Abstract
The single-stranded DNA-binding protein (SSB) of Escherichia coli is involved in all aspects of DNA metabolism: replication, repair, and recombination. In solution, the protein exists as a homotetramer of 18,843-kilodalton subunits. As it binds tightly and cooperatively to single-stranded DNA, it has become a prototypic model protein for studying protein-nucleic acid interactions. The sequences of the gene and protein are known, and the functional domains of subunit interaction, DNA binding, and protein-protein interactions have been probed by structure-function analyses of various mutations. The ssb gene has three promoters, one of which is inducible because it lies only two nucleotides from the LexA-binding site of the adjacent uvrA gene. Induction of the SOS response, however, does not lead to significant increases in SSB levels. The binding protein has several functions in DNA replication, including enhancement of helix destabilization by DNA helicases, prevention of reannealing of the single strands and protection from nuclease digestion, organization and stabilization of replication origins, primosome assembly, priming specificity, enhancement of replication fidelity, enhancement of polymerase processivity, and promotion of polymerase binding to the template. E. coli SSB is required for methyl-directed mismatch repair, induction of the SOS response, and recombinational repair. During recombination, SSB interacts with the RecBCD enzyme to find Chi sites, promotes binding of RecA protein, and promotes strand uptake.
Full text
PDF


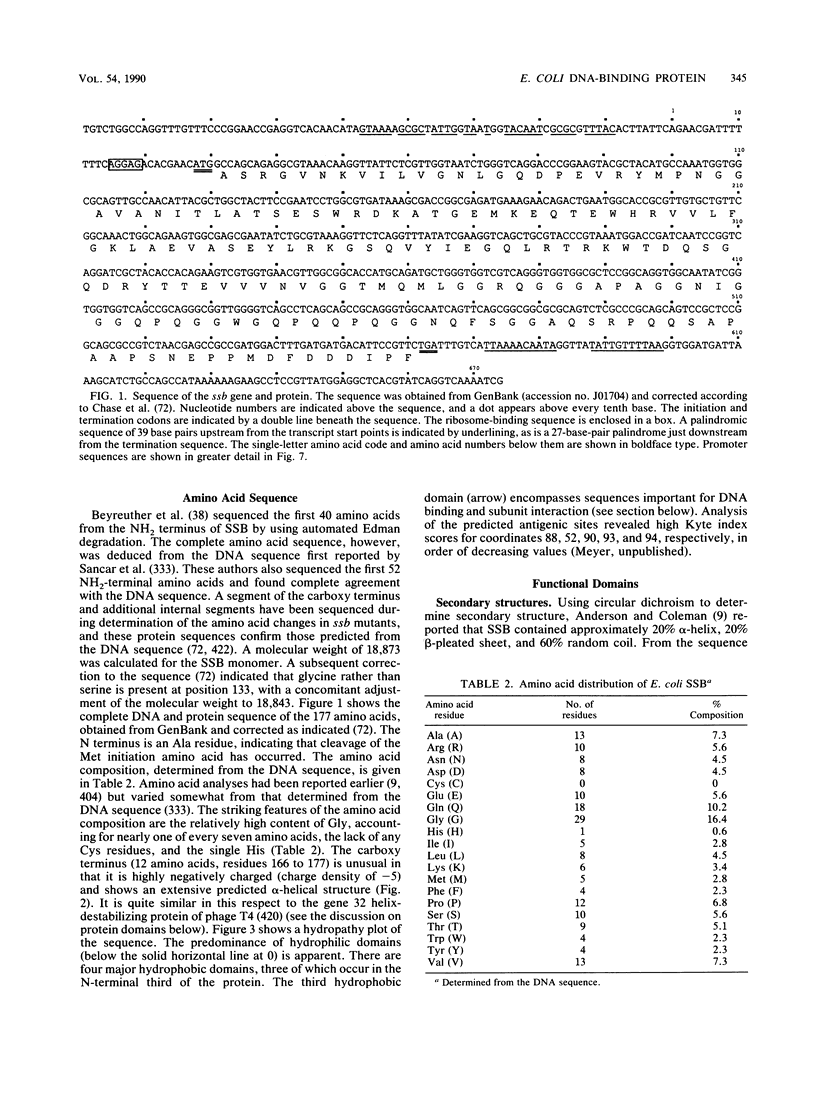
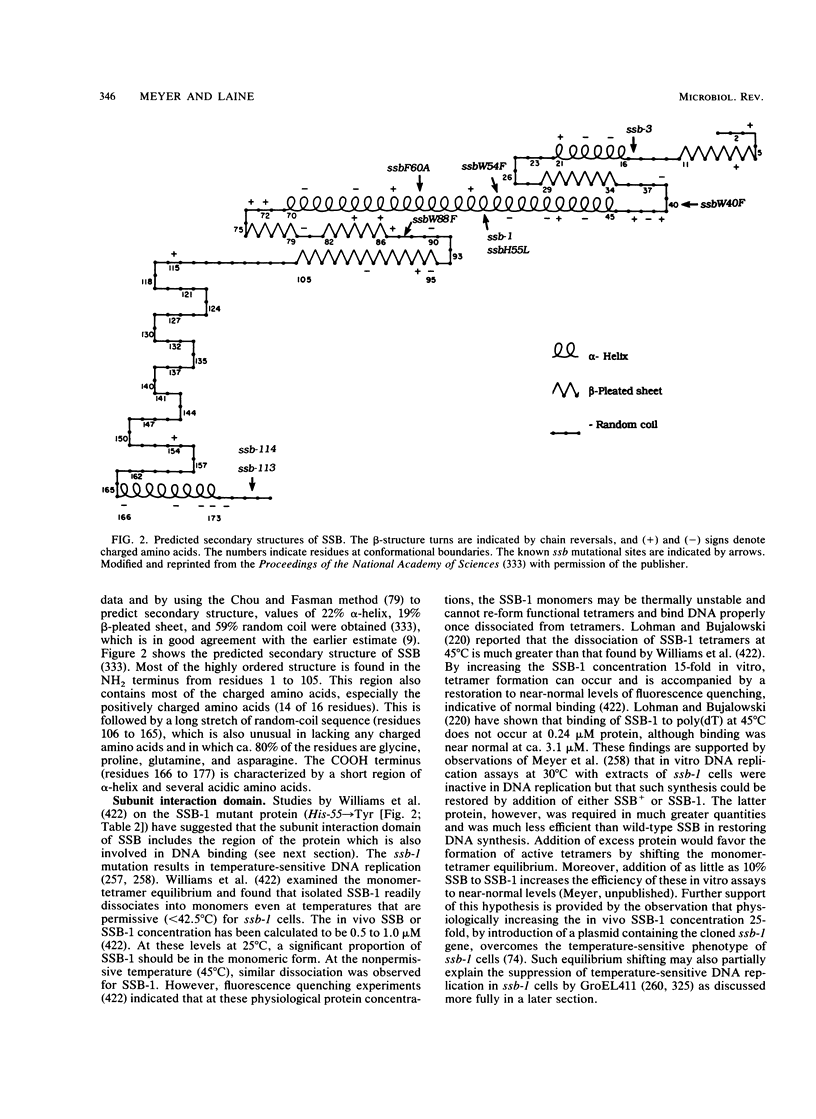
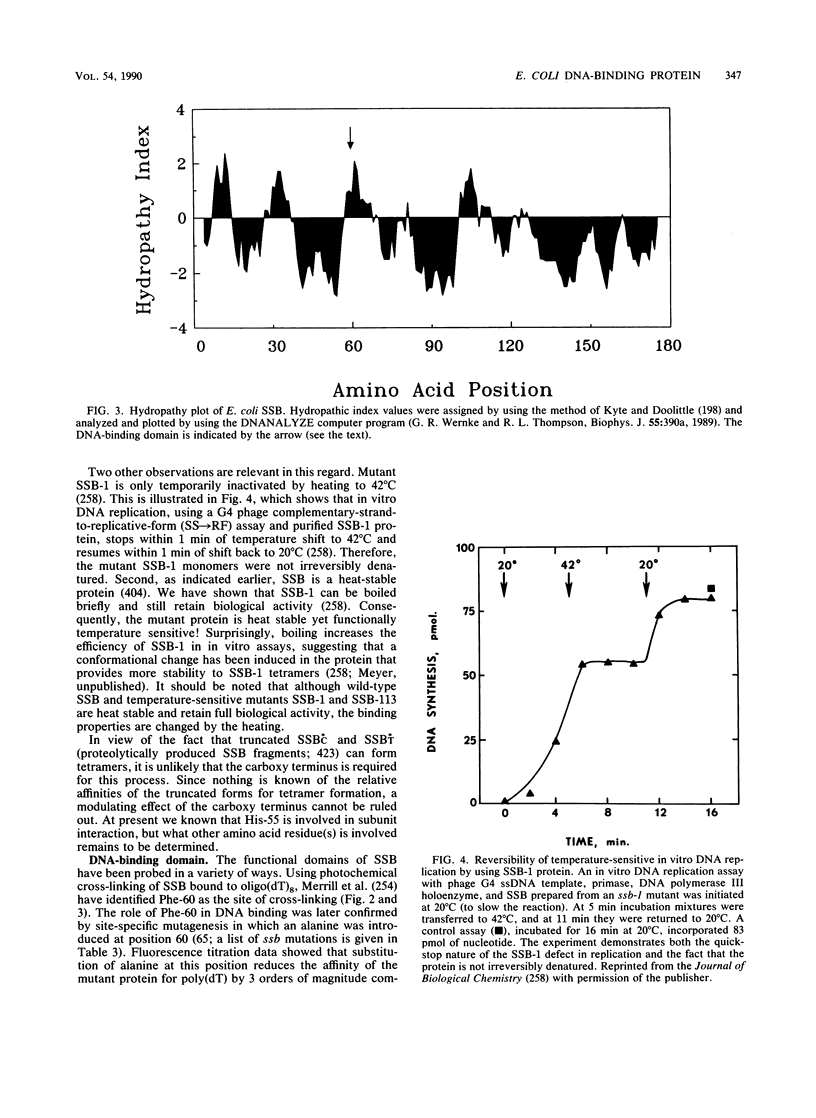







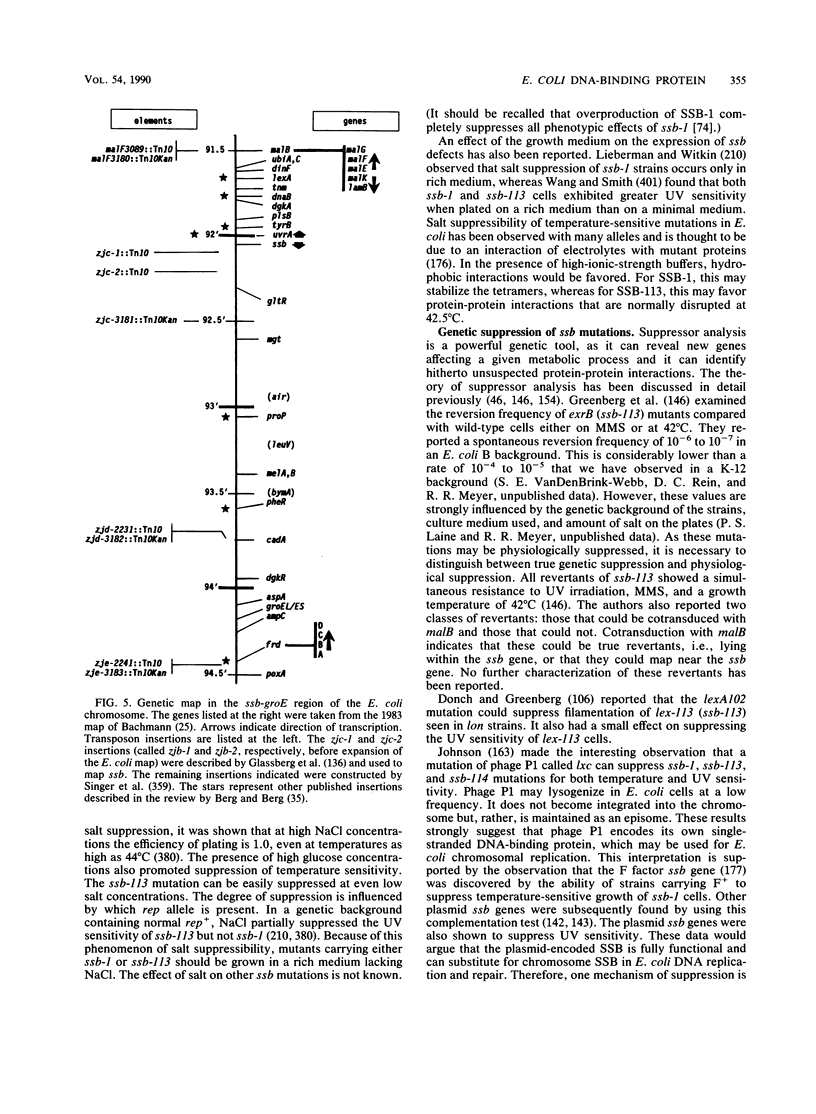





















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abarzúa P., Soeller W., Marians K. J. Mutational analysis of primosome assembly sites. I. Distinct classes of mutants in the pBR322 Escherichia coli factor Y DNA effector sequences. J Biol Chem. 1984 Nov 25;259(22):14286–14292. [PubMed] [Google Scholar]
- Alazard R. J. Study of the expression of UVRA and SSB proteins in vivo in lambda hybrid phages containing the uvrA and ssbA genes of Escherichia coli. Mutat Res. 1983 May;109(2):155–168. doi: 10.1016/0027-5107(83)90043-x. [DOI] [PubMed] [Google Scholar]
- Alberts B. M., Barry J., Bedinger P., Formosa T., Jongeneel C. V., Kreuzer K. N. Studies on DNA replication in the bacteriophage T4 in vitro system. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):655–668. doi: 10.1101/sqb.1983.047.01.077. [DOI] [PubMed] [Google Scholar]
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Alberts B., Sternglanz R. Recent excitement in the DNA replication problem. Nature. 1977 Oct 20;269(5630):655–661. doi: 10.1038/269655a0. [DOI] [PubMed] [Google Scholar]
- Amundsen S. K., Taylor A. F., Chaudhury A. M., Smith G. R. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. A., Coleman J. E. Physiochemical properties of DNA binding proteins: gene 32 protein of T4 and Escherichia coli unwinding protein. Biochemistry. 1975 Dec 16;14(25):5485–5491. doi: 10.1021/bi00696a017. [DOI] [PubMed] [Google Scholar]
- Arai K., Arai N., Shlomai J., Kobori J., Polder L., Low R., Hübscher U., Bertsch L., Kornberg A. Enzyme studies of phi X174 DNA replication. Prog Nucleic Acid Res Mol Biol. 1981;26:9–32. [PubMed] [Google Scholar]
- Arai K., Kornberg A. A general priming system employing only dnaB protein and primase for DNA replication. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4308–4312. doi: 10.1073/pnas.76.9.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K., Kornberg A. Mechanism of dnaB protein action. II. ATP hydrolysis by dnaB protein dependent on single- or double-stranded DNA. J Biol Chem. 1981 May 25;256(10):5253–5259. [PubMed] [Google Scholar]
- Arai K., Kornberg A. Unique primed start of phage phi X174 DNA replication and mobility of the primosome in a direction opposite chain synthesis. Proc Natl Acad Sci U S A. 1981 Jan;78(1):69–73. doi: 10.1073/pnas.78.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K., Low R. L., Kornberg A. Movement and site selection for priming by the primosome in phage phi X174 DNA replication. Proc Natl Acad Sci U S A. 1981 Feb;78(2):707–711. doi: 10.1073/pnas.78.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K., Low R., Kobori J., Shlomai J., Kornberg A. Mechanism of dnaB protein action. V. Association of dnaB protein, protein n', and other repriming proteins in the primosome of DNA replication. J Biol Chem. 1981 May 25;256(10):5273–5280. [PubMed] [Google Scholar]
- Arai K., McMacken R., Yasuda S., Kornberg A. Purification and properties of Escherichia coli protein i, a prepriming protein in phi X174 DNA replication. J Biol Chem. 1981 May 25;256(10):5281–5286. [PubMed] [Google Scholar]
- Arai N., Arai K., Kornberg A. Complexes of Rep protein with ATP and DNA as a basis for helicase action. J Biol Chem. 1981 May 25;256(10):5287–5293. [PubMed] [Google Scholar]
- Arai N., Kornberg A. Rep protein as a helicase in an active, isolatable replication fork of duplex phi X174 DNA. J Biol Chem. 1981 May 25;256(10):5294–5298. [PubMed] [Google Scholar]
- Arai N., Polder L., Akai K., Kornberg A. Replication of phi X174 DNA with purified enzymes. II. Multiplication of the duplex form by coupling of continuous and discontinuous synthetic pathways. J Biol Chem. 1981 May 25;256(10):5239–5246. [PubMed] [Google Scholar]
- Araki H., Ogawa H. A T7 amber mutant defective in DNA-binding protein. Mol Gen Genet. 1981;183(1):66–73. doi: 10.1007/BF00270140. [DOI] [PubMed] [Google Scholar]
- Argos P., Tucker A. D., Philipson L. Primary structural relationships may reflect similar DNA replication strategies. Virology. 1986 Mar;149(2):208–216. doi: 10.1016/0042-6822(86)90122-4. [DOI] [PubMed] [Google Scholar]
- Arnold G. F., Phillips T. A., Tessman I. Levels of DNA topoisomerases, single-stranded-DNA-binding protein, and DNA polymerase I in rho+ and rho-15 strains of Escherichia coli. J Bacteriol. 1989 Sep;171(9):5183–5186. doi: 10.1128/jb.171.9.5183-5186.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach J. I., Howard-Flanders P. Identification of wild-type or mutant alleles of bacterial genes cloned on a bacteriophage lambda vector: isolation of uvrC(am) and other mutants. J Bacteriol. 1981 May;146(2):713–717. doi: 10.1128/jb.146.2.713-717.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backendorf C., Brandsma J. A., Kartasova T., van de Putte P. In vivo regulation of the uvrA gene: role of the "-10" and "-35" promoter regions. Nucleic Acids Res. 1983 Sep 10;11(17):5795–5810. doi: 10.1093/nar/11.17.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. A., Funnell B. E., Kornberg A. Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J Biol Chem. 1987 May 15;262(14):6877–6885. [PubMed] [Google Scholar]
- Baker T. A., Sekimizu K., Funnell B. E., Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986 Apr 11;45(1):53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- Baluch J., Chase J. W., Sussman R. Synthesis of recA protein and induction of bacteriophage lambda in single-strand deoxyribonucleic acid-binding protein mutants of Escherichia coli. J Bacteriol. 1980 Nov;144(2):489–498. doi: 10.1128/jb.144.2.489-498.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay P. K., Wu C. W. Fluorescence and chemical studies on the interaction of Escherichia coli DNA-binding protein with single-stranded DNA. Biochemistry. 1978 Sep 19;17(19):4078–4085. doi: 10.1021/bi00612a032. [DOI] [PubMed] [Google Scholar]
- Barat M., Mignotte B. A DNA binding protein from Xenopus laevis oocyte mitochondria. Chromosoma. 1981;82(4):583–593. doi: 10.1007/BF00295014. [DOI] [PubMed] [Google Scholar]
- Barbour S. D., Nagaishi H., Templin A., Clark A. J. Biochemical and genetic studies of recombination proficiency in Escherichia coli. II. Rec+ revertants caused by indirect suppression of rec- mutations. Proc Natl Acad Sci U S A. 1970 Sep;67(1):128–135. doi: 10.1073/pnas.67.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict R. C., Kowalczykowski S. C. Increase of the DNA strand assimilation activity of recA protein by removal of the C terminus and structure-function studies of the resulting protein fragment. J Biol Chem. 1988 Oct 25;263(30):15513–15520. [PubMed] [Google Scholar]
- Benz E. W., Jr, Reinberg D., Vicuna R., Hurwitz J. Initiation of DNA replication by the dnaG protein. J Biol Chem. 1980 Feb 10;255(3):1096–1106. [PubMed] [Google Scholar]
- Bernstein C. Deoxyribonucleic acid repair in bacteriophage. Microbiol Rev. 1981 Mar;45(1):72–98. doi: 10.1128/mr.45.1.72-98.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold V., Thielmann H. W., Geider K. Carcinogens inhibit DNA synthesis with isolated DNA polymerases from Escherichia coli. FEBS Lett. 1978 Feb 1;86(1):81–84. doi: 10.1016/0014-5793(78)80103-3. [DOI] [PubMed] [Google Scholar]
- Beyreuther K., Berthold-Schmidt V., Geider K. Biological activity and a partial amino-acid sequence of Escherichia coli DNA-binding protein I isolated from overproducing cells. Eur J Biochem. 1982 Apr 1;123(2):415–420. doi: 10.1111/j.1432-1033.1982.tb19784.x. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Radding C. M. Insertions, deletions and mismatches in heteroduplex DNA made by recA protein. Cell. 1983 Dec;35(2 Pt 1):511–520. doi: 10.1016/0092-8674(83)90185-x. [DOI] [PubMed] [Google Scholar]
- Bianchi M., Riboli B., Magni G. E. coli recA protein possesses a strand separating activity on short duplex DNAs. EMBO J. 1985 Nov;4(11):3025–3030. doi: 10.1002/j.1460-2075.1985.tb04039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanar M. A., Sandler S. J., Armengod M. E., Ream L. W., Clark A. J. Molecular analysis of the recF gene of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4622–4626. doi: 10.1073/pnas.81.15.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobst A. M., Langemeier P. W., Warwick-Koochaki P. E., Bobst E. V., Ireland J. C. Nucleic binding affinity of bacteriophage T4 gene 32 protein in the cooperative binding mode. J Biol Chem. 1982 Jun 10;257(11):6184–6193. [PubMed] [Google Scholar]
- Bobst E. V., Bobst A. M., Perrino F. W., Meyer R. R., Rein D. C. Variability in the nucleic acid binding site size and the amount of single-stranded DNA-binding protein in Escherichia coli. FEBS Lett. 1985 Feb 11;181(1):133–137. doi: 10.1016/0014-5793(85)81128-5. [DOI] [PubMed] [Google Scholar]
- Boidot-Forget M., Saison-Behmoaras T., Toulmé J. J., Hélène C. Single-strand binding proteins from phage T4 and E. coli form higher order structures with poly(dT). Biochimie. 1986 Sep;68(9):1129–1134. doi: 10.1016/s0300-9084(86)80188-2. [DOI] [PubMed] [Google Scholar]
- Bonner C. A., Randall S. K., Rayssiguier C., Radman M., Eritja R., Kaplan B. E., McEntee K., Goodman M. F. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J Biol Chem. 1988 Dec 15;263(35):18946–18952. [PubMed] [Google Scholar]
- Botstein D., Maurer R. Genetic approaches to the analysis of microbial development. Annu Rev Genet. 1982;16:61–83. doi: 10.1146/annurev.ge.16.120182.000425. [DOI] [PubMed] [Google Scholar]
- Bouché J. P., Rowen L., Kornberg A. The RNA primer synthesized by primase to initiate phage G4 DNA replication. J Biol Chem. 1978 Feb 10;253(3):765–769. [PubMed] [Google Scholar]
