Abstract
The drug-metabolizing enzyme thiopurine methyltransferase (TPMT) has become one of the best examples of pharmacogenomics to be translated into routine clinical practice. TPMT metabolizes the thiopurines 6-mercaptopurine, 6-thioguanine, and azathioprine, drugs that are widely used for treatment of acute leukemias, inflammatory bowel diseases, and other disorders of immune regulation. Since the discovery of genetic polymorphisms in the TPMT gene, many sequence variants that cause a decreased enzyme activity have been identified and characterized. Increasingly, to optimize dose, pretreatment determination of TPMT status before commencing thiopurine therapy is now routine in many countries. Novel TPMT sequence variants are currently numbered sequentially using PubMed as a source of information; however, this has caused some problems as exemplified by two instances in which authors’ articles appeared on PubMed at the same time, resulting in the same allele numbers given to different polymorphisms. Hence, there is an urgent need to establish an order and consensus to the numbering of known and novel TPMT sequence variants. To address this problem, a TPMT nomenclature committee was formed in 2010, to define the nomenclature and numbering of novel variants for the TPMT gene. A website (http://www.imh.liu.se/tpmtalleles) serves as a platform for this work. Researchers are encouraged to submit novel TPMT alleles to the committee for designation and reservation of unique allele numbers. The committee has decided to renumber two alleles: nucleotide position 106 (G > A) from TPMT*24 to TPMT*30 and position 611 (T > C, rs79901429) from TPMT*28 to TPMT*31. Nomenclature for all other known alleles remains unchanged.
Keywords: allele, nomenclature, pharmacogenetics, thiopurine methyltransferase
Introduction
The thiopurines 6-thioguanine, 6-mercaptopurine (6-MP), and its pro-drug azathioprine are purine based analogues. These drugs were synthesized in the early 1950’s by Elion and Hitchings [1], and within a few years 6-MP was used to successfully treat children with childhood acute lymphoblastic leukemia (ALL) [2]. Today, thiopurines continue to be used extensively in clinical practice as anticancer and immunosuppressive agents despite having a narrow therapeutic index with potential life-threatening drug induced toxicity.
One of the main causes of toxicity is the way in which these prodrugs are metabolized to their active metabolites inside the cell [3,4]. They undergo extensive metabolism to form both active and inactive metabolites causing cell death by several different mechanisms [4]. The main causes of cytotoxicity are through the incorporation of thioguanine nucleotides (TGNs) into the DNA as base analogues [5], inhibition of de novo purine synthesis [6,7], and disturbances in intracellular signaling pathways. The latter of these contributes to the immunosuppressive properties of these agents [8–10]. Thiopurines are S-methylated by the enzyme thiopurine methyltransferase (TPMT, EC 2.1.1.67). This produces methylated metabolites such as S-methylmercaptopurine and S-methylthioguanine, both of which are believed to be inactive, and S-methyl-thioinosine monophosphate, an inhibitor of de novo purine synthesis, emphasizing the importance of TPMT activity in the metabolism of these drugs.
The first study measuring TPMT activity in humans by Weinshilboum and Sladek demonstrated trimodal distribution [11]. They reported that from a cohort of 298 randomly selected Caucasians, 11.1% had intermediate activity, 89.6% had high activity, and 0.3% had no activity. This was critical in helping to understand the role of TPMT activity in patients treated with thiopurines and was highlighted by Lennard [12], who showed that in children treated with 6-MP for ALL, the red blood cell TGN concentrations were inversely correlated to the TPMT activity, indicating that with high TPMT activity more drug was S-methylated to inactive metabolites.
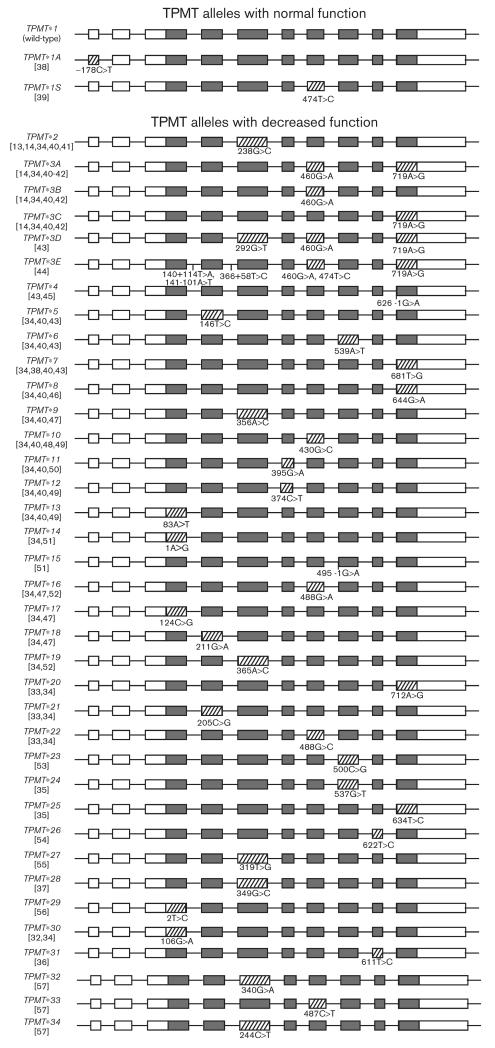
Following the study of TPMT activity, the cloning and characterization of the human TPMT cDNA revealed that these phenotypic variations were primarily due to variation within the coding sequence of the gene itself [13,14]. The human TPMT gene maps to chromosome 6p22.3, which is 34 kb in length and consists of 10 exons. An untranscribed and untranslated pseudogene, homologous to 96% of the TPMT cDNA sequence, has been located to chromosome 18 [15]. To date, about 30 genetic variants have been shown to affect TPMT protein stability and/or enzymatic activity. Most are nonsynonymous single nucleotide polymorphisms (SNPs, Table 1, Fig. 1). The most intensively studied alleles are *2, *3A, and *3C, which represent up to 95% of variant alleles found in most populations [16–18].
