.6.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. T., Cedergren R. Natural selection versus primitive gene structure as determinant of codon usage. Eur J Biochem. 1986 Aug 15;159(1):175–180. doi: 10.1111/j.1432-1033.1986.tb09849.x. [DOI] [PubMed] [Google Scholar]
- Wong J. T. Evolution of the genetic code. Microbiol Sci. 1988 Jun;5(6):174–181. [PubMed] [Google Scholar]
- Yarus M. A specific amino acid binding site composed of RNA. Science. 1988 Jun 24;240(4860):1751–1758. doi: 10.1126/science.3381099. [DOI] [PubMed] [Google Scholar]