- Bouché J. P., Zechel K., Kornberg A. dnaG gene product, a rifampicin-resistant RNA polymerase, initiates the conversion of a single-stranded coliphage DNA to its duplex replicative form. J Biol Chem. 1975 Aug 10;250(15):5995–6001. [PubMed] [Google Scholar]
- Braedt G., Smith G. R. Strand specificity of DNA unwinding by RecBCD enzyme. Proc Natl Acad Sci U S A. 1989 Feb;86(3):871–875. doi: 10.1073/pnas.86.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma J. A., Bosch D., Backendorf C., van de Putte P. A common regulatory region shared by divergently transcribed genes of the Escherichia coli SOS system. Nature. 1983 Sep 15;305(5931):243–245. doi: 10.1038/305243a0. [DOI] [PubMed] [Google Scholar]
- Brandsma J. A., Bosch D., de Ruÿter M., van de Putte P. Analysis of the regulatory region of the ssb gene of Escherichia coli. Nucleic Acids Res. 1985 Jul 25;13(14):5095–5109. doi: 10.1093/nar/13.14.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma J. A., Stoorvogel J., van Sluis C. A., van de Putte P. Effect of lexA and ssb genes, present on a uvrA recombinant plasmid, on the UV survival of Escherichia coli K-12. Gene. 1982 Apr;18(1):77–85. doi: 10.1016/0378-1119(82)90058-0. [DOI] [PubMed] [Google Scholar]
- Brandsma J. A., van Sluis C. A., van de Putte P. Use of transposons in cloning poorly selectable genes of Escherichia coli: cloning of uvrA and adjacent genes. J Bacteriol. 1981 Aug;147(2):682–684. doi: 10.1128/jb.147.2.682-684.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D., Schekman R., Kornberg A. A possible role for RNA polymerase in the initiation of M13 DNA synthesis. Proc Natl Acad Sci U S A. 1971 Nov;68(11):2826–2829. doi: 10.1073/pnas.68.11.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant F. R. Construction of a recombinase-deficient mutant recA protein that retains single-stranded DNA-dependent ATPase activity. J Biol Chem. 1988 Jun 25;263(18):8716–8723. [PubMed] [Google Scholar]
- Bryant F. R., Riddles P. W., Lehman I. R. Studies of the mechanism of DNA pairing by the RecA protein of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:535–539. doi: 10.1101/sqb.1984.049.01.060. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Lohman T. M. A general method of analysis of ligand-macromolecule equilibria using a spectroscopic signal from the ligand to monitor binding. Application to Escherichia coli single-strand binding protein-nucleic acid interactions. Biochemistry. 1987 Jun 2;26(11):3099–3106. doi: 10.1021/bi00385a023. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Lohman T. M. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986 Dec 2;25(24):7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Lohman T. M. Limited co-operativity in protein-nucleic acid interactions. A thermodynamic model for the interactions of Escherichia coli single strand binding protein with single-stranded nucleic acids in the "beaded", (SSB)65 mode. J Mol Biol. 1987 Jun 20;195(4):897–907. doi: 10.1016/0022-2836(87)90493-1. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Lohman T. M. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. I. Evidence and a quantitative model. J Mol Biol. 1989 May 5;207(1):249–268. doi: 10.1016/0022-2836(89)90454-3. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Lohman T. M. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II. Salt, temperature and oligonucleotide length effects. J Mol Biol. 1989 May 5;207(1):269–288. doi: 10.1016/0022-2836(89)90455-5. [DOI] [PubMed] [Google Scholar]
- Bujalowski W., Overman L. B., Lohman T. M. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J Biol Chem. 1988 Apr 5;263(10):4629–4640. [PubMed] [Google Scholar]
- CLARK A. J., MARGULIES A. D. ISOLATION AND CHARACTERIZATION OF RECOMBINATION-DEFICIENT MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1965 Feb;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Finet J. R., Jhon N. I., Khamis M. I., Maki A. H., Ruvolo P. P., Chase J. W. An IncY plasmid-encoded single-stranded DNA-binding protein from Escherichia coli shows the identical pattern of stacked tryptophan residues as the chromosomal ssb gene product. Eur J Biochem. 1988 Dec 1;178(1):101–107. doi: 10.1111/j.1432-1033.1988.tb14434.x. [DOI] [PubMed] [Google Scholar]
- Casas-Finet J. R., Khamis M. I., Maki A. H., Chase J. W. Tryptophan 54 and phenylalanine 60 are involved synergistically in the binding of E. coli SSB protein to single-stranded polynucleotides. FEBS Lett. 1987 Aug 17;220(2):347–352. doi: 10.1016/0014-5793(87)80844-x. [DOI] [PubMed] [Google Scholar]
- Casas-Finet J. R., Khamis M. I., Maki A. H., Ruvolo P. P., Chase J. W. Optically detected magnetic resonance of tryptophan residues in Escherichia coli ssb gene product and E. coli plasmid-encoded single-stranded DNA-binding proteins and their complexes with poly(deoxythymidylic) acid. J Biol Chem. 1987 Jun 25;262(18):8574–8583. [PubMed] [Google Scholar]
- Cassuto E., West S. C., Mursalim J., Conlon S., Howard-Flanders P. Initiation of genetic recombination: homologous pairing between duplex DNA molecules promoted by recA protein. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3962–3966. doi: 10.1073/pnas.77.7.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha T. A., Maki A. H. Close range interactions between nucleotide bases and tryptophan residues in an Escherichia coli single-stranded DNA binding protein-mercurated poly(uridylic acid) complex. A study by optically detected magnetic resonance spectroscopy. J Biol Chem. 1984 Jan 25;259(2):1105–1109. [PubMed] [Google Scholar]
- Chabbert M., Cazenave C., Hélène C. Kinetic studies of recA protein binding to a fluorescent single-stranded polynucleotide. Biochemistry. 1987 Apr 21;26(8):2218–2225. doi: 10.1021/bi00382a022. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Flory J., Ruddle N. H., Murphy J. B., Williams K. R. A monoclonal antibody that recognizes the functional domain of Escherichia coli single-stranded DNA binding protein that includes the ssb-113 mutation. J Biol Chem. 1985 Jun 25;260(12):7214–7218. [PubMed] [Google Scholar]
- Chase J. W., L'Italien J. J., Murphy J. B., Spicer E. K., Williams K. R. Characterization of the Escherichia coli SSB-113 mutant single-stranded DNA-binding protein. Cloning of the gene, DNA and protein sequence analysis, high pressure liquid chromatography peptide mapping, and DNA-binding studies. J Biol Chem. 1984 Jan 25;259(2):805–814. [PubMed] [Google Scholar]
- Chase J. W., Merrill B. M., Williams K. R. F sex factor encodes a single-stranded DNA binding protein (SSB) with extensive sequence homology to Escherichia coli SSB. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5480–5484. doi: 10.1073/pnas.80.18.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase J. W., Murphy J. B., Whittier R. F., Lorensen E., Sninsky J. J. Amplification of ssb-1 mutant single-stranded DNA-binding protein in Escherichia coli. J Mol Biol. 1983 Feb 25;164(2):193–211. doi: 10.1016/0022-2836(83)90075-x. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Whittier R. F., Auerbach J., Sancar A., Rupp W. D. Amplification of single-strand DNA binding protein in Escherichia coli. Nucleic Acids Res. 1980 Jul 25;8(14):3215–3227. doi: 10.1093/nar/8.14.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase J. W., Williams K. R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- Chaudhury A. M., Smith G. R. A new class of Escherichia coli recBC mutants: implications for the role of RecBC enzyme in homologous recombination. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7850–7854. doi: 10.1073/pnas.81.24.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. C., Smith G. R. Cutting of chi-like sequences by the RecBCD enzyme of Escherichia coli. J Mol Biol. 1987 Apr 20;194(4):747–750. doi: 10.1016/0022-2836(87)90252-x. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Chow S. A., Rao B. J., Radding C. M. Reversibility of strand invasion promoted by recA protein and its inhibition by Escherichia coli single-stranded DNA-binding protein or phage T4 gene 32 protein. J Biol Chem. 1988 Jan 5;263(1):200–209. [PubMed] [Google Scholar]
- Christiansen C., Baldwin R. L. Catalysis of DNA reassociation by the Escherichia coli DNA binding protein: A polyamine-dependent reaction. J Mol Biol. 1977 Sep 25;115(3):441–454. doi: 10.1016/0022-2836(77)90164-4. [DOI] [PubMed] [Google Scholar]
- Christiansen G., Griffith J. Visualization of the paranemic joining of homologous DNA molecules catalyzed by the RecA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2066–2070. doi: 10.1073/pnas.83.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysogelos S., Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesla Z., O'Brien P., Clark A. J. Genetic analysis of UV mutagenesis of the Escherichia coli glyU gene. Mol Gen Genet. 1987 Apr;207(1):1–8. doi: 10.1007/BF00331483. [DOI] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Clark A. J., Sandler S. J., Willis D. K., Chu C. C., Blanar M. A., Lovett S. T. Genes of the RecE and RecF pathways of conjugational recombination in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:453–462. doi: 10.1101/sqb.1984.049.01.051. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M., Greipel J., Maass G. Conformation of the DNA undecamer 5'd(A-A-G-T-G-T-G-A-T-A-T) bound to the single-stranded DNA binding protein of Escherichia coli. A time-dependent transferred nuclear Overhauser enhancement study. J Mol Biol. 1986 Jan 5;187(1):119–124. doi: 10.1016/0022-2836(86)90411-0. [DOI] [PubMed] [Google Scholar]
- Cohen S. P., Resnick J., Sussman R. Interaction of single-strand binding protein and RecA protein at the single-stranded DNA site. J Mol Biol. 1983 Jul 15;167(4):901–909. doi: 10.1016/s0022-2836(83)80119-3. [DOI] [PubMed] [Google Scholar]
- Coleman J. E., Oakley J. L. Physical chemical studies of the structure and function of DNA binding (helix-destabilizing) proteins. CRC Crit Rev Biochem. 1980 Jan;7(3):247–289. doi: 10.3109/10409238009105463. [DOI] [PubMed] [Google Scholar]
- Cotterill S. M., Fersht A. R. Direct observation of complexes of ssb and recA proteins with a fluorescent single-stranded deoxyribonucleic acid derivative. Biochemistry. 1983 Dec 6;22(25):5878–5881. doi: 10.1021/bi00294a029. [DOI] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Directionality and polarity in recA protein-promoted branch migration. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Enzymes of general recombination. Annu Rev Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. recA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. recA protein-promoted DNA strand exchange. Stable complexes of recA protein and single-stranded DNA formed in the presence of ATP and single-stranded DNA binding protein. J Biol Chem. 1982 Jul 25;257(14):8523–8532. [PubMed] [Google Scholar]
- Cox M. M., Morrical S. W., Neuendorf S. K. Unidirectional branch migration promoted by nucleoprotein filaments of RecA protein and DNA. Cold Spring Harb Symp Quant Biol. 1984;49:525–533. doi: 10.1101/sqb.1984.049.01.059. [DOI] [PubMed] [Google Scholar]
- Cox M. M., Soltis D. A., Lehman I. R., DeBrosse C., Benkovic S. J. ADP-mediated dissociation of stable complexes of recA protein and single-stranded DNA. J Biol Chem. 1983 Feb 25;258(4):2586–2592. [PubMed] [Google Scholar]
- Cox M. M., Soltis D. A., Livneh Z., Lehman I. R. On the role of single-stranded DNA binding protein in recA protein-promoted DNA strand exchange. J Biol Chem. 1983 Feb 25;258(4):2577–2585. [PubMed] [Google Scholar]