Table 1.
TPMT allele nomenclature
| Allele | dbSNP rsID and corresponding nucleotides on the positive chromosomal strand (for standardization) |
Nucleotide changes in the TPMT gene (given on the negative chromosomal strand, NCBI reference sequence NM_000367.2) |
Gene location | Amino acid change (NCBI reference sequence NP_000358.1) |
References |
|---|---|---|---|---|---|
| TPMT*1 | rs2842934 allele Aa | Wild type 474Ta | – | – | – |
| TPMT*1A | ND, G> A | – 178C>T | Exon I | – | [38] |
| TPMT*1S | A >G at rs2842934 | 474T >C | Exon VII | Ile158Ile | [39] |
| TPMT*2 | C>G at rs1800462 | 238G>C | Exon V | Ala80Pro | [13,14,34,40,41] |
| TPMT*3A | C> T at rs1800460 | 460G>A | Exon VII | Ala154Thr | [14,34,40–42] |
| T >C at rs1142345 | 719A >G | Exon X | Tyr240Cys | – | |
| TPMT*3B | C> T at rs1800460 | 460G>A | Exon VII | Ala154Thr | [14,34,40,42] |
| TPMT*3C | T >C at rs1142345 | 719A >G | Exon X | Tyr240Cys | [14,34,40,42] |
| TPMT*3D | C>A at s72552739 | 292G > T | Exon V | Glu98Sto | [43] |
| C> T at rs1800460 | 460G>A | Exon VII | Ala154Thr | – | |
| T >C at rs1142345 | 719A >G | Exon X | Tyr240Cys | – | |
| TPMT*3E | A > T at rs3931660 | 140+ 114T > A | Intron III | – | [44] |
| T>A at rs12529220 | 141–101A> T | Intron III | – | – | |
| A >G at rs2518463 | 366 + 58T >C | Intron IV | – | – | |
| C> T at rs1800460 | 460G>A | Exon VII | Ala154Thr | – | |
| A >G at rs2842934 | 474T >C | Exon VII | Ile158Ile | – | |
| T >C at rs1142345 | 719A >G | Exon X | Tyr240Cys | – | |
| TPMT*4 | C>T at rs1800584 | 626-1G> A | Intron IX/exon X in splice junction | – | [43,45] |
| TPMT*5 | A >G at rs72552740 | 146T >C | Exon IV | Leu49Ser | [34,40,43] |
| TPMT*6 | T >A at rs75543815 | 539A > T | Exon VIII | Tyr180Phe | [34,40,43] |
| TPMT*7 | A >C at rs72552736 | 681T >G | Exon X | His227Gln | [34,38,40,43] |
| TPMT*8 | C>T at rs56161402 | 644G>A | Exon X | Arg215His | [34,40,46] |
| TPMT*9 | T>G at rs151149760 | 356A >C | Exon V | Lys119Thr | [34,40,47] |
| TPMT*10 | C>G at rs72552737 | 430G>C | Exon VII | Gly144Arg | [34,40,48,49] |
| TPMT*11 | C>T at rs72552738 | 395G > A | Exon VI | Cys132Tyr | [34,40,50] |
| TPMT*12 | ND, G> A | 374C >T | Exon VI | Ser125Leu | [34,40,49] |
| TPMT*13 | T> A at rs72552742 | 83A >T | Exon III | Glu28Val | [34,40,49] |
| TPMT*14 | T>C at rs9333569 | 1A >G | Exon III | Met1Val | [34,51] |
| TPMT*15 | C>T at rs9333570 | 495-1G > A | Intron VII/exon VIII in splice junction | – | [51] |
| TPMT*16 | C> T at rs144041067 | 488G>A | Exon VII | Arg163His | [34,47,52] |
| TPMT*17 | ND, G>C | 124C >G | Exon III | Gln42Glu | [34,47] |
| TPMT*18 | ND, C> T | 211G >A | Exon IV | Gly71Arg | [34,47] |
| TPMT*19 | ND, T>G | 365A>C | Exon V | Lys122Thr | [34,52] |
| TPMT*20 | T>C at rs150900439 | 712A >G | Exon X | Lys238Glu | [33,34] |
| TPMT*21 | G>C at rs200591577 | 205C >G | Exon IV | Leu69Val | [33,34] |
| TPMT*22 | ND, C>G | 488G>C | Exon VII | Arg163Pro | [33,34] |
| TPMT*23 | G>C at rs74423290 | 500C >G | Exon VIII | Ala167Gly | [53] |
| TPMT*24 | C> A at rs6921269 | 537G> T | Exon VIII | Gln179His | [35] |
| TPMT*25 | ND, A >G | 634T >C | Exon X | Cys212Arg | [35] |
| TPMT*26 | A >G at rs72556347 | 622T>C | Exon IX | Phe208Leu | [54] |
| TPMT*27 | ND, A >C | 319T >G | Exon V | Tyr107Asp | [55] |
| TPMT*28 | ND, C>G | 349G>Cb | Exon V | Gly117Arg | [37] |
| TPMT*29 | A >G at rs267607275 | 2T >C | Exon III | Met1Thr | [56] |
| TPMT*30 | Old TPMT*20/*24, ND, C> T | 106G> A | Exon III | Gly36Ser | [32,34] |
| TPMT*31 | Old TPMT*28 A >G at rs79901429 | 611T>C | Exon IX | Ile204Thr | [36] |
| TPMT*32 | C> T at rs115106679 | 340G>A | Exon V | Glu114Lys | [57] |
| TPMT*33 | G> A at rs112339338 | 487C>T | Exon VII | Arg163Cys | [57] |
| TPMT*34 | G> A at rs111901354 | 244C>T | Exon V | Arg82Trp | [57] |
The table defines all the single nucleotide polymorphisms (SNPs) in TPMT as of January 2013.