- Craig N. L., Roberts J. W. E. coli recA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature. 1980 Jan 3;283(5742):26–30. doi: 10.1038/283026a0. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Roberts J. W. Function of nucleoside triphosphate and polynucleotide in Escherichia coli recA protein-directed cleavage of phage lambda repressor. J Biol Chem. 1981 Aug 10;256(15):8039–8044. [PubMed] [Google Scholar]
- Cuozzo M., Silverman P. M. Characterization of the F plasmid TraJ protein synthesized in F' and Hfr strains of Escherichia coli K-12. J Biol Chem. 1986 Apr 15;261(11):5175–5179. [PubMed] [Google Scholar]
- Delius H., Mantell N. J., Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J Mol Biol. 1972 Jun 28;67(3):341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- Dodson M., Echols H., Wickner S., Alfano C., Mensa-Wilmot K., Gomes B., LeBowitz J., Roberts J. D., McMacken R. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: localized unwinding of duplex DNA by a six-protein reaction. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7638–7642. doi: 10.1073/pnas.83.20.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M., Roberts J., McMacken R., Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: complexes with lambda O protein and with lambda O, lambda P, and Escherichia coli DnaB proteins. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4678–4682. doi: 10.1073/pnas.82.14.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donch J. J., Greenberg J. Suppression of filamentation in a new lex mutant by a linked (lexA) mutation in Escherichia coli. Mutat Res. 1976 Mar;34(3):533–538. doi: 10.1016/0027-5107(76)90228-1. [DOI] [PubMed] [Google Scholar]
- Egner C., Azhderian E., Tsang S. S., Radding C. M., Chase J. W. Effects of various single-stranded-DNA-binding proteins on reactions promoted by RecA protein. J Bacteriol. 1987 Aug;169(8):3422–3428. doi: 10.1128/jb.169.8.3422-3428.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Griffith J., Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Scott J. F., Kornberg A. An enzyme system for replication of duplex circular DNA: the replicative form of phage phi X174. Proc Natl Acad Sci U S A. 1976 May;73(5):1594–1597. doi: 10.1073/pnas.73.5.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis D. G., Amundsen S. K., Smith G. R. Genetic functions promoting homologous recombination in Escherichia coli: a study of inversions in phage lambda. Genetics. 1987 Jan;115(1):11–24. doi: 10.1093/genetics/115.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler J. S., Tessman I., Tessman E. S. Lethality of the double mutations rho rep and rho ssb in Escherichia coli. J Bacteriol. 1985 Feb;161(2):609–614. doi: 10.1128/jb.161.2.609-614.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay P. J., Johanson K. O., McHenry C. S., Bambara R. A. Size classes of products synthesized processively by DNA polymerase III and DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1981 Jan 25;256(2):976–983. [PubMed] [Google Scholar]
- Fay P. J., Johanson K. O., McHenry C. S., Bambara R. A. Size classes of products synthesized processively by two subassemblies of Escherichia coli DNA polymerase III holoenzyme. J Biol Chem. 1982 May 25;257(10):5692–5699. [PubMed] [Google Scholar]
- Fiddes J. C., Barrell B. G., Godson G. N. Nucleotide sequences of the separate origins of synthesis of bacteriophage G4 viral and complementary DNA strands. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1081–1085. doi: 10.1073/pnas.75.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firshein W. Role of the DNA/membrane complex in prokaryotic DNA replication. Annu Rev Microbiol. 1989;43:89–120. doi: 10.1146/annurev.mi.43.100189.000513. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., James A. A., Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981 Nov 12;294(5837):184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Fishel R., Kolodner R. Gene conversion in Escherichia coli: the recF pathway for resolution of heteroduplex DNA. J Bacteriol. 1989 Jun;171(6):3046–3052. doi: 10.1128/jb.171.6.3046-3052.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory J., Radding C. M. Visualization of recA protein and its association with DNA: a priming effect of single-strand-binding protein. Cell. 1982 Apr;28(4):747–756. doi: 10.1016/0092-8674(82)90054-x. [DOI] [PubMed] [Google Scholar]
- Flory J., Tsang S. S., Muniyappa K. Isolation and visualization of active presynaptic filaments of recA protein and single-stranded DNA. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory S. S., Tsang J., Muniyappa K., Bianchi M., Gonda D., Kahn R., Azhderian E., Egner C., Shaner S., Radding C. M. Intermediates in homologous pairing promoted by RecA protein and correlations of recombination in vitro and in vivo. Cold Spring Harb Symp Quant Biol. 1984;49:513–523. doi: 10.1101/sqb.1984.049.01.058. [DOI] [PubMed] [Google Scholar]
- Formosa T., Burke R. L., Alberts B. M. Affinity purification of bacteriophage T4 proteins essential for DNA replication and genetic recombination. Proc Natl Acad Sci U S A. 1983 May;80(9):2442–2446. doi: 10.1073/pnas.80.9.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradkin G. E., Rabinkova E. V., Torosian M. V., Aizenberg O. A., Naumova L. A. Rol' vzaimodeistviia in vivo belkov SSB s DNK-polimerazoi II. Mol Biol (Mosk) 1988 Jan-Feb;22(1):111–116. [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R., Georgopoulos C., Tilly K., Herskowitz I., Banuett F. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol Rev. 1984 Dec;48(4):299–325. doi: 10.1128/mr.48.4.299-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller C. W., Richardson C. C. Initiation of DNA replication at the primary origin of bacteriophage T7 by purified proteins. Initiation of bidirectional synthesis. J Biol Chem. 1985 Mar 10;260(5):3197–3206. [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Kaguni J. M., Kornberg A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7370–7374. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell B. E., Baker T. A., Kornberg A. Complete enzymatic replication of plasmids containing the origin of the Escherichia coli chromosome. J Biol Chem. 1986 Apr 25;261(12):5616–5624. [PubMed] [Google Scholar]
- Funnell B. E., Baker T. A., Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987 Jul 25;262(21):10327–10334. [PubMed] [Google Scholar]
- Geider K., Beck E., Schaller H. An RNA transcribed from DNA at the origin of phage fd single strand to replicative form conversion. Proc Natl Acad Sci U S A. 1978 Feb;75(2):645–649. doi: 10.1073/pnas.75.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geider K., Hoffmann-Berling H. Proteins controlling the helical structure of DNA. Annu Rev Biochem. 1981;50:233–260. doi: 10.1146/annurev.bi.50.070181.001313. [DOI] [PubMed] [Google Scholar]
- Geider K. Interaction of DNA with DNA-binding proteins: protein exchange and complex stability. Eur J Biochem. 1978 Jul 3;87(3):617–622. doi: 10.1111/j.1432-1033.1978.tb12414.x. [DOI] [PubMed] [Google Scholar]
- Geider K., Kornberg A. Conversion of the M13 viral single strand to the double-stranded replicative forms by purified proteins. J Biol Chem. 1974 Jul 10;249(13):3999–4005. [PubMed] [Google Scholar]
- Gillen J. R., Willis D. K., Clark A. J. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J Bacteriol. 1981 Jan;145(1):521–532. doi: 10.1128/jb.145.1.521-532.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassberg J., Meyer R. R., Kornberg A. Mutant single-strand binding protein of Escherichia coli: genetic and physiological characterization. J Bacteriol. 1979 Oct;140(1):14–19. doi: 10.1128/jb.140.1.14-19.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G. C., Gargiulo G., Rena-Descalzi L., Worcel A. Escherichia coli single-strand binding protein stabilizes specific denatured sites in superhelical DNA. Nature. 1983 Jun 30;303(5920):770–774. doi: 10.1038/303770a0. [DOI] [PubMed] [Google Scholar]
- Goetz G. S., Hurwitz J. Studies on the role of the phi X174 gene A protein in phi X viral strand synthesis. I. Replication of DNA containing an alteration in position 1 of the 30-nucleotide icosahedral bacteriophage origin. J Biol Chem. 1988 Nov 5;263(31):16421–16432. [PubMed] [Google Scholar]
- Goldmark P. J., Linn S. Purification and properties of the recBC DNase of Escherichia coli K-12. J Biol Chem. 1972 Mar 25;247(6):1849–1860. [PubMed] [Google Scholar]
- Golub E. I., Low K. B. Conjugative plasmids of enteric bacteria from many different incompatibility groups have similar genes for single-stranded DNA-binding proteins. J Bacteriol. 1985 Apr;162(1):235–241. doi: 10.1128/jb.162.1.235-241.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E. I., Low K. B. Derepression of single-stranded DNA-binding protein genes on plasmids derepressed for conjugation, and complementation of an E. coli ssb- mutation by these genes. Mol Gen Genet. 1986 Sep;204(3):410–416. doi: 10.1007/BF00331017. [DOI] [PubMed] [Google Scholar]
- Golub E. I., Low K. B. Indirect stimulation of genetic recombination. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1401–1405. doi: 10.1073/pnas.80.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E., Bailone A., Devoret R. A gene encoding an SOS inhibitor is present in different conjugative plasmids. J Bacteriol. 1988 Sep;170(9):4392–4394. doi: 10.1128/jb.170.9.4392-4394.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J. H., Marians K. J. Mutational analysis of primosome assembly sites. Evidence for alternative DNA structures. J Biol Chem. 1985 Oct 5;260(22):12266–12272. [PubMed] [Google Scholar]
- Greenberg J., Berends L. J., Donch J., Green M. H. exrB: a malB-linked gene in Escherichia coli B involved in sensitivity to radiation and filament formation. Genet Res. 1974 Apr;23(2):175–184. doi: 10.1017/s0016672300014798. [DOI] [PubMed] [Google Scholar]
- Greenberg J., Berends L., Donch J., Johnson B. Reversion studies with exrB in Escherichia coli. Genet Res. 1975 Apr;25(2):109–117. doi: 10.1017/s0016672300015512. [DOI] [PubMed] [Google Scholar]
- Greenberg J., Donch J., Berends L. The dominance of exrB over exrB+ in heterodiploids of Escherichia coli. Genet Res. 1975 Feb;25(1):39–44. doi: 10.1017/s001667230001541x. [DOI] [PubMed] [Google Scholar]
- Greenberg J., Donch J. Sensitivity to elevated temperatures in exrB strains of Escherichia coli. Mutat Res. 1974 Dec;25(3):403–405. doi: 10.1016/0027-5107(74)90070-0. [DOI] [PubMed] [Google Scholar]
- Greipel J., Maass G., Mayer F. Complexes of the single-stranded DNA-binding protein from Escherichia coli (Eco SSB) with poly(dT). An investigation of their structure and internal dynamics by means of electron microscopy and NMR. Biophys Chem. 1987 May 9;26(2-3):149–161. doi: 10.1016/0301-4622(87)80018-2. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Harris L. D. DNA strand exchanges. CRC Crit Rev Biochem. 1988;23 (Suppl 1):S43–S86. [PubMed] [Google Scholar]
- Griffith J. D., Harris L. D., Register J., 3rd Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb Symp Quant Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- Grilley M., Welsh K. M., Su S. S., Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989 Jan 15;264(2):1000–1004. [PubMed] [Google Scholar]
- Hanawalt P. C., Cooper P. K., Ganesan A. K., Smith C. A. DNA repair in bacteria and mammalian cells. Annu Rev Biochem. 1979;48:783–836. doi: 10.1146/annurev.bi.48.070179.004031. [DOI] [PubMed] [Google Scholar]
- Hartman P. E., Roth J. R. Mechanisms of suppression. Adv Genet. 1973;17:1–105. doi: 10.1016/s0065-2660(08)60170-4. [DOI] [PubMed] [Google Scholar]
- Helene C., Maurizot J. C. Interactions of oligopeptides with nucleic acids. CRC Crit Rev Biochem. 1981;10(3):213–258. doi: 10.3109/10409238109113600. [DOI] [PubMed] [Google Scholar]
- Heuser J., Griffith J. Visualization of RecA protein and its complexes with DNA by quick-freeze/deep-etch electron microscopy. J Mol Biol. 1989 Dec 5;210(3):473–484. doi: 10.1016/0022-2836(89)90124-1. [DOI] [PubMed] [Google Scholar]
- Hickson I. D., Arthur H. M., Bramhill D., Emmerson P. T. The E. coli uvrD gene product is DNA helicase II. Mol Gen Genet. 1983;190(2):265–270. doi: 10.1007/BF00330649. [DOI] [PubMed] [Google Scholar]
- Honigberg S. M., Gonda D. K., Flory J., Radding C. M. The pairing activity of stable nucleoprotein filaments made from recA protein, single-stranded DNA, and adenosine 5'-(gamma-thio)triphosphate. J Biol Chem. 1985 Sep 25;260(21):11845–11851. [PubMed] [Google Scholar]
- Horii Z., Clark A. J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973 Oct 25;80(2):327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- Imber R., Low R. L., Ray D. S. Identification of a primosome assembly site in the region of the ori 2 replication origin of the Escherichia coli mini-F plasmid. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7132–7136. doi: 10.1073/pnas.80.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. F. Genetic mapping of the lexC-113 mutation. Mol Gen Genet. 1977 Nov 29;157(1):91–97. doi: 10.1007/BF00268691. [DOI] [PubMed] [Google Scholar]
- Johnson B. F. Suppression of the lexC (ssbA) mutation of Escherichia coli by a mutant of bacteriophage P1. Mol Gen Genet. 1982;186(1):122–126. doi: 10.1007/BF00422923. [DOI] [PubMed] [Google Scholar]
- Johnson B. F. Two-dimensional electrophoretic analysis of the regulation of SOS proteins in three ssb mutants. Arch Microbiol. 1984 Jun;138(2):106–112. doi: 10.1007/BF00413009. [DOI] [PubMed] [Google Scholar]
- Joseph J. W., Kolodner R. Exonuclease VIII of Escherichia coli. II. Mechanism of action. J Biol Chem. 1983 Sep 10;258(17):10418–10424. [PubMed] [Google Scholar]
- Julin D. A., Riddles P. W., Lehman I. R. On the mechanism of pairing of single- and double-stranded DNA molecules by the recA and single-stranded DNA-binding proteins of Escherichia coli. J Biol Chem. 1986 Jan 25;261(3):1025–1030. [PubMed] [Google Scholar]
- Kaguni J. M., Kornberg A. The rho subunit of RNA polymerase holoenzyme confers specificity in priming M13 viral DNA replication. J Biol Chem. 1982 May 25;257(10):5437–5443. [PubMed] [Google Scholar]
- Kahn R., Radding C. M. Separation of the presynaptic and synaptic phases of homologous pairing promoted by recA protein. J Biol Chem. 1984 Jun 25;259(12):7495–7503. [PubMed] [Google Scholar]
- Kaiser K., Murray N. E. Physical characterisation of the "Rac prophage" in E. coli K12. Mol Gen Genet. 1979 Sep;175(2):159–174. doi: 10.1007/BF00425532. [DOI] [PubMed] [Google Scholar]
- Kenyon C. J., Walker G. C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980 May;77(5):2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamis M. I., Casas-Finet J. R., Maki A. H., Murphy J. B., Chase J. W. Investigation of the role of individual tryptophan residues in the binding of Escherichia coli single-stranded DNA binding protein to single-stranded polynucleotides. A study by optical detection of magnetic resonance and site-selected mutagenesis. J Biol Chem. 1987 Aug 15;262(23):10938–10945. [PubMed] [Google Scholar]
- Khamis M. I., Casas-Finet J. R., Maki A. H., Murphy J. B., Chase J. W. Role of tryptophan 54 in the binding of E. coli single-stranded DNA-binding protein to single-stranded polynucleotides. FEBS Lett. 1987 Jan 26;211(2):155–159. doi: 10.1016/0014-5793(87)81427-8. [DOI] [PubMed] [Google Scholar]
- Khamis M. I., Casas-Finet J. R., Maki A. H., Ruvolo P. P., Chase J. W. Optically detected magnetic resonance of tryptophan residues in complexes formed between a bacterial single-stranded DNA binding protein and heavy atom modified poly(uridylic acid). Biochemistry. 1987 Jun 16;26(12):3347–3354. doi: 10.1021/bi00386a015. [DOI] [PubMed] [Google Scholar]
- Khamis M. I., Casas-Finet J. R., Maki A. H. Stacking interactions of tryptophan residues and nucleotide bases in complexes formed between Escherichia coli single-stranded DNA binding protein and heavy atom-modified poly(uridylic) acid. A study by optically detected magnetic resonance spectroscopy. J Biol Chem. 1987 Feb 5;262(4):1725–1733. [PubMed] [Google Scholar]
- Kohno T., Roth J. Electrolyte effects on the activity of mutant enzymes in vivo and in vitro. Biochemistry. 1979 Apr 3;18(7):1386–1392. doi: 10.1021/bi00574a041. [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Capage M. A., Golub E. I., Low K. B. F sex factor of Escherichia coli K-12 codes for a single-stranded DNA binding protein. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4422–4426. doi: 10.1073/pnas.80.14.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Fishel R. A., Howard M. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985 Sep;163(3):1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Helinski D. R. Regulation of initiation of DNA replication. Annu Rev Genet. 1979;13:355–391. doi: 10.1146/annurev.ge.13.120179.002035. [DOI] [PubMed] [Google Scholar]
- Konforti B. B., Davis R. W. 3' homologous free ends are required for stable joint molecule formation by the RecA and single-stranded binding proteins of Escherichia coli. Proc Natl Acad Sci U S A. 1987 Feb;84(3):690–694. doi: 10.1073/pnas.84.3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A. DNA replication. J Biol Chem. 1988 Jan 5;263(1):1–4. [PubMed] [Google Scholar]
- Kornberg A. Enzyme studies of replication of the Escherichia coli chromosome. Adv Exp Med Biol. 1984;179:3–16. doi: 10.1007/978-1-4684-8730-5_1. [DOI] [PubMed] [Google Scholar]
- Kornberg A., Scott J. F., Bertsch L. L. ATP utilization by rep protein in the catalytic separation of DNA strands at a replicating fork. J Biol Chem. 1978 May 10;253(9):3298–3304. [PubMed] [Google Scholar]
- Kowalczykowski S. C., Clow J., Somani R., Varghese A. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA. Demonstration of competitive binding and the lack of a specific protein-protein interaction. J Mol Biol. 1987 Jan 5;193(1):81–95. doi: 10.1016/0022-2836(87)90629-2. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Krupp R. A. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J Mol Biol. 1987 Jan 5;193(1):97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- Krauss G., Sindermann H., Schomburg U., Maass G. Escherichia coli single-strand deoxyribonucleic acid binding protein: stability, specificity, and kinetics of complexes with oligonucleotides and deoxyribonucleic acid. Biochemistry. 1981 Sep 1;20(18):5346–5352. doi: 10.1021/bi00521a040. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Exonucleolytic proofreading. Cell. 1988 Jun 17;53(6):837–840. doi: 10.1016/s0092-8674(88)90189-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Meyer R. R., Loeb L. A. Single-strand binding protein enhances fidelity of DNA synthesis in vitro. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6331–6335. doi: 10.1073/pnas.76.12.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Schaaper R. M., Loeb L. A. Depurination-induced infidelity of deoxyribonucleic acid synthesis with purified deoxyribonucleic acid replication proteins in vitro. Biochemistry. 1983 May 10;22(10):2378–2384. doi: 10.1021/bi00279a012. [DOI] [PubMed] [Google Scholar]
- Kushner S. R., Nagaishi H., Templin A., Clark A. J. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci U S A. 1971 Apr;68(4):824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Shin O., Bodner J. B., McHenry C. S., Bambara R. A. Properties of initiation complexes formed between Escherichia coli DNA polymerase III holoenzyme and primed DNA in the absence of ATP. J Biol Chem. 1987 Feb 15;262(5):2121–2130. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LEHMAN I. R., NUSSBAUM A. L. THE DEOXYRIBONUCLEASES OF ESCHERICHIA COLI. V. ON THE SPECIFICITY OF EXONUCLEASE I (PHOSPHODIESTERASE). J Biol Chem. 1964 Aug;239:2628–2636. [PubMed] [Google Scholar]
- LaDuca R. J., Crute J. J., McHenry C. S., Bambara R. A. The beta subunit of the Escherichia coli DNA polymerase III holoenzyme interacts functionally with the catalytic core in the absence of other subunits. J Biol Chem. 1986 Jun 5;261(16):7550–7557. [PubMed] [Google Scholar]
- LaDuca R. J., Fay P. J., Chuang C., McHenry C. S., Bambara R. A. Site-specific pausing of deoxyribonucleic acid synthesis catalyzed by four forms of Escherichia coli DNA polymerase III. Biochemistry. 1983 Oct 25;22(22):5177–5188. doi: 10.1021/bi00291a018. [DOI] [PubMed] [Google Scholar]
- Lahue R. S., Au K. G., Modrich P. DNA mismatch correction in a defined system. Science. 1989 Jul 14;245(4914):160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Langowski J., Benight A. S., Fujimoto B. S., Schurr J. M., Schomburg U. Change of conformation and internal dynamics of supercoiled DNA upon binding of Escherichia coli single-strand binding protein. Biochemistry. 1985 Jul 16;24(15):4022–4028. doi: 10.1021/bi00336a033. [DOI] [PubMed] [Google Scholar]
- Lasken R. S., Kornberg A. The primosomal protein n' of Escherichia coli is a DNA helicase. J Biol Chem. 1988 Apr 25;263(12):5512–5518. [PubMed] [Google Scholar]
- LeBowitz J. H., McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986 Apr 5;261(10):4738–4748. [PubMed] [Google Scholar]
- Lee M. S., Marians K. J. Escherichia coli replication factor Y, a component of the primosome, can act as a DNA helicase. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8345–8349. doi: 10.1073/pnas.84.23.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire G., Gold L., Yarus M. Autogenous translational repression of bacteriophage T4 gene 32 expression in vitro. J Mol Biol. 1978 Nov 25;126(1):73–90. doi: 10.1016/0022-2836(78)90280-2. [DOI] [PubMed] [Google Scholar]
- Lers N., Salaj-Smic E., Trgovcević Z. Overproduction of SSB protein enhances the capacity for photorepair in Escherichia coli recA cells. Photochem Photobiol. 1989 Feb;49(2):225–227. doi: 10.1111/j.1751-1097.1989.tb04100.x. [DOI] [PubMed] [Google Scholar]
- Lieberman H. B., Witkin E. M. DNA degradation, UV sensitivity and SOS-mediated mutagenesis in strains of Escherichia coli deficient in single-strand DNA binding protein: effects of mutations and treatments that alter levels of Exonuclease V or recA protein. Mol Gen Genet. 1983;190(1):92–100. doi: 10.1007/BF00330329. [DOI] [PubMed] [Google Scholar]
- Lieberman H. B., Witkin E. M. Variable expression of the ssb--1 allele in different strains of Escherichia coli K12 and B: differential suppression of its effects on DNA replication, DNA repair and ultraviolet mutagenesis. Mol Gen Genet. 1981;183(2):348–355. doi: 10.1007/BF00270639. [DOI] [PubMed] [Google Scholar]