dbSNP, single nucleotide polymorphism database; ND, not reported to dbSNP; TPMT, thiopurine methyltransferase.
dbSNP reports G> A at this position: however, the TPMT nomenclature committee has defined wild type as having allele A at this position (positive chromosomal strand) and the *1S allele as having allele G at this position (positive chromosomal strand).
Incorrect nucleotide substitutions are given in Landy et al.’s [37] study: the corrected nucleotide substitution is included in the table (T. Marinaki, 14 September 2012, personal communication).
Fig. 1.
Schematic representation of thiopurine methyltransferase (TPMT) alleles showing affected exons in stripe pattern, unaffected exons in dark gray, and untranslated parts of exons in white. The sizes of the exons are proportional to base pair length, whereas the introns are not. Nucleotide changes in the TPMT gene (given on the negative chromosomal strand, NCBI reference sequence NM_000367.2) are numbered such that the A in the ATG is + 1.
However, large interethnic differences exist in the frequencies of these alleles, and in the prevalence of more rare alleles. Examples are TPMT*8 and TPMT*6, which occur at frequencies between 1.5 and 3.5% in some African and Asian populations [19,20], but very rarely, if at all, in other investigated populations. More detailed information about specific TPMT alleles can be found in Wang et al.’s [21] study. In the future, the application of routine exome or whole genome sequencing is likely to unveil many more rare TPMT sequence variants [22].
Patients inheriting two nonfunctional TPMT alleles are at the highest risk of hematopoietic toxicity if treated with conventional doses of thiopurines (essentially 100% risk), and convert more parent drug into active TGNs because they lack the methylation pathway, whereas patients who inherit one wild-type allele and one nonfunctional allele are at a significantly higher risk of hematopoietic toxicity (~35% cumulative incidence in one study of children with ALL) [23,24].
In contrast to this, ALL patients with high TPMT activity may have a higher risk of relapse if treated with conventional doses of 6-MP (depending on what other ALL therapy they receive); thus, they should be treated on full-dose thiopurine. Those with intermediate activity often require treatment with a lower dose to avoid toxicity. It has been shown that TPMT heterozygous ALL patients do not have a higher risk of ALL relapse if treated with a lower dose of 6-MP [25], and that heterozygous ALL patients have a significantly lower rate of minimal residual disease positivity for early treatment response to 6-MP, at least in the ALL Berlin-Frankfurt-Münster (BFM) trial [26].
Clearly, as this enzyme has such a profound effect on drug metabolism, it is critical that the TPMT status of a patient should be taken into account before commencing thiopurine treatment [27].
In 2011, a guideline from the Clinical Pharmacogenetics Implementation Consortium was published, with the purpose of providing information with which to interpret clinical TPMT genotype tests so that the results can be used successfully to guide the dosing of thiopurines [28].
Although there is still debate on genotype versus phenotype in assessing the TPMT status, it remains both timely and critical that a repository for the naming of these polymorphisms is created. This will have appropriate links to existing resources: thus, investigators in the field can easily identify polymorphisms that have been described previously and their effects on TPMT activity. It is to this end that a new TPMT nomenclature website (http://www.imh.liu.se/tpmtalleles) was launched by representatives from a worldwide group of researchers from the field of TPMT pharmacogenetic research.