- Lin L. L., Little J. W. Autodigestion and RecA-dependent cleavage of Ind- mutant LexA proteins. J Mol Biol. 1989 Dec 5;210(3):439–452. doi: 10.1016/0022-2836(89)90121-6. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Wood R. D. DNA repair and recombination. Curr Opin Cell Biol. 1989 Jun;1(3):475–480. doi: 10.1016/0955-0674(89)90008-2. [DOI] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Livneh Z., Lehman I. R. Recombinational bypass of pyrimidine dimers promoted by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1982 May;79(10):3171–3175. doi: 10.1073/pnas.79.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Picksley S. M., Prescott C. Inducible expression of a gene specific to the RecF pathway for recombination in Escherichia coli K12. Mol Gen Genet. 1983;190(1):162–167. doi: 10.1007/BF00330340. [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Kunkel T. A. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., Bujalowski W. Negative cooperativity within individual tetramers of Escherichia coli single strand binding protein is responsible for the transition between the (SSB)35 and (SSB)56 DNA binding modes. Biochemistry. 1988 Apr 5;27(7):2260–2265. doi: 10.1021/bi00407a002. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., Bujalowski W., Overman L. B. E. coli single strand binding protein: a new look at helix-destabilizing proteins. Trends Biochem Sci. 1988 Jul;13(7):250–255. [PubMed] [Google Scholar]
- Lohman T. M., Green J. M., Beyer R. S. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986 Jan 14;25(1):21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., Overman L. B., Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J Mol Biol. 1986 Feb 20;187(4):603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., Overman L. B. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985 Mar 25;260(6):3594–3603. [PubMed] [Google Scholar]
- Lovett S. T., Clark A. J. Genetic analysis of regulation of the RecF pathway of recombination in Escherichia coli K-12. J Bacteriol. 1983 Mar;153(3):1471–1478. doi: 10.1128/jb.153.3.1471-1478.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S. T., Kolodner R. D. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S. T., Luisi-DeLuca C., Kolodner R. D. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics. 1988 Sep;120(1):37–45. doi: 10.1093/genetics/120.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low R. L., Shlomai J., Kornberg A. Protein n, a primosomal DNA replication protein of Escherichia coli. Purification and characterization. J Biol Chem. 1982 Jun 10;257(11):6242–6250. [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Welsh K., Clark S., Su S. S., Modrich P. Repair of DNA base-pair mismatches in extracts of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:589–596. doi: 10.1101/sqb.1984.049.01.066. [DOI] [PubMed] [Google Scholar]
- Lu C., Scheuermann R. H., Echols H. Capacity of RecA protein to bind preferentially to UV lesions and inhibit the editing subunit (epsilon) of DNA polymerase III: a possible mechanism for SOS-induced targeted mutagenesis. Proc Natl Acad Sci U S A. 1986 Feb;83(3):619–623. doi: 10.1073/pnas.83.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., Kleckner N. Mismatch repair mutations of Escherichia coli K12 enhance transposon excision. Genetics. 1985 Jan;109(1):3–19. doi: 10.1093/genetics/109.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längle-Rouault F., Maenhaut-Michel G., Radman M. GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. EMBO J. 1987 Apr;6(4):1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay V., Linn S. The mechanism of degradation of duplex deoxyribonucleic acid by the recBC enzyme of Escherichia coli K-12. J Biol Chem. 1974 Jul 10;249(13):4286–4294. [PubMed] [Google Scholar]
- Mackay V., Linn S. Selective inhibition of the dnase activity of the recBC enzyme by the DNA binding protein from Escherichia coli. J Biol Chem. 1976 Jun 25;251(12):3716–3719. [PubMed] [Google Scholar]
- Madiraju M. V., Templin A., Clark A. J. Properties of a mutant recA-encoded protein reveal a possible role for Escherichia coli recF-encoded protein in genetic recombination. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6592–6596. doi: 10.1073/pnas.85.18.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S. K., Datta A. R. Mechanisms of recombination by the RecBC and the RecF pathways following conjugation in Escherichia coli K12. Mol Gen Genet. 1979 Jan 16;169(1):67–78. doi: 10.1007/BF00267547. [DOI] [PubMed] [Google Scholar]
- Mahoungou C., Ghrir R., Lecaer J. P., Mignotte B., Barat-Gueride M. The amino-terminal sequence of the Xenopus laevis mitochondrial SSB is homologous to that of the Escherichia coli protein. FEBS Lett. 1988 Aug 1;235(1-2):267–270. doi: 10.1016/0014-5793(88)81276-6. [DOI] [PubMed] [Google Scholar]
- Maki H., Maki S., Kornberg A. DNA Polymerase III holoenzyme of Escherichia coli. IV. The holoenzyme is an asymmetric dimer with twin active sites. J Biol Chem. 1988 May 15;263(14):6570–6578. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. II. A novel complex including the gamma subunit essential for processive synthesis. J Biol Chem. 1988 May 15;263(14):6555–6560. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. III. Distinctive processive polymerases reconstituted from purified subunits. J Biol Chem. 1988 May 15;263(14):6561–6569. [PubMed] [Google Scholar]
- Maples V. F., Kushner S. R. DNA repair in Escherichia coli: identification of the uvrD gene product. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5616–5620. doi: 10.1073/pnas.79.18.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marians K. J. Enzymology of DNA in replication in prokaryotes. CRC Crit Rev Biochem. 1984;17(2):153–215. doi: 10.3109/10409238409113604. [DOI] [PubMed] [Google Scholar]
- Marians K. J., Soeller W., Zipursky S. L. Maximal limits of the Escherichia coli replication factor Y effector site sequences in pBR322 DNA. J Biol Chem. 1982 May 25;257(10):5656–5662. [PubMed] [Google Scholar]
- Matson S. W. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3' to 5' direction. J Biol Chem. 1986 Aug 5;261(22):10169–10175. [PubMed] [Google Scholar]
- Matson S. W., George J. W. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J Biol Chem. 1987 Feb 15;262(5):2066–2076. [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc Natl Acad Sci U S A. 1980 Feb;77(2):857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme of Escherichia coli. Annu Rev Biochem. 1988;57:519–550. doi: 10.1146/annurev.bi.57.070188.002511. [DOI] [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme of Escherichia coli: components and function of a true replicative complex. Mol Cell Biochem. 1985 Feb;66(1):71–85. doi: 10.1007/BF00231826. [DOI] [PubMed] [Google Scholar]
- McMacken R., Ueda K., Kornberg A. Migration of Escherichia coli dnaB protein on the template DNA strand as a mechanism in initiating DNA replication. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4190–4194. doi: 10.1073/pnas.74.10.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensa-Wilmot K., Seaby R., Alfano C., Wold M. C., Gomes B., McMacken R. Reconstitution of a nine-protein system that initiates bacteriophage lambda DNA replication. J Biol Chem. 1989 Feb 15;264(5):2853–2861. [PubMed] [Google Scholar]
- Merrill B. M., Williams K. R., Chase J. W., Konigsberg W. H. Photochemical cross-linking of the Escherichia coli single-stranded DNA-binding protein to oligodeoxynucleotides. Identification of phenylalanine 60 as the site of cross-linking. J Biol Chem. 1984 Sep 10;259(17):10850–10856. [PubMed] [Google Scholar]
- Meyer R. R., Brown C. L., Rein D. C. A new DNA-dependent ATPase from Escherichia coli. Purification and characterization of ATPase IV. J Biol Chem. 1984 Apr 25;259(8):5093–5099. [PubMed] [Google Scholar]
- Meyer R. R., Glassberg J., Kornberg A. An Escherichia coli mutant defective in single-strand binding protein is defective in DNA replication. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1702–1705. doi: 10.1073/pnas.76.4.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. R., Glassberg J., Scott J. V., Kornberg A. A temperature-sensitive single-stranded DNA-binding protein from Escherichia coli. J Biol Chem. 1980 Apr 10;255(7):2897–2901. [PubMed] [Google Scholar]
- Meyer R. R. On the evolutionary origin of mitochondrial DNA. J Theor Biol. 1973 Mar;38(3):647–653. doi: 10.1016/0022-5193(73)90264-6. [DOI] [PubMed] [Google Scholar]
- Meyer R. R., Rein D. C., Glassberg J. The product of the lexC gene of Escherichia coli is single-stranded DNA-binding protein. J Bacteriol. 1982 Apr;150(1):433–435. doi: 10.1128/jb.150.1.433-435.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. R., Shlomai J., Kobori J., Bates D. L., Rowen L., McMacken R., Ueda K., Kornberg A. Enzymatic conversion of single-stranded phiX174 and G4 circles to duplex forms: discontinuous replication. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):289–293. doi: 10.1101/sqb.1979.043.01.035. [DOI] [PubMed] [Google Scholar]
- Meyer R. R., Thomas D. C., Koerner T. J., Rein D. C. Interaction of DNA accessory proteins with DNA polymerase beta of the Novikoff hepatoma. Adv Exp Med Biol. 1984;179:355–361. doi: 10.1007/978-1-4684-8730-5_37. [DOI] [PubMed] [Google Scholar]
- Meyer R. R., Voegele D. W., Ruben S. M., Rein D. C., Trela J. M. Influence of single-stranded DNA-binding protein on recA induction in Escherichia coli. Mutat Res. 1982 Jun;94(2):299–313. doi: 10.1016/0027-5107(82)90293-7. [DOI] [PubMed] [Google Scholar]
- Michaels M. L., Lee M. S., Romano L. J. Contrasting effects of Escherichia coli single-stranded DNA binding protein on synthesis by T7 DNA polymerase and Escherichia coli DNA polymerase I (large fragment). Evidence that binding protein inhibits trans-lesion synthesis by polymerase I. J Biol Chem. 1986 Apr 15;261(11):4847–4854. [PubMed] [Google Scholar]
- Mignotte B., Barat M., Mounolou J. C. Characterization of a mitochondrial protein binding to single-stranded DNA. Nucleic Acids Res. 1985 Mar 11;13(5):1703–1716. doi: 10.1093/nar/13.5.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignotte B., Marsault J., Barat-Gueride M. Effects of the Xenopus laevis mitochondrial single-stranded DNA-binding protein on the activity of DNA polymerase gamma. Eur J Biochem. 1988 Jun 15;174(3):479–484. doi: 10.1111/j.1432-1033.1988.tb14123.x. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Modrich P. Methyl-directed DNA mismatch correction. J Biol Chem. 1989 Apr 25;264(12):6597–6600. [PubMed] [Google Scholar]
- Mok M., Marians K. J. The Escherichia coli preprimosome and DNA B helicase can form replication forks that move at the same rate. J Biol Chem. 1987 Dec 5;262(34):16644–16654. [PubMed] [Google Scholar]
- Molineux I. J., Friedman S., Gefter M. L. Purification and properties of the Escherichia coli deoxyribonucleic acid-unwinding protein. Effects on deoxyribonucleic acid synthesis in vitro. J Biol Chem. 1974 Oct 10;249(19):6090–6098. [PubMed] [Google Scholar]
- Molineux I. J., Gefter M. L. Properties of the Escherichia coli DNA-binding (unwinding) protein interaction with nucleolytic enzymes and DNA. J Mol Biol. 1975 Nov 15;98(4):811–825. doi: 10.1016/s0022-2836(75)80012-x. [DOI] [PubMed] [Google Scholar]