The TPMT nomenclature website
Since the publication of the first TPMT variant (allele *2) in 1995 [13,14], many sequence variants in the TPMT gene have been identified and been given star (*) allele numbers (summarized in Table 1, Fig. 1). Currently, authors number alleles sequentially using PubMed as a source of information about the last published allele; however, this has caused some problems, as exemplified by two instances in which authors’ articles appeared on PubMed at the same time, resulting in the same number being given to different polymorphisms.
To circumvent this in the future, there is a need for a unified nomenclature system, and therefore, in 2010, a TPMT nomenclature committee of representatives from around the world who work in the field was formed with the aim of creating a platform for a full description of known TPMT alleles and for the numbering of subsequent alleles. The TPMT nomenclature website (http://www.imh.liu.se/tpmtalleles) was launched in 2011 with the purpose of managing allele designations and providing a summary of published TPMT alleles. The nomenclature system chosen was based on the established star allele numbering system and on recommended genetic variant nomenclature guidelines [29–31].
Currently, the website covers the nomenclature for published TPMT alleles linked to a designated star allele number, as well as the functionality of the allele. In addition, links to the National Center for Biotechnology Information single nucleotide polymorphism database (dbSNP), as well as publications describing the original identification and/or the characterization of the allele are presented. Researchers are encouraged to submit their own validated novel genetic variants to dbSNP to obtain a reference SNP identification (rsID) to aid consistency in genetic variant mapping.
Inclusion criteria for TPMT alleles
The main function of the website is to encourage researchers worldwide to be confident of which TPMT allele they are referring and to avoid confusion in the literature in the future. The TPMT nomenclature committee has decided upon inclusion criteria that will help to maintain this goal, listed 1–8 in Table 2.
Table 2.
Inclusion criteria for TPMT alleles to be assigned a unique identity and to be included on the TPMT websitea
| (1) On the TPMT nomenclature website, only human TPMT alleles are considered |
| (2) The gene and allele are separated by an asterisk followed by Arabic numerals (e.g. TPMT*1, TPMT*3) |
| (3) Additional nucleotide changes and combinations of nucleotide changes, including silent mutations in the gene, will be assigned letters (e.g. *1A, *1S) |
| (4) To be assigned as a unique allele, it should contain nucleotide changes that have been shown to affect transcription, splicing, translation, post-transcriptional or post-translational modifications or result in at least one amino acid change |
| (5) Numbering of nucleotides in the allele should be as described in Antonarakis and the NomenclatureWorking Group [30]. In the cDNA sequence, the base A in the initiation codon ATG is denoted as + 1 and the base before A is numbered as − 1 |
| (6) Submission of new alleles should be done with information sufficient to fulfill the criteria to be assigned a unique allele (as under criterion 4 above) or letter (as described under criterion 3 above). For incorporation into the website as a unique allele, all exons and exon–intron borders should have been sequenced. If a new allele has been detected on the cDNA level, verification of the mutation(s) on the genomic level is necessary. For acceptance of a new SNP given a separate letter (criterion 3), evidence for its presence on the genomic level is necessary |
| (7) No temporary allelic numbers or letters are provided, and information about any new allele submitted will continuously be published on the website. In case an author does not want to release the information on the website before publication, the webmaster can usually provide him or her with an allelic designation but not release the information on the website until the manuscript has been accepted or published |
| (8) Any novel SNPs should be submitted to dbSNP (NCBI) after TPMT allele designation to obtain a unique rsID for marker mapping – this rsID should then be submitted to the website to be added to the table |
dbSNP, single nucleotide polymorphism database; TPMT, thiopurine methyltransferase.
Renaming of existing TPMT alleles
To improve the existing nomenclature, the committee reviewed all existing allele numbers, and for those that are duplicated, the committee has agreed on the renumbering of two earlier identified and published alleles.
Renaming of TPMT*20/*24 (106G > A in exon III)
The 106G > A SNP in exon 3 was identified during denaturating HPLC screening of the TPMT gene in 200 Japanese individuals [32] and was numbered TPMT*20. At the same time, Schaeffeler et al. [33] identified a SNP at position 712 (A > G, rs150900439), and also numbered it TPMT*20. To address this problem, in 2008 the 106 SNP was renumbered TPMT*24 [34]. However, at the same time in 2008, Garat et al. [35] presented yet another SNP at position 537(G > T), which was numbered TPMT*24. The committee has decided that the SNP at position 106G > A should be designated by the unique star allele number TPMT*30 (Table 1).
Renaming of TPMT*28 (611T > C in exon IX, rs79901429)
The 611T > C (rs79901429) SNP was identified in a Swedish family with Italian ancestries, and was numbered TPMT*28 in 2010 [36]. At the same time, Landy et al. [37] described the identification of a SNP at position 349 (G > C, see Table 1 for comments), which they also numbered TPMT*28. The committee has decided that the SNP at position 611 should be assigned a novel star allele number – TPMT*31 (Table 1).