- Molineux I. J., Gefter M. L. Properties of the Escherichia coli in DNA binding (unwinding) protein: interaction with DNA polymerase and DNA. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3858–3862. doi: 10.1073/pnas.71.10.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux I. J., Pauli A., Gefter M. L. Physical studies of the interaction between the Escherichia coli DNA binding protein and nucleic acids. Nucleic Acids Res. 1975 Oct;2(10):1821–1837. doi: 10.1093/nar/2.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzingo A. F., Christiansen C. Crystallization of single-strand DNA-binding protein. J Mol Biol. 1983 Nov 5;170(3):801–801. doi: 10.1016/s0022-2836(83)80135-1. [DOI] [PubMed] [Google Scholar]
- Moreau P. L. Effects of overproduction of single-stranded DNA-binding protein on RecA protein-dependent processes in Escherichia coli. J Mol Biol. 1987 Apr 20;194(4):621–634. doi: 10.1016/0022-2836(87)90239-7. [DOI] [PubMed] [Google Scholar]
- Moreau P. L. Mutagénèse et réponses induites par l'endommagement de l'ADN chez Escherichia coli: principe des tests bactériens pour la détection des substances cancérogènes ou antitumorales. Bull Cancer. 1988;75(2):147–166. [PubMed] [Google Scholar]
- Moreau P. L. Overproduction of single-stranded-DNA-binding protein specifically inhibits recombination of UV-irradiated bacteriophage DNA in Escherichia coli. J Bacteriol. 1988 Jun;170(6):2493–2500. doi: 10.1128/jb.170.6.2493-2500.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P. L., Roberts J. W. RecA protein--promoted lambda repressor cleavage: complementation between RecA441 and RecA430 proteins in vitro. Mol Gen Genet. 1984;198(2):25–34. doi: 10.1007/BF00328696. [DOI] [PubMed] [Google Scholar]
- Morrical S. W., Cox M. M. Stabilization of recA protein-ssDNA complexes by the single-stranded DNA binding protein of Escherichia coli. Biochemistry. 1990 Jan 23;29(3):837–843. doi: 10.1021/bi00455a034. [DOI] [PubMed] [Google Scholar]
- Morrical S. W., Lee J., Cox M. M. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of recA protein and single-stranded DNA. Biochemistry. 1986 Apr 8;25(7):1482–1494. doi: 10.1021/bi00355a003. [DOI] [PubMed] [Google Scholar]
- Muniyappa K., Shaner S. L., Tsang S. S., Radding C. M. Mechanism of the concerted action of recA protein and helix-destabilizing proteins in homologous recombination. Proc Natl Acad Sci U S A. 1984 May;81(9):2757–2761. doi: 10.1073/pnas.81.9.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskavitch K. M., Linn S. A unified mechanism for the nuclease and unwinding activities of the recBC enzyme of Escherichia coli. J Biol Chem. 1982 Mar 10;257(5):2641–2648. [PubMed] [Google Scholar]
- Myers T. W., Romano L. J. Mechanism of stimulation of T7 DNA polymerase by Escherichia coli single-stranded DNA binding protein (SSB). J Biol Chem. 1988 Nov 15;263(32):17006–17015. [PubMed] [Google Scholar]
- Nakai H., Richardson C. C. The effect of the T7 and Escherichia coli DNA-binding proteins at the replication fork of bacteriophage T7. J Biol Chem. 1988 Jul 15;263(20):9831–9839. [PubMed] [Google Scholar]
- Neidhardt F. C., VanBogelen R. A., Vaughn V. The genetics and regulation of heat-shock proteins. Annu Rev Genet. 1984;18:295–329. doi: 10.1146/annurev.ge.18.120184.001455. [DOI] [PubMed] [Google Scholar]
- Neuendorf S. K., Cox M. M. Exchange of recA protein between adjacent recA protein-single-stranded DNA complexes. J Biol Chem. 1986 Jun 25;261(18):8276–8282. [PubMed] [Google Scholar]
- Niyogi S. K., Ratrie H., 3rd, Datta A. K. Effect of Escherichia coli DNA binding protein on the transcription of single-stranded phage M13 DNA by Escherichia coli RNA polymerase. Biochem Biophys Res Commun. 1977 Sep 9;78(1):343–349. doi: 10.1016/0006-291x(77)91260-8. [DOI] [PubMed] [Google Scholar]
- Nomura N., Low R. L., Ray D. S. Identification of ColE1 DNA sequences that direct single strand-to-double strand conversion by a phi X174 type mechanism. Proc Natl Acad Sci U S A. 1982 May;79(10):3153–3157. doi: 10.1073/pnas.79.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N., Ray D. S. Expression of a DNA strand initiation sequence of ColE1 plasmid in a single-stranded DNA phage. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6566–6570. doi: 10.1073/pnas.77.11.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossal N. G. Prokaryotic DNA replication systems. Annu Rev Biochem. 1983;52:581–615. doi: 10.1146/annurev.bi.52.070183.003053. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Okazaki T. Discontinuous DNA replication. Annu Rev Biochem. 1980;49:421–457. doi: 10.1146/annurev.bi.49.070180.002225. [DOI] [PubMed] [Google Scholar]
- Ollis D., Brick P., Abdel-Meguid S. S., Murthy K., Chase J. W., Steitz T. A. Crystals of Escherichia coli single-strand DNA-binding protein show that the tetramer has D2 symmetry. J Mol Biol. 1983 Nov 5;170(3):797–800. doi: 10.1016/s0022-2836(83)80134-x. [DOI] [PubMed] [Google Scholar]
- Overman L. B., Bujalowski W., Lohman T. M. Equilibrium binding of Escherichia coli single-strand binding protein to single-stranded nucleic acids in the (SSB)65 binding mode. Cation and anion effects and polynucleotide specificity. Biochemistry. 1988 Jan 12;27(1):456–471. doi: 10.1021/bi00401a067. [DOI] [PubMed] [Google Scholar]
- Perrino F. W., Meyer R. R., Bobst A. M., Rein D. C. Interaction of a folded chromosome-associated protein with single-stranded DNA-binding protein of Escherichia coli, identified by affinity chromatography. J Biol Chem. 1988 Aug 25;263(24):11833–11839. [PubMed] [Google Scholar]
- Perrino F. W., Rein D. C., Bobst A. M., Meyer R. R. The relative rate of synthesis and levels of single-stranded DNA binding protein during induction of SOS repair in Escherichia coli. Mol Gen Genet. 1987 Oct;209(3):612–614. doi: 10.1007/BF00331171. [DOI] [PubMed] [Google Scholar]
- Phizicky E. M., Roberts J. W. Kinetics of RecA protein-directed inactivation of repressors of phage lambda and phage P22. J Mol Biol. 1980 May 25;139(3):319–328. doi: 10.1016/0022-2836(80)90133-3. [DOI] [PubMed] [Google Scholar]
- Ponticelli A. S., Schultz D. W., Taylor A. F., Smith G. R. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985 May;41(1):145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- Porter R. D., Black S., Pannuri S., Carlson A. Use of the Escherichia coli SSB gene to prevent bioreactor takeover by plasmidless cells. Biotechnology (N Y) 1990 Jan;8(1):47–51. doi: 10.1038/nbt0190-47. [DOI] [PubMed] [Google Scholar]
- Prasad B. V., Chiu W. Sequence comparison of single-stranded DNA binding proteins and its structural implications. J Mol Biol. 1987 Feb 5;193(3):579–584. doi: 10.1016/0022-2836(87)90268-3. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Cox M. M. recA protein binding to the heteroduplex product of DNA strand exchange. J Biol Chem. 1987 Jan 25;262(3):1337–1343. [PubMed] [Google Scholar]
- Quiñones A., Piechocki R. Differential suppressor effects of the ssb-1 and ssb-113 alleles on uvrD mutator of Escherichia coli in DNA repair and mutagenesis. J Basic Microbiol. 1987;27(5):263–273. doi: 10.1002/jobm.3620270508. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Helical RecA nucleoprotein filaments mediate homologous pairing and strand exchange. Biochim Biophys Acta. 1989 Jul 7;1008(2):131–145. doi: 10.1016/0167-4781(80)90001-9. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing & strand exchange mediated by rec A nucleoprotein filaments & networks. Prog Clin Biol Res. 1986;218:77–94. [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Recombination activities of E. coli recA protein. Cell. 1981 Jul;25(1):3–4. doi: 10.1016/0092-8674(81)90224-5. [DOI] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Ramdas J., Mythili E., Muniyappa K. RecA protein promoted homologous pairing in vitro. Pairing between linear duplex DNA bound to HU Protein (nucleosome cores) and nucleoprotein filaments of recA protein-single-stranded DNA. J Biol Chem. 1989 Oct 15;264(29):17395–17400. [PubMed] [Google Scholar]
- Register J. C., 3rd, Christiansen G., Griffith J. Electron microscopic visualization of the RecA protein-mediated pairing and branch migration phases of DNA strand exchange. J Biol Chem. 1987 Sep 15;262(26):12812–12820. [PubMed] [Google Scholar]
- Register J. C., 3rd, Griffith J. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J Biol Chem. 1985 Oct 5;260(22):12308–12312. [PubMed] [Google Scholar]
- Reinberg D., Zipursky S. L., Hurwitz J. Separate requirements for leading and lagging strand DNA synthesis during phi X A protein-dependent RF goes to RF DNA replication in vitro. J Biol Chem. 1981 Dec 25;256(24):13143–13151. [PubMed] [Google Scholar]
- Resnick J., Sussman R. Escherichia coli single-strand DNA binding protein from wild type and lexC113 mutant affects in vitro proteolytic cleavage of phage lambda repressor. Proc Natl Acad Sci U S A. 1982 May;79(9):2832–2835. doi: 10.1073/pnas.79.9.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben R. C., Gefter M. L. A DNA-binding protein induced by bacteriophage T7. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1846–1850. doi: 10.1073/pnas.70.6.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben R. C., Gefter M. L. A deoxyribonucleic acid-binding protein induced by bacteriophage T7. Purification and properties of the protein. J Biol Chem. 1974 Jun 25;249(12):3843–3850. [PubMed] [Google Scholar]
- Richardson C. C., Romano L. J., Kolodner R., LeClerc J. E., Tamanoi F., Engler M. J., Dean F. B., Richardson D. S. Replication of bacteriophage T7 DNA by purified proteins. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):427–440. doi: 10.1101/sqb.1979.043.01.049. [DOI] [PubMed] [Google Scholar]
- Riddles P. W., Lehman I. R. The formation of paranemic and plectonemic joints between DNA molecules by the recA and single-stranded DNA-binding proteins of Escherichia coli. J Biol Chem. 1985 Jan 10;260(1):165–169. [PubMed] [Google Scholar]
- Roberts J. W., Phizicky E. M., Burbee D. G., Roberts C. W., Moreau P. L. A brief consideration of the SOS inducing signal. Biochimie. 1982 Aug-Sep;64(8-9):805–807. doi: 10.1016/s0300-9084(82)80133-8. [DOI] [PubMed] [Google Scholar]
- Roman L. J., Kowalczykowski S. C. Relationship of the physical and enzymatic properties of Escherichia coli recA protein to its strand exchange activity. Biochemistry. 1986 Nov 18;25(23):7375–7385. doi: 10.1021/bi00371a020. [DOI] [PubMed] [Google Scholar]
- Romano L. J., Richardson C. C. Characterization of the ribonucleic acid primers and the deoxyribonucleic acid product synthesized by the DNA polymerase and gene 4 protein of bacteriophage T7. J Biol Chem. 1979 Oct 25;254(20):10483–10489. [PubMed] [Google Scholar]
- Romano L. J., Richardson C. C. Requirements for synthesis of ribonucleic acid primers during lagging strand synthesis by the DNA polymerase and gene 4 protein of bacteriophage T7. J Biol Chem. 1979 Oct 25;254(20):10476–10482. [PubMed] [Google Scholar]
- Rosamond J., Telander K. M., Linn S. Modulation of the action of the recBC enzyme of Escherichia coli K-12 by Ca2+. J Biol Chem. 1979 Sep 10;254(17):8646–8652. [PubMed] [Google Scholar]
- Ruben S. M., VanDenBrink-Webb S. E., Rein D. C., Meyer R. R. Suppression of the Escherichia coli ssb-1 mutation by an allele of groEL. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3767–3771. doi: 10.1073/pnas.85.11.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyechan W. T., Wetmur J. G. Studies on the cooperative binding of the Escherichia coli DNA unwinding protein to single-stranded DNA. Biochemistry. 1975 Dec 16;14(25):5529–5534. doi: 10.1021/bi00696a023. [DOI] [PubMed] [Google Scholar]
- Ruyechan W. T., Wetmur J. G. Studies on the noncooperative binding of the Escherichia coli DNA unwinding protein to single-stranded nucleic acids. Biochemistry. 1976 Nov 16;15(23):5057–5064. doi: 10.1021/bi00668a017. [DOI] [PubMed] [Google Scholar]
- Römer R., Schomburg U., Krauss G., Maass G. Escherichia coli single-stranded DNA binding protein is mobile on DNA: 1H NMR study of its interaction with oligo- and polynucleotides. Biochemistry. 1984 Dec 4;23(25):6132–6137. doi: 10.1021/bi00320a036. [DOI] [PubMed] [Google Scholar]
- Salaj-Smic E., Lers N., Trgovcević Z. Overproduction of single-stranded DNA-binding protein increases UV-induced mutagenesis in Escherichia coli. Mutat Res. 1988 Jul;208(3-4):179–182. doi: 10.1016/0165-7992(88)90057-7. [DOI] [PubMed] [Google Scholar]
- Salles B., Paoletti C., Villani G. Lack of single-strand DNA-binding protein amplification under conditions of SOS induction in E. coli. Mol Gen Genet. 1983;189(1):175–177. doi: 10.1007/BF00326074. [DOI] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. Cloning of uvrA, lexC and ssb genes of Escherichia coli. Biochem Biophys Res Commun. 1979 Sep 12;90(1):123–129. doi: 10.1016/0006-291x(79)91598-5. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B., Rupp W. D., Little J. W., Mount D. W. LexA protein inhibits transcription of the E. coli uvrA gene in vitro. Nature. 1982 Jul 1;298(5869):96–98. doi: 10.1038/298096a0. [DOI] [PubMed] [Google Scholar]
- Sancar A., Williams K. R., Chase J. W., Rupp W. D. Sequences of the ssb gene and protein. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4274–4278. doi: 10.1073/pnas.78.7.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller H., Uhlmann A., Geider K. A DNA fragment from the origin of single-strand to double-strand DNA replication of bacteriophage fd. Proc Natl Acad Sci U S A. 1976 Jan;73(1):49–53. doi: 10.1073/pnas.73.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R., Weiner J. H., Weiner A., Kornberg A. Ten proteins required for conversion of phiX174 single-stranded DNA to duplex form in vitro. Resolution and reconstitution. J Biol Chem. 1975 Aug 10;250(15):5859–5865. [PubMed] [Google Scholar]
- Scherzinger E., Klotz G. Studies on bacteriophage T7 DNA synthesis in vitro. II. Reconstitution of the T7 replication system using purified proteins. Mol Gen Genet. 1975 Dec 1;141(3):233–249. doi: 10.1007/BF00341802. [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Litfin F., Jost E. Stimulation of T7 DNA polymerase by a new phage-coded protein. Mol Gen Genet. 1973 Jul 2;123(3):247–262. doi: 10.1007/BF00271243. [DOI] [PubMed] [Google Scholar]
- Scheuermann R. H., Echols H. A separate editing exonuclease for DNA replication: the epsilon subunit of Escherichia coli DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7747–7751. doi: 10.1073/pnas.81.24.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmellik-Sandage C. S., Tessman E. S. Signal strains that can detect certain DNA replication and membrane mutants of Escherichia coli: isolation of a new ssb allele, ssb-3. J Bacteriol. 1990 Aug;172(8):4378–4385. doi: 10.1128/jb.172.8.4378-4385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R. J., Wetmur J. G. Kinetics of transfer of Escherichia coli single strand deoxyribonucleic acid binding protein between single-stranded deoxyribonucleic acid molecules. Biochemistry. 1982 Feb 16;21(4):608–615. doi: 10.1021/bi00533a002. [DOI] [PubMed] [Google Scholar]
- Scholtissek S., Grosse F. A plasmid vector system for the expression of a triprotein consisting of beta-galactosidase, a collagenase recognition site and a foreign gene product. Gene. 1988;62(1):55–64. doi: 10.1016/0378-1119(88)90579-3. [DOI] [PubMed] [Google Scholar]
- Schutte B. C., Cox M. M. Homology-dependent changes in adenosine 5'-triphosphate hydrolysis during recA protein promoted DNA strand exchange: evidence for long paranemic complexes. Biochemistry. 1987 Sep 8;26(18):5616–5625. doi: 10.1021/bi00392a006. [DOI] [PubMed] [Google Scholar]
- Schutte B. C., Cox M. M. Homology-dependent underwinding of duplex DNA in recA protein generated paranemic complexes. Biochemistry. 1988 Oct 4;27(20):7886–7894. doi: 10.1021/bi00420a046. [DOI] [PubMed] [Google Scholar]
- Scott J. F., Eisenberg S., Bertsch L. L., Kornberg A. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc Natl Acad Sci U S A. 1977 Jan;74(1):193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevastopoulos C. G., Wehr C. T., Glaser D. A. Large-scale automated isolation of Escherichia coli mutants with thermosensitive DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3485–3489. doi: 10.1073/pnas.74.8.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Huang H. V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986 Mar;112(3):441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman L. A., Gefter M. L. Studies on the mechanism of enzymatic DNA elongation by Escherichia coli DNA polymerase II. J Mol Biol. 1976 May 5;103(1):61–76. doi: 10.1016/0022-2836(76)90052-8. [DOI] [PubMed] [Google Scholar]
- Shibata T., DasGupta C., Cunningham R. P., Radding C. M. Homologous pairing in genetic recombination: formation of D loops by combined action of recA protein and a helix-destabilizing protein. Proc Natl Acad Sci U S A. 1980 May;77(5):2606–2610. doi: 10.1073/pnas.77.5.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., DasGupta C., Cunningham R. P., Radding C. M. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto N., Ikushima N., Utiyama H., Tachibana H., Horie K. Specific and cooperative binding of E. coli single-stranded DNA binding protein to mRNA. Nucleic Acids Res. 1987 Jul 10;15(13):5241–5250. doi: 10.1093/nar/15.13.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. A prepriming DNA replication enzyme of Escherichia coli. II. Actions of protein n': a sequence-specific, DNA-dependent ATPase. J Biol Chem. 1980 Jul 25;255(14):6794–6798. [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc Natl Acad Sci U S A. 1980 Feb;77(2):799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J., Polder L., Arai K., Kornberg A. Replication of phi X174 dna with purified enzymes. I. Conversion of viral DNA to a supercoiled, biologically active duplex. J Biol Chem. 1981 May 25;256(10):5233–5238. [PubMed] [Google Scholar]
- Shwartz H., Livneh Z. Dynamics of termination during in vitro replication of ultraviolet-irradiated DNA with DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1987 Aug 5;262(22):10518–10523. [PubMed] [Google Scholar]
- Shwartz H., Shavitt O., Livneh Z. The role of exonucleolytic processing and polymerase-DNA association in bypass of lesions during replication in vitro. Significance for SOS-targeted mutagenesis. J Biol Chem. 1988 Dec 5;263(34):18277–18285. [PubMed] [Google Scholar]
- Sigal N., Delius H., Kornberg T., Gefter M. L., Alberts B. A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3537–3541. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims J., Benz E. W., Jr Initiation of DNA replication by the Escherichia coli dnaG protein: evidence that tertiary structure is involved. Proc Natl Acad Sci U S A. 1980 Feb;77(2):900–904. doi: 10.1073/pnas.77.2.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims J., Dressler D. Site-specific initiation of a DNA fragment: nucleotide sequence of the bacteriophage G4 negative-strand initiation site. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3094–3098. doi: 10.1073/pnas.75.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slilaty S. N., Rupley J. A., Little J. W. Intramolecular cleavage of LexA and phage lambda repressors: dependence of kinetics on repressor concentration, pH, temperature, and solvent. Biochemistry. 1986 Nov 4;25(22):6866–6875. doi: 10.1021/bi00370a020. [DOI] [PubMed] [Google Scholar]
- Smith G. R., Amundsen S. K., Chaudhury A. M., Cheng K. C., Ponticelli A. S., Roberts C. M., Schultz D. W., Taylor A. F. Roles of RecBC enzyme and chi sites in homologous recombination. Cold Spring Harb Symp Quant Biol. 1984;49:485–495. doi: 10.1101/sqb.1984.049.01.055. [DOI] [PubMed] [Google Scholar]
- Smith G. R. Homologous recombination in E. coli: multiple pathways for multiple reasons. Cell. 1989 Sep 8;58(5):807–809. doi: 10.1016/0092-8674(89)90929-x. [DOI] [PubMed] [Google Scholar]
- Smith G. R. Homologous recombination in procaryotes. Microbiol Rev. 1988 Mar;52(1):1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Kunes S. M., Schultz D. W., Taylor A., Triman K. L. Structure of chi hotspots of generalized recombination. Cell. 1981 May;24(2):429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- Smith G. R. Mechanism and control of homologous recombination in Escherichia coli. Annu Rev Genet. 1987;21:179–201. doi: 10.1146/annurev.ge.21.120187.001143. [DOI] [PubMed] [Google Scholar]
- Soeller W. C., Marians K. J. Deletion mutants defining the Escherichia coli replication factor Y effector site sequences in pBR322 DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7253–7257. doi: 10.1073/pnas.79.23.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller W., Abarzúa P., Marians K. J. Mutational analysis of primosome assembly sites. II. Role of secondary structure in the formation of active sites. J Biol Chem. 1984 Nov 25;259(22):14293–14300. [PubMed] [Google Scholar]
- Soltis D. A., Lehman I. R. An unpaired 3' terminus stimulates recA protein-promoted DNA strand exchange. J Biol Chem. 1983 Dec 10;258(23):14073–14075. [PubMed] [Google Scholar]
- Soltis D. A., Lehman I. R. recA protein promoted DNA strand exchange. J Biol Chem. 1983 May 25;258(10):6073–6077. [PubMed] [Google Scholar]
- Srivenugopal K. S., Morris D. R. Modulation of the relaxing activity of Escherichia coli topoisomerase I by single-stranded DNA binding proteins. Biochem Biophys Res Commun. 1986 Jun 13;137(2):795–800. doi: 10.1016/0006-291x(86)91149-6. [DOI] [PubMed] [Google Scholar]
- Stasiak A., Stasiak A. Z., Koller T. Visualization of RecA-DNA complexes involved in consecutive stages of an in vitro strand exchange reaction. Cold Spring Harb Symp Quant Biol. 1984;49:561–570. doi: 10.1101/sqb.1984.049.01.063. [DOI] [PubMed] [Google Scholar]
- Stayton M. M., Kornberg A. Complexes of Escherichia coli primase with the replication origin of G4 phage DNA. J Biol Chem. 1983 Nov 10;258(21):13205–13212. [PubMed] [Google Scholar]
- Su S. S., Grilley M., Thresher R., Griffith J., Modrich P. Gap formation is associated with methyl-directed mismatch correction under conditions of restricted DNA synthesis. Genome. 1989;31(1):104–111. doi: 10.1139/g89-020. [DOI] [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Huber H. E., Richardson C. C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987 Nov 25;262(33):16212–16223. [PubMed] [Google Scholar]
- Taylor A. F., Schultz D. W., Ponticelli A. S., Smith G. R. RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation-dependence of the cutting. Cell. 1985 May;41(1):153–163. doi: 10.1016/0092-8674(85)90070-4. [DOI] [PubMed] [Google Scholar]
- Taylor A. F., Smith G. R. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol. 1985 Sep 20;185(2):431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- Taylor A., Smith G. R. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 1980 Nov;22(2 Pt 2):447–457. doi: 10.1016/0092-8674(80)90355-4. [DOI] [PubMed] [Google Scholar]