Submission of new alleles
To avoid any confusion in the future, authors are encouraged to submit novel TPMT alleles (sequence and functionality if available), preferably after a manuscript has been accepted but before final proofing, to the nomenclature committee through the website for confidential designation and reservation of a novel allele number by the nomenclature committee, or to contact the editor of the committee (corresponding author of the current paper) by ordinary mail.
To submit a novel TPMT allele to the committee, authors should fill in the form available on the TPMT nomenclature website, with information regarding, for example, the position of the variant, gene location (exon/intron, etc.), nucleotide position (e.g. 238), and SNP flanking sequence. Usage of star allele designations that have not been approved by the nomenclature committee is strongly discouraged, because of the risk of confusion when two different alleles are given the same star number. The editors of the TPMT nomenclature committee (and if appropriate the advisory board, details of which are found on the TPMT nomenclature website) will review the submission to evaluate whether there are enough data to support a new allele designation, and will await publishing before making the novel allele available, ensuring that only peer-reviewed data are published on the TPMT nomenclature website.
Conclusion
To maintain a common nomenclature system within the field, fellow scientists investigating TPMT polymorphisms are strongly encouraged to submit novel TPMT allelic variants to the TPMT nomenclature committee (http://www.imh.liu.se/tpmtalleles) by contacting the webmaster through the website for the designation and reservation of novel TPMT allele numbers confidentially.
The authors of this mini-review, some of whom have themselves identified novel TPMT alleles, are supportive of this new nomenclature system, and will use this system in their future work. This new nomenclature will also be used at www.pharmgkb.org for associations between TPMT alleles and drug responses reported in the literature [58] and at www.LOVD.nl/TPMT, a gene variant database, collecting and displaying all reported TPMT DNA variants.
Acknowledgements
This study was supported by the Swedish Children’s Cancer Foundation and the Swedish Cancer Society (M.L.A.); NHMRC and The Gutsy Group (J.D.); NIH R37 CA 36401, U01 GM 92666, U01 HL 105918, P30 CA 21765, and ALSAC (W.E.); the Jim and Mary Carney Charitable Trust, Whangarei, New Zealand (M.A.K.); Leukaemia and Lymphoma Research, London, UK (L.L.); Guy’s and St Thomas’ Charity (T.M.); NIH Grants UL1 RR025747 and P01CA142538 (H.L.M.); NIH R37 CA 36401, NIH GM 92666, HL 105918, U01 GM 92666, U01 HL 105918, P30 CA 21765, and ALSAC (M.V.R.); Robert Bosch Foundation, Stuttgart (E.S.); Robert Bosch Foundation, Stuttgart and Deutsche Forschungsgemeinschaft SFB685, Germany (M.S.); NIH Grants U19 GM61388 (the Pharmacogenomics Research Network), RO1 GM28157 and RO1 CA132780 (R.W.); the Children’s Cancer Foundation, Singapore (A.E.J.Y.); NIH R24 GM61374 (E.M.M.); NIH R24 GM61374 (J.M.H.); NIH R24 GM61374 (T.E.K.); the Newcastle Healthcare Charity and the Newcastle upon Tyne Hospitals NHS Charity (S.A.C.).
Footnotes
Conflicts of interest M.V.R. and W.E. receive a portion of the income that St Jude Children’s Research Hospital receives from licensing patent rights related to TPMT polymorphisms. E.S. and M.S. are holders of patent rights related to TPMT genetic variants. M.K. is a coinventor on a patent relating to a trinucleotide repeat polymorphism in the TPMT promoter. For the remaining authors there are no conflicts of interest.
References
- 1.Elion GB, Hitchings GH, Vanderwerff H. Antagonists of nucleic acid derivatives. VI. Purines. J Biol Chem. 1951;192:505–518. [PubMed] [Google Scholar]
- 2.Burchenal JH, Murphy ML, Ellison RR, Sykes MP, Tan TC, Leone LA, et al. Clinical evaluation of a new antimetabolite, 6-mercaptopurine, in the treatment of leukemia and allied diseases. Blood. 1953;8:965–999. [PubMed] [Google Scholar]
- 3.Fotoohi AK, Coulthard SA, Albertioni F. Thiopurines: factors influencing toxicity and response. Biochem Pharmacol. 2010;79:1211–1220. doi: 10.1016/j.bcp.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Coulthard S. Mechanism of thiopurine action. J Braz Med Assoc. 2012;58:18–23. [Google Scholar]
- 5.Karran P. Thiopurines, DNA damage, DNA repair and therapy-related cancer. Br Med Bull. 2006;79-80:153–170. doi: 10.1093/bmb/ldl020. [DOI] [PubMed] [Google Scholar]
- 6.Dervieux T, Blanco JG, Krynetski EY, Vanin EF, Roussel MF, Relling MV. Differing contribution of thiopurine methyltransferase to mercaptopurine versus thioguanine effects in human leukemic cells. Cancer Res. 2001;61:5810–5816. [PubMed] [Google Scholar]
- 7.Coulthard SA, Hogarth LA, Little M, Matheson EC, Redfern CP, Minto L, et al. The effect of thiopurine methyltransferase expression on sensitivity to thiopurine drugs. Mol Pharmacol. 2002;62:102–109. doi: 10.1124/mol.62.1.102. [DOI] [PubMed] [Google Scholar]
- 8.Tiede I, Fritz G, Strand S, Poppe D, Dvorsky R, Strand D, et al. CD28-dependent Rac1 activation is the molecular target of azathioprine in primary human CD4 + T lymphocytes. J Clin Invest. 2003;111:1133–1145. doi: 10.1172/JCI16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poppe D, Tiede I, Fritz G, Becker C, Bartsch B, Wirtz S, et al. Azathioprine suppresses ezrin-radixin-moesin-dependent T cell-APC conjugation through inhibition of Vav guanosine exchange activity on Rac proteins. J Immunol. 2006;176:640–651. doi: 10.4049/jimmunol.176.1.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourgine J, Garat A, Allorge D, Crunelle-Thibaut A, Lo-Guidice JM, Colombel JF, et al. Evidence for a functional genetic polymorphism of the Rho-GTPase Rac1. Implication in azathioprine response? Pharmacogenet Genomics. 2011;21:313–324. doi: 10.1097/FPC.0b013e3283449200. [DOI] [PubMed] [Google Scholar]
- 11.Weinshilboum R, Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet. 1980;32:651–662. [PMC free article] [PubMed] [Google Scholar]
- 12.Lennard L, Lilleyman JS, Van Loon J, Weinshilboum RM. Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet. 1990;336:225–229. doi: 10.1016/0140-6736(90)91745-v. [DOI] [PubMed] [Google Scholar]
- 13.Krynetski EY, Schuetz JD, Galpin AJ, Pui CH, Relling MV, Evans WE. A single point mutation leading to loss of catalytic activity in human thiopurine S-methyltransferase. Proc Natl Acad Sci USA. 1995;92:949–953. doi: 10.1073/pnas.92.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai HL, Krynetski EY, Yates CR, Loennechen T, Fessing MY, Krynetskaia NF, et al. Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet. 1996;58:694–702. [PMC free article] [PubMed] [Google Scholar]
- 15.Lee D, Szumlanski C, Houtman J, Honchel R, Rojas K, Overhauser J, et al. Thiopurine methyltransferase pharmacogenetics. Cloning of human liver cDNA and a processed pseudogene on human chromosome 18q21.1. Drug Metab Dispos. 1995;23:398–405. [PubMed] [Google Scholar]
- 16.Spire-Vayron de la Moureyre C, Debuysere H, Mastain B, Vinner E, Marez D, Lo Guidice JM, et al. Genotypic and phenotypic analysis of the polymorphic thiopurine S-methyltransferase gene (TPMT) in a European population. Br J Pharmacol. 1998;125:879–887. doi: 10.1038/sj.bjp.0702152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ameyaw MM, Collie-Duguid ES, Powrie RH, Ofori-Adjei D, McLeod HL. Thiopurine methyltransferase alleles in British and Ghanaian populations. Hum Mol Genet. 1999;8:367–370. doi: 10.1093/hmg/8.2.367. [DOI] [PubMed] [Google Scholar]
- 18.McLeod HL, Pritchard SC, Githang’a J, Indalo A, Ameyaw MM, Powrie RH, et al. Ethnic differences in thiopurine methyltransferase pharmacogenetics: evidence for allele specificity in Caucasian and Kenyan individuals. Pharmacogenetics. 1999;9:773–776. doi: 10.1097/00008571-199912000-00012. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira E, Quental S, Alves S, Amorim A, Prata MJ. Do the distribution patterns of polymorphisms at the thiopurine S-methyltransferase locus in sub-Saharan populations need revision? Hints from Cabinda and Mozambique. Eur J Clin Pharmacol. 2007;63:703–706. doi: 10.1007/s00228-007-0310-8. [DOI] [PubMed] [Google Scholar]
- 20.Schaeffeler E, Zanger UM, Eichelbaum M, Asante-Poku S, Shin JG, Schwab M. Highly multiplexed genotyping of thiopurine s-methyltransferase variants using MALD-TOF mass spectrometry: reliable genotyping in different ethnic groups. Clin Chem. 2008;54:1637–1647. doi: 10.1373/clinchem.2008.103457. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Pelleymounter L, Weinshilboum R, Johnson JA, Hebert JM, Altman RB, et al. Very important pharmacogene summary: thiopurine S-methyltransferase. Pharmacogenet Genomics. 2010;20:401–405. doi: 10.1097/FPC.0b013e3283352860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans WE, Horner M, Chu YQ, Kalwinsky D, Roberts WM. Altered mercaptopurine metabolism, toxic effects, and dosage requirement in a thiopurine methyltransferase-deficient child with acute lymphocytic leukemia. J Pediatr. 1991;119:985–989. doi: 10.1016/s0022-3476(05)83063-x. [DOI] [PubMed] [Google Scholar]