- Tessman E. S., Peterson P. K. Suppression of the ssb-1 and ssb-113 mutations of Escherichia coli by a wild-type rep gene, NaCl, and glucose. J Bacteriol. 1982 Nov;152(2):572–583. doi: 10.1128/jb.152.2.572-583.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler D. S., Stahl F. W. DNA double-chain breaks in recombination of phage lambda and of yeast. Annu Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- Thomas D. C., Meyer R. R. Deoxyribonucleic acid dependent adenosinetriphosphatases from the Novikoff hepatoma. Characterization of a homogeneous adenosinetriphosphatase that stimulates DNA polymerase beta. Biochemistry. 1982 Sep 28;21(20):5060–5068. doi: 10.1021/bi00263a033. [DOI] [PubMed] [Google Scholar]
- Thomas D. C., Rein D. C., Meyer R. R. Purification and enzymological characterization of DNA-dependent ATPase IV from the Novikoff hepatoma. Nucleic Acids Res. 1988 Jul 25;16(14A):6447–6464. doi: 10.1093/nar/16.14.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Selzer G. Initiation of DNA synthesis in Escherichia coli. Annu Rev Biochem. 1979;48:999–1034. doi: 10.1146/annurev.bi.48.070179.005031. [DOI] [PubMed] [Google Scholar]
- Trgovcević Z., Lers N., Brcić-Kostić K., Salaj-Smic E. Post-ultraviolet DNA synthesis in the absence of repair: role of the single-strand DNA-binding protein. Int J Radiat Biol. 1989 May;55(5):739–745. doi: 10.1080/09553008914550791. [DOI] [PubMed] [Google Scholar]
- Tsang S. S., Muniyappa K., Azhderian E., Gonda D. K., Radding C. M., Flory J., Chase J. W. Intermediates in homologous pairing promoted by recA protein. Isolation and characterization of active presynaptic complexes. J Mol Biol. 1985 Sep 20;185(2):295–309. doi: 10.1016/0022-2836(85)90405-x. [DOI] [PubMed] [Google Scholar]
- Uhlmann A., Geider K. Interaction of DNA with DNA binding proteins. III. Infectivity of protein-complexed phage fd DNA in Escherichia coli spheroplasts. Biochim Biophys Acta. 1977 Feb 16;474(4):639–645. doi: 10.1016/0005-2787(77)90083-1. [DOI] [PubMed] [Google Scholar]
- Vales L. D., Chase J. W., Murphy J. B. Effect of ssbA1 and lexC113 mutations on lambda prophage induction, bacteriophage growth, and cell survival. J Bacteriol. 1980 Aug;143(2):887–896. doi: 10.1128/jb.143.2.887-896.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicuna R., Hurwitz J., Wallace S., Girard M. Selective inhibition of in vitro DNA synthesis dependent on phiX174 compared with fd DNA. I. Protein requirements for selective inhibition. J Biol Chem. 1977 Apr 25;252(8):2524–2533. [PubMed] [Google Scholar]
- Villani G., Pierre A., Salles B. Quantification of SSB protein in E. coli and its variation during RECA protein induction. Biochimie. 1984 Jun;66(6):471–476. doi: 10.1016/0300-9084(84)90082-8. [DOI] [PubMed] [Google Scholar]
- Volkert M. R., Hartke M. A. Suppression of Escherichia coli recF mutations by recA-linked srfA mutations. J Bacteriol. 1984 Feb;157(2):498–506. doi: 10.1128/jb.157.2.498-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Lasken R. S., Kornberg A. The dnaB-dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J Biol Chem. 1989 Feb 15;264(5):2469–2475. [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Walker G. C., Marsh L., Dodson L. A. Genetic analyses of DNA repair: inference and extrapolation. Annu Rev Genet. 1985;19:103–126. doi: 10.1146/annurev.ge.19.120185.000535. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. C., Smith K. C. Effects of the ssb-1 and ssb-113 mutations on survival and DNA repair in UV-irradiated delta uvrB strains of Escherichia coli K-12. J Bacteriol. 1982 Jul;151(1):186–192. doi: 10.1128/jb.151.1.186-192.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. V., Smith K. C. Rich growth medium enhances ultraviolet radiation sensitivity and inhibits cell division in ssb mutants of Escherichia coli K-12. Photochem Photobiol. 1984 Jun;39(6):793–797. doi: 10.1111/j.1751-1097.1984.tb08861.x. [DOI] [PubMed] [Google Scholar]
- Watt V. M., Ingles C. J., Urdea M. S., Rutter W. J. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigle J. J. Induction of Mutations in a Bacterial Virus. Proc Natl Acad Sci U S A. 1953 Jul;39(7):628–636. doi: 10.1073/pnas.39.7.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J. H., Bertsch L. L., Kornberg A. The deoxyribonucleic acid unwinding protein of Escherichia coli. Properties and functions in replication. J Biol Chem. 1975 Mar 25;250(6):1972–1980. [PubMed] [Google Scholar]
- Weinstock G. M., McEntee K. RecA protein-dependent proteolysis of bacteriophage lambda repressor Characterization of the reaction and stimulation by DNA-binding proteins. J Biol Chem. 1981 Nov 10;256(21):10883–10888. [PubMed] [Google Scholar]
- West S. C., Cassuto E., Howard-Flanders P. Role of SSB protein in RecA promoted branch migration reactions. Mol Gen Genet. 1982;186(3):333–338. doi: 10.1007/BF00729451. [DOI] [PubMed] [Google Scholar]
- West S. C., Cassuto E., Howard-Flanders P. recA protein promotes homologous-pairing and strand-exchange reactions between duplex DNA molecules. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2100–2104. doi: 10.1073/pnas.78.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Countryman J. K., Howard-Flanders P. Purification and properties of the recA protein of Proteus mirabilis. Comparison with Escherichia coli recA protein; specificity of interaction with single strand binding protein. J Biol Chem. 1983 Apr 10;258(7):4648–4654. [PubMed] [Google Scholar]
- West S. C. Protein-DNA interactions in genetic recombination. Trends Genet. 1988 Jan;4(1):8–13. doi: 10.1016/0168-9525(88)90121-7. [DOI] [PubMed] [Google Scholar]
- Whittier R. F., Chase J. W. DNA repair in E. coli strains deficient in single-strand DNA binding protein. Mol Gen Genet. 1981;183(2):341–347. doi: 10.1007/BF00270638. [DOI] [PubMed] [Google Scholar]
- Whittier R. F., Chase J. W. DNA repair properties of Escherichia coli tif-1, recAo281 and lexA1 strains deficient in single-strand DNA binding protein. Mol Gen Genet. 1983;190(1):101–111. doi: 10.1007/BF00330330. [DOI] [PubMed] [Google Scholar]
- Whittier R. F., Chase J. W., Masker W. E. Repair resynthesis in Escherichia coli mutants deficient in single-stranded DNA-binding protein. Mutat Res. 1983 Oct;112(5):275–286. doi: 10.1016/0167-8817(83)90003-2. [DOI] [PubMed] [Google Scholar]
- Wickner R. B., Wright M., Wickner S., Hurwitz J. Conversion of phiX174 and fd single-stranded DNA to replicative forms in extracts of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3233–3237. doi: 10.1073/pnas.69.11.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. H. DNA replication proteins of Escherichia coli. Annu Rev Biochem. 1978;47:1163–1191. doi: 10.1146/annurev.bi.47.070178.005503. [DOI] [PubMed] [Google Scholar]
- Wickner S. DNA or RNA priming of bacteriophage G4 DNA synthesis by Escherichia coli dnaG protein. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2815–2819. doi: 10.1073/pnas.74.7.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Hurwitz J. Conversion of phiX174 viral DNA to double-stranded form by purified Escherichia coli proteins. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4120–4124. doi: 10.1073/pnas.71.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Hurwitz J. Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc Natl Acad Sci U S A. 1975 Mar;72(3):921–925. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. R., Konigsberg W. DNA helix-destabilizing proteins. Gene Amplif Anal. 1981;2:475–508. [PubMed] [Google Scholar]
- Williams K. R., LoPresti M. B., Setoguchi M., Konigsberg W. H. Amino acid sequence of the T4 DNA helix-destabilizing protein. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4614–4617. doi: 10.1073/pnas.77.8.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. R., LoPresti M. B., Setoguchi M. Primary structure of the bacteriophage T4 DNA helix-destabilizing protein. J Biol Chem. 1981 Feb 25;256(4):1754–1762. [PubMed] [Google Scholar]
- Williams K. R., Murphy J. B., Chase J. W. Characterization of the structural and functional defect in the Escherichia coli single-stranded DNA binding protein encoded by the ssb-1 mutant gene. Expression of the ssb-1 gene under lambda pL regulation. J Biol Chem. 1984 Oct 10;259(19):11804–11811. [PubMed] [Google Scholar]
- Williams K. R., Spicer E. K., LoPresti M. B., Guggenheimer R. A., Chase J. W. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J Biol Chem. 1983 Mar 10;258(5):3346–3355. [PubMed] [Google Scholar]
- Williams R. C., Spengler S. J. Fibers of RecA protein and complexes of RecA protein and single-stranded phi X174 DNA as visualized by negative-stain electron microscopy. J Mol Biol. 1986 Jan 5;187(1):109–118. doi: 10.1016/0022-2836(86)90410-9. [DOI] [PubMed] [Google Scholar]
- Wright M., Buttin G., Hurwitz J. The isolation and characterization from Escherichia coli of an adenosine triphosphate-dependent deoxyribonuclease directed by rec B, C genes. J Biol Chem. 1971 Nov;246(21):6543–6555. [PubMed] [Google Scholar]
- Yarranton G. T., Das R. H., Gefter M. L. Enzyme-catalyzed DNA unwinding. Mechanism of action of helicase III. J Biol Chem. 1979 Dec 10;254(23):12002–12006. [PubMed] [Google Scholar]
- Yoakum G. H., Kushner S. R., Grossman L. Isolation of plasmids carrying either the uvrC or uvrC uvrA and ssb genes of Escherichia coli K-12. Gene. 1980 Dec;12(3-4):243–248. doi: 10.1016/0378-1119(80)90106-7. [DOI] [PubMed] [Google Scholar]
- Zang L. H., Maki A. H., Murphy J. B., Chase J. W. Triplet state sublevel kinetics of tryptophan 54 in the complex of Escherichia coli single-stranded DNA binding protein with single-stranded poly(deoxythymidylic) acid. Biophys J. 1987 Nov;52(5):867–872. doi: 10.1016/S0006-3495(87)83280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechel K., Bouché J. P., Kornberg A. Replication of phage G4. A novel and simple system for the initiation of deoxyribonucleic acid synthesis. J Biol Chem. 1975 Jun 25;250(12):4684–4689. [PubMed] [Google Scholar]
- Zentgraf H., Berthold V., Geider K. Interaction of DNA with DNA binding proteins. II. Displacement of Escherichia coli DNA unwinding protein and the condensed structure of DNA complexed with protein HD. Biochim Biophys Acta. 1977 Feb 16;474(4):629–638. doi: 10.1016/0005-2787(77)90082-x. [DOI] [PubMed] [Google Scholar]
- Zipursky S. L., Marians K. J. Identification of two Escherichia coli factor Y effector sites near the origins of replication of the plasmids (ColE1 and pBR322. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6521–6525. doi: 10.1073/pnas.77.11.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A., Brenner S. L. An alpha-helical peptide model for electrostatic interactions of proteins with DNA. The N terminus of RecA. J Mol Biol. 1989 Oct 5;209(3):447–457. doi: 10.1016/0022-2836(89)90009-0. [DOI] [PubMed] [Google Scholar]
- van Mansfeld A. D., van Teeffelen H. A., Fluit A. C., Baas P. D., Jansz H. S. Effect of SSB protein on cleavage of single-stranded DNA by phi X gene A protein and A* protein. Nucleic Acids Res. 1986 Feb 25;14(4):1845–1861. doi: 10.1093/nar/14.4.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ende A., Baker T. A., Ogawa T., Kornberg A. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: primase as the sole priming enzyme. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3954–3958. doi: 10.1073/pnas.82.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]