- 24.Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, Krynetski EY, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 25.Relling MV, Pui CH, Cheng C, Evans WE. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood. 2006;107:843–844. doi: 10.1182/blood-2005-08-3379. [DOI] [PubMed] [Google Scholar]
- 26.Stanulla M, Schaeffeler E, Flohr T, Cario G, Schrauder A, Zimmermann M, et al. Thiopurine methyltransferase (TPMT) genotype and early treatment response to mercaptopurine in childhood acute lymphoblastic leukemia. Jama. 2005;293:1485–1489. doi: 10.1001/jama.293.12.1485. [DOI] [PubMed] [Google Scholar]
- 27.Lennard L, Gibson BE, Nicole T, Lilleyman JS. Congenital thiopurine methyltransferase deficiency and 6-mercaptopurine toxicity during treatment for acute lymphoblastic leukaemia. Arch Dis Child. 1993;69:577–579. doi: 10.1136/adc.69.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther. 2011;89:387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White JA, McAlpine PJ, Antonarakis S, Cann H, Eppig JT, Frazer K, et al. Guidelines for human gene nomenclature (1997). HUGO Nomenclature Committee. Genomics. 1997;45:468–471. doi: 10.1006/geno.1997.4979. [DOI] [PubMed] [Google Scholar]
- 30.Antonarakis SE. Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 31.Den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki T, Goto E, Konno Y, Hiratsuka M, Mizugaki M. Three novel single nucleotide polymorphisms of the human thiopurine S-methyltransferase gene in Japanese individuals. Drug Metab Pharmacokinet. 2006;21:332–336. doi: 10.2133/dmpk.21.332. [DOI] [PubMed] [Google Scholar]
- 33.Schaeffeler E, Eichelbaum M, Reinisch W, Zanger UM, Schwab M. Three novel thiopurine S-methyltransferase allelic variants (TPMT*20, *21, *22)-association with decreased enzyme function. Hum Mutat. 2006;27:976. doi: 10.1002/humu.9450. [DOI] [PubMed] [Google Scholar]
- 34.Ujiie S, Sasaki T, Mizugaki M, Ishikawa M, Hiratsuka M. Functional characterization of 23 allelic variants of thiopurine S-methyltransferase gene (TPMT*2-*24) Pharmacogenet Genomics. 2008;18:887–893. doi: 10.1097/FPC.0b013e3283097328. [DOI] [PubMed] [Google Scholar]
- 35.Garat A, Cauffiez C, Renault N, Lo-Guidice JM, Allorge D, Chevalier D, et al. Characterisation of novel defective thiopurine S-methyltransferase allelic variants. Biochem Pharmacol. 2008;76:404–415. doi: 10.1016/j.bcp.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Appell ML, Wennerstrand P, Peterson C, Hertervig E, Martensson LG. Characterization of a novel sequence variant, TPMT*28, in the human thiopurine methyltransferase gene. Pharmacogenet Genomics. 2010;20:700–707. doi: 10.1097/FPC.0b013e3283402ee4. [DOI] [PubMed] [Google Scholar]
- 37.Landy J, Bhuva N, Marinaki A, Mawdsley J. Novel thiopurine methyltransferase variant TPMT*28 results in a misdiagnosis of TPMT deficiency. Inflamm Bowel Dis. 2010;17:1441–1442. doi: 10.1002/ibd.21505. [DOI] [PubMed] [Google Scholar]
- 38.Spire-Vayron de la Moureyre C, Debuysere H, Sabbagh N, Marez D, Vinner E, Chevalier ED, et al. Detection of known and new mutations in the thiopurine S-methyltransferase gene by single-strand conformation polymorphism analysis. Hum Mutat. 1998;12:177–185. doi: 10.1002/(SICI)1098-1004(1998)12:3<177::AID-HUMU5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 39.Yates CR, Krynetski EY, Loennechen T, Fessing MY, Tai HL, Pui CH, et al. Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997;126:608–614. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 40.Salavaggione OE, Wang L, Wiepert M, Yee VC, Weinshilboum RM. Thiopurine S-methyltransferase pharmacogenetics: variant allele functional and comparative genomics. Pharmacogenet Genomics. 2005;15:801–815. doi: 10.1097/01.fpc.0000174788.69991.6b. [DOI] [PubMed] [Google Scholar]
- 41.Tai HL, Krynetski EY, Schuetz EG, Yanishevski Y, Evans WE. Enhanced proteolysis of thiopurine S-methyltransferase (TPMT) encoded by mutant alleles in humans (TPMT*3A, TPMT*2): mechanisms for the genetic polymorphism of TPMT activity. Proc Natl Acad Sci USA. 1997;94:6444–6449. doi: 10.1073/pnas.94.12.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szumlanski C, Otterness D, Her C, Lee D, Brandriff B, Kelsell D, et al. Thiopurine methyltransferase pharmacogenetics: human gene cloning and characterization of a common polymorphism. DNA Cell Biol. 1996;15:17–30. doi: 10.1089/dna.1996.15.17. [DOI] [PubMed] [Google Scholar]
- 43.Otterness D, Szumlanski C, Lennard L, Klemetsdal B, Aarbakke J, Park-Hah JO, et al. Human thiopurine methyltransferase pharmacogenetics: gene sequence polymorphisms. Clin Pharmacol Ther. 1997;62:60–73. doi: 10.1016/S0009-9236(97)90152-1. [DOI] [PubMed] [Google Scholar]
- 44.Colleoni L, Kapetis D, Maggi L, Camera G, Canioni E, Cavalcante P, et al. A new thiopurine S-methyltransferase haplotype associated with intolerance to azathioprine. J Clin Pharmacol. 2012 doi: 10.1177/0091270011435989. doi: 10.1177/0091270011435989. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 45.Otterness D, Szumlanski C, Weinshilboum R. Human thiopurine methyltransferase pharmacogenetics: identification of a novel variant allele. J Invest Med. 1996;44:248A. [Google Scholar]
- 46.Hon YY, Fessing MY, Pui CH, Relling MV, Krynetski EY, Evans WE. Polymorphism of the thiopurine S-methyltransferase gene in African-Americans. Hum Mol Genet. 1999;8:371–376. doi: 10.1093/hmg/8.2.371. [DOI] [PubMed] [Google Scholar]
- 47.Schaeffeler E, Fischer C, Brockmeier D, Wernet D, Moerike K, Eichelbaum M, et al. Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German-Caucasians and identification of novel TPMT variants. Pharmacogenetics. 2004;14:407–417. doi: 10.1097/01.fpc.0000114745.08559.db. [DOI] [PubMed] [Google Scholar]
- 48.Colombel JF, Ferrari N, Debuysere H, Marteau P, Gendre JP, Bonaz B, et al. Genotypic analysis of thiopurine S-methyltransferase in patients with Crohn’s disease and severe myelosuppression during azathioprine therapy. Gastroenterology. 2000;118:1025–1030. doi: 10.1016/s0016-5085(00)70354-4. [DOI] [PubMed] [Google Scholar]
- 49.Hamdan-Khalil R, Allorge D, Lo-Guidice JM, Cauffiez C, Chevalier D, Spire C, et al. In vitro characterization of four novel non-functional variants of the thiopurine S-methyltransferase. Biochem Biophys Res Commun. 2003;309:1005–1010. doi: 10.1016/j.bbrc.2003.08.103. [DOI] [PubMed] [Google Scholar]
- 50.Schaeffeler E, Stanulla M, Greil J, Schrappe M, Eichelbaum M, Zanger UM, et al. A novel TPMT missense mutation associated with TPMT deficiency in a 5-year-old boy with ALL. Leukemia. 2003;17:1422–1424. doi: 10.1038/sj.leu.2402981. [DOI] [PubMed] [Google Scholar]
- 51.Lindqvist M, Haglund S, Almer S, Peterson C, Taipalensu J, Hertervig E, et al. Identification of two novel sequence variants affecting thiopurine methyltransferase enzyme activity. Pharmacogenetics. 2004;14:261–265. doi: 10.1097/00008571-200404000-00006. [DOI] [PubMed] [Google Scholar]
- 52.Hamdan-Khalil R, Gala JL, Allorge D, Lo-Guidice JM, Horsmans Y, Houdret N, et al. Identification and functional analysis of two rare allelic variants of the thiopurine S-methyltransferase gene, TPMT*16 and TPMT*19. Biochem Pharmacol. 2005;69:525–529. doi: 10.1016/j.bcp.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 53.Lindqvist M, Skoglund K, Karlgren A, Soderkvist P, Peterson C, Kidhall I, et al. Explaining TPMT genotype/phenotype discrepancy by haplotyping of TPMT*3A and identification of a novel sequence variant, TPMT*23. Pharmacogenet Genomics. 2007;17:891–895. doi: 10.1097/FPC.0b013e3282ef642b. [DOI] [PubMed] [Google Scholar]
- 54.Kham SK, Soh CK, Aw DC, Yeoh AE. TPMT*26 (208F→L), a novel mutation detected in a Chinese. Br J Clin Pharmacol. 2009;68:120–123. doi: 10.1111/j.1365-2125.2009.03405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng Q, Vannaprasaht S, Peng Y, Angsuthum S, Avihingsanon Y, Yee VC, et al. Thiopurine S-methyltransferase pharmacogenetics: functional characterization of a novel rapidly degraded variant allozyme. Biochem Pharmacol. 2010;79:1053–1061. doi: 10.1016/j.bcp.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee CK, Loh TP, Wong ST, Lee HK, Huan PT, Chiu LL, et al. Detection of a novel single nucleotide polymorphism of the human thiopurine s-methyltransferase gene in a Chinese individual. Drug Metab Pharmacokinet. 2012;27:559–561. doi: 10.2133/dmpk.dmpk-12-sc-008. [DOI] [PubMed] [Google Scholar]
- 57.Lennard L, Cartwright CS, Wade R, Richards SM, Vora A. Thiopurine methyltransferase genotype-phenotype discordance, and thiopurine active metabolite formation, in childhood acute lymphoblastic leukaemia. Br J Clin Pharmacol. 2012 doi: 10.1111/bcp.12066. doi: 10.1111/bcp.12066. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]